Background: HIF-1 is a transcription factor with a well documented role in cancer progression, whereas FHL proteins can suppress tumor growth.
Results: Three FHL proteins inhibit HIF-1 transcriptional activity via distinct mechanisms.
Conclusion: FHL proteins function to inhibit HIF-1 activity.
Significance: Inhibition of HIF-1 by FHL proteins may partially explain their ability to suppress tumor growth.
Keywords: Hypoxia, Hypoxia-inducible Factor (HIF), Transcription Factors, Transcription Regulation
Abstract
Hypoxia-inducible factor 1 (HIF-1) is a transcription factor that promotes angiogenesis, metabolic reprogramming, and other critical aspects of cancer biology. The four-and-a-half LIM domain (FHL) proteins are a family of LIM domain-only proteins implicated in transcriptional regulation and suppression of tumor growth. Here we describe functional interactions between the FHL proteins and HIF-1. FHL1–3 inhibit HIF-1 transcriptional activity and HIF-1α transactivation domain function by oxygen-independent mechanisms. FHL2 directly interacts with HIF-1α to repress transcriptional activity. FHL1 binds to the p300/CBP co-activators and disrupts binding with HIF-1α. FHL3 does not bind to HIF-1α or p300, indicating that it regulates transactivation by a novel molecular mechanism. Expression of the FHL proteins increased upon HIF-1α induction, suggesting the existence of a feedback loop. These results identify FHL proteins as negative regulators of HIF-1 activity, which may provide a mechanism by which they suppress tumor growth.
Introduction
The four-and-a-half LIM domain (FHL)4 proteins have a domain architecture consisting of an amino-terminal half LIM domain followed by four full LIM domains in tandem (1). Multiple sequence alignments demonstrate that FHL1–3 share roughly 50% sequence identity. The FHL proteins have been shown to regulate a variety of transcription factors including SMAD proteins, β-catenin, FOXO1, SRF, AP-1, NFAT, MyoD, and the androgen receptor (2). Recently, the FHL proteins have been implicated in cancer (3, 4). FHL1 and FHL2 expression is down-regulated in several types of human cancers (3). Overexpression of FHL1–3 suppressed hepatocellular tumor xenograft formation in nude mice, whereas knockdown of FHL1–3 had the opposite effect (4). The FHL proteins have also been implicated in other biological processes, particularly muscle growth and differentiation (5–9).
Hypoxia-inducible factor 1 (HIF-1) is a heterodimeric transcription factor consisting of HIF-1α and HIF-1β subunits (10). HIF-1 coordinates adaptive responses to hypoxia by regulating the expression of hundreds of target genes mediating changes in metabolism, angiogenesis, autophagy, and cell growth (11, 12). The HIF-2α protein has a more restricted tissue distribution than HIF-1α but shares sequence similarity and functional overlap (13). HIF target genes include Glut1 encoding glucose transporter 1 (14), PDK1 encoding pyruvate dehydrogenase kinase 1 (15), VegfA encoding vascular endothelial growth factor A (16), Epo encoding erythropoietin (17), and Sod2 encoding manganese superoxide dismutase (18). In recent years HIF-1 has emerged as a promising target for cancer therapeutics (12, 19). HIF-1α overexpression is a common feature of human cancers (20, 21), where it mediates adaptation to the hypoxic tumor microenvironment. Numerous tumor suppressors including p53, PTEN, and the von Hippel Lindau (VHL) protein inhibit HIF-1 activity, whereas viral oncoproteins increase HIF-1 activity (12, 21).
HIF-1α protein stability and transcriptional activity are modulated according to the cellular O2 concentration through the hydroxylation of key amino acid residues. Hydroxylation at proline 402 and proline 564 by prolyl hydroxylase domain proteins allows the binding of the VHL protein and subsequent ubiquitination and degradation of HIF-1α (22–24). The HIF-1α interacting protein OS-9 promotes prolyl hydroxylation of HIF-1α (25). Two other HIF-1α interacting proteins, SSAT2 (26) and MCM7 (27), promote VHL-dependent ubiquitination of HIF-1α. HIF-1α transactivation domain (TAD) function is regulated by FIH-1 (factor inhibiting HIF-1) (28), which hydroxylates asparagine 803, thereby disrupting interaction between the CH1 domain of p300 and the carboxyl-terminal TAD (residues 786–826) of HIF-1α (C-TAD) (29, 30).
Recent work has revealed that HIF-1 activity is also regulated by O2-independent pathways. RACK1 was identified as a negative regulator of HIF-1α protein stability (31). RACK1-dependent ubiquitination is modulated by calcineurin signaling (32), Hsp90 inhibitors (31), and the proteins SSAT1 (33) and Sept9-v1 (34). Other O2-independent regulators of HIF-1α stability include the E3 ubiquitin protein ligases hypoxia-associated factor (35) and ChIP/Hsp70 (36). Reptin was recently described as an O2-independent regulator of HIF-1α transactivation function (37), whereas hypoxia-associated factor (38) and NEMO (39) have been shown to selectively regulate HIF-2α transactivation function. Here we report that all three FHL family members negatively regulate HIF-1 transactivation function in an O2-independent manner.
EXPERIMENTAL PROCEDURES
Tissue Culture and Cells
HEK293, HEK293T, HeLa, and Hep3B cells were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. The cells were maintained at 37 °C in a 5% CO2, 95% air incubator. Hypoxia was induced by exposing cells to 1% O2, 5% CO2, balance N2 at 37 °C in a modular incubator chamber (Billups-Rothenberg).
Immunoprecipitation (IP) and Western Blot (WB) Assays
The cells were lysed in PBS with 0.1% Tween 20, 1 mm DTT, protease inhibitor mixture, sodium orthovanadate, and sodium fluoride, followed by gentle sonication. For IP assays, 30 μl of anti-V5-agarose beads (Sigma) were added to 2.5 mg of cell lysate overnight at 4 °C. The beads were washed four times in lysis buffer. The proteins were eluted in SDS sample buffer and fractionated by SDS-PAGE. Antibodies used in WB assays were: GST (GE Healthcare); V5 (Invitrogen); FLAG (Sigma); β-actin (Santa Cruz); Myc epitope, CBP, FHL1, FHL2, and HIF-2α (Novus Biologicals); and HIF-1α and p300 (BD Biosciences).
GST Pulldown Assays
GST fusion proteins were purified as described (26). [35S]Methionine-labeled proteins were generated in reticulocyte lysates using a T7-coupled in vitro transcription/translation system (Promega). For in vitro GST pulldown experiments, 10 μl of programmed reticulocyte lysate was incubated with 2 μg of GST fusion protein in 500 μl of PBS-T binding buffer (Dulbecco's PBS, pH 7.4, 0.1% Tween 20) at 4 °C for 4 h, followed by the addition of 30 μl of glutathione-Sepharose 4B beads for 2 h. For GST pulldown from cell lysates, 2 μg of GST fusion protein was added to 2 mg of whole cell lysate and incubated overnight at 4 °C, followed by the addition of 30 μl of glutathione-Sepharose 4B beads for 2 h. The beads were washed four times with PBS-T. The proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE followed by autoradiography using Molecular Imager FX (Bio-Rad) or by WB assay.
Reporter Assays
20,000 HEK293 cells were seeded onto 24-well plates and 48 h after seeding were transfected with plasmid DNA using FuGENE 6 (Roche Applied Science). Control reporter pSV-Renilla (10 ng), HIF-1-dependent reporter p2.1 (120 ng), and expression vectors were used. For transactivation assays, pSV-Renilla (10 ng), pG5-E1b-Luc (100 ng), and expression vectors were used. For HIF-1α-CH1 2-hybrid interaction assays, pSV-Renilla (10 ng), pG5-E1b-Luc (100 ng), VP16-CH1 plasmid (150 ng), and other expression vectors were used. For HIF target gene promoters, VegfA-Luc, Sod2-Luc, or Epo-Luc (100 ng) and expression vectors (400 ng) were used. Hep3B cells were transfected using Lipofectamine 2000 (Invitrogen). The cells were lysed, and luciferase activities were determined with a multiwell luminescence reader (Perkin-Elmer Life Science) using a dual luciferase reporter assay system (Promega).
Quantitative Real Time Reverse Transcriptase-PCR Assays
Total RNA was extracted from HEK293T cells using TRIzol (Invitrogen) and treated with DNase I (Ambion). Total RNA (1 μg) was used as template for first strand cDNA synthesis with the iScript cDNA Synthesis system (Bio-Rad). PCR was performed using IQ SYBR Green Supermix and the iCycler Real-Time PCR Detection System (Bio-Rad). Expression of target mRNA relative to 18 S rRNA was calculated based on the threshold cycle (CT) for amplification as 2−Δ(CT), where ΔCT = CT,target − CT,18S. The primer sequences used were: 18 S rRNA, CGG CGA CGA CCC ATT CGA AC and GAA TCG AAC CCT GAT TCC CCG TC; FHL1, GCT GTG GAG GAC CAG TAT TAC and CAG TAG TCG TGC CAG GAT TG; FHL2, ACT GCT TCT GTG ACT TGT ATG C and GTT ATG CCA CTG CCG TTG C; FHL3, GGA GTG ACA TAC CGT GAT C and GCA GGA GAA GCA GTT GTG; GLUT1, CGG GCC AAG AGT GTG CTA AA and TGA CGA TAC CGG AGC CAA TG; PDK1, ACC AGG ACA GCC AAT ACA AG and CCT CGG TCA CTC ATC TTC AC; and VEGF, CTT GCC TTG CTG CTC TAC and TGG CTT GAA GAT GTA CTG G.
shRNA Assays
The vector pSR.retro.GFP.Neo.circular.stuffer (Oligo Engine) was used for shRNA expression. Oligonucleotides were annealed and ligated into BglII- and HindIII-digested vector. The sequences used were CCA AGG AGG TGC ACT ATA A for shFHL1, CTG CTT CTG TGA CTT GTA T for shFHL2 and CAG TGG CTG TGA ACA GCC A for shFHL3. shRNA vectors against HIF-1α and HIF-2α were previously described (40).
Statistical Analysis
The data are presented as the means ± S.D. except where otherwise indicated. Differences between conditions were analyzed for statistical significance using Student's t test. Differences between three or more experimental conditions were analyzed by two-way analysis of variance with Bonferroni correction.
RESULTS
FHL2 Is a Novel HIF-1α Interacting Protein
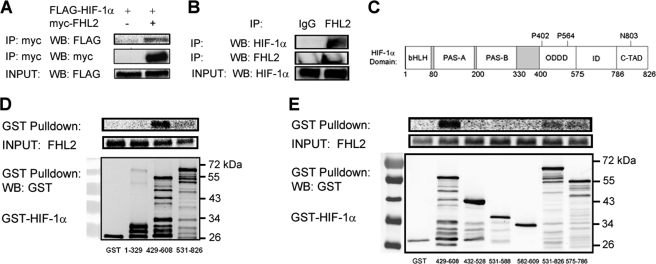
We have utilized several experimental approaches to identify novel HIF-1α interacting proteins (25–28, 31, 36, 41). A yeast two-hybrid screen (26) identified FHL2 as a putative HIF-1α interacting protein. Co-IP experiments verified that FHL2 interacted with FLAG-tagged HIF-1α in 293T cells (Fig. 1A). In Hep3B cells, endogenous FHL2 co-immunoprecipitated with endogenous HIF-1α induced by exposure to hypoxia (Fig. 1B). Binding assays with in vitro transcribed and translated FHL2, and various GST-HIF-1α deletion constructs were used to confirm the initial interaction and localize the binding site on HIF-1α (Fig. 1, C and D). FHL2 bound to residues 429–608, which encompass the O2-dependent degradation domain of HIF-1α and showed weak binding to the overlapping TAD (residues 531–826), with no detectable binding to the amino-terminal basic helix-loop-helix and PAS (Per-Arnt-Sim homology) domains (residues 1–329) of HIF-1α (Fig. 1D). Further attempts to narrow down the interaction site showed that residues 429–608 of HIF-1α comprised the smallest region that retained strong binding to FHL2 (Fig. 1E).
FIGURE 1.
FHL2 interacts with HIF-1α. A, FLAG-HIF-1α co-immunoprecipitates with FHL2. 293T cells were co-transfected with expression vectors encoding FLAG-HIF-1α and either EV or vector encoding Myc epitope-tagged FHL2. At 24 h post-transfection, the cells were lysed and the lysates were immunoprecipitated with anti-Myc antibody. IP products and cell lysates were subjected to WB with anti-Myc or anti-FLAG antibody. B, endogenous FHL2 co-immunoprecipitates with endogenous HIF-1α. Hep3B cells were exposed to 1% O2 for 6 h, lysed, and subjected to immunoprecipitation with anti-IgG or anti-FHL2 antibody. IP products were subjected to WB with anti-FHL2 or anti-HIF-1α antibody. C, the location of the basic helix-loop-helix domain (bHLH), Per-Arnt-Sim homology domain (PAS), O2-dependent degradation domain (ODDD), inhibitory domain (ID), and C-TAD of HIF-1α are shown. Sites of prolyl (P) and asparaginyl (N) hydroxylation are indicated. D and E, FHL2 interacts specifically and directly with HIF-1α(429–608). Purified GST and GST fusion proteins containing the indicated amino acid residues of HIF-1α were incubated with in vitro transcribed, in vitro translated, and 35S-labeled FHL2, captured with glutathione-Sepharose beads and analyzed by SDS-PAGE and autoradiography (top panels) or by WB with GST antibody (bottom panel).
FHL2 Inhibits HIF-1 Transcriptional Activity
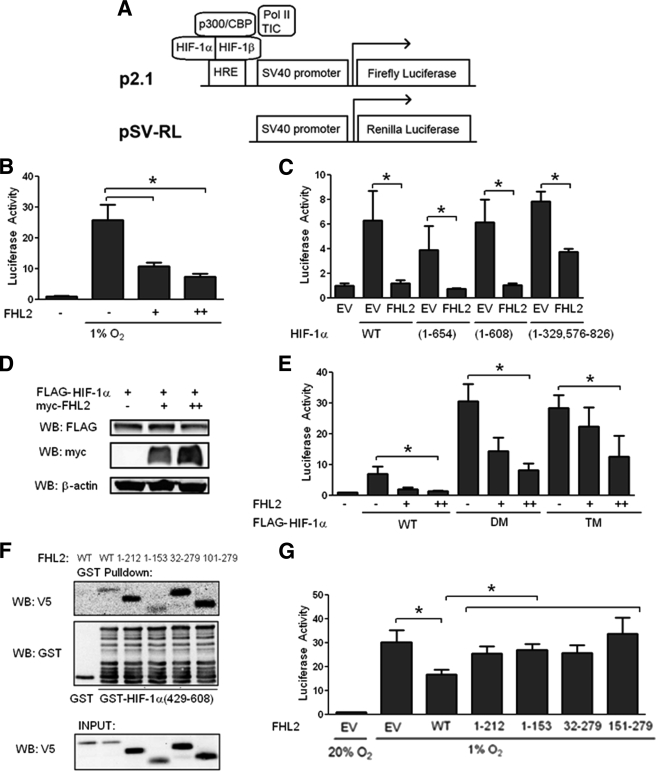
To determine whether FHL2 affected HIF-1 transcriptional activity, we took advantage of a previously described HIF-1 reporter assay (42). 293 cells were co-transfected with two reporter plasmids, p2.1 and pSV-RL. p2.1 contains the firefly luciferase-coding sequence downstream of a HIF-1-dependent hypoxia-response element from the ENO1 gene and SV40 promoter sequences, whereas pSV-RL contains Renilla luciferase coding sequences downstream of the SV40 promoter only (Fig. 2A). The ratio of firefly to Renilla luciferase is a quantitative measure of HIF-1 transcriptional activity. FHL2 inhibited HIF-1 activity induced by hypoxia in a dose-dependent manner (Fig. 2B). Utilizing expression vectors encoding various HIF-1α deletion constructs, we found that the carboxyl-terminal region of HIF-1α (residues 609–826) was dispensable for the effect of FHL2 (Fig. 2C). FHL2 had a greatly reduced effect on the activity of a HIF-1α(1–329/576–826) deletion construct, consistent with the impaired binding to residues 576–826 of HIF-1α.
FIGURE 2.
FHL2 inhibits HIF-1 transcriptional activity in an O2-independent manner. A, schematic of HIF reporter assay. B, FHL2 inhibits hypoxia-induced HIF activity. 293 cells were co-transfected with control Renilla luciferase reporter pSV-RL; HIF-1-dependent firefly luciferase reporter p2.1; and either EV or FHL2 expression vector. At 24 h post-transfection, the cells were exposed to either nonhypoxic (20% O2) or hypoxic (1% O2) conditions as indicated for another 24 h and then lysed, and the ratio of firefly:Renilla luciferase activity was determined. The results are normalized to cells transfected with EV under nonhypoxic conditions. C, effect of FHL2 on HIF-1α deletion mutants. 293 cells were co-transfected with pSV-RL; p2.1; vector encoding the indicated HIF-1α deletion mutant; and either EV or FHL2 vector. 48 h post-transfection, the cells were lysed, and the ratio of firefly:Renilla activity was measured. D, FHL2 does not affect HIF-1α protein levels. 293T cells were co-transfected with 1 μg of FLAG-HIF-1α expression vector and either 0 (−), 1 (+), or 2 (++) μg of Myc-FHL2 expression vector. 48 h post-transfection, the cells were lysed and probed with anti-FLAG, anti-V5, and anti-β-actin antibodies. E, the effect of FHL2 is independent of HIF-1α hydroxylation. 293 cells were co-transfected with pSV-RL; p2.1; vector encoding the wild-type FLAG-HIF-1α (WT), P402A/P564A double-mutant (DM) HIF-1α, or P402A/P564A/N803A triple mutant (TM) HIF-1α; and either EV or FHL2 vector. 24 h post-transfection, the cells were lysed to determine firefly:Renilla activity. F, binding of FHL2 deletion mutants to HIF-1α. Purified GST or GST-HIF-1α(429–608) fusion protein was incubated with in vitro transcribed, in vitro translated, and 35S-labeled full-length FHL2 or FHL2 deletion construct as indicated. GST proteins were captured with glutathione-Sepharose beads and analyzed by SDS-PAGE and autoradiography or by WB with anti-GST antibody. G, full-length (WT) FHL2 is required to inhibit HIF-1 activity. 293 cells were co-transfected with pSV-RL; p2.1; and EV or vector encoding the indicated FHL2 deletion construct. 24 h post-transfection, the cells were exposed to 1% O2 for an additional 24 h and then lysed, and the ratio of firefly:Renilla activity was determined. Total plasmid transfected was kept constant in all experiments. The results are shown as the means ± S.D. *, p < 0.01 compared with EV.
Although FHL2 binds to the O2-dependent degradation domain of HIF-1α, overexpression of FHL2 had no effect on FLAG-HIF-1α protein levels (Fig. 2D). The effects of FHL2 on wild-type HIF-1α, double-mutant HIF-1α (DM) harboring proline-to-alanine (P402A/P564A) mutations, and triple-mutant HIF-1α (TM) with an additional N803A mutation showed that all three hydroxylation sites are dispensable for negative regulation of HIF-1α by FHL2 (Fig. 2E). Based on these results, we conclude that FHL2 inhibits HIF-1 transcriptional activity in a hydroxylation-independent manner.
Full-length FHL2 Is Required to Inhibit HIF-1
We compared the binding of a series of in vitro transcribed and translated FHL2 deletion constructs to GST-HIF-1α(429–608). We observed binding of all deletion constructs tested to HIF-1α (Fig. 2F). However, in the HIF-1 reporter assay only full-length FHL2 inhibited HIF-1 activity (Fig. 2G). These results demonstrate that domains of FHL2 that are dispensable for interaction with HIF-1α are nonetheless essential for inhibition of HIF-1 transcriptional activity.
Other FHL Family Members Inhibit HIF-1
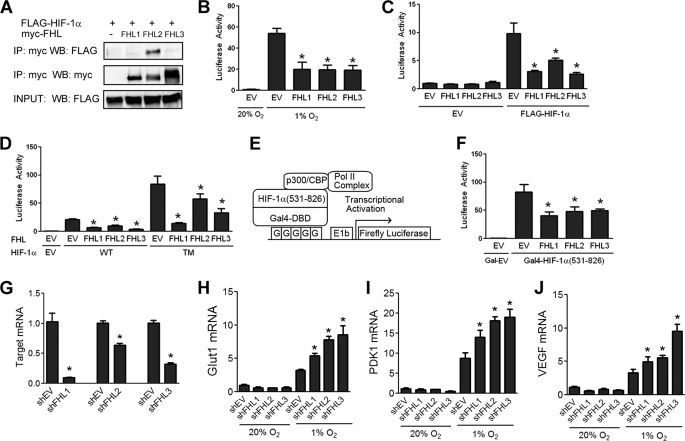
FHL1 and FHL3 belong to the same protein family as FHL2, sharing the same domain structure, as well as a high degree of sequence similarity. To determine whether other FHL family members can interact with HIF-1α, 293T cells were co-transfected with expression vectors encoding FLAG-HIF-1α and Myc-tagged FHL proteins. Co-IP experiments revealed that FHL2 is the only FHL family member with detectable binding to HIF-1α (Fig. 3A).
FIGURE 3.
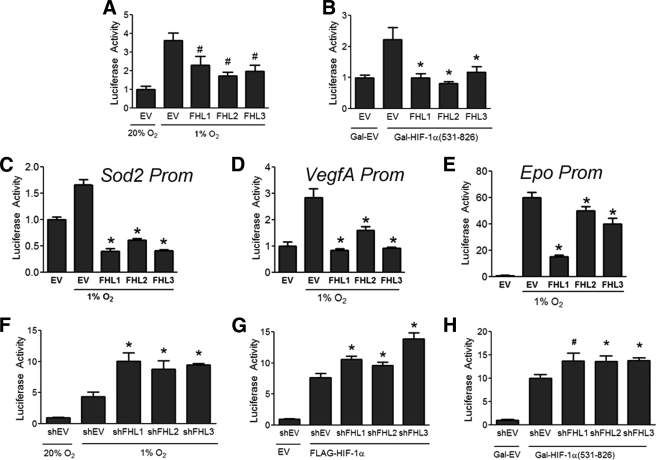
All three FHL proteins inhibit HIF-1 transcriptional activity. A, FHL2 is the only FHL family member that binds HIF-1α. 293T cells were co-transfected with FLAG-HIF-1α and either EV or vector encoding Myc-FHL1, Myc-FHL2, or Myc-FHL3. Lysates were subjected to immunoprecipitation (IP) with anti-Myc antibody and probed for FLAG-HIF-1α and Myc-FHL by WB. B, FHL1–3 inhibit HIF-1 activity induced by hypoxia. 293 cells were co-transfected with p2.1, pSV-RL, and EV or vector encoding FHL1, FHL2, or FHL3. At 24 h post-transfection, cells were exposed to either 20% O2 or 1% O2 as indicated for another 24 h, then lysed and the ratio of firefly:Renilla activity was determined. (C) FHL1–3 inhibit HIF-1 activity induced by HIF-1α overexpression under nonhypoxic conditions. 293 cells were co-transfected with p2.1, pSV-RL, FLAG-HIF-1α vector or EV, and either EV, FHL1, FHL2, or FHL3 vector. 24 h post-transfection, the cells were lysed, and the ratio of firefly:Renilla activity was determined. D, FHL1–3 inhibit HIF-1 in a hydroxylation-independent manner. 293 cells were co-transfected with pSV-RL; p2.1; vector encoding wild-type FLAG-HIF-1α (WT) or P402A/P564A/N803A triple mutant (TM); and either EV, FHL1, FHL2, or FHL3 vector. 24 h post-transfection, the cells were lysed to determine firefly:Renilla activity. E, schematic of HIF-1α TAD assay. F, FHL1–3 inhibit HIF-1α TAD function. 293 cells were co-transfected with pSV-RL; pG5-E1b-Luc; either Gal4-HIF-1α(531–826) or Gal4-EV; and either EV, FHL1, FHL2, or FHL3 vector. 24 h post-transfection, the cells were exposed to hypoxia for an additional 24 h. The cells were harvested, and the ratio of firefly:Renilla luciferase was determined. The amount of total plasmid transfected was kept constant in all experiments. The results are shown as the means ± S.D. G, HeLa cells were transfected with either empty shRNA vector (shEV) or shRNA targeting FHL1, FHL2, or FHL3. 48 h post-transfection RNA was isolated, and quantitative RT-PCR of the target mRNA was performed. H–J, HeLa cells were transfected with either shEV or shRNA targeting FHL1, FHL2, or FHL3. 24 h post-transfection the cells were exposed to either 20% O2 or 1% O2 for an additional 24 h. RNA was isolated, and quantitative RT-PCR was performed against GLUT1 (H), PDK1 (I), and VEGF (J). The results are shown as the means ± S.E. *, p < 0.01 compared with EV or shEV.
However, despite the lack of physical interaction with HIF-1α, overexpression of FHL1 or FHL3 inhibited HIF-1 transcriptional activity that was induced by exposure to hypoxia (Fig. 3B) or by overexpression of FLAG-HIF-1α (Fig. 3C). The effects of FHL1 and FHL3 were also hydroxylation-independent, because FHL1–3 inhibited the activity of HIF-1α(TM), which harbors mutations at all three hydroxylation sites (Fig. 3D).
To examine whether the FHL proteins regulate HIF-1α transactivation, 293 cells were co-transfected with reporter plasmid pG5-E1b-Luc, which contains five Gal4 binding sites upstream of the E1b gene promoter and firefly luciferase coding sequences and an expression vector encoding the Gal4 DNA-binding domain either alone (Gal-EV) or fused to HIF-1α(531–826) (Fig. 3E). In this assay, FHL1–3 all inhibited HIF-1α TAD function (Fig. 3F).
To determine whether endogenous levels of the FHL proteins regulate HIF-1 target gene expression, we constructed shRNA expression vectors targeting FHL1, FHL2, or FHL3 (Fig. 3G). Knockdown of the FHL proteins increased expression of three HIF target genes, GLUT1 (Fig. 3H), PDK1 (Fig. 3I), and VEGF (Fig. 3J), under hypoxic conditions but not under nonhypoxic conditions when the HIF-1α protein is not stabilized.
FHL1 Disrupts Binding of HIF-1α to p300
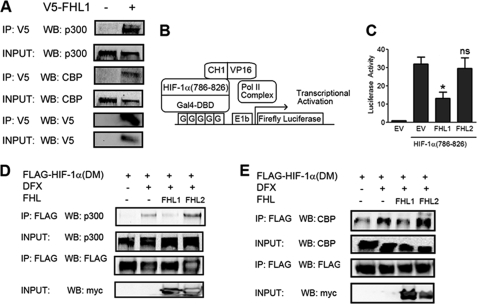
HIF-1α and HIF-2α C-TAD function are regulated by the p300/CBP co-activators. Because FHL1 did not interact with HIF-1α, we hypothesized that FHL1 modulated HIF-1 transcriptional activity by binding to its co-activators instead. To test this hypothesis, 293T cells were transfected with V5-FHL1 expression vector, lysed, and subjected to IP with control IgG or V5 antibody. The results demonstrate that FHL1 interacts with both p300 and CBP (Fig. 4A).
FIGURE 4.
FHL1 disrupts binding of HIF-1α to p300/CBP co-activators. A, FHL1 interacts with p300 and CBP. 293T cells transfected with V5-FHL1 were lysed and immunoprecipitated with a control IgG or V5 antibody. Immunoprecipitates were probed with p300, CBP, and Myc antibodies. B, mammalian two-hybrid assay of HIF-1α TAD-p300 interaction. C, FHL1 inhibits interaction between the HIF-1α C-TAD and p300 CH1 domain. 293 cells were co-transfected with Gal4-HIF-1α(786–826) and VP16-CH1 expression vectors; pG5-E1b-Luc and pSV-RL reporter plasmids; and EV, FHL1, or FHL2 vector. 24 h post-transfection, the cells were exposed to an additional 24 h of hypoxic or nonhypoxic conditions. The cells were harvested, and the ratio of firefly:Renilla luciferase was determined. The results are shown as the means ± S.D. *, p < 0.01 compared with EV. D, FHL1 disrupts HIF-1α-p300 interaction. 293T cells were co-transfected with vector encoding FLAG-tagged double mutant (P402A/P564A) HIF-1α (DM) and either EV, FHL1, or FHL2 vector as indicated. 24 h post-transfection, the cells were left untreated or treated with 100 μm DFX for 6 h, then lysed, and subjected to IP with anti-FLAG antibodies. Immunoprecipitates and cell lysates were subjected to WB with antibodies against p300, FLAG, or Myc. E, FHL1 disrupts HIF-1α-CBP interaction. 293T cells were co-transfected with FLAG-HIF-1α(DM) vector and either EV or vector encoding FHL1 or FHL2 as indicated. 24 h post-transfection, the cells were left untreated or treated with 100 μm DFX for 6 h, then lysed, and subjected to IP with anti-FLAG antibodies. Immunoprecipitates and cell lysates were subjected to WB with antibodies against CBP, FLAG, V5, or Myc.
To determine whether FHL1 affected p300 binding to HIF-1α, we utilized a mammalian two-hybrid assay. 293 cells were transfected with reporter plasmid pG5-E1b-Luc and expression vectors encoding Gal4-HIF-1α(786–826), and the CH1 domain of p300 fused to the VP16 co-activator (Fig. 4B). Hypoxia stimulated an increase in the functional interaction between the C-TAD of HIF-1α and the CH1 domain of p300 in this assay, which was inhibited by FHL1 (Fig. 4C). FHL2 does not interact with either the HIF-1α C-TAD or the p300 CH1 domain and as expected showed no effect in the two-hybrid assay (Fig. 4C).
To verify these results, we analyzed the interaction of HIF-1α with endogenous p300 in a co-IP assay. We utilized an expression vector encoding FLAG-tagged HIF-1α(P402A/P564A) (FLAG-HIF-1α(DM)) so that total levels of HIF-1α protein would not be affected by hydroxylase inhibitors. Treating cells with the hydroxylase inhibitor desferrioxamine (DFX) had no effect on the stability of the mutant construct but, by blocking asparagine hydroxylation, increased the HIF-1α-p300 interaction. Overexpression of FHL1, but not FHL2, blocked this effect (Fig. 4D). These results confirmed that FHL1 functions in a hydroxylation-independent fashion. We observed a similar inhibitory effect of FHL1 overexpression on HIF-1α-CBP interaction (Fig. 4E), whereas FHL2 overexpression had no effect. Thus, the results of both co-IP and two-hybrid assays demonstrate that FHL1 inhibits the transcriptional activity of HIF-1α by competing with it for binding to CBP and p300.
FHL Proteins Regulate HIF-1 Activity in Hepatocellular Carcinoma Cells
The FHL proteins were recently reported to inhibit the growth of human hepatocellular carcinoma xenografts (4). Overexpression of FHL1, FHL2, or FHL3 slowed the rate of tumor growth, whereas knockdown of FHL1, FHL2, or FHL3 expression had the opposite effect (4). To determine whether FHL proteins inhibit HIF-1 activity in human hepatocellular carcinoma, we performed experiments with Hep3B cells. Consistent with the results observed in 293 cells, overexpression of FHL1–3 in Hep3B cells had an inhibitory effect on HIF-1 transcriptional activity (Fig. 5A) and HIF-1α TAD function (Fig. 5B).
FIGURE 5.
FHL proteins inhibit HIF-1 transcriptional activity in hepatocellular carcinoma cells. A, FHL1–3 inhibit HIF-1 activity in Hep3B cells subjected to hypoxia. Hep3B cells were co-transfected with pSV-RL; p2.1; FLAG-HIF-1α; and either EV or vector encoding FHL1, FHL2, or FHL3. 24 h post-transfection, cells were exposed to an additional 24 h under nonhypoxic or hypoxic conditions. Cells were then lysed and the ratio of firefly:Renilla activity was measured. B, FHL1–3 inhibit HIF-1α TAD function. Hep3B cells were co-transfected with pSV-RL; pG5-E1b-Luc; either Gal4-HIF-1α(531–826) or Gal4-EV; and either EV or FHL1, FHL2, or FHL3 vector. 24 h post-transfection, the cells were lysed to determine firefly:Renilla activity. C–E, FHL1–3 inhibit HIF activity at target gene promoters. Hep3B cells were co-transfected with the indicated luciferase promoter constructs and either EV, FHL1, FHL2, or FHL3 vector. 24 h post-transfection the cells were exposed to an additional 24 h under nonhypoxic or hypoxic conditions. The cells were then lysed, and luciferase activity was measured. F, FHL1–3 knockdown increases HIF-1 activity under hypoxic conditions. Hep3B cells were co-transfected with pSV-RL; p2.1; and either empty vector (shEV) or vector encoding shRNA against FHL1, FHL2, or FHL3. 24 h post-transfection the cells were exposed to an additional 24 h at 20% or 1% O2. The cells were lysed, and the ratio of firefly:Renilla activity was determined. G, FHL1–3 knockdown increases HIF-1 activity under nonhypoxic conditions. Hep3B cells were co-transfected with pSV-RL; p2.1; FLAG-HIF-1α vector or EV; and either EV or vector encoding shRNA against FHL1, FHL2, or FHL3. 48 h post-transfection the cells were lysed, and the ratio of firefly:Renilla activity was determined. H, FHL1–3 knockdown up-regulates HIF-1α TAD function. Hep3B cells were co-transfected with pSV-RL; pG5-E1b-Luc; either Gal4-HIF-1α(531–826) or Gal4-EV; and either EV or vector encoding shRNA against FHL1, FHL2, or FHL3. 24 h post-transfection the cells were exposed to 24 h of hypoxia and then lysed to determine firefly:Renilla activity. The amount of total plasmid transfected was kept constant in all experiments. The results are shown as the means ± S.D. *, p < 0.01; #, p < 0.05 compared with EV or shEV.
To examine the effect of the FHL proteins on HIF target gene promoters, we utilized three previously described reporter constructs (43) containing the promoter regions of Sod2, VegfA, and Epo. Activity of the three reporters was induced by exposure to hypoxia in Hep3B cells, and overexpression of FHL1, FHL2, or FHL3 was able to block this effect (Fig. 5, C–E).
We further examined the effect of endogenous FHL proteins on HIF-1 transcriptional activity. Knockdown of FHL1, FHL2, or FHL3 in Hep3B cells led to an increase in HIF-1 reporter activity induced either by exposure to hypoxia (Fig. 5F) or by co-transfection of a FLAG-HIF-1α vector (Fig. 5G). Similarly, knockdown of FHL1, FHL2, or FHL3 led to a significant increase in HIF-1α TAD function (Fig. 5H).
FHL2 Does Not Inhibit HIF-2 Activity
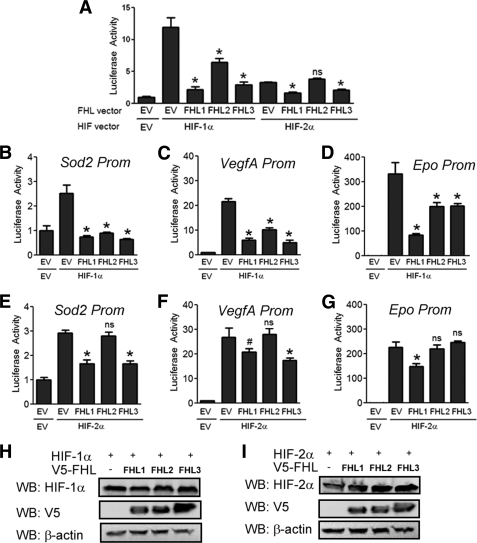
Because of the similarity between the HIF-1α and HIF-2α protein sequences, we examined whether the FHL proteins regulate HIF-2 activity. We could not detect binding of FHL2 or any FHL family member to HIF-2α under conditions where the FHL2-HIF-1α interaction was demonstrated (data not shown). Although we observed inhibition of HIF-1 activity by all three FHL proteins (Fig. 6A), FHL2 was not able to inhibit HIF-2 activity in the same assay (Fig. 6A). However, FHL1 and FHL3 effectively inhibited HIF-2 activity, as well as HIF-1 activity (Fig. 6A).
FIGURE 6.
FHL1 and FHL3, but not FHL2, inhibit HIF-2 transcriptional activity. A, FHL2 inhibits HIF-1, but not HIF-2 activity. 293 cells were co-transfected with p2.1, pSV-RL, HIF-1α or HIF-2α expression vector, and either EV, FHL1, FHL2, or FHL3 vector. 24 h post-transfection, the cells were lysed, and the ratio of firefly:Renilla activity was determined. B-D, FHL1–3 inhibit HIF-1 activity at target gene promoters. Hep3B cells were co-transfected with the indicated luciferase reporter, HIF-1α expression vector, and either EV, FHL1, FHL2, or FHL3 expression vector. 24 h post-transfection cells were lysed, and luciferase activity measured. E–G, FHL1 and FHL3, but not FHL2, inhibit HIF-2 activity at target gene promoters. Hep3B cells were co-transfected with the indicated luciferase reporter, HIF-2α expression vector, and either EV, FHL1, FHL2, or FHL3 expression vector. 24 h post-transfection the cells were lysed, and luciferase activity was measured. The results are shown as the means ± S.D. *, p < 0.01; #, p < 0.05 compared with EV; ns, not significant. H and I, FHL1–3 have no effect on HIF-1α or HIF-2α protein levels. 293T cells were co-transfected with HIF-1α expression vector (H) or HIF-2α expression vector (I) and either EV or V5-tagged FHL1, FHL2, or FHL3 expression vector. 24 h post-transfection the cells were lysed and probed with anti-HIF-1α or anti-HIF-2α, anti-V5, or β-actin antibodies.
To be sure these effects were not an artifact of the reporter plasmid used, we examined the effect of the FHL proteins on the Sod2, VegfA, and Epo gene promoters. All of the FHL proteins were able to inhibit activity at these promoters induced by HIF-1α overexpression (Fig. 6, B–D). By contrast, FHL2 had no effect on activity induced by HIF-2α overexpression on any of these promoters (Fig. 6, E–G). However, FHL1 and FHL3 were able to inhibit HIF-2 activity (Fig. 6, E–G). These results indicate that FHL1 and FHL3 inhibit HIF-1 and HIF-2 through mechanisms independent of direct binding to HIF-α proteins, whereas direct and specific binding is necessary for the effect of FHL2 on HIF-1α.
Just as overexpression of the FHL proteins had no effect on the stability of HIF-1α (Fig. 6H), we found that the FHL proteins had no effect on stability of HIF-2α (Fig. 6I). Thus, inhibition of HIF-1 and HIF-2 by the FHL proteins is independent of effects on HIF-1α or HIF-2α stability.
FHL Expression Is Induced by Hypoxia
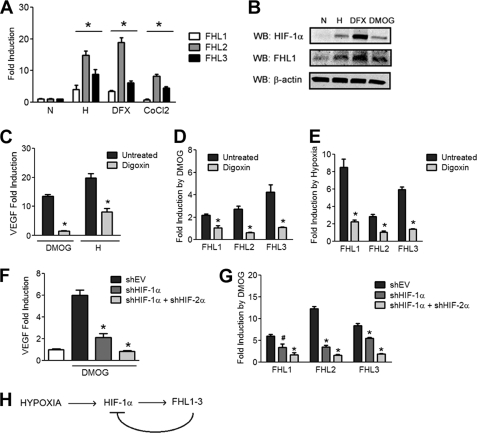
Previous work demonstrated that FHL1 levels were increased in human pulmonary arterial smooth muscle cells exposed to hypoxia (44). To determine whether the expression of other FHL family members is also regulated by hypoxia, we analyzed the expression of FHL1, FHL2, and FHL3 mRNA upon exposure to hypoxia or either of two different chemical inducers of HIF-1α, DFX and cobalt chloride. Expression of FHL mRNAs was significantly increased under all conditions where HIF-1α was induced (Fig. 7A). WB analysis of Hep3B cells exposed to hypoxia or to a hydroxylase inhibitor (DFX or dimethyloxalylglycine (DMOG)) demonstrated a corresponding increase in FHL1 protein levels (Fig. 7B).
FIGURE 7.
Increased FHL expression in hypoxic cells. A, HIF-1α inducers increase expression of FHL1–3 mRNA. Hep3B cells were exposed to 48 h of nonhypoxic (N) or hypoxic (H) conditions or were treated with DFX (100 μm) or cobalt chloride (100 μm). Afterward, RNA was isolated, and quantitative real time RT-PCR of FHL1, FHL2, and FHL3 mRNA was performed. B, HIF-1α inducers increase FHL1 protein levels. Hep3B cells were exposed to 48 h of nonhypoxic (N) or hypoxic conditions (H) or were treated with DFX (100 μm) or DMOG (1 mm). The cells were then lysed, and the lysates were probed with anti-FHL1, anti-HIF-1α, or anti-β-actin antibodies. C, digoxin blocks induction of VEGF by HIF-1. Hep3B cells were left untreated or treated with digoxin for 24 h and treated with DMOG or exposed to hypoxia as indicated. RNA was isolated, and RT-PCR of VEGF mRNA was performed. Fold induction over the untreated and normoxic control is shown. D and E, digoxin blocks induction of FHL family members by HIF-1. Hep3B cells were left untreated or treated with digoxin and then treated with DMOG or vehicle for 24 h (D) or exposed to nonhypoxic or hypoxic conditions for 24 h (E). RNA was isolated, and quantitative RT-PCR of FHL1, FHL2, and FHL3 was performed. Fold induction over the vehicle condition is shown in each case. F and G, Hep3B cells were stably transfected with empty shRNA vector or vector encoding shRNA against HIF-1α or both HIF-1α and HIF-2α. The cells were treated with DMOG or vehicle for 24 h, after which RNA was isolated and quantitative RT-PCR of VEGF (F) and FHL1, FHL2, and FHL3 (G) was performed. The results are shown as the means ± S.E. *, p < 0.01; #, p < 0.05 compared with untreated and nonhypoxic conditions. H, feedback regulation of HIF-1 activity by FHL1–3.
We tested whether HIF-1 activity regulates expression of FHL1, FHL2, and FHL3 mRNA using both pharmacological and genetic tools. Digoxin was recently described as a selective inhibitor of HIF-1α mRNA translation and subsequent HIF-1 activity (45). We confirmed that digoxin blocked induction of VEGF mRNA in cells treated with DMOG or exposed to hypoxia (Fig. 7C). Similarly, digoxin blocked induction of FHL1, FHL2, and FHL3 mRNA upon treatment with DMOG (Fig. 7D) or exposure to hypoxia (Fig. 7E).
We stably infected Hep3B cells with virus encoding either empty shRNA vector, shRNA against HIF-1α, or shRNAs against both HIF-1α and HIF-2α. We confirmed that knockdown of HIF-1 and HIF-2 activity blocked induction of VEGF mRNA in cells treated with DMOG (Fig. 7F). Similarly, knockdown of HIF-1α and HIF-2α blocked induction of FHL1, FHL2, and FHL3 mRNA (Fig. 7G). Thus, activity of both HIF-1 and HIF-2 serves to up-regulate expression of the FHL proteins.
DISCUSSION
In this study, we have identified a novel function of FHL proteins as inhibitors of HIF-1 activity. All three FHL family members inhibit HIF-1α TAD function in a hydroxylation-independent manner. Remarkably, the three proteins utilize three distinct mechanisms of action. FHL2 is the only family member that binds directly and selectively to HIF-1α. FHL2 does not bind HIF-2α, nor does it regulate HIF-2 activity. In contrast, FHL1 and FHL3 inhibited both HIF-1 and HIF-2 activity. FHL1 binds directly to p300 and CBP, whereas the precise molecular mechanism by which FHL3 regulates HIF transcriptional activity remains to be determined.
FHL2 has been shown to regulate the activity of other transcription factors, often by functioning as a scaffold protein. FHL2 promoted the interaction of sirtuin 1 with FOXO1, leading to increased deacetylation and repression of FOXO activity (46), whereas it promoted p300-β-catenin interaction and enhanced β-catenin transcriptional activity (9, 48, 49). Deletion mutants of FHL2 with preserved binding to HIF-1α were ineffective in inhibiting HIF-1 activity (Fig. 2, E and F), suggesting that binding to other (co-repressor) proteins may be essential for FHL2 to repress HIF-1α TAD function.
FHL1 inhibited HIF-1 activity by competing with HIF-1α for binding to p300 and CBP (Fig. 5). FHL1 may thus function in a similar manner as the protein CITED2 (50). Homozygosity for a knock-out allele at the locus encoding CITED2 leads to embryonic lethality in mice because of defects in heart development (51, 52), which are partially rescued by heterozygosity for a knock-out allele at the locus encoding HIF-1α (53). In contrast, FHL1 knock-out mice are viable with comparatively mild defects in cardiac function (7). FHL2 knock-out mice are also viable but display enhanced cardiac hypertrophy in response to chronic infusion of isoproterenol (8). Because the FHL proteins share a redundant tissue distribution, it is possible that inhibition of HIF-1 by other FHL family members may contribute to the mild phenotypes observed in FHL1 and FHL2 knock-out mice.
The observation that FHL family members operate through different molecular mechanisms mirrors findings for other protein families that regulate HIF-1, such as the MCM (27) and SSAT (26, 33) proteins. MCM3 inhibited HIF-1α TAD function, whereas MCM7 enhanced proteasomal degradation of HIF-1α (27). SSAT2 enhanced O2-dependent degradation of HIF-1α mediated through the VHL ubiquitin ligase complex (26), whereas SSAT1 enhanced O2-independent degradation of HIF-1α mediated by RACK1-dependent ubiquitination (33). The existence of multiple FHL family members acting via different mechanisms may allow differential regulation of HIF-1 versus HIF-2 target genes. For example, hypoxia-associated factor and Sirt1 have recently been shown to function as selective HIF-2α co-activators (38, 43), and we demonstrate here that FHL2 is a selective HIF-1α co-repressor, whereas FHL1 and FHL3 negatively regulate both HIF-1α and HIF-2α. FHL1 binding to p300 may also inhibit the activity of other transcription factors that bind to the CH1 domain, including ETS1, HNF4, p53, PIT1, and STAT2 (47). FHL3 is likely to bind to another component of the co-activator complex that interacts directly or indirectly with the TADs of HIF-1α and HIF-2α.
Finally, we detected coordinated up-regulation of the FHL proteins upon the induction of HIF-1α by exposure to hypoxia or treatment with hydroxylase inhibitors. This was blocked by treatment with the HIF inhibitor digoxin or by knockdown of HIF-1α and HIF-2α, implicating the HIF proteins in this process. This result is consistent with previous work suggesting regulation of FHL1 expression by the HIFs in pulmonary arterial smooth muscle cells (44) and indicates the presence of a feedback loop that may serve to limit HIF activity under conditions of prolonged hypoxia (Fig. 7H).
Acknowledgments
We thank P. Coulombe, C. Dang, and G. Tomaselli (Johns Hopkins University) for advice and suggestions; K. Padgett (Novus Biologicals, Inc.) for providing antibodies against Myc epitope, FHL1, FHL2, CBP, and HIF-2α; E. Huang (University of Utah) for providing the CH1-VP16 expression vector; and J. Garcia (University of Texas-Southwestern) for providing the HIF target gene promoter plasmids.
This work was supported in part by National Institutes of Health Public Health Service Contracts N01-HV28180 and HHS-N268201000032C. This work was also supported by funds from the Johns Hopkins Institute for Cell Engineering.
- FHL
- four-and-a-half LIM domain
- CBP
- CREB-binding protein
- C-TAD
- carboxyl-terminal transactivation domain
- DFX
- desferrioxamine
- DMOG
- dimethyloxalylglycine
- DM
- double mutant
- EV
- empty vector
- GLUT1
- glucose transporter-1
- IP
- immunoprecipitation
- HIF
- hypoxia-inducible factor
- MCM
- minichromosome maintenance
- PDK1
- pyruvate dehydrogenase kinase 1
- SSAT
- spermidine/spermine N(1)-acetyltransferase
- TAD
- transactivation domain
- TM
- triple mutant
- VHL
- von Hippel-Lindau protein
- WB
- Western blot.
REFERENCES
- 1. Kadrmas J. L., Beckerle M. C. (2004) The LIM domain. From the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- 2. Johannessen M., Møller S., Hansen T., Moens U., Van Ghelue M. (2006) The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol. Life Sci. 63, 268–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kleiber K., Strebhardt K., Martin B. T. (2007) The biological relevance of FHL2 in tumour cells and its role as a putative cancer target. Anticancer Res. 27, 55–61 [PubMed] [Google Scholar]
- 4. Ding L., Wang Z., Yan J., Yang X., Liu A., Qiu W., Zhu J., Han J., Zhang H., Lin J., Cheng L., Qin X., Niu C., Yuan B., Wang X., Zhu C., Zhou Y., Li J., Song H., Huang C., Ye Q. (2009) Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-β-like signaling pathway. J. Clin. Invest. 119, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cowling B. S., McGrath M. J., Nguyen M. A., Cottle D. L., Kee A. J., Brown S., Schessl J., Zou Y., Joya J., Bönnemann C. G., Hardeman E. C., Mitchell C. A. (2008) Identification of FHL1 as a regulator of skeletal muscle mass. Implications for human myopathy. J. Cell Biol. 183, 1033–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schessl J., Zou Y., McGrath M. J., Cowling B. S., Maiti B., Chin S. S., Sewry C., Battini R., Hu Y., Cottle D. L., Rosenblatt M., Spruce L., Ganguly A., Kirschner J., Judkins A. R., Golden J. A., Goebel H. H., Muntoni F., Flanigan K. M., Mitchell C. A., Bönnemann C. G. (2008) Proteomic identification of FHL1 as the protein mutated in human reducing body myopathy. J. Clin. Invest. 118, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheikh F., Raskin A., Chu P. H., Lange S., Domenighetti A. A., Zheng M., Liang X., Zhang T., Yajima T., Gu Y., Dalton N. D., Mahata S. K., Dorn G. W., 2nd, Heller-Brown J., Peterson K. L., Omens J. H., McCulloch A. D., Chen J. (2008) An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J. Clin. Invest. 118, 3870–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kong Y., Shelton J. M., Rothermel B., Li X., Richardson J. A., Bassel-Duby R., Williams R. S. (2001) Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to β-adrenergic stimulation. Circulation 103, 2731–2738 [DOI] [PubMed] [Google Scholar]
- 9. Martin B., Schneider R., Janetzky S., Waibler Z., Pandur P., Kühl M., Behrens J., von der Mark K., Starzinski-Powitz A., Wixler V. (2002) The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Semenza G. L. (2009) Physiology (Bethesda) 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 12. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 13. Patel S. A., Simon M. C. (2008) Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 15, 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ebert B. L., Firth J. D., Ratcliffe P. J. (1995) Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 270, 29083–29089 [DOI] [PubMed] [Google Scholar]
- 15. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase. A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 16. Forsythe J. A., Jiang B. H., Iyer N. V., Agani F., Leung S. W., Koos R. D., Semenza G. L. (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell Biol. 16, 4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Semenza G. L., Wang G. L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 19. Melillo G. (2007) Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev. 26, 341–352 [DOI] [PubMed] [Google Scholar]
- 20. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 21. Semenza G. L. (2010) Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation. Implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 23. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 24. Yu F., White S. B., Zhao Q., Lee F. S. (2001) HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baek J. H., Mahon P. C., Oh J., Kelly B., Krishnamachary B., Pearson M., Chan D. A., Giaccia A. J., Semenza G. L. (2005) OS-9 interacts with hypoxia-inducible factor 1α and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol. Cell 17, 503–512 [DOI] [PubMed] [Google Scholar]
- 26. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Hubbi M. E., Byun H., Semenza G. L. (2007) Spermidine/spermine-N1-acetyltransferase 2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1α. J. Biol. Chem. 282, 23572–23580 [DOI] [PubMed] [Google Scholar]
- 27. Hubbi M. E., Luo W., Baek J. H., Semenza G. L. (2011) MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol. Cell 42, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mahon P. C., Hirota K., Semenza G. L. (2001) FIH-1. A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 30. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y. V., Hubbi M. E., Pan F., McDonald K. R., Mansharamani M., Cole R. N., Liu J. O., Semenza G. L. (2007) Calcineurin promotes hypoxia-inducible factor 1α expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 282, 37064–37073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Zhang H., Semenza G. L. (2007) Spermidine/spermine N(1)-acetyltransferase-1 binds to hypoxia-inducible factor-1α (HIF-1α) and RACK1 and promotes ubiquitination and degradation of HIF-1α. J. Biol. Chem. 282, 33358–33366 [DOI] [PubMed] [Google Scholar]
- 34. Amir S., Wang R., Simons J. W., Mabjeesh N. J. (2009) SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. J. Biol. Chem. 284, 11142–11151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koh M. Y., Darnay B. G., Powis G. (2008) Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol. Cell. Biol. 28, 7081–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo W., Zhong J., Chang R., Hu H., Pandey A., Semenza G. L. (2010) Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 285, 3651–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J. S., Kim Y., Kim I. S., Kim B., Choi H. J., Lee J. M., Shin H. J., Kim J. H., Kim J. Y., Seo S. B., Lee H., Binda O., Gozani O., Semenza G. L., Kim M., Kim K. I., Hwang D., Baek S. H. (2010) Negative regulation of hypoxic responses via induced Reptin methylation. Mol. Cell 39, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koh M. Y., Lemos R., Jr., Liu X., Powis G. (2011) The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 71, 4015–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bracken C. P., Whitelaw M. L., Peet D. J. (2005) Activity of hypoxia-inducible factor 2α is regulated by association with the NF-kappaB essential modulator. J. Biol. Chem. 280, 14240–14251 [DOI] [PubMed] [Google Scholar]
- 40. Wong C. C., Gilkes D. M., Zhang H., Chen J., Wei H., Chaturvedi P., Fraley S. I., Wong C. M., Khoo U. S., Ng I. O., Wirtz D., Semenza G. L. (2011) Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc. Natl. Acad. Sci. U.S.A. 108, 16369–16374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated co-activator for hypoxia-inducible factor 1. Cell 145, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 43. Dioum E. M., Chen R., Alexander M. S., Zhang Q., Hogg R. T., Gerard R. D., Garcia J. A. (2009) Regulation of hypoxia-inducible factor 2α signaling by the stress-responsive deacetylase sirtuin 1. Science 324, 1289–1293 [DOI] [PubMed] [Google Scholar]
- 44. Kwapiszewska G., Wygrecka M., Marsh L. M., Schmitt S., Trösser R., Wilhelm J., Helmus K., Eul B., Zakrzewicz A., Ghofrani H. A., Schermuly R. T., Bohle R. M., Grimminger F., Seeger W., Eickelberg O., Fink L., Weissmann N. (2008) Fhl-1, a new key protein in pulmonary hypertension. Circulation 118, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 45. Zhang H., Qian D. Z., Tan Y. S., Lee K., Gao P., Ren Y. R., Rey S., Hammers H., Chang D., Pili R., Dang C. V., Liu J. O., Semenza G. L. (2008) Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc. Natl. Acad. Sci. U.S.A. 105, 19579–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Y., Hou H., Haller E. M., Nicosia S. V., Bai W. (2005) Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 24, 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Freedman S. J., Sun Z. Y., Poy F., Kung A. L., Livingston D. M., Wagner G., Eck M. J. (2002) Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor 1 alpha. Proc. Natl. Acad. Sci. U.S.A. 99, 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei Y., Renard C. A., Labalette C., Wu Y., Lévy L., Neuveut C., Prieur X., Flajolet M., Prigent S., Buendia M. A. (2003) Identification of the LIM protein FHL2 as a co-activator of β-catenin. J. Biol. Chem. 278, 5188–5194 [DOI] [PubMed] [Google Scholar]
- 49. Labalette C., Renard C. A., Neuveut C., Buendia M. A., Wei Y. (2004) Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and β-catenin. Mol. Cell Biol. 24, 10689–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhattacharya S., Michels C. L., Leung M. K., Arany Z. P., Kung A. L., Livingston D. M. (1999) Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 13, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bamforth S. D., Bragança J., Eloranta J. J., Murdoch J. N., Marques F. I., Kranc K. R., Farza H., Henderson D. J., Hurst H. C., Bhattacharya S. (2001) Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat. Genet. 29, 469–474 [DOI] [PubMed] [Google Scholar]
- 52. Yin Z., Haynie J., Yang X., Han B., Kiatchoosakun S., Restivo J., Yuan S., Prabhakar N. R., Herrup K., Conlon R. A., Hoit B. D., Watanabe M., Yang Y. C. (2002) The essential role of Cited2, a negative regulator for HIF-1α, in heart development and neurulation. Proc. Natl. Acad. Sci. U.S.A. 99, 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu B., Doughman Y., Turakhia M., Jiang W., Landsettle C. E., Agani F. H., Semenza G. L., Watanabe M., Yang Y. C. (2007) Partial rescue of defects in Cited2-deficient embryos by HIF-1α heterozygosity. Dev. Biol. 301, 130–140 [DOI] [PubMed] [Google Scholar]