Background: We hypothesized that STAT proteins mediate the protumorigenic function of heparanase.
Results: Heparanase enhances the phosphorylation of STAT3 and STAT5b, SRC and EGFR. Notably, cytoplasmic rather than nuclear phospho-STAT3 correlated with poor prognosis.
Conclusion: Heparanase levels are associated with the outcome of head and neck cancer patients.
Significance: A novel feature of head and neck cancer is revealed.
Keywords: Cancer Biology, Cell Signaling, Epidermal Growth Factor Receptor (EGFR), Glycosaminoglycan, Head And Neck Cancer, Heparan Sulfate, Phosphorylation, STAT3, Heparanase, Localization
Abstract
Activity of heparanase is implicated strongly in dissemination of metastatic tumor cells and cells of the immune system. In addition, heparanase enhances the phosphorylation of selected signaling molecules, including SRC and EGFR, in a manner that requires secretion but not enzymatic activity of heparanase and is mediated by its C-terminal domain. Clinically, heparanase staining is associated with larger tumors and increased EGFR phosphorylation in head and neck carcinoma. We hypothesized that signal transducer and activator of transcription (STAT) proteins mediate the protumorigenic function of heparanase downstream of the EGFR. We provide evidence that heparanase enhances the phosphorylation of STAT3 and STAT5b but not STAT5a. Moreover, enhanced proliferation of heparanase transfected cells was attenuated by STAT3 and STAT5b siRNA, but not STAT5a or STAT1 siRNA. Clinically, STAT3 phosphorylation was associated with head and neck cancer progression, EGFR phosphorylation, and heparanase expression and cellular localization. Notably, cytoplasmic rather than nuclear phospho-STAT3 correlated with increased tumor size (T-stage; p = 0.007), number of metastatic neck lymph nodes (p = 0.05), and reduced survival of patients (p = 0.04).
Introduction
Heparanase is an endo-β-d-glucuronidase capable of cleaving heparan sulfate side chains of heparan sulfate proteoglycans at a limited number of sites (1, 2). Heparanase activity correlated with the metastatic potential of tumor-derived cells, attributed to enhanced cell dissemination as a consequence of heparan sulfate cleavage and remodeling of the extracellular matrix barrier (1, 2). Similarly, heparanase activity was implicated in neovascularization, inflammation, and autoimmunity, involving migration of vascular endothelial cells and activated cells of the immune system (1–3). Evidence has shown that heparanase is up-regulated in various primary solid tumors (i.e. carcinomas and sarcomas) and hematological malignancies (4–7). Heparanase up-regulation correlated with increased lymph node and distant metastasis, increased microvessel density, and reduced post-operation survival of cancer patients, thus providing a strong clinical support for the prometastatic and proangiogenic features of the enzyme and encouraging the development of heparanase inhibitors (8–12). In addition, heparanase up-regulation in primary human tumors correlated in some cases with tumors bigger in size (4). Likewise, heparanase overexpression enhanced (13, 14), whereas local delivery of anti-heparanase siRNA inhibited (14) the progression of tumor xenografts. These results imply that heparanase function is not limited to tumor metastasis but is engaged in the progression of primary lesions. The cellular and molecular mechanisms underlying these aspects of heparanase function are not entirely clear but likely involve proangiogenic features (4, 15). In addition, results obtained in recent years indicate that heparanase facilitates the phosphorylation and activity of selected signaling molecules and induces transcription of proangiogenic (i.e. VEGF-A, VEGF-C, COX-2), prothrombotic (i.e. tissue factor), mitogenic (hepatocyte growth factor), and osteolyic (RANKL) genes (4, 13, 15–20). Signaling function requires heparanase secretion but not enzymatic activity and appears to be mediated by its C-terminal domain (21–24).
We have reported previously that heparanase enhances the phosphorylation of EGFR3 in an SRC-dependent manner, leading to increased cell proliferation and colony formation in soft agar (21). Because, in this system, ERK phosphorylation did not appear to be affected by heparanase (23, 25), we hypothesized that STAT proteins mediate the proliferative effect downstream EGFR. We provide evidence that heparanase enhances the phosphorylation of STAT3 and STAT5b but not STAT5a. Enhanced STAT5b phosphorylation by heparanase was attenuated by PP2 and CL-387785 or tyrphostin AG1478 (selective inhibitors of SRC and EGFR, respectively) but not PD98059, a MEK inhibitor. Moreover, enhanced proliferation of heparanase transfected cells was attenuated by STAT3 and STAT5b siRNA but not STAT5a or STAT1 siRNA. Clinically, STAT3 phosphorylation was associated with head and neck cancer progression and with EGFR phosphorylation and heparanase levels. Notably, cytoplasmic rather than nuclear phospho-STAT3 correlated with increased tumor size (T-stage; p = 0.007), number of metastatic neck lymph nodes (p = 0.05), and reduced the survival of patients (p = 0.04).
MATERIALS AND METHODS
Antibodies and Reagents
The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): anti-lamin A/C (sc-7292), anti-SRC (sc-18 and sc-19), anti-phosphotyrosine (sc-7020), anti-AKT (sc-5298), anti-EGFR (sc-03), anti-pEGFR (Tyr1173, sc-12351R), anti-STAT3 (sc-7179), anti-phospho-STAT3 (Tyr705; sc-8059), anti-STAT5a (sc-1081), anti-STAT5b (sc-1656), anti-phospho-ERK (sc-7383), and anti-ERK2 (sc-154). Polyclonal antibodies to phospho-SRC (Tyr416) and phospho-AKT (Ser473) were purchased from Cell Signaling (Beverly, MA). Anti-actin antibody was purchased from Sigma. Anti-heparanase polyclonal antibody (no. 733) has been described previously (21). Bromodeoxyuridine (BrdU) was purchased from GE Healthcare, and anti-BrdU monoclonal antibody-HRP conjugated was purchased from Roche Applied Science. The selective PI3K (LY 294002), MAPK (PD 98059), SRC (PP2), and EGFR (AG1478; CL-387785) inhibitors were purchased from Calbiochem and were dissolved in dimethyl sulfoxide as stock solutions. Dimethyl sulfoxide was added to the cell culture as control.
Cell Culture and Transfection
Mouse embryonic fibroblasts have been described previously (26). FaDu pharynx carcinoma cells were kindly provided by Dr. Eben L. Rosenthal (University of Alabama at Birmingham, Birmingham, AL) (27), SQ-20B laryngeal carcinoma and JSQ3 nasal vestibule carcinoma cells were kindly provided by Dr. Ralph Weichselbaum (University of Chicago, Chicago, IL) (28), and CAG myeloma cells were kindly provided by Dr. Ben-Zion Katz (Tel Aviv Sourasky Medical Center, Tel Aviv, Israel) (29). Human LNCaP prostate carcinoma, U87 glioma, Cal27 tongue carcinoma, and T47D breast carcinoma cells were purchased from the ATCC. Cells were cultured in DMEM supplemented with glutamine, pyruvate, antibiotics, and 10% fetal calf serum in a humidified atmosphere containing 5% CO2 at 37 °C. For stable transfection, cells were transfected with heparanase gene constructs using the FuGENE reagent according to the manufacturer's instructions (Roche Applied Science), selected with Zeocin (Invitrogen) for 2 weeks, expanded, and pooled, as described (17, 21). Cells were passed in culture no more that 3 months after being thawed from authentic stocks.
Cell Fractionation, Immunoprecipitation, and Protein Blotting
Cell fractionation was carried out utilizing NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer's instructions (Pierce). Preparation of cell lysates, immunoprecipitation, and immunoblotting were performed essentially as described (17, 21).
Cell Proliferation
For growth curves, cells (5 × 104) were seeded into 6-cm culture dishes in duplicate. Cells were dissociated with trypsin/EDTA and counted every other day using a Coulter Counter, and cell numbers were further confirmed by counting with a hemacytometer. Additionally, cell proliferation was analyzed by BrdU incorporation using cell proliferation labeling reagent (1:1000, GE Healthcare) as described (21). At least 1000 cells were counted for each cell type.
Gene Silencing and PCR Analysis
Transfection and analysis of cells following siRNA transfection were carried out essentially as described (21). Anti-heparanase, anti-STAT3, anti-STAT5a, anti-STAT5b, anti-STAT1, and control anti-GFP siRNA oligonucleotides (siGENOME ON-TARGETplus SMARTpool duplex) were purchased from Dharmacon (Thermo Fisher Scientific) and transfection was carried out with DharmaFECT3 reagent, according to the manufacturer's instructions (Dharmacon). Total RNA was extracted with TRIzol (Invitrogen), and RNA (1 μg) was amplified using a one-step PCR amplification kit, according to the manufacturer's instructions (ABgene, Epsom, UK). The PCR primer sets were as follows: heparanase F, 5′-AGGTCTGCATATGGAGGCGG-3′ and heparanase R, 5′-TGAACTTCCTGGCCGGAGAG-3′; and GAPDH F, 5′ CCAGCCGAGCCACATCGCTC-3′ and GAPDH R, 5′ATGAGCCCCAGCCTTCTCCAT-3′.
Immunocytochemistry
Immunofluorescent staining was performed essentially as described (21, 30, 31). Staining was observed under a fluorescent confocal microscope.
Immunostaining
Staining of formalin-fixed, paraffin-embedded 5-micron sections was performed essentially as described (17, 32, 33). Briefly, slides were deparaffinized and rehydrated, and endogenous peroxidase activity was quenched (30 min) by 3% hydrogen peroxide in methanol. Slides were then subjected to antigen retrieval by boiling (20 min) in 10 mm citrate buffer, pH 6.0. Following washes with PBS, slides were incubated with 10% normal goat serum in PBS for 60 min to block nonspecific binding and incubated (20 h, 4 °C) with anti-phospho-STAT3, diluted 1:100 in blocking solution. Slides were extensively washed with PBS containing 0.01% Triton X-100 and incubated with a secondary reagent (Envision kit) according to the manufacturer's instructions (Dako). Following additional washes, color was developed with the AEC reagent (Dako), sections were counterstained with hematoxylin and mounted as described (17, 21). Immunostained specimens were examined by a senior pathologist (I. Naroditsky) who was blind to clinical data of the patients. Staining was scored according to the intensity of staining (0, none; +1, weak to moderate; +2, strong), and the percentage (extent staining) of tumor cells that were stained. The extent of staining was further categorized as low (0, <10%), moderate (+1, 10–50%), and high (+2, >50% of the cells). Staining localization was categorized as cytoplasmic or nuclear. Specimens that were similarly stained with preimmune serum, or applying the above procedure but lacking the primary antibody, yielded no detectable staining.
DNA Binding Assay
Binding of STAT5 to β-casein promoter sequences was carried out essentially as described (34). Briefly, nuclear extracts of control (Vo) and heparanase-transfected cells were incubated (18 h, 4 °C) with double-stranded biotinylated oligonucleotides containing the STAT5 binding site of bovine β-casein promoter (5′-GATTTCTAGGAATTCAA-3′). Strepavidin-agarose beads (Sigma) were then added, and after washing, bound material was subjected to immunoblotting as described above.
Statistics
Univariate association between STAT parameters (intensity, extent, and localization of staining) and between clinical parameters, pathological parameters, and the outcome of patients, as well as heparanase and EGFR staining intensity, extent, and localization, were analyzed using Chi square tests (Pearson, Fisher's exact test) or analysis of variance. Univariate and multivariate ordinal logistic fits were performed to detect independent parameters that affect disease stage (21). All experiments were repeated at least three times with similar results.
RESULTS
Heparanase Enhances STAT Protein Phosphorylation
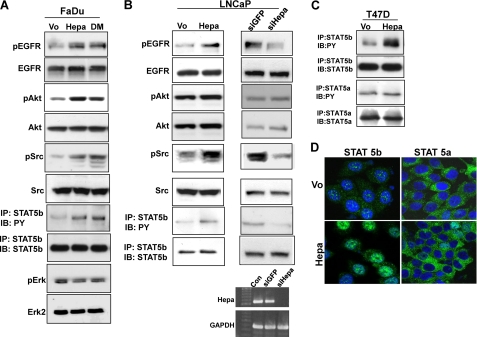
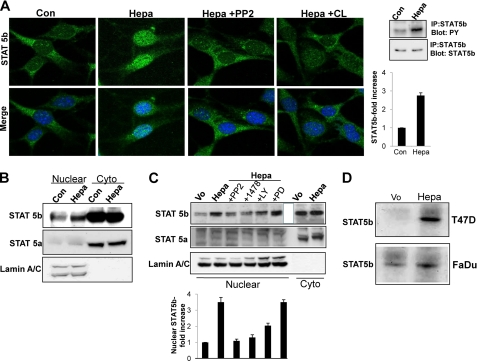
Overexpression of wild type (Hepa) or inactive, double-mutated (Glu225 and Glu343) (21) heparanase gene constructs in FaDu pharyngeal carcinoma cells resulted in a marked increase in the phosphorylation levels of EGFR (pEGFR; Fig. 1A, upper panel), AKT (pAKT, Fig. 1A, third panel), and SRC (pSrc; Fig. 1A, fifth panel) (supplemental Fig. 1, A–C). Similarly, heparanase overexpression enhanced, whereas heparanase gene silencing (Fig. 1B, right lower panels) attenuated the phosphorylation of EGFR (Fig. 1B, upper panels) and SRC (Fig. 1B, fifth panels) in LNCaP prostate carcinoma cells (supplemental Fig. 1, E, F, H, and I). In contrast with FaDu cells, however, AKT phosphorylation was not significantly affected by heparanase in LNCaP cells (Fig. 1B, third panels; supplemental Fig. 1, G and J). Because ERK phosphorylation does not appear to be regulated by heparanase (Fig. 1A, lower panels; supplemental Fig. 1D) (23, 25), we hypothesized that STAT proteins mediate the proliferative effect of heparanase noted in LNCaP cells (21). Of the seven mammalian STAT proteins, STAT3 and STAT5 are often associated with neoplasia, are found downstream the EGFR signaling pathway, and can also be phosphorylated directly by SRC (35–37). Indeed, STAT5b phosphorylation was increased nearly 3-fold in heparanase-transfected FaDu (Fig. 1A, seventh panel; supplemental Fig. 2A) and LNCaP (Fig. 1B, left, seventh panel; supplemental Fig. 2B) cells, whereas heparanase gene silencing resulted in a comparable, 3-fold decrease in STAT5b phosphorylation (Fig. 1B, right, seventh panel; supplemental Fig. 2C). A similar increase in STAT5b phosphorylation was observed in T47D breast carcinoma cells overexpressing heparanase (Fig. 1C; supplemental Fig. 2D), accompanied by a marked increase in nuclear STAT5b (Fig. 1D, left panels). In contrast, the phosphorylation status (Fig. 1C, third panel; supplemental Fig. 2E) and cellular localization (Fig. 1D, right panels) of STAT5a was not altered by heparanase, indicating a specific effect on STAT5b. Accordingly, exogenous addition of recombinant heparanase to mouse embryonic fibroblasts resulted in efficient phosphorylation (Fig. 2A, right most panels) and translocation of STAT5b to the cell nucleus (Fig. 2A, Hepa). Enhanced nuclear translocation was similarly observed in LNCaP prostate carcinoma and CAG myeloma cells overexpressing heparanase (supplemental Fig. 3A, left and right panels, respectively). Enhanced nuclear translocation of STAT5b was further confirmed biochemically, clearly revealing increased amounts of STAT5b (Fig. 2B, upper panel) but not STAT5a (Fig. 2B, second panel) in the cell nuclei following exogenous addition of latent heparanase. Notably, nuclear translocation following addition of heparanase was markedly attenuated by PP2 and CL-387785, selective inhibitors of SRC and EGFR, respectively. This was observed by immunofluorescent staining (Fig. 2A) and biochemically, subjecting nuclear extracts to immunoblotting applying anti-STAT5b (Fig. 2C, upper and lower panels) antibodies. These results indicate that enhanced STAT5b protein phosphorylation and nuclear translocation by heparanase are mediated by SRC and EGFR, whereas MAPK does not seem to be involved.
FIGURE 1.
Heparanase enhances STAT5b phosphorylation. FaDu pharynx carcinoma (A) and LNCaP prostate carcinoma (B, left panels) cells were stably transfected with control empty vector (Vo), wild type (Hepa), or mutated inactive (double mutated (DM)) heparanase gene constructs and cell lysates were subjected to immunoblotting applying anti-pEGFR (upper panels), anti-EGFR (second panels), anti-pAKT (third panels), anti-AKT (fourth panels), anti-pSrc (fifth panels), anti-Src (sixth panels), anti-pErk (ninth panel), and anti-Erk 2 (tenth panel) antibodies. Corresponding lysates were subjected to immunoprecipitation (IP) with anti-STAT5b antibody, followed by immunoblotting (IB) with anti-phosphotyrosine (PY, seventh panels) or anti STAT5b (eighth panels) antibodies. LNCaP prostate carcinoma cells were left untreated (Con) or were transfected with anti-GFP or anti-heparanase (siHepa) siRNA oligonucleotides. Total RNA was extracted 2 days following transfection, and heparanase expression was examined by RT-PCR analysis. GAPDH transcript was used as an internal control for RNA loading (B, right lower panels). Cell lysates were prepared from corresponding LNCaP cells and subjected to immunoblotting applying the antibodies described above (B, right panels). C, T47D breast carcinoma cells were stably transfected with heparanase gene construct (Hepa) or an empty vector (Vo), and cell lysates were immunoprecipitated with antibodies directed against STAT5b (upper panel) or STAT5a (third panel) followed by immunoblotting with anti-phosphotyrosine antibody (PY). Membranes were then striped and reprobed with anti-STAT5b (second panel) or anti-STAT5a (fourth panel) antibodies. D, immunofluorescent staining. Control (Vo, upper panels) and heparanase-transfected (Hepa, lower panels) T47D cells were stained with anti-STAT5b (left panels) or anti-STAT5a (right panels) antibodies (green). Nuclei counterstaining (DAPI) appears in blue. Note the nuclear translocation of STAT5b but not STAT5a in heparanase overexpressing cells. Original magnification, ×63.
FIGURE 2.
STAT5b phosphorylation and nuclear translocation by heparanase is mediated by SRC and EGFR. A, inhibition study. Mouse embryonic fibroblasts were left untreated (Con) or were incubated with heparanase without (Hepa) or after the cells were preincubated with PP2 (5 μm) or CL-387785 (CL; 0.1 μm) for 30 min. Following a 1-h incubation with heparanase, cells were fixed and stained with anti-STAT5b antibody (upper panels). Merge images with DAPI counter staining (blue) are shown in the lower panels. Original magnification, ×63. Corresponding control (Vo) and heparanase-transfected cell lysates were immunoprecipitated (IP) with anti-STAT5b antibody followed by immunoblotting with anti-phosphotyrosine (PY) or anti STAT5b antibodies (rightmost panels). Densitometry analysis of pSTAT5b is shown in the right lower panel. B, control (Con) and heparanase-treated (Hepa) cells from corresponding cultures were fractionated into nuclear and cytoplasmic (Cyto) fractions and subjected to immunoblotting applying antibodies directed against STAT5b (upper panel), STAT5a (middle panel), and lamin A/C to validate fraction purity and protein loading (lower panel). C, heparanase-transfected FaDu cells were left untreated (Hepa) or incubated (4 h) with SRC (PP2, 5 μm), EGFR (1478, 5 μm), PI3K (LY, 15 μm), or MEK (PD, 10 μm) inhibitors. Nuclear fractions were then prepared and subjected to immunoblotting applying anti-STAT5b (upper panel), anti-STAT5a (second panel) and anti-lamin A/C (third panel) antibodies. Nuclear fraction of control (Vo) cells and cytoplasmic fractions (Cyto) of control (Vo) and heparanase-transfected (Hepa) cells were included as control. Densitometry analysis of nuclear STAT5b levels is shown in the lower panel. D, DNA binding. Nuclear extracts of control (Vo) and heparanase-transfected (Hepa) T47D (upper panel) and FaDu (second panel) cells were incubated (20 h, 4 °C) with biotinylated oligonucleotides containing the STAT5 binding site of the bovine β-casein promoter. Strepavidin-agarose beads were then added for 60 min, and, after washing, agarose-bound material was subjected to immunoblotting with anti-STAT5b antibody. Note the increased STAT5b association with the casein gene promoter following heparanase overexpression.
To confirm that increased phosphorylation (Fig. 1) and nuclear translocation (Figs. 1D and 2, A–C) of STAT5b is associated with enhanced gene transcription, nuclear extracts of control (Vo) and heparanase-transfected T47D (Hepa; Fig. 2D, upper panel) and FaDu (Fig. 2D, lower panel) cells were incubated with biotinylated oligonucleotides corresponding to the β-casein promoter (34) that is regulated by STAT5 (38). Association of STAT5b with the β-casein promoter was markedly increased in heparanase overexpressing cells (Fig. 2D and supplemental Fig. 2, F and G), indicating that heparanase facilitates the transcriptional activity of selected STAT proteins.
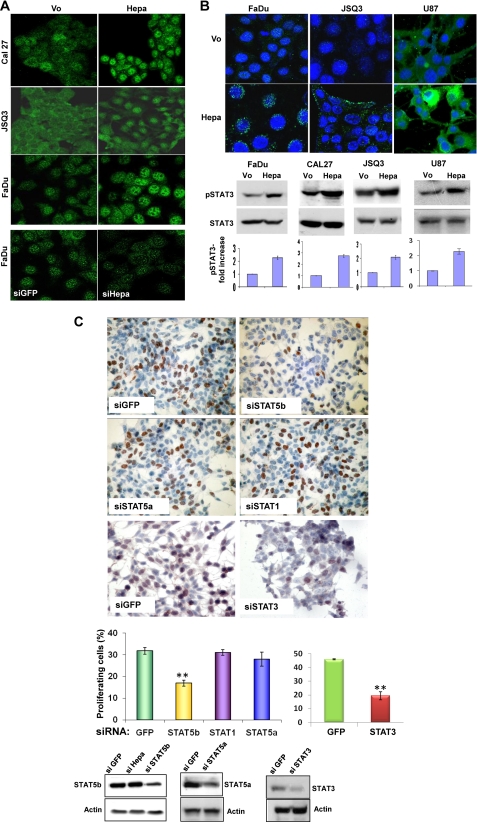
Immunofluorescent staining further revealed increased nuclear translocation of STAT5b in heparanase overexpressing Cal27 tongue carcinoma, JSQ3 nasal vestibule carcinoma, and FaDu cells (Fig. 3A, upper panels, respectively), whereas heparanase gene silencing results in reduced levels of nuclear STAT5b (Fig. 3A, lower panels), indicating that endogenous heparanase modulates STAT5b localization. Notably, not only STAT5b but also STAT3 phosphorylation is increased following heparanase overexpression in FaDu (Fig. 3B, lower left panel), Cal27 (Fig. 3B, lower second lefthand panel), JSQ3 (Fig. 3B, lower second righthand panel) and U87 glioma (Fig. 3B, right panels) cells. In FaDu cells as well as in JSQ3 and U87 cells, phospho-STAT3 evidently was not translocated to the cell nucleus but rather resided perinuclearly (FaDu; Fig. 3B, upper left panel), at the cell periphery (JSQ3; Fig. 3B, upper middle panels), possibly at sites of focal adhesions (39), or diffusely distributed in the cell cytoplasm (U87; upper right panels).
FIGURE 3.
Heparanase modulates STAT5b and STAT3 phosphorylation. A, heparanase overexpression and gene silencing. Cal27 (upper panels), JSQ3 (second panels), and FaDu cells (third panels) were transfected with an empty vector (Vo) or heparanase gene construct (Hepa), and stably transfected cells were subjected to immunofluorescent staining with anti-STAT5b antibody. FaDu cells were transfected similarly with anti-GFP (siGFP) or anti-heparanase (siHepa) siRNA oligonucleotides. Three days thereafter, cells were subjected to immunofluorescent staining with anti-STAT5b antibody (lower panels). Note increased nuclear STAT5b in heparanase-overexpressing cells and decreased nuclear STAT5b following heparanase gene silencing. Original magnification, ×40. B, STAT3 phosphorylation. FaDu (lower left panels), Cal27 (second left panel), JSQ3 (second right panel), and U87 (lower right panels) cells were transfected with empty control (Vo) or heparanase expression vector (Hepa), and lysate samples were subjected to immunoblotting applying anti-pSTAT3 or anti-STAT3 antibodies. STAT3 phosphorylation index calculated by densitometry analysis (Vo cells arbitrary set to a value of 1) of at least five independent experiments is shown in the bottom panels. Control (Vo) and heparanase-transfected (Hepa) FaDu (upper left panel), JSQ3 (middle panels), and U87 (upper right panels) cells were also subjected to immunofluorescent staining applying anti-pSTAT3 antibody (green); nuclei counterstaining (DAPI) is shown in blue. Original magnification, ×63. C, BrdU incorporation. Subconfluent heparanase-transfected LNCaP cells were transfected with control (siGFP), anti-STAT5b, anti-STAT5a, anti-STAT1, or anti-STAT3 siRNA oligonucleotides. After recovery for 2 days in growth medium, cells were kept in serum-free medium for 20 h followed by incubation with BrdU (1:1000) for 2 h. Cells were then fixed and immunostained with anti BrdU monoclonal antibodies. Positively stained, red-brown nuclei were counted versus blue, hematoxylin counterstained nuclei (upper panel). At least 1000 cells were counted for each cell type, and the percentage of positively stained cells is noted in each bar. Decreased STAT3, STAT5b and STAT5a levels following siRNA transfection are shown in the lower panels.
To reveal whether STAT protein mediates the proliferative function of heparanase, LNCaP cells were transfected with siRNAs directed against STAT, and cell proliferation was evaluated by BrdU incorporation. Anti-STAT5b siRNA resulted in a significant decrease in STAT5b levels (Fig. 3C, lower left panel) and a 2-fold decrease in BrdU incorporation (Fig. 3C, siSTAT5b; p = 0.003). Likewise, a 2-fold decrease in BrdU incorporation was noted following STAT3 gene silencing (Fig. 3C, siSTAT3). In contrast, BrdU incorporation was not significantly altered by siRNAs directed against STAT5a or STAT1 (Fig. 3C, siSTAT5a, STAT1), suggesting that STAT3 and STAT5b mediate cell proliferation elicited by heparanase in LNCaP cells.
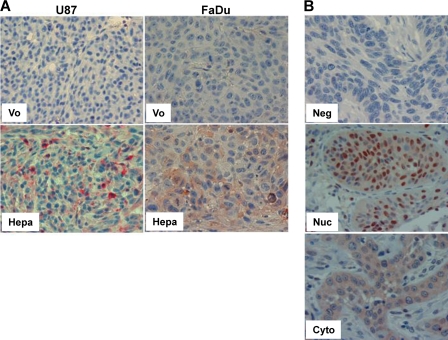
We next applied immunohistochemistry to reveal STAT phosphorylation levels in the course of tumor xenograft development. We could not obtain satisfactory staining for pSTAT5 applying polyclonal and monoclonal antibodies from various vendors. Anti-pSTAT3 antibody nonetheless yielded reproducible specific staining. STAT3 phosphorylation was markedly increased in tumor xenografts produced by heparanase-transfected U87 glioma cells (Fig. 4A, left panels), a model system in which heparanase overexpression resulted in significant increase in tumor development (22) and in line with the protumorigenic properties of STAT3 in glioma (40). A similar increase in pSTAT3 was evident in tumor xenografts produced by FaDu cells overexpressing heparanase (Fig. 4A, right panels). In these tumor models, phospho-STAT3 was not translocated to the cell nuclei but was localized in the cytoplasm, in agreement with the in vitro phenotype (Fig. 3B). Only modest increases in pSTAT3 levels were observed following heparanase overexpression by CAG myeloma cells (supplemental Fig. 3B). In contrast, STAT3 phosphorylation was markedly augmented in endothelial cells lining blood vessels in xenografts produced by heparanase overexpressing CAG cells compared with control mock-transfected cells (supplemental Fig. 3B), altogether suggesting that, depending on the cell type, heparanase may activate STAT3 in cells of the tumor and/or its microenvironment.
FIGURE 4.
Immunohistochemistry analysis of pSTAT3. A, tumor xenografts. Formalin-fixed, paraffin-embedded 5-micron sections of xenografts produced by control (Vo) and heparanase-transfected U87 (left panels) and FaDu (right panels) cells were stained with anti-pSTAT3 antibody. Note increased reactivity in heparanase-transfected cells, assuming cytoplasmic localization. B, tumor biopsies. Formalin-fixed, paraffin-embedded 5-micron sections of head and neck tumor biopsies were subjected to immunostaining applying anti-pSTAT3 antibody, as described under “Materials and Methods.” Shown are representative photomicrographs of pSTAT3-negative (Neg; upper panel) and positively stained specimens exhibiting prevalent nuclear (Nuc; middle panel) or cytoplasmic (Cyto; lower panel) localization. Original magnification, ×40.
Clinical Significance of Phospho-STAT3 in Head and Neck Squamous Cell Carcinoma
We applied immunostaining to reveal pSTAT3 levels in biopsies of head and neck squamous cell carcinoma (HNSCC), a cohort utilized previously in a number of studies (15, 17, 21, 32). Clinical description of the patients included in the study is summarized in Table 1. Positive pSTAT3 staining was found in 94% (81/86) of the tumor specimens, whereas 6% (5/86) of the specimens were found negative for pSTAT3 (Fig. 4B, upper panel). In most of the specimens (83%; 67/81), pSTAT3 staining localized preferentially to the cell nuclei (Fig. 4B, middle panel). In the other cases (27%; 14/81), pSTAT3 was predominantly localized in the cell cytoplasm (Fig. 4B, lower panel). We analyzed the clinical significance of nuclear versus cytoplasmic pSTAT3, omitting the small number of cases (5) found negative for pSTAT3. Importantly, cytoplasmic pSTAT3 was associated with tumor larger in size (T-stage). Hence, all 14 cases with cytoplasmic pSTAT3 were diagnosed with large tumors (T-stage 3–4) compared with 64% of such tumors where pSTAT3 assumed nuclear localization (p = 0.007; Table 2, “T-Stage”). One of the most crucial prognostic factors for the survival of patients with HNSCC is tumor metastasis to neck lymph nodes. Notably, the number of metastatic lymph nodes found in the pathological specimens of neck dissections was twice as much in head and neck cancer patients having cytoplasmic versus nuclear pSTAT3 (3 versus 1.56 for cytoplasmic and nuclear pSTAT3, respectively; supplemental Fig. 3C; p = 0.05). Accordingly, the status of patients was worse when pSTAT3 was localized to the cytoplasm: 79% of the patients with cytoplasmic pSTAT3 died by the end of the study, compared with 46% of the patients exhibiting nuclear pSTAT3 (p = 0.04; Table 2, “Status”).
TABLE 1.
Clinical description of the patients included in this study
| Site of tumor | No. of patients | Percent |
|---|---|---|
| Larynx | 65 | 76 |
| Pharynx | 9 | 10 |
| Oropharynx | 5 | |
| Hypopharynx | 4 | |
| Skin (metastatic) | 5 | 6 |
| Oral cavity | 3 | 3 |
| Unknown origin | 2 | 2 |
| Other | 2 | 2 |
| T-stage | ||
| T0–2 | 25 | 29 |
| T3 | 35 | 40 |
| T4 | 26 | 30 |
| N-stage | ||
| N0 | 45 | 52 |
| N1 | 13 | 15 |
| N2–3 | 28 | 33 |
TABLE 2.
Cytoplasmic pSTAT3 associates with tumor size (T-stage), patient status, EGFR phosphorylation, heparanase expression, and cellular localization
Values designates the number of patients, and values in parentheses designate the percent of the total in that group.
| pSTAT3 localization | Cytoplasmic pSTAT3 | Nuclear pSTAT3 | Total |
|---|---|---|---|
| T-Stage (p = 0.007) | |||
| 0–2 | 0 | 24 (36) | 24 |
| 3–4 | 14 (100) | 43 (64) | 57 |
| Total | 14 | 67 | 81 |
| Status (p = 0.04) | |||
| Dead | 11 (79) | 31 (46) | 42 |
| Alive | 3 (21) | 36 (54) | 39 |
| Total | 14 | 67 | 81 |
| pEGFR (p = 0.006) | |||
| Negative | 0 | 18 (37) | 18 |
| Positive | 14 (100) | 30 (63) | 44 |
| Total | 14 | 48 | 62a |
| Heparanase extent (p = 0.03) | |||
| Low | 1 (7) | 21 (36) | 22 |
| High | 13 (93) | 37 (64) | 50 |
| Total | 14 | 58 | 72b |
| Heparanase localization (p = 0.03) | |||
| Cytoplasmic | 10 (71) | 21 (36) | 31 |
| Nuclear | 4 (29) | 37 (64) | 41 |
| Total | 14 | 58 | 72b |
a Data of 19 patients were missing.
b Data of nine patients were missing.
We have reported previously that in this cohort of patients, staining for pEGFR associates with tumor size (21). Notably, pEGFR levels were associated with pSTAT3 localization (Table 2, “pEGFR”). Thus, all cases (100%) exhibiting cytoplasmic localization of pSTAT3 were also positive for pEGFR compared with 63% of the specimens positive for pEGFR and assuming nuclear localization of pSTAT3, differences that are statistically highly significance (p = 0.006). Even a higher significance of association is obtained when pEGFR staining extent is analyzed in comparison with pSTAT3 cellular localization (supplemental Fig. 3D). In this analysis, mean pEGFR staining extent was 58.9% ± 7.7 in specimens harboring cytoplasmic pSTAT3 compared with 27.5% ± 4.1 in specimens where pSTAT3 assumed nuclear localization (p = 0.0007). These results strongly imply that pSTAT3 is found downstream EGFR, associating with disease progression in HNSCC. Most importantly, heparanase staining extent was associated with pSTAT3 localization (Table 2, “Heparanase extent”). Thus, 93% of the specimens that exhibit cytoplasmic localization of pSTAT3 displayed high levels of heparanase staining extent compared with 64% of the cases in which pSTAT3 assumed nuclear localization (p = 0.03). Furthermore, pSTAT3 localization was associated with heparanase localization (Table 2, “Heparanase localization”). Remarkably, nuclear heparanase was associated with nuclear pSTAT3, both of which predict favorable outcome (32, 41), whereas cytoplasmic heparanase, which correlates with increased EGFR phosphorylation (21), was linked with cytoplasmic pSTAT3 (p = 0.03) and enhanced tumor progression (Table 2, “T-stage” and “Status”).
Ordinal logistic fit subsequently was performed to identify parameters associating with HNSCC tumor size (T-stage). In univariate analysis, both heparanase and pSTAT3 localization were found significant (p = 0.01 and 0.0009 for heparanase and pSTAT3, respectively). However, when using multivariate logistic fit that included both parameters, pSTAT3 localization remained significant (p = 0.004), whereas heparanase became insignificant (p = 0.11), indicating that the two parameters are interconnected. Thus, the protumorigenic function of heparanase in HNSCC (32) likely involves STAT3 phosphorylation.
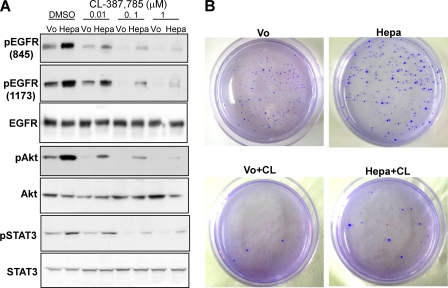
To further reveal the significance of heparanase in HNSCC, we next examined whether cells overexpressing heparanase acquire resistance toward EGFR inhibitors. For this purpose, we subjected FaDu pharynx carcinoma cells overexpressing heparanase to increasing concentrations of CL-387785, an irreversible EGFR inhibitor, compared with control cells harboring an empty vector (Vo). Heparanase overexpression resulted in a marked increase in the phosphorylation levels of EGFR (Fig. 5A, upper and second panels, dimethyl sulfoxide), AKT (fourth row), and STAT3 (sixth row), in agreement with previous results (Fig. 1). Phosphorylation of EGFR, AKT, and STAT3 was markedly attenuated in cells treated with CL-387785 in a dose-dependent manner (Fig. 5A), as would be expected. Notably, cells overexpressing heparanase were significantly less sensitive to CL-387785 treatment and phosphorylation of all signaling molecules was still evident even at the highest concentration of the drug (1 μm; Fig. 5A, Hepa; supplemental Fig. 4). The consequence of this increase in signaling is translated to colony formation. Hence, cells overexpressing heparanase produced more and larger colonies in soft agar than control cells in the presence of CL-387785 (Fig. 5B, Hepa+CL). These results suggest that heparanase levels not only associates with the outcome of patients (32) but might also influence the response to treatment.
FIGURE 5.
Cells overexpressing heparanase are less sensitive to EGFR inhibitor. A, control (Vo) and heparanase-transfected (Hepa) FaDu cells were incubated with the indicated concentration (μm) of CL-387785 for 2 h. Vehicle (dimethyl sulfoxide (DMSO)) was used as control. Total cell lysates were subjected to immunoblotting with antibodies directed against the phosphorylated state of EGFR (pEGFR, upper and second panels), AKT (pAKT, fourth panel), and STAT3 (pSTAT3, sixth panel). Total amounts of EGFR, AKT, and STAT3 are shown in the third, fifth, and seventh panels, respectively. Note the sustained EGFR signaling in heparanase overexpressing cells even in the presence of high doses (1 μm) of CL-387785. B, colony formation. Control (Vo) and heparanase-transfected (Hepa) FaDu cells (5 × 103) were mixed with soft agar and cultured for 3 weeks in the absence or presence of EGFR inhibitor, CL-387785 (CL, 0.1 μmol/liter). Shown are photomicrographs of colonies at low (×10) magnification. Note more and larger colonies produced by heparanase overexpressing cells following EGFR inhibition compared with control cells.
DISCUSSION
Approximately 45,000 new cases of head and neck cancer are diagnosed in the United States each year, and the estimated worldwide incidence is 500,000 (42). Despite of the introduction of novel therapeutics and improved techniques in surgery, radiation, and chemotherapy, locoregional and distant recurrence remain common and is almost always fatal. Development of new therapeutics is thus needed urgently.
We have reported previously that heparanase expression is induced in HNSCC and is associated with tumors larger in size, increased invasiveness (extracapsular extension), and reduced the survival of patients (32). In addition, we have found that heparanase facilitates the phosphorylation of SRC, leading to augmented EGFR phosphorylation, enhanced cell proliferation, and larger colonies in soft agar (21). Clinically, increased EGFR phosphorylation was associated with larger tumors and with heparanase expression and cytoplasmic localization in HNSCC (21), yet molecular determinants that mediate heparanase function downstream to EGFR have not been so far characterized. The results presented in this study suggest a linear association by which heparanase, when localized in the cell cytoplasm, facilitates the phosphorylation of SRC and EGFR which, in turn, facilitates STAT3 phosphorylation, leading to cancer progression. Nuclear heparanase, in contrast, does not activate EGFR or STAT3 and predicts favorable outcome of head and neck cancer patients (21).
Although expression and phosphorylation of STAT3 are elevated in HNSCC as compared with normal epithelial cells, and its activation is thought to represent an early event in head and neck carcinogenesis (43–46), the prognostic value of STAT3 is not entirely clear. Seethala et al. (47) reported that pSTAT3 levels did not correlate with clinical outcome in two cohorts of head and neck cancer patients. Likewise, we could not find significant correlations between the staining intensity or extent of nuclear phospho-STAT3 and clinical (i.e. T-stage, N-stage) or molecular (i.e. heparanase, EGFR) parameters (data not shown). Utilizing tissue micro array and automated quantitative analysis, Pectasides et al. (41) recently reported that high levels of nuclear STAT3 are associated with a favorable outcome, predicting a lower risk of progression and death of head and neck cancer patients. Notably, we found that cytoplasmic pSTAT3 (Fig. 4B, lower panel) is associated with tumors larger in size (Table 2, “T-stage”), increased number of metastatic lymph nodes (supplemental Fig. 3C), and reduced survival of patients (Table 2, “Status”) compared with nuclear STAT3. To the best of our knowledge, this is the first systematic evaluation of the clinical significance of cytoplasmic versus nuclear pSTAT3. Cytoplasmic localization of pSTAT3 may therefore be considered a valuable diagnostic parameter in HNSCC.
Previous reports have shown that pSTAT3 is localized not only in cell nuclei but also in cytoplasmic protrusions and focal adhesions of migrating cells and in endocytic vesicles (i.e. endosomes) (48, 49), in a SRC-dependent manner (39). Indeed, immunofluorescent staining clearly revealed increased pSTAT3 levels in heparanase overexpressing cells, which was localized in the cell periphery (JSQ3; Fig. 3B) and cytoplasmic vesicles residing perinuclearly (FaDu; Fig. 3B), indicating that pSTAT3 is retained in the cytoplasm of some head and neck cancer cell lines. Retention of pSTAT3 in the cytoplasm, possibly due to impaired interaction with importins (36), may sequester pSTAT3 from the nucleus, preventing the induction of proapoptotic/differentiation genes (41), or governing transcription-independent function of pSTAT3 leading to head and neck tumor development (48, 49). Mechanisms(s) underlying protumorigenic function of cytoplasmic pSTAT3 are yet to be defined. Increased STAT3 expression and activation has been associated with amplified EGFR signaling, which occurs in most epithelial malignancies including HNSCC, where anti-EGFR therapeutics are implemented clinically (i.e. Erbitux) or are being evaluated in phase III clinical trials (43). STAT3 activation also occurs through SRC, an event considered sufficient to induce cell transformation (50). Although we were unable to obtain reproducible staining for pSrc, staining for pEGFR was found to be tightly associated with head and neck tumor progression (21) and cytoplasmic localization of pSTAT3 (Table 2, “pEGFR”; supplemental Fig. 3D), further strengthening the validity and significance of the obtained results.
A key occurrence in the above signaling events is induction of heparanase and its cellular localization. We have reported previously that heparanase expression is increased in the majority (86%) of head and neck cancer patients and, when localized in the cytoplasm, correlates with poor prognosis (32) and increased EGFR phosphorylation (21). The current study further links between cytoplasmic heparanase and cytoplasmic pSTAT3, first by showing that 93% of the cases in which pSTAT3 assumes cytoplasmic localization also exhibit high extent of heparanase staining (Table 2, “Heparanase extent”), and second, by demonstrating that heparanase and pSTAT3 coincide in the cytoplasm in 71% of the cases (Table 2, “Heparanase localization”), and last, by double staining demonstrating that heparanase and pSTAT3 co-localize in the cytoplasm of the very same cells (data not shown). In tumor models, pSTAT3 staining was increased in xenografts produced by U87 glioma and FaDu cells overexpressing heparanase (Fig. 4A), corresponding to accelerated tumor growth (22). In CAG myeloma, pSTAT3 staining was augmented most prominently in endothelial cells (supplemental Fig. 3B). Thus, heparanase, secreted by tumor cells or other cells in the tumor milieu, may enhance STAT3 phosphorylation in the tumor and/or its microenvironment, including the tumor vasculature, in agreement with the proangiogenic properties of STAT3 (51). It should be noted that cytoplasmic heparanase in tumor biopsies represents a secretable heparanase protein responsible for eliciting SRC and EGFR phosphorylation (21); nuclear translocation simply may sequester heparanase and prevent its function extracellularly or play a direct role in gene regulation (4). Noteworthy, increased STAT3 phosphorylation was found in inflamed colons of transgenic mice overexpressing heparanase compared with control mice, associated with highly proliferating colon epithelial cells (52). This suggests that STAT3 activation by heparanase is not limited to cancer progression but rather is common in different biological settings.
Although STAT3 phosphorylation and its clinical relevance appear striking, heparanase function is not restricted to this family member. We found that phosphorylation of STAT5b is increased following heparanase overexpression or exogenous addition, whereas heparanase gene silencing results in decreased STAT5b phosphorylation. This is shown by IP, immunoblotting, and nuclear translocation applying immunofluorescent staining and cell fractionation in a number of cell lines (Figs. 1, 2, and 3A). Notably, comparable increase in STAT5b phosphorylation was observed in FaDu cells transfected with mutated, enzymatically inactive heparanase (Fig. 1A, DM), thus supporting the notion that heparanase exerts both enzymatic and nonenzymatic effects (13, 15, 18). In contrast, phosphorylation of STAT5a was not altered by heparanase, clearly revealing the specificity of this regulatory mechanism. Increased STAT5b phosphorylation by heparanase is mediated by SRC and EGFR because its phosphorylation and nuclear translocation (Fig. 2, A and B) were prevented by selective inhibitors of SRC (PP2) and EGFR (CL-387785). This is in agreement with STAT5b activation by receptor (i.e. EGFR) and non-receptor (i.e. SRC) tyrosine kinases (44, 53). Importantly, heparanase overexpression facilitated the interaction of STAT5b with the β-casein promoter region (Fig. 2D; supplemental Fig. 2, F and G), implying its biological functionality. Furthermore, STAT5b gene silencing was associated with reduced proliferation of heparanase overexpressing LNCaP cells, whereas silencing of STAT5a or STAT1 genes had no such effect (Fig. 3C), in line with a protumorigenic function of STAT5b in prostate cancer (54). STAT5b activation is noted in several other human malignancies, including head and neck carcinoma (55), resulting in resistance to cisplatin-mediated apoptosis and to growth inhibition by anti-EGFR small molecule inhibitor (erlotinib) (56). Acquired resistance of heparanase overexpressing cells toward EGFR inhibitor (Fig. 5; supplemental Fig. 4) is in line with this notion and further supports the clinical significance of heparanase expression in head and neck and possibly other carcinomas. Studies examining STAT3 and STAT5 phosphorylation and its association with heparanase staining and clinical outcome in several other patient cohorts are currently in progress.
Supplementary Material
This work was supported, in whole or in part, by NCI, National Institutes of Health Grant CA106456. This work was also supported by Israel Science Foundation Grant 593/10; the Israel Cancer Research Fund; the US-Israel Binational Science Foundation, and the Rappaport Family Institute Fund (to I. V.).

This article contains supplemental Figs. 1–4.
- EGFR
- epidermal growth factor receptor
- HNSCC
- head and neck squamous cell carcinoma
- Hepa
- heparanase-treated
- Vo
- empty vector.
REFERENCES
- 1. Parish C. R., Freeman C., Hulett M. D. (2001) Heparanase: a key enzyme involved in cell invasion. Biochim. Biophys. Acta 1471, M99–108 [DOI] [PubMed] [Google Scholar]
- 2. Vlodavsky I., Friedmann Y. (2001) Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J. Clin. Invest. 108, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dempsey L. A., Brunn G. J., Platt J. L. (2000) Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem. Sci. 25, 349–351 [DOI] [PubMed] [Google Scholar]
- 4. Ilan N., Elkin M., Vlodavsky I. (2006) Regulation, function, and clinical significance of heparanase in cancer metastasis and angiogenesis. Int. J. Biochem. Cell Biol. 38, 2018–2039 [DOI] [PubMed] [Google Scholar]
- 5. Sanderson R. D., Yang Y., Kelly T., MacLeod V., Dai Y., Theus A. (2005) Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies. J. Cell. Biochem. 96, 897–905 [DOI] [PubMed] [Google Scholar]
- 6. Shafat I., Bem-Arush M. W., Issakov J., Meller I., Naroditsky I., Tortoteto M., Cassinelli G., Lanzi C., Pisano C., Ilan N., Vlodavsky I., Zunino F. (2011) Preclinical and clinical significance of heparanase in Ewing sarcoma. J. Cell. Mol. Med. 115, 1857–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vreys V., David G. (2007) Mammalian heparanase: What is the message? J. Cell Mol. Med. 11, 427–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casu B., Vlodavsky I., Sanderson R. D. (2008) Non-anticoagulant heparins and inhibition of cancer. Pathophysiol. Haemost. Thromb. 36, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dredge K., Hammond E., Davis K., Li C. P., Liu L., Johnstone K., Handley P., Wimmer N., Gonda T. J., Gautam A., Ferro V., Bytheway I. (2010) The PG500 series: Novel heparan sulfate mimetics as potent angiogenesis and heparanase inhibitors for cancer therapy. Invest. New Drugs 28, 276–283 [DOI] [PubMed] [Google Scholar]
- 10. McKenzie E. A. (2007) Heparanase: A target for drug discovery in cancer and inflammation. Br. J. Pharmacol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miao H. Q., Liu H., Navarro E., Kussie P., Zhu Z. (2006) Development of heparanase inhibitors for anti-cancer therapy. Curr. Med. Chem. 13, 2101–2111 [DOI] [PubMed] [Google Scholar]
- 12. Vlodavsky I., Ilan N., Naggi A., Casu B. (2007) Heparanase: Structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr. Pharm. Des. 13, 2057–2073 [DOI] [PubMed] [Google Scholar]
- 13. Levy-Adam F., Ilan N., Vlodavsky I. (2010) Tumorigenic and adhesive properties of heparanase. Semin. Cancer Biol. 20, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lerner I., Baraz L., Pikarsky E., Meirovitz A., Edovitsky E., Peretz T., Vlodavsky I., Elkin M. (2008) Function of heparanase in prostate tumorigenesis: Potential for therapy. Clin. Cancer Res. 14, 668–676 [DOI] [PubMed] [Google Scholar]
- 15. Barash U., Cohen-Kaplan V., Dowek I., Sanderson R. D., Ilan N., Vlodavsky I. (2010) Proteoglycans in health and disease: New concepts for heparanase function in tumor progression and metastasis. FEBS J. 277, 3890–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okawa T., Naomoto Y., Nobuhisa T., Takaoka M., Motoki T., Shirakawa Y., Yamatsuji T., Inoue H., Ouchida M., Gunduz M., Nakajima M., Tanaka N. (2005) Heparanase is involved in angiogenesis in esophageal cancer through induction of cyclooxygenase-2. Clin. Cancer Res. 11, 7995–8005 [DOI] [PubMed] [Google Scholar]
- 17. Cohen-Kaplan V., Naroditsky I., Zetser A., Ilan N., Vlodavsky I., Doweck I. (2008) Heparanase induces VEGF-C and facilitates tumor lymphangiogenesis. Int. J. Cancer 123, 2566–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fux L., Ilan N., Sanderson R. D., Vlodavsky I. (2009) Heparanase: Busy at the cell surface. Trends Biochem. Sci. 34, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramani V. C., Yang Y., Ren Y., Nan L., Sanderson R. D. (2011) Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J. Biol. Chem. 286, 6490–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y., Ren Y., Ramani V. C., Nan L., Suva L. J., Sanderson R. D. (2010) Heparanase enhances local and systemic osteolysis in multiple myeloma by up-regulating the expression and secretion of RANKL. Cancer Res. 70, 8329–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen-Kaplan V., Doweck I., Naroditsky I., Vlodavsky I., Ilan N. (2008) Heparanase augments epidermal growth factor receptor phosphorylation: Correlation with head and neck tumor progression. Cancer Res. 68, 10077–10085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fux L., Feibish N., Cohen-Kaplan V., Gingis-Velitski S., Feld S., Geffen C., Vlodavsky I., Ilan N. (2009) Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 69, 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zetser A., Bashenko Y., Edovitsky E., Levy-Adam F., Vlodavsky I., Ilan N. (2006) Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and SRC activation. Cancer Res. 66, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 24. Gingis-Velitski S., Zetser A., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) Heparanase induces endothelial cell migration via protein kinase B/AKT activation. J. Biol. Chem. 279, 23536–23541 [DOI] [PubMed] [Google Scholar]
- 25. Barash U., Cohen-Kaplan V., Arvatz G., Gingis-Velitski S., Levy-Adam F., Nativ O., Shemesh R., Ayalon-Sofer M., Ilan N., Vlodavsky I. (2010) A novel human heparanase splice variant, T5, endowed with protumorigenic characteristics. Faseb J. 24, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ben-Zaken O., Gingis-Velitski S., Vlodavsky I., Ilan N. (2007) Heparanase induces AKT phosphorylation via a lipid raft receptor. Biochem. Biophys. Res. Commun. 361, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosenthal E. L., Kulbersh B. D., Duncan R. D., Zhang W., Magnuson J. S., Carroll W. R., Zinn K. (2006) In vivo detection of head and neck cancer orthotopic xenografts by immunofluorescence. Laryngoscope 116, 1636–1641 [DOI] [PubMed] [Google Scholar]
- 28. Weichselbaum R. R., Dunphy E. J., Beckett M. A., Tybor A. G., Moran W. J., Goldman M. E., Vokes E. E., Panje W. R. (1989) Epidermal growth factor receptor gene amplification and expression in head and neck cancer cell lines. Head Neck 11, 437–442 [DOI] [PubMed] [Google Scholar]
- 29. Nadav L., Katz B. Z., Baron S., Cohen N., Naparstek E., Geiger B. (2006) The generation and regulation of functional diversity of malignant plasma cells. Cancer Res. 66, 8608–8616 [DOI] [PubMed] [Google Scholar]
- 30. Levy-Adam F., Feld S., Suss-Toby E., Vlodavsky I., Ilan N. (2008) Heparanase facilitates cell adhesion and spreading by clustering of cell surface heparan sulfate proteoglycans. PLoS ONE 3, e2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zetser A., Levy-Adam F., Kaplan V., Gingis-Velitski S., Bashenko Y., Schubert S., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) Processing and activation of latent heparanase occurs in lysosomes. J. Cell Sci. 117, 2249–2258 [DOI] [PubMed] [Google Scholar]
- 32. Doweck I., Kaplan-Cohen V., Naroditsky I., Sabo E., Ilan N., Vlodavsky I. (2006) Heparanase localization and expression by head and neck cancer: Correlation with tumor progression and patient survival. Neoplasia 8, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy-Adam F., Feld S., Cohen-Kaplan V., Shteingauz A., Gross M., Arvatz G., Naroditsky I., Ilan N., Doweck I., Vlodavsky I. (2010) Heparanase 2 interacts with heparan sulfate with high affinity and inhibits heparanase activity. J. Biol. Chem. 285, 28010–28019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valgeirsdóttir S., Ruusala A., Heldin C. H. (1999) MEK is a negative regulator of Stat5b in PDGF-stimulated cells. FEBS Lett. 450, 1–7 [DOI] [PubMed] [Google Scholar]
- 35. Frank D. A. (2007) STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 251, 199–210 [DOI] [PubMed] [Google Scholar]
- 36. Reich N. C., Liu L. (2006) Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 37. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paukku K., Silvennoinen O. (2004) STATs as critical mediators of signal transduction and transcription: Lessons learned from STAT5. Cytokine Growth Factor Rev. 15, 435–455 [DOI] [PubMed] [Google Scholar]
- 39. Silver D. L., Naora H., Liu J., Cheng W., Montell D. J. (2004) Activated signal transducer and activator of transcription (STAT) 3: Localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 64, 3550–3558 [DOI] [PubMed] [Google Scholar]
- 40. Atkinson G. P., Nozell S. E., Benveniste E. T. (2010) NF-κB and STAT3 signaling in glioma: Targets for future therapies. Expert Rev. Neurother. 10, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pectasides E., Egloff A. M., Sasaki C., Kountourakis P., Burtness B., Fountzilas G., Dafni U., Zaramboukas T., Rampias T., Rimm D., Grandis J., Psyrri A. (2010) Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin. Cancer Res. 16, 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009) Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- 43. Fung C., Grandis J. R. (2010) Emerging drugs to treat squamous cell carcinomas of the head and neck. Expert Opin. Emerg. Drugs 15, 355–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lai S. Y., Johnson F. M. (2010) Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: Implications for future therapeutic approaches. Drug Resist. Updat. 13, 67–78 [DOI] [PubMed] [Google Scholar]
- 45. Leeman R. J., Lui V. W., Grandis J. R. (2006) STAT3 as a therapeutic target in head and neck cancer. Expert Opin. Biol. Ther. 6, 231–241 [DOI] [PubMed] [Google Scholar]
- 46. Song J. I., Grandis J. R. (2000) STAT signaling in head and neck cancer. Oncogene 19, 2489–2495 [DOI] [PubMed] [Google Scholar]
- 47. Seethala R. R., Gooding W. E., Handler P. N., Collins B., Zhang Q., Siegfried J. M., Grandis J. R. (2008) Immunohistochemical analysis of phosphotyrosine signal transducer and activator of transcription 3 and epidermal growth factor receptor autocrine signaling pathways in head and neck cancers and metastatic lymph nodes. Clin. Cancer Res. 14, 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Germain D., Frank D. A. (2007) Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin. Cancer Res. 13, 5665–5669 [DOI] [PubMed] [Google Scholar]
- 49. Sehgal P. B. (2008) Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev. Biol. 19, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schlessinger K., Levy D. E. (2005) Malignant transformation but not normal cell growth depends on signal transducer and activator of transcription 3. Cancer Res. 65, 5828–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Z., Han Z. C. (2008) STAT3: A critical transcription activator in angiogenesis. Med. Res. Rev. 28, 185–200 [DOI] [PubMed] [Google Scholar]
- 52. Lerner I., Hermano E., Zcharia E., Rodkin D., Bulvik R., Doviner V., Rubinstein A. M., Ishai-Michaeli R., Atzmon R., Sherman Y., Meirovitz A., Peretz T., Vlodavsky I., Elkin M. (2011) Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J. Clin. Invest. 121, 1709–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu H., Jove R. (2004) The STATs of cancer–new molecular targets come of age. Nat. Rev. Cancer 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 54. Li H., Zhang Y., Glass A., Zellweger T., Gehan E., Bubendorf L., Gelmann E. P., Nevalainen M. T. (2005) Activation of signal transducer and activator of transcription-5 in prostate cancer predicts early recurrence. Clin. Cancer Res. 11, 5863–5868 [DOI] [PubMed] [Google Scholar]
- 55. Xi S., Zhang Q., Gooding W. E., Smithgall T. E., Grandis J. R. (2003) Constitutive activation of Stat5b contributes to carcinogenesis in vivo. Cancer Res. 63, 6763–6771 [PubMed] [Google Scholar]
- 56. Koppikar P., Lui V. W., Man D., Xi S., Chai R. L., Nelson E., Tobey A. B., Grandis J. R. (2008) Constitutive activation of signal transducer and activator of transcription 5 contributes to tumor growth, epithelial-mesenchymal transition, and resistance to epidermal growth factor receptor targeting. Clin. Cancer Res. 14, 7682–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.