Background: Ethanol-related metabolic activities induce the changes in the small molecule metabolome.
Results: Metabolomic analysis revealed that the level of N-acetyltaurine (NAT) in urine increases dramatically after ethanol consumption.
Conclusion: Ethanol-induced NAT biosynthesis is mainly caused by a novel reaction between taurine and excessive acetate produced by ethanol metabolism.
Significance: NAT is a novel metabolite of ethanol that can function as the biomarker of hyperacetatemia.
Keywords: Alcohol, Liver Metabolism, Mass spectrometry (MS), Metabolism, Metabolomics, N-Acetyltaurine, Acetate, Ethanol, Stable Isotope Labeling, Taurine
Abstract
The influence of ethanol on the small molecule metabolome and the role of CYP2E1 in ethanol-induced hepatotoxicity were investigated using liquid chromatography-mass spectrometry (LC-MS)-based metabolomics platform and Cyp2e1-null mouse model. Histological and biochemical examinations of ethanol-exposed mice indicated that the Cyp2e1-null mice were more resistant to ethanol-induced hepatic steatosis and transaminase leakage than the wild-type mice, suggesting CYP2E1 contributes to ethanol-induced toxicity. Metabolomic analysis of urinary metabolites revealed time- and dose-dependent changes in the chemical composition of urine. Along with ethyl glucuronide and ethyl sulfate, N-acetyltaurine (NAT) was identified as a urinary metabolite that is highly responsive to ethanol exposure and is correlated with the presence of CYP2E1. Subsequent stable isotope labeling analysis using deuterated ethanol determined that NAT is a novel metabolite of ethanol. Among three possible substrates of NAT biosynthesis (taurine, acetyl-CoA, and acetate), the level of taurine was significantly reduced, whereas the levels of acetyl-CoA and acetate were dramatically increased after ethanol exposure. In vitro incubation assays suggested that acetate is the main precursor of NAT, which was further confirmed by the stable isotope labeling analysis using deuterated acetate. The incubations of tissues and cellular fractions with taurine and acetate indicated that the kidney has the highest NAT synthase activity among the tested organs, whereas the cytosol is the main site of NAT biosynthesis inside the cell. Overall, the combination of biochemical and metabolomic analysis revealed NAT is a novel metabolite of ethanol and a potential biomarker of hyperacetatemia.
Introduction
Alcohol abuse is an important health issue in the United States and around the world (1). Ethanol, because of its high permeability across the cellular membrane, can inflict adverse effects on various parts of the body, including the central nervous system, cardiovascular system, and hepatogastrointestinal organs (2–4). The most common ailment associated with alcohol abuse is alcoholic liver disease, a type of hepatic dysfunction caused by ethanol-induced liver injury (5). Over the past decades, numerous efforts have been undertaken to understand the molecular mechanism of these complex pathophysiological events (6–8). However, many aspects of ethanol-induced pathogenesis remain largely unknown.
Based on current available knowledge on ethanol-induced toxicities, the most important contributing factor in these events is the metabolism of ethanol itself (9). The central route of ethanol metabolism is the biotransformation of ethanol into acetyl coenzyme A (acetyl-CoA) through a three-step reaction process, which includes the consecutive oxidation of ethanol to acetaldehyde and then to acetate, followed by the esterification of CoA. The formation of a large quantity of acetyl-CoA and reduced NADH from this process directly disrupts the nutritional and metabolic homeostasis (10). In addition, ethanol metabolism also produces reactive species, including acetaldehyde and free radicals, which can directly attack proteins, lipids, and many other cellular components (11). As the consequence of ethanol-induced protein adduction (12), lipid peroxidation, and oxidative stress (13), the functions of proteins and the structure of cellular membrane are altered, leading to mitochondrial damage, pro-inflammatory responses, and impairment of the antioxidant system (14, 15).
Enzymes responsible for initiating ethanol metabolism, which is the oxidation of ethanol to acetaldehyde, have been identified as alcohol dehydrogenases (16), catalase (17), and cytochrome P450 2E1 (CYP2E1) (18). The metabolism of ethanol from intestinal flora or moderate alcohol consumption in the liver is mainly conducted by alcohol dehydrogenases because of their relatively lower Km values (19, 20), and catalase-mediated acetaldehyde production is potentially important in the extrahepatic tissues, especially in the brain (21). CYP2E1, a microsomal enzyme that is inducible after repeated ethanol exposure, was identified as a key component of the microsomal ethanol-oxidizing system (22), which is capable of generating reactive oxygen species during the conversion of ethanol to acetaldehyde. A pathogenic role of CYP2E1 in ethanol-induced toxicities has been suggested based on its distribution, expression pattern, and enzymatic properties (23) but was not well validated due to the inconsistent results obtained from several animal studies using the Cyp2e1-null mice, which are deficient in CYP2E1 activity. In two recent studies, ethanol-induced hepatosteatosis and oxidative stress were significantly higher in the wild-type and the CYP2E1-humanized mice than the Cyp2e1-null mice, suggesting a contributing role for CYP2E1 in ethanol-induced liver injury (24, 25). However, in two other studies, no clear difference between the wild-type and the Cyp2e1-null mice was observed with regard to the adverse effects of ethanol treatment (26, 27). Therefore, additional studies are needed to determine whether CYP2E1 can significantly contribute to the ethanol-induced liver injury.
One prominent feature of ethanol-induced chronic toxicities is the gradual development of disease, i.e. alcoholic liver disease always starts from a reversible stage of hepatosteatosis, and then gradually progresses to the irreversible stages of hepatic fibrosis and cirrhosis and liver cancer in certain cases (28). As the modulations of biochemical pathways and metabolic reactions occur at each stage of ethanol-elicited diseases, examining these events in cellular and molecular levels provides an excellent venue for discovering specific and sensitive biomarkers that can indicate the extent of ethanol abuse and the scope of tissue damage. Many analytical approaches have been adopted for this purpose. For instance, metabolite analyses have identified EtG2 and ethyl sulfate, two minor metabolites of ethanol, as the indicators of ethanol exposure (29, 30), whereas enzymatic assays have established γ-glutamyltransferase and transaminases as the markers of liver injury (31, 32). In addition, carbohydrate-deficient transferrin in the serum, mean corpuscular volume of red blood cell, and acetaldehyde adducts have also been used for detecting ethanol abuse (31, 33, 34). Recent arrival of metabolomics, a global system biology methodology for measuring small molecule metabolite profiles and fluxes in biological matrices (35), offers a new and powerful tool to characterize the metabolic changes induced by ethanol and to identify the small molecule biomarkers of ethanol-induced toxicities. Mass spectrometry (MS)-based and nuclear magnetic resonance (NMR)-based metabolomic analyses of urine, blood, and tissue samples have revealed the changes of organic acids (36), amino acids (37), and their derivatives (38), as well as fatty acids and associated lipid species (39) in the biofluids following ethanol treatment.
In this study, the metabolic events elicited by ethanol exposure and the role of CYP2E1 in ethanol-induced hepatotoxicity were investigated using LC-MS-based metabolomics as the analytical platform and the Cyp2e1-null mice as the animal model. NAT, a urinary metabolite that is formed mainly by the enzymatic reaction between acetate and taurine, was identified as a novel metabolite of ethanol in vivo.
EXPERIMENTAL PROCEDURES
Reagents
Ethanol, [2H6]ethanol, [2H4]acetate, cesium chloride, magnesium chloride, EDTA, diazinon, α-alanine, cysteine, and creatinine were purchased from Sigma. N-Acetylneuraminic acid, tri-phenylphosphine, 2-picolylamine, mercury(II) chloride, butyric acid, propionic acid, and β-alanine were purchased from Alfa Aesar (Ward Hill, MA). Dehydrated agar, 4-hydroxyphenylacetic acid, p-chloro-l-phenylalanine, taurine, and dansyl chloride LC-MS-grade water, acetonitrile, formic acid, calcium chloride, ferrous(II) chloride, and acetate were purchased from Fisher. EtG was purchased from Toronto Research Chemical (North York, Ontario, Canada); acetyl-CoA was from Roche Applied Science; 2,2′-dipyridyl disulfide and phenylmethylsulfonyl fluoride (PMSF) were from MP Biomedicals (Solon, OH).
Animals and Ethanol Treatments
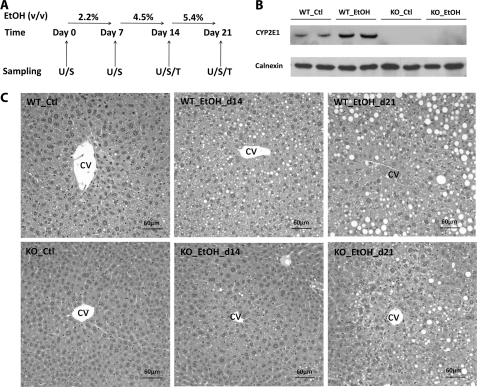
Male wild-type and Cyp2e1-null mice on the 129/Sv strain background (40), 10–12 weeks old, were used in this study. All animals were maintained in a University of Minnesota animal facility under a standard 12-h light/12-h dark cycle with food and water ad libitum. Handling and treatment procedures were in accordance with animal study protocols approved by the University of Minnesota Animal Care and Use Committee. A modified semi-solid diet that was formulated based on Lieber-DeCarli liquid ethanol diet was supplied to mice using customized feeding tubes (41, 42). Details of diet preparation and the feeding tube are described in the supplemental “Experimental Procedures”. To acclimate to the semi-solid diet, both wild-type and Cyp2e1-null mice were initially fed the control semi-solid dextrose diet for 3 days before the treatment. Afterward, the mice were fed either the ethanol diet or the control dextrose diet for 21 days. The concentration of ethanol was increased weekly from 2.2% (v/v) to 4.5% (v/v) and finally 5.4% (v/v) (Fig. 1A). The amount of food consumption of each group was monitored and was determined as statistically comparable.
FIGURE 1.
Influence of ethanol treatment on CYP2E1 protein level and liver histology of wild-type (WT) and Cyp2e1-null (KO) mice. A, schedule of ethanol treatment and sample collection. Both WT and KO mice were fed with either control (Ctl) or ethanol (EtOH) diet for 21 days. The concentration of ethanol was increased weekly from 2.2% (v/v) to 4.5% (v/v) and finally 5.4% (v/v). Urine (U) and serum (S) samples were collected once a week. Tissues (T) were harvested on days 14 and 21. B, Western blot of CYP2E1 protein expression after 21 days feeding of control or ethanol diet. C, histology of liver from WT and KO mice fed with control or ethanol diet. The location of hepatic central vein (CV) was marked.
Sample Collection and Liver Histology
Serum samples were collected by submandibular bleeding. Urine samples were collected by housing the mice in metabolic cages for 24 h. The liver and other tissue samples were harvested after the mice were euthanized by carbon dioxide. All tissue samples were stored at −80 °C before further analysis except for 1 aliquot of liver tissue that was immediately fixed in 10% formalin solution after dissection, then embedded in paraffin, sectioned, and stained with hematoxylin and eosin for general histology.
Preparation of Tissue Homogenates and Intracellular Fractions
Tissue homogenates from liver, kidney, heart, muscle, and brain were prepared by the homogenization in a buffer containing 320 mm sucrose, 50 mm phosphate-buffered saline (PBS) solution, 1 mm EDTA, and protease inhibitor and then centrifuged at 600 × g for 10 min to remove the nuclear pellet. Intracellular fractions were obtained by stepwise centrifugation. Briefly, the tissue homogenates of liver and kidney were centrifuged at 9000 × g for 20 min. Mitochondrial fraction was prepared by washing, centrifuging, and then reconstituting the pellet in a suspension buffer containing 100 mm PBS, 20% (v/v) glycerol, 1 mm EDTA, and protease inhibitor. The supernatant from 9000 × g centrifugation was further centrifuged at 100,000 × g for 1 h. The resulting supernatant was the cytosolic fraction, and the precipitate was reconstituted in the suspension buffer as the microsomal fraction.
Biochemical Assays
Serum alanine aminotransferase activity, liver and serum triacylglycerol (TAG) level, and blood urea nitrogen level were measured using the colorimetric assay kits from Pointe Scientific (Canton, MI). The lipid fraction of the liver was prepared by chloroform/methanol extraction (43).
Western Blotting
The expression level of CYP2E1 in the mouse liver microsome was measured using a monoclonal antibody (1-98-1) against mouse CYP2E1 (44). Calnexin was used as the loading control of microsomal proteins.
Urine Sample Preparation and LC-MS Analysis
Urine samples were prepared by mixing 50 μl of urine with 200 μl of 50% aqueous acetonitrile and centrifuging at 21,000 × g for 10 min to remove protein and particulates. Supernatants were injected into a Waters AcquityTM ultra-performance liquid chromatography system (Milford, MA) and separated by a gradient of mobile phase ranging from water to 95% aqueous acetonitrile containing 0.1% formic acid over a 10-min run. LC eluant was introduced into a Waters SYNAPT QTOF mass spectrometer (QTOF-MS) for accurate mass measurement and MS/MS analysis. Capillary voltage and cone voltage for electrospray ionization was maintained at 3 kV and 30 V for positive mode detection, and at −3 kV and −35 V for negative mode detection, respectively. Source temperature and desolvation temperature were set at 120 and 350 °C, respectively. Nitrogen was used as both cone gas (50 liters/h) and desolvation gas (600 liters/h) and argon as collision gas. For accurate mass measurement, the mass spectrometer was calibrated with sodium formate solution (range m/z 50–1000) and monitored by the intermittent injection of the lock mass leucine enkephalin ([M + H]+ = 556.2771 m/z and [M − H]− = 554.2615 m/z) in real time. After data acquisition in QTOF-MS, chromatograms and spectra of urine samples were processed by MassLynxTM software (Waters).
Chemometric Analysis and Biomarker Identification
Chromatographic and spectral data of urine samples from the wild-type and Cyp2e1-null mice were deconvoluted by MarkerLynxTM software (Waters). A multivariate data matrix containing information on sample identity, ion identity (RT and m/z), and ion abundance was generated through centroiding, de-isotoping, filtering, peak recognition, and integration. The intensity of each ion was calculated by normalizing the single ion counts versus the total ion counts in the whole chromatogram. The data matrix was further exported into SIMCA-P+TM software (Umetrics, Kinnelon, NJ), and transformed by mean-centering and Pareto scaling, a technique that increases the importance of low abundance ions without significant amplification of noise. Both unsupervised and supervised multivariate data analyses (MDA), including principal components analysis, projection to latent structures-discriminant analysis (PLS-DA), and orthogonal projection to latent structures (OPLS) analysis, were applied to classify the measured urine samples (45). Principal components were generated by MDA to represent the major latent variables in the data matrix and were described in a scores scatter plot. Potential urinary biomarkers were identified by analyzing ions contributing to the principal components and to the separation of sample groups in the loading scatter plot.
Structural Elucidation and Synthesis of NAT
The structures of urinary biomarkers identified by metabolomic analysis were determined by accurate mass-based elemental composition analysis, MS/MS fragmentation, and the comparisons with authentic standards. The MS/MS fragmentation was conducted by using collision energy ramping from 10 to 40 eV in SYNAPT QTOF-MS.
NAT was synthesized based on a method described by Johnson et al. (46). Briefly, 0.5 g of taurine was dissolved in a mixture of 7 ml of water and 2.5 ml of pyridine, and then 2 ml of acetic anhydride was added dropwise. The reaction mixture was stirred at 4 °C overnight. After the solvent was removed under vacuum, the product was recrystallized using a mixture of ethanol and dichloromethane (1:1) and dried under vacuum for 10 h. A yield of 286 mg of NAT (43%) was obtained, m.p. 99–101 °C.
Stable Isotope Labeling Analysis of Ethanol Metabolism and Biosynthesis of NAT
Semi-solid diets containing 2.2% unlabeled ethanol, 2.2% [2H6]ethanol, or 1.1% [2H4]acetate were fed to the wild-type mice for 7 days. 24-Hour urine samples were collected and then analyzed by ultra-performance liquid chromatography-QTOF-MS. Data from [2H6]ethanol or [2H4]acetate treatment were compared with those from unlabeled ethanol treatment through a OPLS-based metabolomic analysis (47). Deuterated metabolites of ethanol and acetate were identified in the S-plots of OPLS models and further confirmed by comparing extracted ion chromatograms of unlabeled metabolites with those of deuterated metabolites and by MS/MS fragmentation.
Quantitation of Urinary Biomarkers
Urinary biomarkers were quantified by accurate mass-based ion extraction chromatograms. Standard curves of creatinine, EtG, and NAT ranging from 10 to 500 μm were prepared using dynamic range enhancement function of SYNAPT QTOF-MS (30). Urinary concentrations of EtG and NAT were determined by the integration of peak area and fitting with the standard curve using QuanLynxTM software (Waters) and expressed as the molar ratio to creatinine.
Quantitation of the Precursors of NAT in Liver and Serum
To determine the effects on ethanol overdose on the levels of potential precursors of NAT in vivo, a 4 g/kg dose of ethanol was administered to both wild-type and Cyp2e1-null mice by intraperitoneal (i.p.) injection (48). Serum samples were collected prior to the injection and at 0.5, 1, 2, 4, and 6 h after the injection. Liver samples were harvested prior to the injection and at 1 and 6 h after the injection. To measure the amount of taurine in the liver, the sample mixture was prepared by mixing 5 μl of liver homogenate with 5 μl of 100 μm p-chloro-l-phenylalanine (internal standard) and then followed by the addition of 40 μl of 10 mm Na2CO3 (pH 11). The sample mixture then reacted with 100 μl of freshly prepared dansyl chloride in acetone (3 mg/ml) by vortexing and heating at 60 °C for 10 min (49). The reaction mixture was centrifuged at 21,000 × g for 10 min at 4 °C. Supernatant was transferred to an HPLC vial and injected into the LC/MS system for quantitation. The peak area of dansyl taurine derivative ([M + H]+ = 395.0730) was monitored for determining the concentration of taurine. The standard curve ranging from 50 to 1000 μm was prepared using the same procedure. To measure the hepatic content of acetyl-CoA, 100 μl of liver homogenate was added with sulfadimethoxine (internal standard) and extracted twice by 250 μl of 50% aqueous acetonitrile. The extract was dried by vacuum and reconstituted by 50 μl of phosphate-buffered saline containing 20% acetonitrile. Acetyl-CoA ([M + H]+ = 810.1336) concentration was determined by LC-MS with a standard curve from 5 to 200 μm. To measure the amount of acetate in the liver and serum, 10 μl of liver homogenate or serum sample was diluted by 90 μl of acetonitrile and then mixed with 10 μl of 100 μm [2H4]acetate (internal standard). The derivatization reagents, including 10 μl of 10 mm triphenylphosphine, 10 μl of 10 mm 2,2′-dipyridyl disulfide, and 10 μl of 10 mm 2-picolylamine, were added into the sample mixture consecutively (50). The reaction mix was vortexed and then heated at 60 °C for 15 min. After centrifuging at 21,000 × g for 10 min, the supernatant was transferred to an HPLC vial and injected 5 μl into the LC/MS system for analysis. The peak area of 2-picolylamine-acetate derivative ([M + H]+ = 151.0866) was monitored for determining the concentration of acetate. The standard curve ranging from 50 to 1000 μm was prepared using the same procedure.
Enzymatic Kinetics of NAT Biosynthesis Reactions
In vitro enzyme kinetics experiment was conducted by incubating liver homogenate, 20 mm taurine, and acetyl-CoA or acetate ranging from 100 μm to 6 mm in a phosphate-buffered saline solution at 37 °C for 10 min. Reaction was terminated by adding equal volume of acetonitrile. NAT as the reaction product was further analyzed by LC-MS. Km and Vmax values of the enzyme for acetyl-CoA and acetate were calculated based on Michaelis-Menten equation.
Sites of NAT Biosynthesis
To determine the activity of NAT biosynthesis in the metabolically active organs, tissue homogenates of liver, kidney, heart, brain, and muscle were incubated with 20 mm taurine and 2.5 mm acetate for 30 min, and the yield of NAT was measured. Similarly, the intracellular location of NAT biosynthesis was determined by incubating intracellular fractions of liver and kidney, including cytosol, mitochondria, and microsome, with 20 mm taurine and 2.5 mm acetate, followed by the quantitation of NAT.
Statistics
Experimental values are expressed as mean ± S.D. Statistical analysis was performed with two-tailed Student's t tests for unpaired data, and a p value of <0.05 was considered as statistically significant.
RESULTS
Different Responses to Ethanol Feeding from Wild-type and Cyp2e1-null Mice
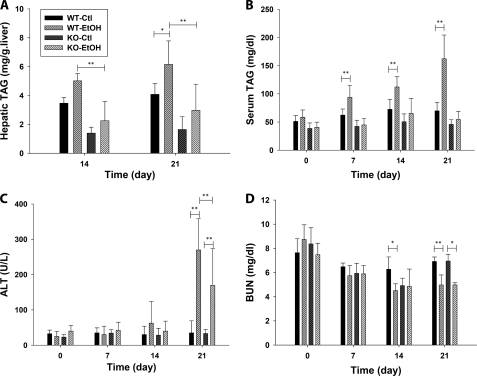
To define the role of CYP2E1 in ethanol-induced hepatotoxicity, the wild-type and Cyp2e1-null mice were fed with control or ethanol diet for 21 days (Fig. 1A). To avoid the urine and fetal contamination associated with liquid ethanol diet, a modified semi-solid ethanol diet was formulated (detailed in the supplemental “Experimental Procedures”) and well received by the mice used in this study. After a 21-day ethanol feeding, CYP2E1 protein was dramatically increased in the wild-type mice but remained absent in the Cyp2e1-null mice (Fig. 1B), confirming that CYP2E1 expression is responsive to ethanol treatment (25). No significant difference in the change of body weight was observed between the two mouse lines fed with the control diet. However, the body weight of wild-type and Cyp2e1-null mice fed with ethanol diet was slightly decreased after starting the 5.4% ethanol diet at day 15 (data not shown). Because hepatic steatosis is a well established acute response to ethanol treatment, microscopic examination of liver histology was conducted. The results showed that the development of both microvesicular and macrovesicular steatosis around the central vein was delayed in the Cyp2e1-null mice compared with the wild-type mice (Fig. 1C). Consistent with the histological analysis, the triacylglycerol (TAG) level in the liver and serum of wild-type mice was increased in a time- and dose-dependent pattern although its level in the Cyp2e1-null mice was not significantly altered by the ethanol exposure (Fig. 2, A and B). It is noteworthy that the basal level of TAG in the liver of Cyp2e1-null mice was significantly lower than its level in the wild-type mice (Fig. 2A). This difference between the two mouse lines has also been observed in a separate study (24), but its cause remains to be determined.
FIGURE 2.
General biochemical parameters of wild-type (WT) and Cyp2e1-null (KO) mice after fed with control or ethanol diet for 21 days. A, TAG levels in the liver. B, TAG levels in the serum. C, serum alanine aminotransferase (ALT) activity. D, blood urea nitrogen (BUN) levels. Values were presented as mean ± S.D. (n = 8). (*, p < 0.05, and **, p < 0.01).
The ethanol-elicited liver injury was evaluated by measuring the activity of serum alanine aminotransferase. The increase of alanine aminotransferase activity occurred in both mouse lines after a 21-day ethanol feeding, but the increase in the Cyp2e1-null mice was significantly less than that in the wild-type mice (Fig. 2C). Furthermore, the effect of ethanol treatment on the kidney function of wild-type and Cyp2e1-null mice was determined by measuring BUN level. During the 21-day ethanol feeding, the decrease of BUN level in the wild-type mice occurred after a 14-day treatment, but did not happen in the Cyp2e1-null mice until day 21 of the treatment (Fig. 2D). Overall, both histological and biochemical analysis of the responses to ethanol feeding from the wild-type and Cyp2e1-null mice indicated that deficiency of CYP2E1, a protein that is involved in ethanol metabolism and also inducible by ethanol exposure, reduces the toxic effects of ethanol, suggesting a contributing role of CYP2E1 in the ethanol-induced toxicity.
Metabolomic Investigation of Ethanol-induced Metabolic Events in Wild-type and Cyp2e1-null Mice
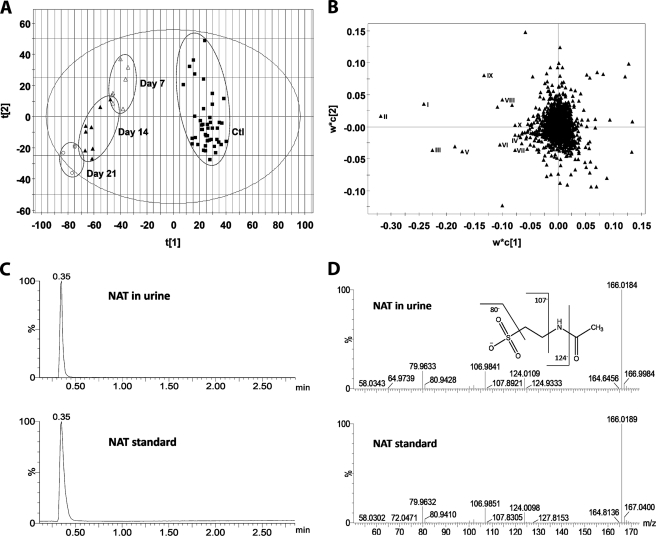
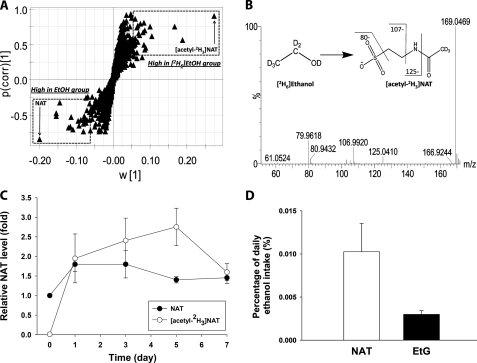
To further explore the mechanism underlying general toxic effects of ethanol and different responses of the wild-type and Cyp2e1-null mice to ethanol challenge, metabolic events induced by ethanol treatment were examined through LC-MS-based metabolomic analysis of urine samples from the two mouse lines. After the data acquired from the chromatograms and mass spectra of LC-MS analysis were processed by the PLS-DA, a type of supervised MDA, a two-component multivariate model, represented by a scores scatter plot, was constructed to illustrate the relationship among sample groups (Fig. 3A and supplemental Fig. S1). The distinctive separation of urine samples collected at the different time points of ethanol feeding in the wild-type mice (circled and labeled in Fig. 3A) suggested that ethanol treatment dramatically altered the chemical composition of the urinary metabolome in a time- and dose-dependent pattern, implying the possibility of identifying the urinary metabolites that can function as the biomarkers of ethanol exposure. Similarly, the ethanol-exposed Cyp2e1-null mice were also clearly separated from their untreated controls in a PLS-DA model, even though the genotype-dependent differences between wild-type and Cyp2e1-null mice, represented by the separation of two mouse lines along the principal component 2 of the model, preexist prior to the ethanol treatment (supplemental Fig. S1). The urinary ions contributing to the classification of sample groups were further characterized in a loadings scatter plot (Fig. 3B), and the chemical identities of ions that are highly induced by ethanol treatment were further determined by accurate mass measurement, elemental composition analysis, MS/MS fragmentation, and comparisons with authentic standards if available (Table 1). Among them (I–X), EtG (III), and ethyl sulfate (IV) are the minor metabolites of ethanol (29, 30); 4-hydroxyphenylacetic acid sulfate (II) as well as its in-source fragment (IX) have been shown as the metabolites of intestinal flora that are highly responsive to ethanol treatment (36); and N-acetylneuraminic acid (X), a functional component of glycoproteins and glycolipids, has been identified as a biomarker of alcohol abuse in the saliva and serum (51). The observation of these known markers of ethanol exposure indicated that the ethanol feeding method in this study was effective in altering the urinary metabolome, and the multivariate analysis was efficient in identifying small molecule markers. More importantly, NAT (I) was identified as a novel marker that is highly responsive to ethanol treatment based on its high value in the loading scatter plot. Its chemical identity was confirmed by the comparisons of its chromatographic peak and MS/MS spectra with synthesized standard (Fig. 3, C and D).
FIGURE 3.
Identification of urinary metabolites induced by ethanol treatment through LC-MS-based metabolomics. Conditions of LC-MS measurement and procedures of data processing and analysis were described under “Experimental Procedures.” A, scores plot of a PLS-DA model on urine samples from the wild-type mice fed with control and ethanol diets. All samples from the mice with no ethanol exposure were labeled as Ctl (■), whereas the samples from ethanol treatment were labeled according to the time points of sample collection, which are day 7 (Δ), 14 (▴), and 21 (○). The t[1] and t[2] values represent the scores of each sample in the principal component 1 and 2, respectively. Fitness (R2) and prediction power (Q2) of this PLS-DA model are 0.64 and 0.43, respectively. The model was validated through the recalculation of R2 and Q2 values after the permutation of sample identities. B, loadings plot of urinary ions contributing to the classification of urine samples from the wild-type mice treated with control and ethanol diet. The w*c[1] and w*c[2] values represent the contributing weights of each ion to the principal components 1 and 2 of the PLS-DA model, respectively. Major urinary ions (I–X) induced by ethanol treatment in the wild-type mice were labeled. C, extracted chromatograms of NAT in urine (I) and NAT standard. D, representative MS/MS fragmentation spectra of NAT in urine (I) and NAT standard. The fragmentation pattern of NAT was interpreted in the inlaid diagram.
TABLE 1.
Urinary metabolite markers of ethanol treatment
The process of identifying urinary metabolites responsive to ethanol treatment was described under “Experimental Procedures” and “Results.” The most prominent markers revealed by the loadings plot of PLS-DA model (labeled in Fig. 3B) were further investigated by accurate mass measurement, MS/MS fragmentation. Retention time (RT) of each marker was its elution time from a 10-min run in a C18 UPLC column. The identities of markers I, II, III, IX, and X were confirmed by the comparisons with authentic standards. ND means not determined.
| Markers | RT | [M − H]− | Formula | Identity |
|---|---|---|---|---|
| min | ||||
| I | 0.35 | 166.0174 | C4H8NO4S− | NAT |
| II | 1.97 | 230.9963 | C8H7O6S− | 4-Hydroxyphenylacetic acid sulfate |
| III | 0.68 | 221.0661 | C8H13O7− | Ethyl glucuronide |
| IV | 0.33 | 124.9909 | C2H5O4S− | Ethyl sulfate |
| V | 5.39 | 317.1592 | C15H25O7− | ND |
| VI | 5.30 | 329.1600 | C16H25O7− | ND |
| VII | 5.62 | 319.1756 | C15H27O7− | ND |
| VIII | 5.24 | 317.1598 | C15H25O7− | ND |
| IX | 1.97 | 151.0395 | C8H7O3− | In-source fragment of II |
| X | 2.32 | 308.0982 | C11H18NO9− | N-Acetylneuraminic acid |
Quantitation of Urinary Biomarkers of Ethanol Exposure
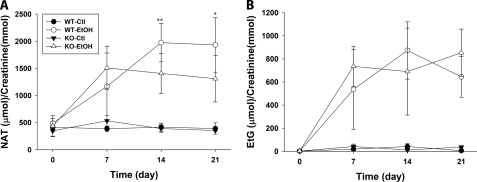
To validate the observation of metabolomics analysis (Fig. 3), the levels of urinary NAT (I) and EtG (III) in the wild-type and Cyp2e1-null mice during the 21-day ethanol treatment were compared through two different normalization approaches. One approach was to calculate the molar ratio to creatinine, which is commonly used to quantify urinary metabolites (52); the other approach was based on the relative abundance of the individual ion in the total MS signals, which were used to construct the multivariate model on urine samples (detailed in the supplemental “Experimental Procedures”). The result from creatinine-based normalization showed that comparable amounts of NAT existed in the urine of both wild-type and Cyp2e1-null mice prior to ethanol exposure, and its level remained stable in the control group, suggesting NAT is already a constitutive component of mouse urine even without ethanol treatment (Fig. 4A). After a 7-day exposure of 2.2% ethanol, the amount of NAT increased in both mouse lines to a comparable level. However, subsequent treatments of 7-day 4.5% ethanol and 7-day 5.4% ethanol led to a significantly higher level of NAT in the wild-type mice than its level in the Cyp2e1-null mice (Fig. 4A). In contrast, urinary EtG was undetectable prior to the ethanol treatment, confirming that EtG is not an endogenous compound but an ethanol metabolite (27). In addition, even though the amount of EtG increased following ethanol exposure, no significant difference between ethanol-treated wild-type and Cyp2e1-null mice was observed (Fig. 4B). Compared with the result from creatinine-based normalization, the result from total MS signal-based normalizations showed an even clearer difference between the two mouse lines on the urinary level of NAT at all three time points, including day 7 of ethanol exposure (supplemental Fig. S2). The discrepancy between the results from these two analytical approaches is likely caused by the fluctuation of creatinine excretion, which has been observed in previous alcohol-related studies (53, 54). Taken together, the quantitation of urinary NAT and EtG confirmed their identities as the biomarkers of ethanol exposure because both of them were dramatically induced by ethanol. The main difference between them is that NAT, an endogenous compound, can also reflect the different responses of the two mouse lines to ethanol treatment.
FIGURE 4.
Quantitation of urinary biomarkers of ethanol exposure. Urinary concentrations of NAT and EtG in both wild-type (WT) and Cyp2e1-null (KO) mice during the 21-day ethanol treatment were calculated as the molar ratio to creatinine. A, urinary concentration of NAT. B, urinary concentration of EtG. Values were presented as mean ± S.D. (n = 8). *, p < 0.05 and **, p < 0.01 indicate statistical significance between WT and KO samples at the same time point.
Identification of NAT as a Metabolite of Ethanol through Stable Isotope Labeling Analysis
NAT has been identified as a chemical component of mouse urine in a recent metabolomics study on the radiation-induced toxicity (46). However, its relation with ethanol has not been shown previously. To examine the source of newly synthesized NAT after ethanol exposure, deuterated ethanol ([2H6]ethanol) was fed to the wild-type mice for 7 days, and the chemical composition of urine samples from both unlabeled and labeled ethanol treatments was compared through the LC-MS-based metabolomic analysis. As shown in a loadings S-plot of detected urinary ions from an OPLS analysis, NAT and its isotopic counterpart, [acetyl-2H3]NAT, were identified as the most prominent ions corresponding to the unlabeled and labeled ethanol treatments, respectively (Fig. 5A). The migration of the deuterium atoms from labeled ethanol into the acetyl group of NAT was further confirmed by the MS/MS fragmentation of [acetyl-2H3]NAT in the urine, indicating ethanol can be the metabolic precursor of NAT (Fig. 5B). To determine the direct contribution of exogenous ethanol to the NAT biosynthesis, the relative abundance of unlabeled NAT and [acetyl-2H3]NAT was further compared. The result showed that the level of unlabeled NAT, which did not originate from deuterated ethanol, increased slightly after ethanol exposure, but more importantly, the level of [acetyl-2H3]NAT in urine, which was from deuterated ethanol, increased dramatically, suggesting that ethanol is the major source of newly synthesized NAT after ethanol exposure (Fig. 5C). Furthermore, the amount of ethanol that was converted in vivo into the urinary NAT was calculated as the percentage of daily ethanol intake through an equal molar conversion (1 m NAT is equivalent to 1 m ethanol) and was compared with that of EtG. The results showed that the urinary contents of NAT and EtG are only equal to a small amount of ethanol consumed by mice (Fig. 5D), indicating that NAT, similar to EtG, is a minor metabolite of ethanol. Overall, the stable isotope labeling analysis defined NAT as both an endogenous metabolite that is constitutively synthesized and an exogenous metabolite formed by ethanol metabolism.
FIGURE 5.
Identification of NAT as a metabolite of ethanol through stable isotope labeling. The diet containing 2.2% unlabeled ethanol or deuterated ethanol ([2H6]ethanol) was fed to the wild-type mice for 7 days as described under “Experimental Procedures.” A, S-plot of urinary ions from an OPLS analysis of LC-MS data from the wild-type mice treated with unlabeled and deuterated ethanol. NAT and its isotopic counterpart, [acetyl-2H3]NAT, are marked. B, representative MS/MS spectrum of [acetyl-2H3]NAT in the urine of wild-type mice treated with deuterated ethanol. The fragmentation pattern of [acetyl-2H3]NAT from deuterated ethanol was interpreted in the inlaid diagram. C, relative abundance of NAT and [acetyl-2H3]NAT during the 7-day exposure of deuterated ethanol. The ion intensity of unlabeled NAT in the urine samples of wild-type mice prior to the exposure of deuterated ethanol was arbitrarily set as 1. D, NAT as a minor metabolite of ethanol. The amounts of daily excretion of NAT and EtG in urine were calculated as the percentage of daily ethanol intake through an equal molar conversion.
Identification of NAT as the Product of the Enzymatic Reaction between Taurine and Acetate
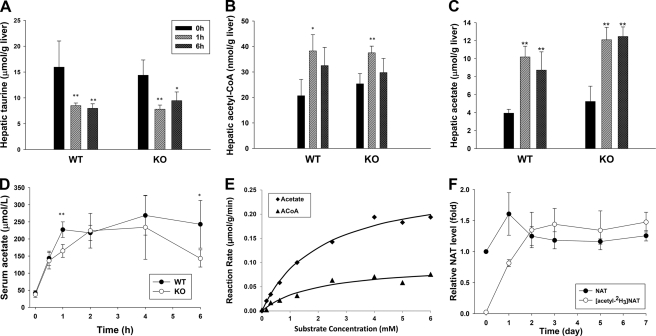
As a taurine ester, NAT is likely formed by the esterification reaction with taurine. Because stable isotope labeling analysis revealed that ethanol can donate an acetyl group for NAT biosynthesis (Fig. 5B), acetyl-CoA and acetate, two ethanol metabolites containing acetyl groups, were considered as possible candidates of acetyl donors. To test this hypothesis, the levels of taurine, acetyl-CoA, and acetate in the wild-type and Cyp2e1-null mice were measured after administering a bolus dose of ethanol (intraperitoneal injection of 4 g/kg ethanol). As taurine and acetyl-CoA are highly enriched inside the cells due to their specific transport and biosynthesis mechanisms, the influences of ethanol were evaluated by measuring their levels in the liver. In contrast, acetate is highly diffusive and much more evenly distributed in the body. Hence, the acetate levels in both liver and serum were examined to determine the impact of ethanol on its turnover. Results from quantitative LC-MS analysis showed that all three metabolites were affected by ethanol treatment (Fig. 6, A–D). Among them, the hepatic taurine level was significantly reduced in both wild-type and Cyp2e1-null mice (Fig. 6A), suggesting that the utilization of taurine occurred quickly after ethanol challenge. In contrast to the decrease of hepatic taurine level, the concentrations of acetyl-CoA and acetate in the liver of both mouse lines were significantly increased by ethanol challenge (Fig. 6, B and C), as the metabolism of excessive ethanol is expected to elevate their levels in the liver (55, 56). One clear difference between hepatic acetate and acetyl-CoA is that the concentration of hepatic acetate was over 200-fold higher than the concentration of hepatic acetyl-CoA. In addition, the dramatic increase of serum acetate levels was observed in both wild-type and Cyp2e1-null mice following ethanol challenge (Fig. 6D). The high level of serum acetate was sustained in the wild-type mice for 6 h, but it was gradually reduced in the Cyp2e1-null mice after 4 h of ethanol treatment (Fig. 6D). Overall, the changes of three metabolites after ethanol challenge, i.e. the decrease of taurine and the increase of acetyl-CoA and acetate, were consistent with the increase of NAT, suggesting these changes potentially contribute to ethanol-induced NAT biosynthesis.
FIGURE 6.
Acetate as a main substrate for NAT biosynthesis in vivo. Levels of potential precursors of NAT in the wild-type (WT) and Cyp2e1-null (KO) mice after intraperitoneal injection of 4 g/kg ethanol were measured by the LC-MS methods described under “Experimental Procedures.” A, hepatic taurine level. B, hepatic acetyl-CoA level. C, hepatic acetate level. D, serum acetate level. E, enzyme kinetics of NAT biosynthesis in vitro. NAT biosynthesis was conducted by incubating liver homogenate with 20 mm taurine and various concentrations of acetate or acetyl-CoA. F, relative abundance of NAT and [acetyl-2H3]NAT during the 7-day exposure of deuterated acetate. The WT mice were fed with the diet containing 1.1% deuterated acetate ([2H4]acetate) for 7 days. The ion intensity of unlabeled NAT in the urine samples of WT mice prior to the exposure of deuterated acetate was arbitrarily set as 1. Values were presented as mean ± S.D. (n = 4). *, p < 0.05, and **, p < 0.01, indicate statistical significance between post- and before-ethanol treatments in A–C and statistical significance between WT and KO samples at the same time point in D.
Because acetyl-CoA is the direct downstream metabolite of acetate in ethanol metabolism, two possible routes of NAT biosynthesis from acetate exist. One is the direct reaction of acetate with taurine, while the other is the reaction of acetyl-CoA with taurine after acetate was converted to acetyl-CoA. To examine these two routes of NAT biosynthesis, the in vitro incubations of liver homogenate of the wild-type mice with taurine and various concentrations of acetate or acetyl-CoA were performed, and the kinetics parameters of enzyme reactions were further determined. The 20 mm taurine concentration was chosen based on the concentration of hepatic taurine in the wild-type mouse (Fig. 6A) and the reported physiological concentration of taurine (57). The results showed that both acetyl-CoA and acetate can react with taurine to produce NAT in vitro with kinetic features of enzyme reaction (Fig. 6E). The Km and Vmax values for the reaction of acetyl-CoA and taurine are 1.96 mm and 0.10 μmol/g of liver/min, respectively, whereas the Km and Vmax values for the reaction of acetate and taurine are 2.10 mm and 0.27 μmol/g liver/min, respectively. Considering the correlation between the Km value of in vitro reactions (Fig. 6E) and the metabolite concentrations in the liver (Fig. 6, B and C), it is evident that the direct reaction of acetate and taurine, instead of the reaction of acetyl-CoA and taurine, should be the main route of NAT biosynthesis in vivo (detailed under “Discussion”). To further determine the role of acetate in NAT biosynthesis, a stable isotope labeling analysis was conducted by feeding the wild-type mice with the diet containing 1.1% deuterated acetate ([2H4]acetate) for 7 days. The results from monitoring both unlabeled NAT and [acetyl-2H3]NAT in the urine clearly demonstrated that the exposure of [2H4]acetate not only quickly led to the biosynthesis of [acetyl-2H3]NAT but also maintained its level during the 7-day treatment (Fig. 6F), confirming that the excessive acetate is the source of newly synthesized NAT after ethanol overdose.
A preliminary investigation on the enzymatic properties of NAT biosynthesis has been conducted through the in vitro assay using the compounds that can potentially affect the reaction, including metal ions (Ca2+, Cs2+, Fe2+, Hg2+, and Mg2+), chelator (EDTA), substrate analogs of taurine (β-alanine, α-alanine, and cysteine) and acetate (propionic acid and butyric acid), and known inhibitors of esterases (PMSF and diazinon). The results indicated that the metal ion is likely required as a cofactor because EDTA dramatically suppresses the enzymatic reaction, and various metal ions have either stimulatory or inhibitory effects on the reaction. Furthermore, the effects of substrate competition were observed using structural analogs of acetate or taurine, suggesting that acetate and taurine might not be the exclusive substrates of the proposed NAT synthase. Finally, two common esterase inhibitors had suppressive effects on the enzymatic activity (supplemental Table S1).
Sites of NAT Biosynthesis
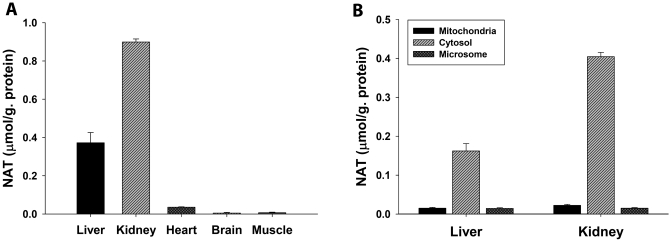
Although the liver is the major site of ethanol metabolism, the high level of acetate in serum makes acetate available for further metabolism in extrahepatic tissues after ethanol exposure. To determine the tissues/organs that are capable of NAT biosynthesis, the in vitro incubations of taurine and acetate were conducted using the homogenates of tissues/organs that are metabolically active. The result clearly showed that the kidney has the highest enzymatic activity of NAT biosynthesis from taurine and acetate, even much higher than the activity in the liver (Fig. 7A). The contributions of heart, muscle, and brain to NAT biosynthesis in vivo are likely minimal based on their low activities even though their taurine contents are reportedly high (57). Furthermore, the intracellular locations of NAT biosynthesis were examined using subcellular fractions of kidney and liver, including mitochondria, cytosol, and microsome. It is clear from the results of in vitro incubations that the cytosolic fractions have much higher activity of NAT biosynthesis than the mitochondrial and microsomal fractions (Fig. 7B). In summary, in vitro study of NAT biosynthesis suggested that the cytosolic enzymes in kidney and liver are mainly responsible for NAT biosynthesis in vivo.
FIGURE 7.
NAT biosynthesis in tissues and intracellular fractions. Incubations of taurine and acetate with tissue homogenates or intracellular fractions were described under “Experimental Procedures.” A, NAT biosynthesis by liver, kidney, heart, brain, and muscle homogenates. B, NAT biosynthesis by mitochondrial, cytosolic, and microsomal factions of mouse liver and kidney.
DISCUSSION
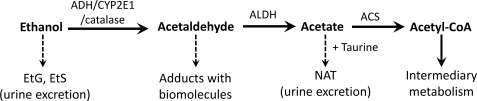
Ethanol metabolism has been studied extensively due to the ubiquitous presence of ethanol in nature and its association with hepatotoxicity and neurotoxicity in humans. A three-step route of ethanol metabolism has been established (9), in which acetaldehyde and acetate are the oxidized intermediates in the metabolic process of converting ethanol to acetyl-CoA, a central metabolite in the intermediary metabolism (Fig. 8). Besides these major metabolic reactions, minor metabolic pathways of ethanol and acetaldehyde have also been identified and characterized as important factors in ethanol-related toxicities. Glucuronidation and sulfation, two conjugation reactions, function as a minor detoxification mechanism to convert ethanol to EtG and ethyl sulfate, two unreactive metabolites that have been used as the biomarkers of ethanol consumption (29, 30). In contrast to the detoxification feature of ethanol conjugation reactions, nonenzymatic adduction reactions between acetaldehyde and biomolecules (proteins and DNAs) have been suggested as a major contributing factor in ethanol-induced toxicities (58). In this study, the formation of NAT was identified and characterized as a novel metabolic pathway of acetate, which may serve as a protective mechanism by removing excessive acetate from blood to urine (Fig. 8).
FIGURE 8.
Role of NAT and CYP2E1 in ethanol metabolism. Major metabolic pathways of ethanol (arrow with a solid line), encompassing three enzymatic steps and two intermediates, are responsible for converting ethanol into acetyl-CoA, a central metabolite of intermediary metabolism, whereas minor metabolic pathways (arrow with a dashed line) affect the toxic effects of ethanol, i.e. the adductions of acetaldehyde with biomolecules contribute to the toxicity, but the formations of EtG, ethyl sulfate (EtS), and NAT facilitate the excretion of ethanol and acetate. CYP2E1 contributes to the formation of acetaldehyde, which is further oxidized to acetate, the substrate of NAT biosynthesis. ALDH, aldehyde dehydrogenase; ACS, acetyl-CoA synthetase.
NAT, a highly water-soluble and hygroscopic compound, was previously found in nature as a major component in the sticky droplet of orb spider web (59). Its presence in the biofluids of mammals has not been described until a recent study on the urinary metabolome of radiation-exposed rat, in which the urinary NAT level was increased by γ-irradiation (46). This study, for the first time, showed that NAT is a constitutive component of mouse urine, and its level dramatically increased after ethanol exposure. Stable isotope labeling analysis using deuterated ethanol further revealed that NAT is indeed a novel metabolite of ethanol (Fig. 5C). As a taurine ester, NAT is likely formed by one or multiple N-acetylation reactions between taurine and ethanol metabolites. To test this hypothesis, the possibilities of acetyl-CoA and acetate, two ethanol metabolites containing acetyl groups, as the donor of the acetyl group of NAT were investigated both in vitro and in vivo. Hyperacetatemia, instead of high levels of acetyl-CoA, was defined as the major contributor of NAT induction after ethanol exposure based on several facts. 1) The quantitation data showed that the concentration of acetate is much higher than the concentration of acetyl-CoA in vivo both before and after ethanol exposure (Fig. 6, B and C). Hence, the concentrations of acetate in the liver and serum are comparable with the determined Km value of the reaction between acetate and taurine, but the concentration of hepatic acetyl-CoA is far below the Km value of the reaction between acetyl-CoA and taurine (Fig. 6E), suggesting that the reaction between acetate and taurine is much more likely to happen in vivo. 2) Incubations of cellular fractions of the liver and kidney indicated that the intracellular site of NAT biosynthesis is the cytosol, not the mitochondria or microsome (Fig. 7B). Because acetyl-CoA concentration in the cytosol is much lower than its concentration in the mitochondria (60), it is unlikely that active NAT biosynthesis from acetyl-CoA occurs in the cytosol. Furthermore, this proposed NAT biosynthesis pathway was supported by observing the decrease of taurine content in the liver after ethanol challenge (Fig. 6A) and detecting the formation of deuterated NAT after the treatment of deuterated acetate (Fig. 6F), suggesting the consumption of taurine and acetate for the formation of NAT in vivo. The enzymatic feature of NAT biosynthesis from acetate and taurine was further investigated and can summarized as follows. 1) The reaction between acetate and taurine is enzymatic because boiled liver and kidney homogenates fail to generate NAT from acetate and taurine but can yield a small amount of NAT from acetyl-CoA and taurine (data not shown). 2) The reaction between acetate and taurine is likely a one-step direct reaction that does not require the conversion of acetate to acetyl-CoA before reacting with taurine. This conclusion is largely based on the fact that ATP and CoA, two essential cofactors in acetyl-CoA synthetase-mediated acetyl-CoA synthesis from acetate (61), are not required for the NAT biosynthesis from acetate and taurine. 3) Metal ion is likely required for the catalytic activity of NAT synthase based on the inhibitory effects of chelator and the modulatory effects of various metal ions (supplemental Table S1). 4) The enzyme responsible for NAT synthesis is mainly located in the cytosol of kidney and liver, as suggested by the NAT synthesis through in vitro incubation of various organ homogenates and intracellular fractions (Fig. 7). In summary, NAT synthase is likely a cytosolic metalloenzyme in the kidney and liver that can directly catalyze the esterification reaction between taurine and acetate without the involvement of ATP and CoA. At present, the exact protein identity of NAT synthase is still unknown. We expect the purification and characterization of this enzyme in future studies will reveal more information on the cofactor requirement, catalytic property, and its relation with ethanol treatment.
Acetate and taurine, as the sources of NAT biosynthesis, are two biochemically important metabolites. Therefore, the formation of NAT is expected to affect the metabolic pathways related to acetate and taurine, as evidenced by the increased acetate level and the decreased taurine level after ethanol treatment in this study (Fig. 6, A–D). Acetate in the body originates from both endogenous and exogenous sources, including the hydrolysis of acetyl-CoA, the metabolism of gut flora, and ethanol metabolism (62). When these acetate-producing routes are activated, such as in the diabetic humans and animals (63, 64) and after excessive ethanol consumption, hyperacetatemia, the high level of acetate in blood, can be readily induced. In addition, hyperacetatemia also commonly occurs in long term hemodialysis patients after using dialysis fluid containing sodium acetate as buffering agent (65). Hyperacetatemia has been associated with the development of dyslipoproteinemia and atherosclerosis, especially in some hemodialysis patients (66), and has also been recently implicated as a main cause of alcohol hangover headache (67). Moreover, increased serum acetate has been shown as a better marker of problem drinking among drunken driver than ethanol (68). At present, the acetate level in human patients is mainly monitored by measuring its level in blood. However, the results from this study suggested that urinary NAT can potentially become an effective biomarker of hyperacetatemia based on the correlation between serum acetate level and urinary NAT level, as well as the reaction mechanism of NAT biosynthesis. Because of the noninvasive nature of urine collection and the convenience of NAT detection (which does not require the derivatization procedure used in the measurement of acetate), measuring NAT might provide an alternative approach for evaluating the status of acetate metabolism and enable the early diagnosis of diseases and toxic effects associated with hyperacetatemia. This hypothesis will require further validation in various hyperacetatemic situations.
Taurine, with its numerous physiological functions, is a highly abundant free amino acid in the body. Its intracellular concentration ranges between 5 and 50 mm in many mammalian tissues (57). Because taurine is an end product of methionine metabolism pathway (69), its level can partially reflect the status of sulfur-containing amino acids in vivo. The protective effects of taurine against the ethanol-induced toxicities, such as hepatic steatosis and lipid peroxidation, have been revealed in several studies on taurine supplementation (70, 71) and have been further proven by the studies of taurine depletion, in which the increased susceptibility to ethanol-induced hepatic dysfunction was observed (72). The mechanism behind these observations has been largely attributed to the indirect interaction between taurine and ethanol through the membrane protection and antioxidant activities of taurine. Our results from this study provide the first evidence that taurine reacts directly with acetate, a major metabolite of ethanol, at physiologically relevant concentrations. Therefore, besides its indirect interaction with ethanol, taurine indeed can directly interfere with ethanol metabolism through the formation of NAT to remove the excessive acetate. This conclusion is further supported by the observation of much higher activity of NAT biosynthesis in the kidney than other organs, which can facilitate the excretion of NAT into urine. Further studies will demonstrate whether the supplementation or the depletion of taurine will significantly affect the NAT production after ethanol exposure and whether taurine could be an effective antidote against the hyperacetatemia in the pathological conditions.
One of the aims of this study was to examine the role of CYP2E1 in the development of ethanol-induced steatosis because the previous studies yielded different conclusions on this issue (24–27). Both histological and biochemical analyses of the responses of the wild-type and Cyp2e1-null mice to ethanol treatment in this study (Figs. 1 and 2) indicated that deficiency of CYP2E1 reduces the toxic effects of ethanol. The correlation of the Cyp2e1 genotype with higher NAT levels in urine (Fig. 4A and supplemental Fig. S2A) and higher acetate levels in serum (Fig. 6D) further supports the role of CYP2E1 in ethanol metabolism as an enzyme participating in the oxidation of ethanol to acetaldehyde, especially in ethanol overdose. Interestingly, our conclusion on a contributing role of CYP2E1 in the ethanol-induced toxicity is consistent with the results from previous studies using a comparable oral feeding method (24, 25), but it is different from other studies using the intragastric infusion method, in which CYP2E1 deficiency failed to make a difference in terms of steatosis (26, 27). Even though the causes leading to these inconsistent observations on ethanol and CYP2E1 remain largely unknown, it has been suggested that the endotoxemia and the increased TNFα level associated with the intragastric infusion model might contribute to the differences among these studies (25).
The identification of NAT as a metabolite of ethanol and a biomarker of hyperacetatemia was mainly facilitated by the adoption of untargeted LC-MS-based metabolomic analysis in this study because the traditional bioanalysis approaches focusing on examining a single molecule or a defined cluster of molecules lack the capacity to detect unexpected or novel metabolites in complex biological matrices. Compared with previous metabolomics studies on ethanol intoxication (36–39), this study has further expanded the power of metabolomics in characterizing biochemical events using stable isotope labeling analysis. Because of the mass difference between unlabeled compound and its stable isotope-labeled counterpart, the metabolic fates of exogenous compounds can be effectively tracked through the MS-based metabolomic analysis of biological samples in the loadings plot of the MDA model, as shown by the identification of ethanol metabolites in this study (Fig. 5A) and the identification of novel acetaminophen metabolites in our recent study on the toxicity-related acetaminophen metabolites (73). In this study, the stable isotope labeling approach also conveniently distinguished the endogenous NAT from the exogenous NAT after deuterated ethanol and acetate treatment (Figs. 5C and 6F). Because of its effectiveness in the identification of metabolites and the characterization of the metabolic pathway, stable isotope labeling will have broad application in the MS-based metabolomics.
Overall, the combination of LC-MS-based metabolomics, stable isotope labeling, animal modeling, and in vivo and in vitro biochemical analysis in this study enabled the unambiguous identification of NAT as a novel metabolite of ethanol formed by the unreported enzymatic reaction between acetate and taurine. Ethanol-induced hyperacetatemia, which is partially contributed by the CYP2E1-mediated ethanol metabolism, is likely the main cause of increased NAT levels in urine. The value of NAT as the biomarker of diseases, such as ethanol-induced tissue injury or hemodialysis-related toxicity, requires further investigation.
Supplementary Material
Acknowledgment
We thank Dr. Frank J. Gonzalez (Laboratory of Metabolism, NCI, National Institutes of Health) for providing Cyp2e1−/− mice and CYP2E1 antibody.
This work was supported by a University of Minnesota startup fund (to C. C.).

This article contains supplemental “Experimental Procedures,” Table S1, and Figs. S1 and S2.
- EtG
- ethyl glucuronide
- NAT
- N-acetyltaurine
- MDA
- multivariate data analysis
- PLS-DA
- projection to latent structures-discriminant analysis
- OPLS
- orthogonal projection to latent structures
- MS/MS
- tandem mass spectrometry
- LC-MS
- liquid chromatography-mass spectrometry
- EtG
- ethyl glucuronide
- BUN
- blood urea nitrogen
- TAG
- triacylglycerol
- QTOF
- quadrupole TOF
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl
- PLS-DA
- projection to latent structures-discriminant analysis.
REFERENCES
- 1. Lieber C. S. (1995) Medical disorders of alcoholism. N. Engl. J. Med. 333, 1058–1065 [DOI] [PubMed] [Google Scholar]
- 2. Sable H. J., Rodd Z. A., Bell R. L., Schultz J. A., Lumeng L., McBride W. J. (2005) Effects of ethanol drinking on central nervous system functional activity of alcohol-preferring rats. Alcohol 35, 129–135 [DOI] [PubMed] [Google Scholar]
- 3. Mukamal K. J., Chung H., Jenny N. S., Kuller L. H., Longstreth W. T., Jr., Mittleman M. A., Burke G. L., Cushman M., Beauchamp N. J., Jr., Siscovick D. S. (2005) Alcohol use and risk of ischemic stroke among older adults. The cardiovascular health study. Stroke 36, 1830–1834 [DOI] [PubMed] [Google Scholar]
- 4. Rao R. K., Seth A., Sheth P. (2004) Recent advances in alcoholic liver disease. I. Role of intestinal permeability and endotoxemia in alcoholic liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 286, G881–G884 [DOI] [PubMed] [Google Scholar]
- 5. Mann R. E., Smart R. G., Govoni R. (2003) The epidemiology of alcoholic liver disease. Alcohol Res. Health 27, 209–219 [PMC free article] [PubMed] [Google Scholar]
- 6. McClain C. J., Song Z., Barve S. S., Hill D. B., Deaciuc I. (2004) Recent advances in alcoholic liver disease. IV. Dysregulated cytokine metabolism in alcoholic liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G497–G502 [DOI] [PubMed] [Google Scholar]
- 7. Hines I. N., Wheeler M. D. (2004) Recent advances in alcoholic liver disease. III. Role of the innate immune response in alcoholic hepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G310–G314 [DOI] [PubMed] [Google Scholar]
- 8. You M., Crabb D. W. (2004) Recent advances in alcoholic liver disease. II. Minireview. Molecular mechanisms of alcoholic fatty liver. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G1–G6 [DOI] [PubMed] [Google Scholar]
- 9. Lieber C. S. (2005) Metabolism of alcohol. Clin. Liver Dis. 9, 1–35 [DOI] [PubMed] [Google Scholar]
- 10. Lieber C. S. (2000) Alcohol. Its metabolism and interaction with nutrients. Annu. Rev. Nutr. 20, 395–430 [DOI] [PubMed] [Google Scholar]
- 11. Nakamura K., Iwahashi K., Furukawa A., Ameno K., Kinoshita H., Ijiri I., Sekine Y., Suzuki K., Iwata Y., Minabe Y., Mori N. (2003) Acetaldehyde adducts in the brain of alcoholics. Arch. Toxicol. 77, 591–593 [DOI] [PubMed] [Google Scholar]
- 12. Worrall S., de Jersey J., Nicholls R., Wilce P. (1993) Acetaldehyde/protein interactions. Are they involved in the pathogenesis of alcoholic liver disease? Dig. Dis. 11, 265–277 [DOI] [PubMed] [Google Scholar]
- 13. Nordmann R., Ribière C., Rouach H. (1992) Implication of free radical mechanisms in ethanol-induced cellular injury. Free Radic. Biol. Med. 12, 219–240 [DOI] [PubMed] [Google Scholar]
- 14. Cederbaum A. I. (2001) Introduction-serial review. Alcohol, oxidative stress, and cell injury. Free Radic. Biol. Med. 31, 1524–1526 [DOI] [PubMed] [Google Scholar]
- 15. Arteel G. E. (2003) Oxidants and antioxidants in alcohol-induced liver disease. Gastroenterology 124, 778–790 [DOI] [PubMed] [Google Scholar]
- 16. Theorell H., McKee J. S. (1961) Mechanism of action of liver alcohol dehydrogenase. Nature 192, 47–50 [DOI] [PubMed] [Google Scholar]
- 17. Keilin D., Hartree E. F. (1945) Properties of catalase. Catalysis of coupled oxidation of alcohols. Biochem. J. 39, 293–301 [PMC free article] [PubMed] [Google Scholar]
- 18. Tsutsumi M., Lasker J. M., Shimizu M., Rosman A. S., Lieber C. S. (1989) The intralobular distribution of ethanol-inducible P450IIE1 in rat and human liver. Hepatology 10, 437–446 [DOI] [PubMed] [Google Scholar]
- 19. Dalziel K., Dickinson F. M. (1966) The kinetics and mechanism of liver alcohol dehydrogenase with primary and secondary alcohols as substrates. Biochem. J. 100, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norberg A., Jones A. W., Hahn R. G., Gabrielsson J. L. (2003) Role of variability in explaining ethanol pharmacokinetics. Research and forensic applications. Clin. Pharmacokinet. 42, 1–31 [DOI] [PubMed] [Google Scholar]
- 21. Aragon C. M., Stotland L. M., Amit Z. (1991) Studies on ethanol-brain catalase interaction. Evidence for central ethanol oxidation. Alcohol Clin. Exp. Res. 15, 165–169 [DOI] [PubMed] [Google Scholar]
- 22. Lieber C. S., DeCarli L. M. (1968) Ethanol oxidation by hepatic microsomes. Adaptive increase after ethanol feeding. Science 162, 917–918 [DOI] [PubMed] [Google Scholar]
- 23. Caro A. A., Cederbaum A. I. (2004) Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu. Rev. Pharmacol. Toxicol. 44, 27–42 [DOI] [PubMed] [Google Scholar]
- 24. Lu Y., Zhuge J., Wang X., Bai J., Cederbaum A. I. (2008) Cytochrome P450 2E1 contributes to ethanol-induced fatty liver in mice. Hepatology 47, 1483–1494 [DOI] [PubMed] [Google Scholar]
- 25. Lu Y., Wu D., Wang X., Ward S. C., Cederbaum A. I. (2010) Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic. Biol. Med. 49, 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kono H., Bradford B. U., Yin M., Sulik K. K., Koop D. R., Peters J. M., Gonzalez F. J., McDonald T., Dikalova A., Kadiiska M. B., Mason R. P., Thurman R. G. (1999) CYP2E1 is not involved in early alcohol-induced liver injury. Am. J. Physiol. 277, G1259–G1267 [DOI] [PubMed] [Google Scholar]
- 27. Isayama F., Froh M., Bradford B. U., McKim S. E., Kadiiska M. B., Connor H. D., Mason R. P., Koop D. R., Wheeler M. D., Arteel G. E. (2003) The CYP inhibitor 1-aminobenzotriazole does not prevent oxidative stress associated with alcohol-induced liver injury in rats and mice. Free Radic. Biol. Med. 35, 1568–1581 [DOI] [PubMed] [Google Scholar]
- 28. MacSween R. N., Burt A. D. (1986) Histologic spectrum of alcoholic liver disease. Semin. Liver Dis. 6, 221–232 [DOI] [PubMed] [Google Scholar]
- 29. Wurst F. M., Skipper G. E., Weinmann W. (2003) Ethyl glucuronide—the direct ethanol metabolite on the threshold from science to routine use. Addiction 98, Suppl. 2, 51–61 [DOI] [PubMed] [Google Scholar]
- 30. Wurst F. M., Dresen S., Allen J. P., Wiesbeck G., Graf M., Weinmann W. (2006) Ethyl sulfate. A direct ethanol metabolite reflecting recent alcohol consumption. Addiction 101, 204–211 [DOI] [PubMed] [Google Scholar]
- 31. Anton R. F., Lieber C., Tabakoff B. (2002) Carbohydrate-deficient transferrin and γ-glutamyltransferase for the detection and monitoring of alcohol use. Results from a multisite study. Alcohol Clin. Exp. Res. 26, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 32. Rosman A. S., Lieber C. S. (1994) Diagnostic utility of laboratory tests in alcoholic liver disease. Clin. Chem. 40, 1641–1651 [PubMed] [Google Scholar]
- 33. Wymer A., Becker D. M. (1990) Recognition and evaluation of red blood cell macrocytosis in the primary care setting. J. Gen. Intern. Med. 5, 192–197 [DOI] [PubMed] [Google Scholar]
- 34. Freeman T. L., Tuma D. J., Thiele G. M., Klassen L. W., Worrall S., Niemelä O., Parkkila S., Emery P. W., Preedy V. R. (2005) Recent advances in alcohol-induced adduct formation. Alcohol Clin. Exp. Res. 29, 1310–1316 [DOI] [PubMed] [Google Scholar]
- 35. Chen C., Gonzalez F. J., Idle J. R. (2007) LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 39, 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manna S. K., Patterson A. D., Yang Q., Krausz K. W., Li H., Idle J. R., Fornace A. J., Jr., Gonzalez F. J. (2010) Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J. Proteome Res. 9, 4176–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masuo Y., Imai T., Shibato J., Hirano M., Jones O. A., Maguire M. L., Satoh K., Kikuchi S., Rakwal R. (2009) Omic analyses unravels global molecular changes in the brain and liver of a rat model for chronic Sake (Japanese alcoholic beverage) intake. Electrophoresis 30, 1259–1275 [DOI] [PubMed] [Google Scholar]
- 38. Bradford B. U., O'Connell T. M., Han J., Kosyk O., Shymonyak S., Ross P. K., Winnike J., Kono H., Rusyn I. (2008) Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease. Toxicol. Appl. Pharmacol. 232, 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zivkovic A. M., Bruce German J., Esfandiari F., Halsted C. H. (2009) Quantitative lipid metabolomic changes in alcoholic micropigs with fatty liver disease. Alcohol Clin. Exp. Res. 33, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee S. S., Buters J. T., Pineau T., Fernandez-Salguero P., Gonzalez F. J. (1996) Role of CYP2E1 in the hepatotoxicity of acetaminophen. J. Biol. Chem. 271, 12063–12067 [DOI] [PubMed] [Google Scholar]
- 41. Lieber C. S., DeCarli L. M. (1989) Liquid diet technique of ethanol administration. 1989 update. Alcohol Alcohol. 24, 197–211 [PubMed] [Google Scholar]
- 42. Bykov I., Palmén M., Piirainen L., Lindros K. O. (2004) Oral chronic ethanol administration to rodents by agar gel diet. Alcohol Alcohol. 39, 499–502 [DOI] [PubMed] [Google Scholar]
- 43. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 44. Park S. S., Ko I. Y., Patten C., Yang C. S., Gelboin H. V. (1986) Monoclonal antibodies to ethanol-induced cytochrome P-450 that inhibit aniline and nitrosamine metabolism. Biochem. Pharmacol. 35, 2855–2858 [DOI] [PubMed] [Google Scholar]
- 45. van Velzen E. J., Westerhuis J. A., van Duynhoven J. P., van Dorsten F. A., Hoefsloot H. C., Jacobs D. M., Smit S., Draijer R., Kroner C. I., Smilde A. K. (2008) Multilevel data analysis of a crossover designed human nutritional intervention study. J. Proteome Res. 7, 4483–4491 [DOI] [PubMed] [Google Scholar]
- 46. Johnson C. H., Patterson A. D., Krausz K. W., Lanz C., Kang D. W., Luecke H., Gonzalez F. J., Idle J. R. (2011) Radiation metabolomics. 4. UPLC-ESI-QTOFMS-based metabolomics for urinary biomarker discovery in γ-irradiated rats. Radiat. Res. 175, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westerhuis J. A., van Velzen E. J., Hoefsloot H. C., Smilde A. K. (2010) Multivariate paired data analysis. Multilevel PLSDA versus OPLSDA. Metabolomics 6, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vasiliou V., Ziegler T. L., Bludeau P., Petersen D. R., Gonzalez F. J., Deitrich R. A. (2006) CYP2E1 and catalase influence ethanol sensitivity in the central nervous system. Pharmacogenet. Genomics 16, 51–58 [DOI] [PubMed] [Google Scholar]
- 49. Márquez F. J., Quesada A. R., Sánchez-Jiménez F., Núñez de Castro I. (1986) Determination of 27 dansyl amino acid derivatives in biological fluids by reversed-phase high performance liquid chromatography. J. Chromatogr. 380, 275–283 [DOI] [PubMed] [Google Scholar]
- 50. Higashi T., Ichikawa T., Inagaki S., Min J. Z., Fukushima T., Toyo'oka T. (2010) Simple and practical derivatization procedure for enhanced detection of carboxylic acids in liquid chromatography-electrospray ionization-tandem mass spectrometry. J. Pharm. Biomed. Anal. 52, 809–818 [DOI] [PubMed] [Google Scholar]
- 51. Pönniö M., Alho H., Heinälä P., Nikkari S. T., Sillanaukee P. (1999) Serum and saliva levels of sialic acid are elevated in alcoholics. Alcohol. Clin. Exp. Res. 23, 1060–1064 [DOI] [PubMed] [Google Scholar]
- 52. Warrack B. M., Hnatyshyn S., Ott K. H., Reily M. D., Sanders M., Zhang H., Drexler D. M. (2009) Normalization strategies for metabonomic analysis of urine samples. J. Chromatogr B. Analyt. Technol. Biomed Life Sci. 877, 547–552 [DOI] [PubMed] [Google Scholar]
- 53. Chung F. M., Yang Y. H., Shieh T. Y., Shin S. J., Tsai J. C., Lee Y. J. (2005) Effect of alcohol consumption on estimated glomerular filtration rate and creatinine clearance rate. Nephrol. Dial. Transplant. 20, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 54. Schaeffner E. S., Kurth T., de Jong P. E., Glynn R. J., Buring J. E., Gaziano J. M. (2005) Alcohol consumption and the risk of renal dysfunction in apparently healthy men. Arch. Intern. Med. 165, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 55. Kondrup J., Grunnet N. (1973) The effect of acute and prolonged ethanol treatment on the contents of coenzyme A, carnitine, and their derivatives in rat liver. Biochem. J. 132, 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bode C., Stähler E., Kono H., Goebell H. (1970) Effects of ethanol on free coenzyme A, free carnitine, and their fatty acid esters in rat liver. Biochim. Biophys. Acta 210, 448–455 [DOI] [PubMed] [Google Scholar]
- 57. Hayes K. C., Sturman J. A. (1981) Taurine in metabolism. Annu. Rev. Nutr. 1, 401–425 [DOI] [PubMed] [Google Scholar]
- 58. Brooks P. J., Theruvathu J. A. (2005) DNA adducts from acetaldehyde. Implications for alcohol-related carcinogenesis. Alcohol 35, 187–193 [DOI] [PubMed] [Google Scholar]
- 59. Mayer J., Denger K., Smits T. H., Hollemeyer K., Groth U., Cook A. M. (2006) N-Acetyltaurine dissimilated via taurine by Delftia acidovorans NAT. Arch. Microbiol. 186, 61–67 [DOI] [PubMed] [Google Scholar]
- 60. Siess E. A., Brocks D. G., Wieland O. H. (1978) Distribution of metabolites between the cytosolic and mitochondrial compartments of hepatocytes isolated from fed rats. Hoppe-Seylers Z. Physiol. Chem. 359, 785–798 [DOI] [PubMed] [Google Scholar]
- 61. Jogl G., Tong L. (2004) Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry 43, 1425–1431 [DOI] [PubMed] [Google Scholar]
- 62. Buckley B. M., Williamson D. H. (1977) Origins of blood acetate in the rat. Biochem. J. 166, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smith R. F., Humphreys S., Hockaday T. D. (1986) The measurement of plasma acetate by a manual or automated technique in diabetic and nondiabetic subjects. Ann. Clin. Biochem. 23, 285–291 [DOI] [PubMed] [Google Scholar]
- 64. Murthy V. K., Steiner G. (1972) Hepatic acetic thiokinase. Possible regulatory step in lipogenesis. Metabolism 21, 213–221 [DOI] [PubMed] [Google Scholar]
- 65. Desch G., Oules R., Mion C., Descomps B., De Paulet A. C. (1978) Plasma acetate levels during hemodialysis. Clin. Chim. Acta 85, 231–241 [DOI] [PubMed] [Google Scholar]
- 66. Yalçin A., Kocaolu S., Akçiçek F., Ozyer A. (1999) Effects of acetate or bicarbonate dialysis solutions on serum HDL and HDL subfractions of patients undergoing hemodialysis. Curr. Med. Res. Opin. 15, 310–315 [DOI] [PubMed] [Google Scholar]
- 67. Maxwell C. R., Spangenberg R. J., Hoek J. B., Silberstein S. D., Oshinsky M. L. (2010) Acetate causes alcohol hangover headache in rats. PLoS One 5, e15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roine R. P., Korri U. M., Ylikahri R., Penttila A., Pikkarainen J., Salaspuro M. (1988) Increased serum acetate as a marker of problem drinking among drunken drivers. Alcohol Alcohol. 23, 123–126 [PubMed] [Google Scholar]
- 69. Huxtable R. J. (1992) Physiological actions of taurine. Physiol. Rev. 72, 101–163 [DOI] [PubMed] [Google Scholar]
- 70. Watanabe A., Hobara N., Nagashima H. (1985) Lowering of liver acetaldehyde but not ethanol concentrations by pretreatment with taurine in ethanol-loaded rats. Experientia 41, 1421–1422 [DOI] [PubMed] [Google Scholar]
- 71. Kerai M. D., Waterfield C. J., Kenyon S. H., Asker D. S., Timbrell J. A. (1999) Reversal of ethanol-induced hepatic steatosis and lipid peroxidation by taurine. A study in rats. Alcohol Alcohol. 34, 529–541 [DOI] [PubMed] [Google Scholar]
- 72. Kerai M. D., Waterfield C. J., Kenyon S. H., Asker D. S., Timbrell J. A. (2001) The effect of taurine depletion by βalanine treatment on the susceptibility to ethanol-induced hepatic dysfunction in rats. Alcohol Alcohol. 36, 29–38 [DOI] [PubMed] [Google Scholar]
- 73. Chen C., Krausz K. W., Idle J. R., Gonzalez F. J. (2008) Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J. Biol. Chem. 283, 4543–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.