Background: SCARB2 is the cellular receptor for EV71.
Results: Seven highly variable amino acids on SCARB2 and a region in proximity to residue Gln-172 of VP1 are critical for EV71 infection.
Conclusion: EV71 might adopt the canyon receptor engagement mode for viral entry.
Significance: Elucidation of receptor and viral determinants revealed a possible target for anti-EV71 interventions.
Keywords: Host-Pathogen Interactions, Infectious Diseases, Scavenger Receptor, Viral Protein, Virus Entry, Capsid VP1, EV71, SCARB2
Abstract
Enterovirus 71 (EV71) is one of the major pathogens that cause hand, foot, and mouth disease outbreaks in young children in the Asia-Pacific region in recent years. Human scavenger receptor class B 2 (SCARB2) is the main cellular receptor for EV71 on target cells. The requirements of the EV71-SCARB2 interaction have not been fully characterized, and it has not been determined whether SCARB2 serves as an uncoating receptor for EV71. Here we compared the efficiency of the receptor from different species including human, horseshoe bat, mouse, and hamster and demonstrated that the residues between 144 and 151 are critical for SCARB2 binding to viral capsid protein VP1 of EV71 and seven residues from the human receptor could convert murine SCARB2, an otherwise inefficient receptor, to an efficient receptor for EV71 viral infection. We also identified that EV71 binds to SCARB2 via a canyon of VP1 around residue Gln-172. Soluble SCARB2 could convert the EV71 virions from 160 S to 135 S particles, indicating that SCARB2 is an uncoating receptor of the virus. The uncoating efficiency of SCARB2 significantly increased in an acidic environment (pH 5.6). These studies elucidated the viral capsid and receptor determinants of enterovirus 71 infection and revealed a possible target for antiviral interventions.
Introduction
Enterovirus 71 (EV71)3 belongs to human enterovirus species A of the genus Enterovirus within the Picornaviridae family. It is genetically close to another human enterovirus species A virus, coxsackievirus A16 (CA16) (1). Both EV71 and CA16 are common causes of hand-foot-mouth disease (HFMD) that mostly affect young children. HFMD is often mild, self-limiting, and characterized by symptoms of fever, sore throat, diarrhea, and papulovesicular rash on the hands, feet, and oropharyngeal mucosa. However, unlike CA16, EV71 infection can also cause severe neurological complications such as acute encephalitis, poliomyelitis-like paralysis, and aseptic meningitis with or without pulmonary edema disease. The EV71-associated neurological complications can sometimes be fatal, and neurogenic pulmonary edema is thought to be the main pathogenic process in fatal cases (2–5). EV71 infections manifested with severe neurological diseases with high mortality rates were first reported in Bulgaria in 1975 and Hungary in 1978 (6, 7). Two decades later, large and severe HFMD epidemics reemerged in Malaysia in 1997 and Taiwan in 1998 and 2000 (8, 9). Since then, there have been more frequent large scale EV71-associated epidemics throughout the Asia-Pacific region, and fatal instances of neurotropic infections have become increasingly common, particularly in Southeast and East Asia. Between January 2009 and May 2011, the EV71 epidemics in China have resulted in more than two million HFMD cases and about 1000 deaths.4 As poliovirus, which causes poliomyelitis, is nearly eradicated, EV71 has emerged as an important human enterovirus and raised considerable public health concerns (11, 12). However, effective antiviral agents or vaccines against EV71 are currently unavailable.
EV71 is a nonenveloped virus comprising a positive single-stranded RNA genome (∼7.4 kb in length) that is packed within an outer protein capsid usually composed of four proteins (VP1–VP4) (1). Based on homologous structural modeling of the EV71 capsid proteins, the overall architecture of the virion is believed to be similar to other enteroviruses (e.g. poliovirus) in the picornavirus family (13). The capsid of the virus has an icosahedral symmetry and is ∼30 nm in diameter. It consists of 60 copies of pentameric subunits that are formed by each of the four capsid proteins, VP1, VP2, VP3, and VP4 (14, 15). VP1–VP3 of picornaviruses are exposed on the virion surface and are responsible for host-receptor binding, whereas VP4 is located inside the particle. VP1 is the most external, surface-accessible, and immunodominant protein among the picornavirus capsid proteins (16).
It has been shown that EV71 enters host cells via a receptor-mediated endocytosis pathway that is pH- and clathrin-dependent (17). Two human functional receptors for EV71 have recently been identified: human P-selectin glycoprotein ligand-1 (PSGL-1; CD162) and human scavenger receptor class B 2 (SCARB2; also known as lysosomal membrane protein II or CD36b-like-2) (18, 19). The PSGL-1 is expressed mainly on leukocytes, and it is likely that PSGL-1 primarily serves as a receptor for EV71 infection of leukocytes (19). It has also been shown that tyrosine sulfation, but not O-glycosylation, in the N-terminal region of PSGL-1 may facilitate virus entry and replication of EV71 in leukocytes (20). SCARB2 is a type III multiple transmembrane glycoprotein with both N and C termini on the cytosol side (21) located primarily in the membranes of lysosomes and endosomes and has been demonstrated as a specific receptor for targeting β-glucocerebrosidase to lysosomes (22). SCARB2 is widely expressed on different cell types including neurons; it may be directly involved in EV71 infection of the brain. Moreover, unlike PSGL-1, SCARB2 can be utilized by most EV71 strains as an entry receptor, indicating that SCARB2 may play a critical role in the early steps of EV71 infection (18, 19). The entire exon 4 of SCARB2 has been shown recently to be responsible for the interaction with VP1 protein of EV71 (23). However, the detailed molecular interaction between VP1 and SCARB2 has not been determined. It is still unknown whether the SCARB2 and VP1 interaction contributes to EV71 viral particle uncoating during the entry step of EV71 infection or whether SCARB2 binding mediates capsid alteration during uncoating.
In this study, using a single round pseudotype EV71 virus infection system, we investigated the ability of several SCARB2 molecules from different species including human, mouse, horseshoe bat, and hamster to serve as a receptor for EV71 infection. By comparing SCARB2s from human and horseshoe bat with that of hamster and mouse SCARB2, we identified critical residues required for human SCARB2 (hSCARB2) binding to VP1; this region comprises residues 144–151 in a highly variable region among species. We demonstrated that changing seven residues of mouse SCARB2 (mSCARB2) to corresponding residues of hSCARB2 could convert the mSCARB2 to an efficient receptor for EV71. We further identified that the determinant on VP1 for receptor binding, which comprises Gln-172 and surrounding residues, forms a canyon around the plateau on the virion surface. Finally, we showed that SCARB2, but not PSGL-1, is an uncoating receptor for EV71, and the uncoating efficiency of SCARB2 is significantly increased in an acidic environment (pH 5.6).
MATERIALS AND METHODS
Cell Lines, Antibodies, Plasmids, and Virus
Human embryonic kidney (HEK)-293T cells, human rhabdomyosarcoma (RD) cells, and baby hamster kidney (BHK21) cells were maintained in Dulbecco's modified essential medium (DMEM) containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in 5% CO2. Rabbit anti-actin IgG and horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit IgG antibodies were purchased from Sigma-Aldrich. SCARB2 cDNAs of human, mouse, horseshoe bat, and hamster were cloned from RD cells, L929 cells, primary fibroblasts of horseshoe bat, and BHK21 cells, respectively. These cDNA clones were subsequently cloned into pCAGGS vector fused with a nine-residue C-terminal tag (C9; TETSQVAPA) that is recognized by the mouse monoclonal antibody 1D4 (National Cell Culture Center). The virus enterovirus 71 (Fuyang (FY) strain; GenBankTM accession number EU703812; genotypic subgroup C4) was isolated from a patient in Fuyang City during the HFMD epidemic of 2007 in China. Virus was propagated and titrated in RD cells.
Generation of SCARB2 Knockdown RD Cells (RD/SCARB2 kd)
Short hairpin RNA (shRNA) sequences 5′-CCGGGACCAGAGTATCGAGAAGAAACTCGAGTTTCTTCTCGATACTCTGGTCTTTTTG-3′ and 5′-AATTCAAAAAGACCAGAGTATCGAGAAGAAACTCGAGTTTCTTCTCGATACTCTGGTC-3′ targeting the coding sequence of human SCARB2 mRNA were cloned into pLKO.1-puro vector (Sigma) driven by an hU6 promoter with AgeI and EcoRI sites. For production of the lentiviral particles, 293T cells were co-transfected with pLKO.1-shSCARB2, psPAX2, and pMD2.G (Sigma) at a ratio of 4:3:1. 48 h after transfection, the cell culture supernatant was harvested and filtered using a 0.45-μm filter membrane. RD cells were transduced with lentiviruses carrying the transgene pLKO.1-shSCARB2. After 4 days of incubation, transduced RD cells were screened with 2.0 μg/ml puromycin (Sigma) to obtain shSCARB2-incorporated RD (RD/SCARB2 kd) cells. SCARB2 mRNA levels of RD cells and RD/SCARB2 kd cells were determined by real time RT-PCR using the following primers: hSCARB2-qF (5′-CCAATACGTCAGACAATGCC) and hSCARB2-qR (5′-ACCATTCTTGCAGATGCTGA-3′).
Molecular Cloning of Epidemic EV71(FY) Strain
For cloning the full length of the FY RNA genome, 5 × 106 RD cells were infected with EV71(FY) with a multiplicity of infection of 0.1, and total RNA was isolated with TRIzol reagent when 80–90% cells rounded up. The genome of EV71(FY) was cloned using a PCR-based method. Briefly, three fragments ranging from nucleotides 1 to 2859, 2859 to 5391, and 5391 to 7405 of EV71 were amplified from FY cDNA and cloned into pMD18T vector (Takara). After sequence verification, the three fragments were ligated in vitro. The DNA sequence of T7 promoter was added to the 5′-end of the viral genome cDNA.
Production of Replication-competent EV71(FY)-EGFP Virus
A replication-competent EV71 with an EGFP reporter gene (EV71(FY)-EGFP virus; Fig. 1) was generated on the basis of pSVA-EV71-EGFP (18), which is an infectious clone of EV71 strain SK-EV006 (AB469182) kindly provided by Dr. Satoshi Koike. The coding sequence of EGFP was inserted between the 5′-untranslated region (UTR) and VP4 with an EV71 2A protease recognition site (-AITTL-) in between EGFP and VP4. The structural genes of SK-EV006 in pSVA-EV71-EGFP were replaced with EV71(FY) strain (GenBank accession number EU703812). EV71(FY)-EGFP RNA was prepared by in vitro transcription with a SmaI-linearized plasmid harboring EV71(FY)-EGFP as a template using a RiboMAX large scale RNA production kit (Promega). RD cells were transfected with EV71(FY)-EGFP RNA by Lipofectamine 2000 (Invitrogen). EV71(FY)-EGFP viruses were collected by two rounds of freeze-thaw cycles when the GFP-positive cells reached 80–90% and then filtered through a 0.45-μm filter, aliquoted, and frozen at −80 °C until use.
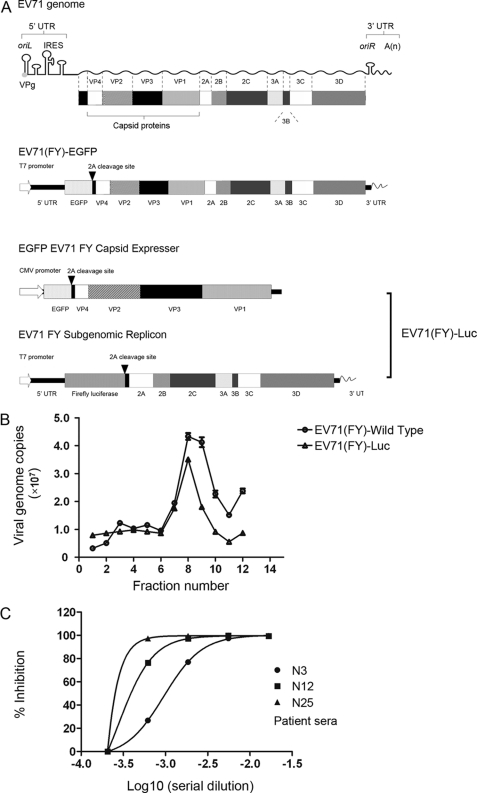
FIGURE 1.
Single round EV71 infection system. A, schematic map of the EV71 viral RNA genome, replication-competent EV71(FY)-EGFP viral genome, and EV71 replicon and capsid expresser for the single round EV71 infection system, EV71(FY)-Luc. EV71(FY)-EGFP is a replication-competent virus with the EGFP reporter gene inserted between the 5′-UTR and VP4. An EV71 2A protease recognition site (-AITTL-) was inserted between EGFP and VP4. For the single round EV71 infection system (EV71(FY)-Luc), the EV71 capsid expresser was used to express all the structural capsid genes in trans, an EGFP gene was inserted upstream of the EV71(FY) capsid gene, and all other EV71 viral genes were deleted. The EGFP was separated from the structural genes by a 2A self-cleavage site. EV71 replicon was produced by replacing the capsid coding region with a firefly luciferase reporter gene in the full-length EV71(FY) genome, and a T7 promoter was placed at the 5′-end for transcription in vitro. B, characterization of EV71 pseudotype virus by ultracentrifugation. Both wild type EV71(FY) and pseudotype virus EV71(FY)-Luc were applied to sucrose gradient ultracentrifugation, and the copy number of viral genomic RNA in each fraction was subsequently determined by quantitative RT-PCR. EV71(FY) virus was quantified with primers located in VP1. Specific primers for firefly luciferase reporter gene were used for quantifying pseudotype EV71(FY)-Luc virus. The data shown are representative of three independent experiments. Error bars represent S.D. between duplicate samples analyzed in the experiment. C, inhibition of EV71(FY)-Luc pseudotype viral infection of RD cells. EV71 pseudotype virus was incubated with patient anti-EV71 serum samples at 4 °C for 1 h prior to infection of RD cells in a 96-well plate in a total volume of 100 μl in quadruplicate. Luciferase activity was measured 24 h postinfection. Non-linear regression analysis of the neutralization curve of anti-EV71 serum was conducted using GraphPad Prism. N3, N12, and N25 are convalescent sera from different patients that showed low, medium, and high neutralization activity, respectively, against EV71 infection in a classic plaque reduction assay. IRES, internal ribosome entry site.
EV71 Single Round Infection System
EV71 Subgenomic Replicon
The EV71 replicon was constructed by replacing the capsid coding region with a firefly luciferase reporter gene and a T7 promoter at the 5′-end for transcription in vitro. The EV71 replicon was linearized, and 1 μg of purified DNA was then used as template for EV71 replicon RNA synthesis with a RiboMAX large scale RNA production kit (Promega). Transcribed RNA was purified by isopropanol precipitation and used for transfection or frozen at −80 °C until use.
EV71 Capsid and Its Mutant Expressers
The EV71 capsid expresser was constructed to express the structural capsid genes in trans. EGFP gene was inserted upstream of the EV71(FY) capsid gene and was separated by a 2A self-cleavage site. The EGFP is used to ensure transfection efficiency of the structural genes. Capsid variants with mutations were prepared by site-directed mutagenesis of VP1 and verified by DNA sequencing.
Preparation of EV71 Pseudotype Virus EV71(FY)-Luc
FY or FY mutant pseudotype was produced by sequential transfection of FY or FY mutant capsid plasmid and EV71 replicon RNA into HEK-293T cells. Briefly, pcDNA6-FY-capsid or mutant capsid plasmid was transfected into HEK-293T cells at 60–80% confluence; 24 h post-capsid transfection, replicon RNA was then transfected using Lipofectamine 2000 (Invitrogen). FY pseudotype was harvested 24 h post-RNA transfection with two rounds of freeze-thaw cycle.
EV71 Single Round Virus Infection
50 μl of EV71(FY)-Luc or mutant pseudotype was mixed with 50 μl of fresh complete medium supplemented with 2% FBS and applied in quadruplicate to cells on a 96-well plate. Firefly luciferase activity was measured following the Luciferase Assay System manual (Promega) at 24 h postinfection. In the case of pH dependence experiments, cells were pretreated with bafilomycin A1 or NH4Cl at 37 °C for 1 h. The cells were infected with EV71(FY)-Luc at 37 °C for 1 h and then changed to fresh culture medium after washing with PBS once.
Quantification of Virions by Measuring Viral Genome RNA
EV71(FY)-Luc pseudotypes were produced on 293T cells and frozen at −80 °C. For viral RNA quantification, 10 μl of each pseudotype was used for RNA extraction by TRIzol reagent (Invitrogen). cDNA was synthesized with reverse transcriptase from Moloney murine leukemia virus (RNase H−, Takara), and virions were quantified by real time PCR (SYBR Premix Ex TaqII (Perfect Real Time), Takara) using the primers fLuc-F (5′-caaatacgatttatctaatttacacga-3′) and fLuc-R (5′-ccggtatccagatccacaac-3′) located in the luciferase gene.
EV71(FY)-Luc Binding Assay on RD Cells
80 μl of EV71(FY)-Luc or mutant pseudotypes (∼108 copies of viral genome) was added to RD cells on a 48-well plate in duplicate and incubated at 4 °C for 1.5–2 h, and unbound virions were washed away with ice-cold PBS. Bound virions were quantified by real time RT-qPCR using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference.
Neutralization Assay with Single Round EV71(FY)-Luc Pseudotype
EV71(FY)-Luc pseudotype virus was incubated with anti-EV71 serum sample for 1 h on ice and then added to RD cells in a 96-well plate in a total volume of 100 μl. Luciferase activity was measured 24 h postinfection.
SCARB2 Binding Site Mapping
Chimeric SCARB2 mutants were constructed using standard PCR-based methods, and all were cloned into pCAGGS-C9 vector. RD/SCARB2 kd or BHK21 cells seeded on a 24-well plate were transfected with SCARB2 or mutants by Lipofectamine 2000. At 20 h post-transfection, cells were infected with EV71(FY)-EGFP or EV71(FY)-Luc. GFP fluorescence images were taken at the times indicated after EV71(FY)-EGFP infection; firefly luciferase activities were measured 24 h postinfection.
Surface Expression Level of Chimeric SCARB2 Mutants
Cells overexpressing SCARB2 or mutants were surface-biotinylated with sulfo-NHS-LC-biotin (sulfosuccinimidyl 6-(biotinamido)hexanoate; Pierce) following the manufacturer's instruction manual. Then cells were lysed in 500 μl of 1% CHAPS in PBS (Sigma) with 1× mixture protease inhibitor (Roche Applied Science). Equal amounts of total proteins were used for the pulldown assay. 1 μg of 1D4 (C9 tag-specific monoclonal antibody) together with 10 μl of mixed protein A and G Dynal beads (Invitrogen) were added to the supernatants to pull down the C9-tagged proteins. After washing three times with 0.3% CHAPS in PBS, bound proteins were eluted by boiling in 1× loading buffer for 5 min and subsequently examined with HRP-streptavidin (Sigma) using a Western blot assay.
Comparative Model of VP1 Structure
VP1 protein of EV71(FY) shares 36% sequence identity with polio VP1 for which the crystal structure is available (Protein Data Bank code 3EPC). We therefore used the polio VP1 to model the EV71 VP1 structure with the homology modeling program MODELER (24). The model with the best discrete optimized protein energy score (25) using the default settings in MODELER was chosen and subsequently validated using PROCHECK. The three-dimensional architecture of EV71 virus capsid was constructed by replacing the VP1 domain of poliovirus with the modeled VP1 domain of EV71 on the cryo-EM structure of poliovirus (Protein Data Bank code 2PLV). The visualization of the virus capsid was built using the multiscale models in Chimera (26). Representative figures of structural models were also generated with Chimera (27).
Purification of Wild Type EV71 Viruses Using Ultracentrifugation
RD cells of five 15-cm dishes were infected with EV71(FY) virus stock at a multiplicity of infection of 0.1 and cultured in DMEM with 2% FBS. When 80–90% of cells showed a cytopathic effect, the supernatant was collected and concentrated by filtration through a 100-kDa-cutoff centrifugal filter (Millipore). The concentrated virus was loaded on top of a CsCl gradient (1.2–1.6 g/ml; discontinuous) followed by ultracentrifugation at 52,000 rpm for 1 h 45 min at 4 °C in a Beckman SW55Ti rotor. After dialysis with PBS, the purified EV71 was stored at −80 °C until use.
Pulldown Assay of EV71 with Soluble SCARB2-mFcs
SCARB2 extracellular regions of human, mouse, and chimeras were fused with the Fc region of mouse IgG2a to make soluble receptors. Proteins were expressed in HEK-293T cells and purified with protein A-Sepharose beads. 1 μg of purified soluble SCARB2-mFc was incubated with 1 μl of purified EV71(FY) wild type virus or EV71(FY)-Luc pseudovirus with VP1 mutant (∼5 × 108 genome copies) in a total volume of 50 μl in PBS at 4 °C for 3 h, and then 10 μl of protein A Dynal beads (Invitrogen) was added followed by further incubation for 1 h at 4 °C. The beads were then incubated with 300 μl of TRIzol lysis buffer to release bound virions after three washes with PBS (containing 0.001% Triton X-100). The bound virions were quantified by real time RT-PCR.
Flotation Assay of Sucrose Density Gradient Centrifugation
20 μl of purified virus (∼5 × 109 genome copies) was incubated with 5 μg of purified hSCARB2-mFc or hPSGL1-mFc in PBS containing 0.5% BSA in a total volume of 100 μl at 4 °C overnight. For low pH treatment, HCl was added to the mixture to bring the pH to 5.6. The mixture was then incubated at 37 °C for 100 min, subsequently applied to a 15–30% discontinuous sucrose gradient, and ultracentrifuged at 40,000 rpm for 70 min at 10 °C in a Beckman MLS-50 rotor. 20 μl of each fraction sample (22 fractions total) was subjected to RT-qPCR. The 135 S particle was made in vitro from native 160 S virions by heating for 3 min in a low salt buffer (4 mm CaCl2, 20 mm HEPES, pH 7.4).
RESULTS
Generation of EV71(FY)-Luc Pseudovirus for Single Round Infection
To dissect the molecular interaction mechanism of EV71 and its receptors, we first established a single round pseudotype EV71 reporter virus system that allowed us to study EV71 infection solely at the entry level. The EV71 replicon and plasmid expressing EV71 capsid proteins were constructed from an EV71 virus (FY, C4 strain) isolated in Fuyang, China. The subgenomic replicon plasmid (EV71 FY Subgenomic Replicon) contains UTRs, firefly luciferase (Luc) reporter gene, and all nonstructural protein regions with a T7 promoter sequence inserted at the 5′-end. The capsid-expressing plasmid (EGFP EV71 FY Capsid) contains all the capsid genes (VP1–VP4) and an EGFP reporter gene that are under the control of a CMV promoter (Fig. 1A). The single round entry virus was produced by sequential transfections of the capsid-expressing plasmid and EV71 FY subgenome RNA that was transcribed from EV71 FY Subgenomic Replicon in vitro (see “Materials and Methods”). The produced virus is designated as EV71(FY)-Luc. Unlike the replication-competent EV71(FY)-EGFP virus, which contains the full-length RNA genome of EV71 with an EGFP reporter inserted between the 5′-UTR and VP4 (Fig. 1A), EV71(FY)-Luc pseudotype viruses can only enter target cells once because they lack structural genes and their capsid proteins were provided in producing cells in trans. In an ultracentrifugation analysis, EV71(FY)-Luc pseudotype viruses migrated the same as the wild type virus in a discontinuous 15–30% sucrose gradient (Fig. 1B). Moreover, convalescent sera of patients recovered from EV71 infection neutralized the infection of EV71(FY)-Luc pseudoviruses in a dose-dependent manner (Fig. 1C). These results demonstrated that the single round EV71(FY)-Luc pseudotype virus system can be used as a surrogate for wild type virus to study viral entry. This system provides a sensitive and quantitative approach to investigate the detailed molecular interaction between EV71 and receptor at the entry level only and is devoid of any ambiguity that might be caused by postentry events.
SCARB2s from Several Animal Species Support Infection by EV71 with Different Efficiencies
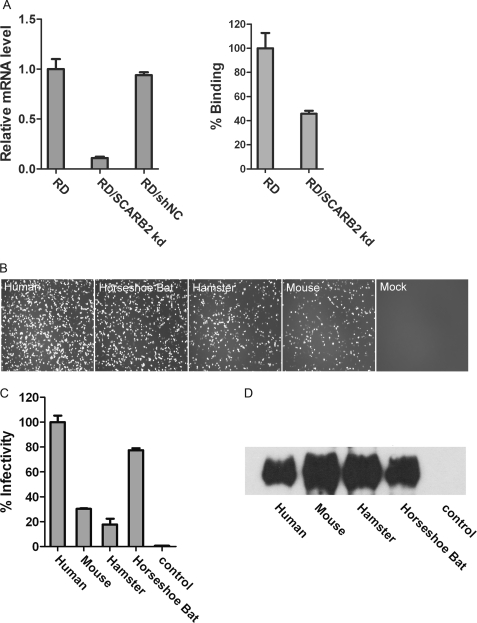
SCARB2, a type III glycoprotein with both N and C termini within the cytoplasm, is endogenously expressed on a wide range of tissues and cells in different species. It is located primarily on the membranes of lysosomes and late endosomes, and its cell surface expression level varies significantly among different cells (28). We established stable SCARB2 knockdown RD cell lines (RD/SCARB2 kd) by using a lentivirus-based system that carries hSCARB2-specific shRNA. The hSCARB2 mRNA expression level was reduced by 90% in the RD/SCARB2 kd cells as compared with RD cells that were transduced with an irrelevant shRNA (RD/shNC cells) (Fig. 2A, left panel). The binding activity of EV71(FY)-Luc pseudotype virus to RD/SCARB2 kd cells was greatly reduced (Fig. 2A, right panel). To compare the susceptibility of RD, RD/SCARB2 kd, and RD/shNC cells to EV71 viral infection, we infected these cell lines with a fully replication-competent EV71(FY)-EGFP virus that was produced by transfection of RD cells with EV71(FY)-EGFP RNA transcribed in vitro (see “Materials and Methods”). As shown in supplemental Fig. 1A, EV71(FY)-EGFP efficiently infected RD and RD/shNC cells but not RD/SCARB2 kd cells. Cytopathic effects were also observed in RD and RD/shNC cells but not in RD/SCARB2 kd cells (supplemental Fig. 1B). To examine whether expression of human SCARB2 can rescue EV71 infection in RD/SCARB2 kd cells, we transfected the cells with an expression construct that encodes hSCARB2 but with a silent nucleotide mutation (hSCARB2smut) that makes the shRNA against SCARB2 ineffective. As expected, transient transfection of RD/SCARB2 kd cells with the hSCARB2smut, but not control vector (mock), resulted in strong restoration of EV71 infection (Fig. 2B, far left panel). These results confirmed that hSCARB2 is sufficient to permit EV71 infection in RD/SCARB2 kd cells.
FIGURE 2.
SCARB2 from diverse species supports EV71 infection with various efficiencies. A, SCARB2 knockdown RD (RD/SCARB2 kd) cell line. Left panel, the SCARB2 mRNA levels in RD cells, stable RD/SCARB2 kd cells, and control RD cells treated with an unrelated shRNA were determined by real time PCR. The SCARB2 mRNA levels for each cell line were normalized using GAPDH as an internal control. Right panel, relative binding efficiency of EV71 pseudotype virus EV71(FY)-Luc to RD and RD/SCARB2 kd cells. The data are shown as averages and S.D. from triplicates analyzed in one experiment, which is representative of three independent experiments. B, SCARB2s from human and horseshoe bat supported more efficient EV71(FY)-EGFP infection than those from mouse and hamster. Human SCARB2 (SCARB2smut that carries nonsense nucleotide mutations to mismatch shRNA target sequence) and SCARB2s of horseshoe bat, mouse, and hamster, respectively, were transiently transfected into a stable SCARB2 kd cell line. 24 h post-transfection, cells were infected with EV71(FY)-EGFP viruses, and infected cells were photographed 24 h postinfection. C, single round EV71(FY)-Luc infection of cells complemented with SCARB2 from human, mouse, hamster, and horseshoe bat. BHK21 cells were transfected with plasmids encoding SCARB2 (fused with a C9 tag at the C terminus) from different species as indicated, and the cells were then infected with EV71(FY)-Luc reporter virus. Luciferase activity was determined 24 h postinfection, and the percentage of infectivity was calculated and normalized to the infectivity of human SCARB2-transfected cells considered as 100%. Infection of EV71(FY)-Luc in untransfected BHK21 cells was used as a control. The data reflect the means with S.D. bars from quadruplicate data points within one experiment. Similar results were obtained from three independent experiments. D, cell surface expression of SCARB2s. BHK21 cells from the same transfections as in C were harvested 24 h post-transfection, cell surface molecules were biotinylated, and cells were lysed. The biotinylated SCARB2s were then pulled down with a C9-specific antibody, 1D4, from the cell lysate and subsequently detected with HRP-streptavidin by Western blot. Untransfected BHK21 cells were used as a control.
It has been shown that cynomolgus monkeys and to a lesser extent suckling mice and nonobese diabetic/severe combined immunodeficient mice can be infected with EV71 bearing adaptive mutations (29). Whether EV71 can infect other animals is currently unknown. We cloned and sequenced SCARB2 genes from horseshoe bat, mouse, and hamster and aligned their predicted amino acid sequences with hSCARB2 (supplemental Fig. 2). The alignment showed that human SCARB2 and SCARB2s from the other three species share ∼86% identity and more than 94% similarity. The functions of these SCARB2s in mediating EV71 infection were tested by receptor complementarity experiments. SCARB2s were transiently transfected into RD/SCARB2 kd cells, the cells were infected with EV71(FY)-EGFP (Fig. 2B) or in separate experiments transfected into BHK21 cells, and then infected with EV71(FY)-Luc (Fig. 2C). EV71(FY)-EGFP infected mouse or hamster SCARB2-expressing cells less efficiently compared with human SCARB2-expressing cells (RD/SCARB2 kd cells transfected with SCARB2smut as described above) (Fig. 2B). This result is consistent with a recently published study showing that mouse SCARB2 does not support robust EV71 infection (23). Interestingly, cells expressing horseshoe bat SCARB2 supported more efficient EV71(FY)-EGFP viral infection (Fig. 2B).
To confirm that the species difference of SCARB2 in supporting EV71 infection was solely at the entry level, we also used the single round EV71(FY)-Luc system that contains firefly luciferase as a reporter to quantify viral infectivity on receptor-complemented cells. SCARB2s from human, mouse, hamster, and horseshoe bat were transfected into BHK21 cells, surface expression of these SCARB2s was determined, and the cells were infected with EV71(FY)-Luc reporter virus. Luciferase activity was determined 24 h postinfection. Consistent with the infection of EV71(FY)-GFP that is replication-competent but with a much clearer difference, infection of single round EV71(FY)-Luc on various SCARB2s from different species is distinct. EV71(FY)-Luc infected human and horseshoe bat SCARB2-expressing cells efficiently; the infectivity on bat SCARB2 cells is ∼80% of that on hSCARB2 cells, and apparently there are no postentry restrictions of BHK21 cells for EV71 infection. However, mouse and hamster SCARB2-expressing cells were infected poorly (up to 32% of the infectivity in hSCARB2 cells; Fig. 2C). Although the surface expression level of SCARB2 from mouse and hamster was actually slightly higher than that from human (Fig. 2D), they only support limited EV71(FY)-Luc infection (Fig. 2C).
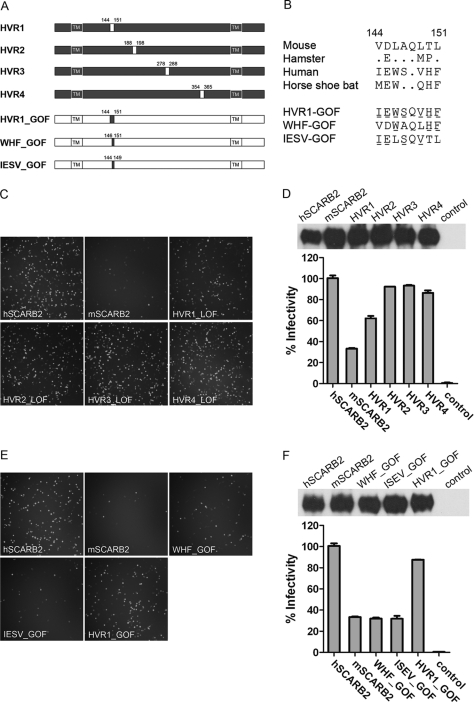
Identifying Critical Residues of SCARB2 for EV71 Infection
The amino acid sequence alignment of SCARB2 from human, horseshoe bat, mouse, and hamster revealed four highly variable regions (HVRs) in the receptor: HVR1 (residues 144–151), HVR2 (residues 188–198), HVR3 (residues 278–288), and HVR4 (residues 354–365) (supplemental Fig. 2). To identify the critical binding region of SCARB2 for mediating virus binding, two mouse/human SCARB2 chimeras and four HVR1-, HVR2-, HVR3-, and HVR4-swapped mutants were constructed. The two chimeras were designated as HM (N-terminal 300 residues from human and the remaining 178 residues from mouse) and MH (N-terminal 300 residues from mouse and the rest from human) (supplemental Fig. 3A), whereas each of the HVR-swapped hSCARB2 mutants has one of the four HVR regions replaced with that of mSCARB2 (Fig. 3A). The receptor activity of these variants was examined by transient transfection into RD/SCARB2 kd and/or BHK21 cells and followed by EV71(FY)-GFP or EV71(FY)-Luc infection. All the SCARB2 variants were expressed at comparable levels on the cell surface (Fig. 3, D and F, upper panels, and supplemental Fig. 3C, right panel). Infection of HM SCARB2-expressing cells was as efficient as that of hSCARB2 cells. In contrast, the reciprocal SCARB2 chimera MH-expressing cells showed a significant reduction in infection (supplemental Fig. 3, B and C), which is consistent with a recent report that the N terminus of SCARB2 is important for EV71 binding and infection (23). Mutant SCARB2 HVR1-LOF (human HVR1 residues 144–151 replaced with mouse HVR1, loss-of-function (LOF) mutant)-expressing cells showed reduced susceptibility to viral infection (∼30% reduction as compared with wild type hSCARB2), whereas HVR2-LOF, HVR3-LOF, and HVR4-LOF mutants did not significantly affect viral infection as shown by EV71(FY)-GFP (Fig. 3C) and EV71(FY)-Luc viral infection assays (Fig. 3D). Sequence comparison of HVR1 of SCARB2 across species showed that only one common amino acid (Gln-148) of the eight residues is shared by all four species (Fig. 3B). Based on HVR1 sequence alignment, SCARB2s can be divided into two groups: group 1 containing mouse and hamster and group 2 containing human and horseshoe bat. For both groups, the two members within each group share five identical residues. Between groups, e.g. human and mouse SCARB2, seven of the eight amino acids are different (Fig. 3B). To confirm that the residues in HVR1 are responsible for the reduced susceptibility of mouse SCARB2-expressing cells to EV71 infection, we altered three of the seven different residues to their human counterpart in the HVR1 of mSCARB2, e.g. from VDLAQLTL to VDWAQLHF (WHF_GOF (gain-of-function mutant)), or the other four different residues to their human counterpart on the HVR1 of mSCARB2 from VDLAQLTL to IELSQVTL (IESV_GOF mutant) and found that both mutants could not restore infectivity. However, remarkably, when all seven different residues between mouse and human SCARB2 were switched from mouse to the human counterpart (IEWSQVHF (HVR1_GOF)), highly efficient EV71 infection was observed and reached ∼85% of the infectivity of hSCARB2 (Fig. 3, E and F), demonstrating a critical role of these seven residues in HVR1 in SCARB2 interaction with the virus.
FIGURE 3.
Identification of critical residues on SCARB2 for EV71 infection. A, schematic diagram of chimeric SCARB2 proteins. Swap mutants of human SCARB2 with residues from mouse SCARB2 at the highly variable regions (HVR1, HVR2, HVR3, and HVR4, respectively) were generated by replacing the human SCARB2 sequence (black) with the murine counterpart (white). The positions of the amino acids exchanged are indicated. Mouse SCARB2 with three (WHF), four (IESV), or seven (HVR1) GOF mutant residues was constructed by replacing the corresponding murine residues with their human counterparts on a mouse SCARB2 expression plasmid. All SCARB2s contain a C9 tag at the C terminus. TM, transmembrane domain. B, amino acid alignment of the HVR1 region of SCARB2s from four different species and GOF mutants of mouse SCARB2. GOF mutants are the three- (WHF), four- (IESV) or seven-residue (HVR1) mutations in the HVR1 region. Residues changed from mouse to human are indicated with underlines. C and D, swapping the HVR1 of human SCARB2 with the mouse counterpart interferes with EV71 infection. RD SCARB2/kd cells transfected with plasmids encoding human SCARB2, mouse SCARB2, or human SCARB2 HVR1–4 LOF mutants were infected with replication-competent EV71(FY)-GFP. Infected cells were photographed 18 h postinfection (C). Infections of EV71(FY)-Luc pseudotype virus were determined on BHK21 cells transfected with various SCARB2s. Firefly luciferase reporter activity was measured 24 h postinfection (D, lower panel), and the surface expression of these SCARB2 variants was also examined (D, upper panel). All SCARB2s contained a C9 tag at the C terminus. Untransfected BHK21 cells were used as a control in D. E and F, introduction of seven residues of human SCARB2 into mouse SCARB2 converts mouse SCARB2 to an efficient EV71 receptor for viral infection. EV71(FY)-GFP infection (E) was recorded as in C. For EV71(FY)-Luc infection (F), surface expression of SCARB2 mutants was determined for BHK21 cells transfected with SCARB2s (F, upper panel). EV71(FY)-Luc infection in these cells was measured 24 h postinfection (F, lower panel). Untransfected BHK21 cells were used as a control in F, and the infectivity of EV71(FY)-Luc in BHK21 cells transfected with hSCARB2 was set as 100%. All the data shown represent three or more experiments. Cell surface expression of SCARB2s was determined similarly to that in Fig. 2D. For both D and F, the data are representative of three independent experiments. Error bars represent S.D. from quadruplicate samples analyzed within the experiment.
EV71 Enters Cells via Canyon around Gln-172 Residue on VP1
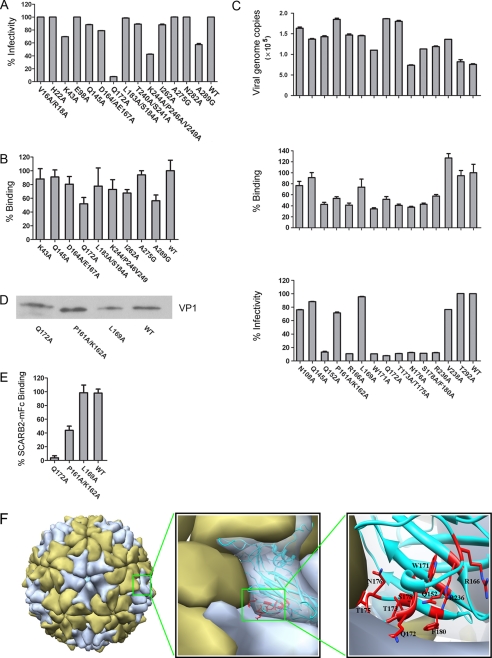
Picornavirus VP1 protein is arrayed around the 5-fold axis on the virion surface, it is highly exposed, and it is the most prominent structural protein. VP1 plays an important role in mediating receptor binding for viral entry, and its sequence has been used for picornavirus classification (30). It has been shown that VP1 of EV71 is involved in binding SCARB2 and is the capsid protein responsible for EV71 viral entry; however, molecular details of the interaction between VP1 and SCARB2 are lacking. We aligned VP1 protein sequences of several EV71 strains including currently circulating strains and found mutation hot spots in VP1 (supplemental Fig. 4). We hypothesized that these mutation hot spots might be a result of evolutionary pressures, such as mutants that can escape host neutralizing antibody and/or mutations for host adaptation with higher receptor binding efficiency. We therefore mutated these residues on VP1 of the EV71(FY) strain to alanine and incorporated them into EV71(FY)-Luc pseudotype viruses. Using the VP1 mutant viruses, we examined the efficiency of their binding to SCARB2 on RD cells and their functions for mediating EV71(FY)-Luc infections. A set of mutations covering ∼20 solvent-exposed residues on VP1 was initially examined by quantifying the pseudovirus particles bound on RD cells using quantitative RT-PCR targeting viral genomic RNA. Among all the VP1 mutants tested, EV71(FY)-Luc bearing the Q172A VP1 mutation showed the most dramatic loss in both binding and infection activity on RD cells (Fig. 4, A and B; not all mutants are shown).
FIGURE 4.
EV71 enters cells via canyon around Gln-172 residue on VP1. A, effect of mutations on surface-exposed VP1 residues for EV71 infection of RD cells. B, effect of VP1 mutants on viral binding activity in RD cells. C, mutagenesis analysis of VP1 around Gln-172 revealed a cluster of residues involved in the interactions between VP1 and SCARB2 as shown by reduction in viral binding and RD cell infectivity. Upper panel, RNA genome copies of each mutant virion were examined with qRT-PCR. Middle panel, mutant virus binding to RD cells. The RD cell-bound EV71(FY)-Luc virions were quantified by RT-qPCR, and binding of mutant virus was normalized to the pseudotype virus bearing wild type VP1. Bottom panel, mutant viral infectivity measurement. EV71(FY)-Luc pseudotype mutant viruses were used to infect RD cells in quadruplicate, and viral infectivity was measured 24 h postinfection by luciferase reporter. Infectivity of mutant virus was normalized to the wild type EV71(FY)-Luc viruses. D, Western blot analysis of VP1 proteins on mutant EV71 pseudovirions. The same amounts of pseudovirions (normalized by viral RNA genome copies quantified by qRT-PCR) bearing mutant VP1 as indicated were examined for VP1 protein levels on the virions by Western blot. Pseudovirions were concentrated by ultracentrifugation, and VP1 proteins were detected by mouse anti-EV71 polyclonal serum. E, SCARB2-mFc pulldown assay on EV71 pseudovirions carrying mutant VP1. 1 μg of purified SCARB2-mFc fusion protein was incubated with ∼5 × 108 pseudovirions bearing wild type or mutant VP1 at 4 °C for 4 h in the presence of protein A beads. After extensive washing with PBS three times, the bound virions were quantified by RT-qPCR. The data shown in A–E are representative of 2–3 independent experiments for each panel. Error bars represent S.D. from triplicate samples analyzed within one experiment. F, the potential binding interfaces between the canyon on EV71 VP1 and SCARB2 were mapped on the three-dimensional architecture of EV71 virus capsid. The capsid architecture shown was built by replacing the VP1 on the cryo-EM structure of poliovirus (Protein Data Bank code 2PLV) with our modeled VP1 structure. The EV71 VP1 residues whose alteration substantially decreased SCARB2 binding are colored in red (Gln-152, Arg-166, Trp-171, Thr-173, Gln-172, Thr-175, Asn-176, Ser-178, Phe-180, and Arg-236, respectively).
We subsequently mutated all the surface residues in the vicinity of Gln-172 based on the structural model of EV71 VP1, which shares high sequence identity with the poliovirus VP1 template structure (Protein Data Bank code 3EPC). A total of 15 additional VP1 capsid mutations were made and examined for their activity for both receptor binding and viral infection. In addition to Q172A, EV71 pseudotypes bearing VP1 mutations at residues Gln-152, Arg-166, Trp-171, Thr-173, Thr-175, Asn-176, Ser-178, Phe-180, and Arg-236 showed greatly decreased viral binding and infectivity in RD cells (Fig. 4C). To ensure that the mutations did not interfere with virus assembly, we quantified the genome copy numbers of viral RNA of these VP1 mutant pseudotype viruses by quantitative RT-PCR. The result showed that virus assembly of most mutants with reduced viral binding and infectivity was not impaired significantly as compared with that of other mutants. Of all residue tested, only N176A showed reduced binding and infection with lower assembly efficiency (Fig. 4C, upper panel). A Western blot was also performed to detect the level of VP1 on purified Q172A mutant pseudotype viruses. As shown in Fig. 4D, the level of VP1 detected by anti-EV71 serum on Q172A VP1 mutant viruses was similar to that on the pseudotype viruses bearing wild type VP1 or the three other VP1 mutants, P161A, K162A, and L169A, that are located in the vicinity of Gln-172 with either a minor or no effect on EV71 pseudovirus binding or infection in RD cells. To demonstrate the direct interaction between VP1 and SCARB2, we examined the binding of wild type and selected mutants including the Q172A mutant viruses with purified, soluble SCARB2 by a pulldown experiment (Fig. 4E). As expected, binding of SCARB2 to mutant viruses was reduced (P161A/K162A) or abolished (Q172A) consistent with the reduced or loss of binding and infectivity of those mutants in RD cells. Collectively, these results demonstrate that a region around residue Gln-172 on VP1 is critically important for EV71 infection mediated by SCARB2.
Next we constructed a three-dimensional structural model of EV71 virion by replacing the VP1 of poliovirus with that of EV71 in the cryo-EM structure of poliovirus (31). As shown in Fig. 4F, left and middle panels, a “canyon” region similar to that on poliovirus was observed on the EV71 virion surface. Interestingly, Gln-172 and other adjacent residues on VP1 that are critical for receptor binding and viral infectivity as revealed by our mutagenesis studies were mapped to the canyon region on the model of EV71 virion. These residues line the wall of the canyon and are mainly on the EF loop region (loop connecting β-strands E and F) (Fig. 4F, right panel, and supplemental Fig. 2). Taken together, it is likely that EV71 binds to SCARB2 via the canyon region on VP1 that is probably composed of residues Gln-152, Arg-166, Trp-171, Gln-172, Thr-173, Thr-175, and Ser-178.
SCARB2 Is Uncoating Receptor of EV71
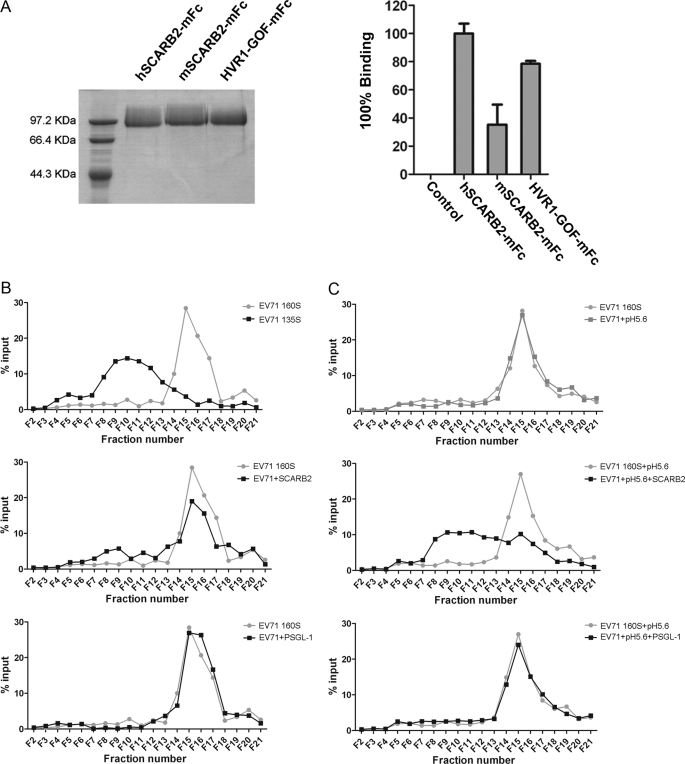
For a productive infection, viral RNA genome must be released into the cytoplasm of the host cells. For some enteroviruses, receptor engagement is the initial trigger of the viral uncoating process that involves capsid alterations by conformational changes. As a result, different virus particles are formed during viral entry: mature enterovirus particles sediment at ∼160 S, whereas the other two forms of viral particles sediment at about 135 and 80 S (32). The 80 S particles represent the final product of the entry process (they are empty particles that have shed genomic RNA), whereas the 135 S particles still retain the full complement of genomic RNA but lack some or all of their content of VP4 and have externalized most of the N-terminal extension of VP1 that is normally inside the virions. Conversion from native mature 160 S particles to the 135 S particles can occur intracellularly following cellular receptor engagement; it can also be induced in some cases with soluble receptors in solution or by simply heating the 160 S virus particles to 50 °C for a short time in a low salt buffer (33). It is unknown whether SCARB2 serves as an uncoating receptor for EV71 entry. In those enteroviruses for which receptor binding triggers virus uncoating (conversion from 160 S to 135 S particles), the receptors usually dock within the canyon on the virus surface. Our data have shown that SCARB2 binds to the canyon area on VP1 of EV71; therefore, we surmised that SCARB2 engagement may trigger virus uncoating, and thus, SCARB2 might serve as an uncoating receptor for EV71 infection. To examine this, we expressed soluble SCARB2 as mouse IgG2a Fc (mFc) fusion proteins by transient transfection of 293T cells with their encoding plasmids (Fig. 5A, left panel). As expected, the hSCARB2-mFc fusion proteins pulled down EV71 virus efficiently as did the HVR1-GOF mutant (HVR1 of mouse SCARB2 replaced with human HVR1), but not the mouse SCARB2 did not (Fig. 5A, right panel). We then prepared EV71 135 S particle by heating purified wild type EV71 virions and used it as a reference for testing whether soluble receptor could induce virus particle conversion from 160 S to 135 S. By ultracentrifugation in a 15–30% discontinuous sucrose gradient, the heat-treated viral particles shifted from the major peak fraction 15–16 (160 S) to fraction 10 (135 S) (Fig. 5B, upper panel). To test whether soluble receptor could induce virus uncoating, 5 μg of purified hSCARB2-mFc was mixed with EV71 particles that were purified by CsCl ultracentrifugation and incubated overnight, then the temperature was shifted to 37 °C, and the mixture was incubated at 37 ºC for 100 min. The mixture of virus and receptor was then subjected to ultracentrifugation at 40,000 rpm for 70 min at 10 °C. As shown in Fig. 5B (middle panel), soluble receptor hSCARB2-mFc was able to uncoat the viruses: ∼15% of intact virions (in fraction 15–16; 160 S) shifted to fractions 8–12, a position identical to that of the 135 S particle induced by heating the virus. Moreover, when the mixture was incubated at 37 °C under low pH (=5.6) treatment, the shift from 160 S to 135 S particles was much more distinct (Fig. 5C, middle panel) compared with that under neutral pH (Fig. 5B, middle panel). We also tested two other mFc fusion proteins: the extracellular domain of human PSGL-1 and the seven HVR1 residues of hSCARB2; both were fused at the N terminus of the mouse IgG2a Fc (supplemental Fig. 5A, left panel). Although soluble PSGL1-mFc could bind EV71 efficiently (supplemental Fig. 5A, right panel), neither PSGL1-mFc (Fig. 5B, bottom panel) nor HVR1-mFc (data not shown) could effectively uncoat EV71 virions. Of note, treatment with low pH alone (Fig. 5C, upper panel) or PSGL-1 plus low pH (Fig. 5C, bottom panel) did not alter the distribution of the virions upon ultracentrifugation. These results support that SCARB2, but not PSGL-1, is an uncoating receptor of EV71 and that low pH can facilitate conformational transition(s) of EV71 viral particles. Consistent with this, it was recently reported that low endosomal pH is required for EV71 infection (17), and we also observed that endosome acidification inhibitors such as bafilomycin A1 and ammonium chloride efficiently inhibited EV71(FY)-GFP and EV71(FY)-Luc entry (supplemental Fig. 5B).
FIGURE 5.
SCARB2 is an uncoating receptor of EV71. A, pulldown assay of EV71 with soluble SCARB2-mFc variants. Left panel, SDS-PAGE of purified SCARB2-mFc fusion proteins with the extracellular region of human or mouse SCARB2 or HVR1-GOF-mFc. Right panel, 1 μg of SCARB2-mFc proteins shown on the left was incubated with 1 μl of purified EV71(FY) wild type virus (∼5 × 108 virions) at 4 °C for 4 h in the presence of protein A beads. After extensive washing with PBS three times, the bound virions were quantified by RT-qPCR. 1 μg of unrelated mFc fusion protein (coronavirus HKU1 S1-mFc) was used as a control. All data points show means and S.D. (error bars) for duplicate samples from a representative experiment (n = 3). B and C, virion flotation assay with sucrose density gradient ultracentrifugation. Purified virus (∼5 × 109) was incubated with 5 μg of hSCARB2-mFc or hPSGL1-mFc followed by incubation at 37 °C for 100 min at neutral pH (B) or pH 5.6 (C) and then loaded onto a 15–30% discontinuous sucrose gradient for ultracentrifugation at 40,000 rpm for 70 min at 10 °C in a Beckman MLS-50 rotor. 20 μl of each fraction of a total 21 fractions was subjected to RT-qPCR to quantify virus in each fraction. The 135 S reference particle was made in vitro from native 160 S virions by heating for 3 min in a low salt buffer (4 mm CaCl2, 20 mm HEPES, pH 7.4).
DISCUSSION
In addition to SCARB2, several other cell surface molecules including human PSGL-1 (18, 19), sialic acid-linked glycan (34), dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN; CD209) (35), and annexin II (36) have also been demonstrated to facilitate EV71 entry. However, SCARB2 is likely an indispensable receptor for EV71 infection. Viral infection in RD cells is greatly reduced when the interaction between EV71 and SCARB2 is blocked by antibodies (18, 19) or in RD cells in which SCARB2 expression is knocked down. SCARB2 is ubiquitously expressed on a wide range of tissues and cells in different species; however, the expression level, particularly on the cell surface, could vary significantly among different cells. It is located primarily on the membranes of lysosomes and late endosomes and has been implicated in participating in maintaining endosomal transport and lysosomal biogenesis (37, 38). Eckhardt et al. (39) showed that only 10–20% of SCARB2 was expressed on the surface of transfected Chinese hamster ovary cells; the majority of the rest was expressed on intracellular membranes. We made RD/SCARB2 kd cell line in which the mRNA SCARB2 expression was significantly knocked down (Fig. 2A) and its function in supporting viral entry is completely knocked out (supplemental Fig. 1). Intriguingly, the viral binding activity of the RD/SCARB2 kd cells remained up to ∼50% as compared with RD cells. As there is no detectable PSGL-1 mRNA in RD cells (data not shown), the viral particle may bind to cells via other attachment factor(s) on RD/SCARB2 kd cells. However, the attachment mediated by other factors is unlikely to be productive in terms of infection.
In this study, we cloned horseshoe bat (Rhinolophidae family) and hamster SCARB2 genes that have not been reported previously. RD/SCARB2 kd or nonsusceptible BHK21 cells transfected with SCARB2s from four different species (horseshoe bat, hamster, mouse, and human) showed various susceptibilities to EV71 infection. Viral infection assays were carried out with both replication-competent EV71 recombinant EGFP reporter virus (EV71(FY)-EGFP) and pseudotype EV71 single round viruses (EV71(FY)-Luc), and similar results were obtained for both viral infection systems. This confirmed that the infectivity difference among these cells is solely attributed to a species difference of the SCARB2 receptor at the viral entry level. The bat SCARB2-expressing cells supported viral infection efficiently (equal to ∼80% of human SCARB2 cells), whereas BHK21 cells expressing hamster or mouse SCARB2 poorly supported EV71 infection (∼30% of human SCARB2 by luciferase assay). Although humans are the only known natural hosts of enteroviruses including EV71, it is intriguing that bat SCARB2-expressing cells can be infected by EV71 efficiently. Bat is a unique and enigmatic group of mammals that account for ∼20% of extant mammalian species (40). A large number of viruses have been isolated from or detected in bats. Highly pathogenic viral diseases due to transmission of viruses from bats have been demonstrated for rabies virus, lyssaviruses, and Nipah and Hendra viruses and can be inferred for severe acute respiratory syndrome coronavirus-like virus. Therefore, bats have been increasingly recognized as reservoir hosts for viruses that can cross species barriers to infect humans and other domestic and wild mammals (41). Very recently, three novel picornaviruses were discovered from diverse bat species (42). The question remains whether horseshoe bats could be infected by EV71 and/or serve as a host or reservoir for EV71 in nature. It would be interesting to investigate whether EV71 or EV71-like viral RNA and/or anti-EV71 antibodies can be detected in horseshoe bats or other species of bats.
It is worth noting that the EV71(FY)-EGFP was able to replicate and spread viruses in RD cells efficiently. Cytopathic effects were observed in RD cells but not in RD/SCARB2 kd cells. This is different from the EV71-GFP reporter virus generated from an infectious clone of EV71 strain SK-EV006 that had a defect in spreading viruses in RD cells for unknown reasons and thus was used as a surrogate for single round virus in infection assays in a recent report (23). Here we produced genuine single round EV71 viruses by sequential transfection of an EV71 structural gene expresser and an EV71 replicon bearing a luciferase reporter in producing cells. The resultant pseudoviruses can only enter the target cell once because they are not able to produce any infectious progenies for a second infection in target cells due to a lack of structure genes on the replicon. Upon ultracentrifugation analysis, the pseudotype virus exhibited a sedimentation pattern similar to that of wide type virus. It could also be neutralized by patient sera in a dose-dependent manner. The single round EV71 system provides a sensitive and quantitative approach to examine EV71 infection at the entry level and is devoid of any ambiguity that might be caused by postentry events. Moreover, this system is safer to work with than replication-competent viruses and is particularly useful for testing antiviral reagents including neutralizing antibodies or small molecule drugs, serological screening of viral infection, and evaluations of vaccine sera.
To identify critical binding residues in SCARB2 responsible for viral binding, amino acid sequences of SCARB2 from four species (two non-efficient receptors (mouse and hamster) and two efficient receptors (bat and human)) were analyzed. Despite that SCARB2s from those four species share high sequence homology, four regions showing a high degree of variance were found among the four species. Cells expressing the HVR1 (residues 144–151) human SCARB2 mutant (human HVR1 replaced with mouse HVR1) had reduced susceptibility to viral infection as compared with wild type hSCARB2, whereas HVR2-, HVR3-, and HVR4-swapped mutants did not affect viral infection. This result indicates that HVR1 of human SCARB2 is a region critical for viral infection. This was further confirmed by three other mouse SCARB2 gain-of-function mutants (WHF-, IESV-, and HVR1-GOF). The WHF and IESV mutants that had three and four of the seven residues, respectively, in the HVR1 mutated to their human counterpart did not show a significantly increased ability to support EV71 infection. However, the HVR1-GOF mutant that had all of the seven different residues between mouse and human SCARB2 mutated to their human counterpart demonstrated a greatly enhanced infectivity level similar to that of human SCARB2. In contrast to a recent study in which the important amino acids for EV71 infection were localized throughout the entire region of amino acids 142–204 (23), our study demonstrated it is the seven residues 144–151 that are critical for the viral capsid binding. Interestingly, these seven residues are adjacent to residues 152–167, which presumably form a coiled coil domain in SCARB2 and have been demonstrated to be necessary for β-glucocerebrosidase binding (22).
EV71 as a member of the Enterovirus genus in the picornavirus family is predicted to have an icosahedral capsid architecture similar to that of other enteroviruses consisting of 60 copies of four capsid proteins, VP1, VP2, VP3, and VP4 (13). The outer surface of the virion is dominated by star-shaped mesas at the 5-fold axes and by a three-bladed propeller-like structure at the 3-fold axes. VP1 components are clustered around the 5-fold axes of symmetry, whereas VP2 and VP3 alternate around the 3-fold axes (1). There is a deep canyon encircling the 5-fold axes below VP1 that separates the VP1 clustered star-shaped mesas from the three-bladed propellers consisting of VP2 and VP3. This canyon has been shown or structurally predicted to be the site of receptor engagement for many viruses in the picornavirus family (1). For EV71, it has only been shown that VP1 is involved in binding to SCARB2 and is the capsid protein responsible for EV71 viral entry (23). To identify the determinants on VP1 for receptor binding and determine whether EV71 adopts a canyon receptor engagement mode, we first aligned amino acid sequences of VP1s from several circulating EV71 strains. Some mutation hot spots, mostly on loop regions according to secondary structure prediction and homology modeling, were identified. Because entry of EV71 is largely mediated by capsid protein VP1, we mutated those residues on VP1 of the EV71(FY) strain to alanine and incorporated them into EV71(FY)-Luc pseudotype viruses to investigate the efficiency of binding and infection of those mutants. Among the residues mutated, one is Gln-145 that has been implicated previously in EV71 adaptation to a murine model (43, 44). In our study, we did not find a significant effect of the Q145A mutation on either viral binding or viral infection in RD cells that express human SCARB2. Importantly, residues Gln-172 and those in its vicinity including Gln-152, Arg-166, Trp-171, Thr-173, Thr-175, Asn-176, Ser-178, Phe-180, and Arg-236 were found to be vital for binding to human SCARB2 and for efficient infection of cells expressing this receptor. To rule out the potential effect of these mutants on virus assembly, we quantified the genome copy numbers of viral RNA of the mutant pseudotype viruses by quantitative RT-PCR and showed that virus assembly of most of those mutants was not impaired significantly as compared with that of other mutants. We also confirmed that the Q172A mutant VP1 protein level on the pseudotype viruses is similar to that on wild type virus or other mutants that did not affect receptor binding and viral infectivity. Our pulldown data on Q172A pseudotype viruses further support a direct interaction of this residue and SCARB2. A previous study using synthetic peptides containing 15 overlapping peptides covering the entire VP1 sequence revealed that a peptide covering residues 163–177 of VP1 is as efficient as the whole virion in eliciting an EV71-specific IgG response (45). Another report also showed that amino acid residues 66–132, a region outside of the binding site identified by our work, may be involved in the dimerization of VP1 capsid protein (46). These observations are consistent with our findings in that the Gln-172 and residues in its immediate vicinity are probably exposed. Mutations on these residues are unlikely to lead to assembly defects of the virus.
It is generally believed that picornavirus particles are primed and undergo viral “breathing” at physiological temperatures with transient exposure of portions of VP4 and the N terminus of VP1 (47, 48). For some viruses, like poliovirus, receptor binding greatly accelerates the stepwise uncoating process by converting the particles from the native 160 S to the 135 S A particles (49). Many cellular receptors identified to date for enterovirus belong to the immunoglobulin superfamily of receptors (50, 51). They contain two to seven immunoglobulin-like domains, and frequently the first (membrane-distal) Ig-like domain protrudes into the deep canyon of the receptor binding site and triggers the disassembling of the particles (52, 53). Interestingly, SCARB2 is significantly different from existing enterovirus receptors. Presumably, both the N and C termini of SCARB2 are embedded in the plasma membrane or more frequently the lysosome membrane. There is no structural information available for this or related molecules. It is difficult to predict the configuration of SCARB2 and exactly how this molecule maneuvers to reach the key binding sites of VP1 on intact virions. It is also unknown whether EV71, like several other picornaviruses, contains a “pocket factor” inside the canyon on the viral surface (54, 55). Nonetheless, our experiments with purified human SCARB2 demonstrate that binding of soluble SCARB2, but not PSGL-1, to EV71 particles could lead to a major shift of the virions from 160 S to 135 S particles, indicating that conformation changes occur upon receptor binding on EV71. Remarkably, consistent with the data that infection of EV71 could be blocked by endosome acidification inhibitor, the uncoating efficiency of SCARB2 is significantly higher in an acidic environment (pH 5.6), indicating that unlike poliovirus entry, which is pH-independent (49, 56), protonation of certain residue(s) of EV71 capsids might play an important role in viral uncoating after receptor binding.
Phylogenetic analysis indicates that coxsackievirus type A 16 is likely the ancestor of EV71 and that EV71 emerged from CA16 during the 1940s (57). Both CA16 and EV71 utilize SCARB2 as an entry receptor (18), but only EV71 can occasionally induce neurological symptoms, whereas CA16 rarely does so. The factors that determine the pathogenic differences of these two viruses are currently unknown (11, 12). Viral entry is frequently a determinant of the virulence of a virus (10). Identification of the viral and receptor determinants of EV71 may help to elucidate the pathogenic differences of these two viruses.
Taken together, in this study we localized both receptor and viral determinants of the virus-receptor interactions. In particular, residues 144–151 of SCARB2 are critically important for viral binding and infection. Altering seven residues of mSCARB2 to the corresponding residues of hSCARB2 converted the mSCARB2 to an efficient receptor for EV71. The viral determinants for receptor binding was mapped to a canyon or “pocket” region on the viral particle surface comprising residue Gln-172 and surrounding residues of the capsid protein VP1. We further demonstrated that human SCARB2, but not PSGL-1, could induce conformational changes of the viral capsid from 160 S to 135 S. Additional conformational changes of viral capsids in an acidic environment would ultimately lead to uncoating of the RNA genome in the cytosol.
Supplementary Material
This work was supported by the Major State Basic Research Development Program of China (973 Program Grant 2010CB530101) and a research and development grant from the Beijing Municipal Government.

This article contains supplemental Figs. 1–5.
Monthly Reports of Nationally Notifiable Infectious Diseases, released by the Ministry of Health of the People's Republic of China, February 2011.
- EV71
- enterovirus 71
- SCARB2
- scavenger receptor class B 2
- CA16
- coxsackievirus A16
- PSGL-1
- P-selectin glycoprotein ligand-1
- hSCARB2
- human SCARB2
- mSCARB2
- mouse SCARB2
- RD
- rhabdomyosarcoma
- HFMD
- hand-foot-mouth disease
- kd
- knockdown
- EGFP
- enhanced GFP
- FY
- Fuyang
- qPCR
- quantitative PCR
- mFc
- mouse IgG2a Fc
- HVR
- highly variable region
- LOF
- loss-of-function
- GOF
- gain-of-function.
REFERENCES
- 1. Racaniello V. R. (2007) in Fields Virology (Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., eds) 5 Ed., pp. 795–838, Vol. 1, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Huang C. C., Liu C. C., Chang Y. C., Chen C. Y., Wang S. T., Yeh T. F. (1999) Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 341, 936–942 [DOI] [PubMed] [Google Scholar]
- 3. Vallet S., Legrand Quillien M. C., Dailland T., Podeur G., Gouriou S., Schuffenecker I., Payan C., Marcorelles P. (2009) Fatal case of enterovirus 71 infection, France, 2007. Emerg. Infect. Dis. 15, 1837–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu M., Meng G., He Y. X., Zheng J., Liao S. L., Zhong Y. F., Zhao X. S., Shao H. Q., Wang Y. P., Gao Z. C., Gao Z. F. (2009) Pathology of enterovirus 71 infection: an autopsy study of 5 cases. Zhonghua Bing Li Xue Za Zhi 38, 81–85 [PubMed] [Google Scholar]
- 5. Lum L. C., Wong K. T., Lam S. K., Chua K. B., Goh A. Y. (1998) Neurogenic pulmonary oedema and enterovirus 71 encephalomyelitis. Lancet 352, 1391. [DOI] [PubMed] [Google Scholar]
- 6. Chumakov M., Voroshilova M., Shindarov L., Lavrova I., Gracheva L., Koroleva G., Vasilenko S., Brodvarova I., Nikolova M., Gyurova S., Gacheva M., Mitov G., Ninov N., Tsylka E., Robinson I., Frolova M., Bashkirtsev V., Martiyanova L., Rodin V. (1979) Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch. Virol. 60, 329–340 [DOI] [PubMed] [Google Scholar]
- 7. Nagy G., Takátsy S., Kukán E., Mihály I., Dömök I. (1982) Virological diagnosis of enterovirus type 71 infections: experiences gained during an epidemic of acute CNS diseases in Hungary in 1978. Arch. Virol. 71, 217–227 [DOI] [PubMed] [Google Scholar]
- 8. Chan L. G., Parashar U. D., Lye M. S., Ong F. G., Zaki S. R., Alexander J. P., Ho K. K., Han L. L., Pallansch M. A., Suleiman A. B., Jegathesan M., Anderson L. J. (2000) Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. For the Outbreak Study Group. Clin. Infect. Dis. 31, 678–683 [DOI] [PubMed] [Google Scholar]
- 9. Ho M., Chen E. R., Hsu K. H., Twu S. J., Chen K. T., Tsai S. F., Wang J. R., Shih S. R. (1999) An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 341, 929–935 [DOI] [PubMed] [Google Scholar]
- 10. Li W., Zhang C., Sui J., Kuhn J. H., Moore M. J., Luo S., Wong S. K., Huang I. C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W. A., Guan Y., Choe H., Farzan M. (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24, 1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu J., Qian Y., Wang S., Serrano J. M., Li W., Huang Z., Lu S. (2010) EV71: an emerging infectious disease vaccine target in the Far East? Vaccine 28, 3516–3521 [DOI] [PubMed] [Google Scholar]
- 12. Ooi M. H., Wong S. C., Lewthwaite P., Cardosa M. J., Solomon T. (2010) Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 9, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 13. Ranganathan S., Singh S., Poh C. L., Chow V. T. (2002) The hand, foot and mouth disease virus capsid: sequence analysis and prediction of antigenic sites from homology modelling. Appl. Bioinformatics 1, 43–52 [PubMed] [Google Scholar]
- 14. Pallansch M., Roos R. (2007) in Fields Virology (Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., eds) 5th Ed., pp. 839–893, Vol. 1, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 15. Rossmann M. G., Johnson J. E. (1989) Icosahedral RNA virus structure. Annu. Rev. Biochem. 58, 533–573 [DOI] [PubMed] [Google Scholar]
- 16. Rossmann M. G., Arnold E., Erickson J. W., Frankenberger E. A., Griffith J. P., Hecht H. J., Johnson J. E., Kamer G., Luo M., Mosser A. G. (1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153 [DOI] [PubMed] [Google Scholar]
- 17. Hussain K. M., Leong K. L., Ng M. M., Chu J. J. (2011) The essential role of clathrin-mediated endocytosis in the infectious entry of human enterovirus 71. J. Biol. Chem. 286, 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamayoshi S., Yamashita Y., Li J., Hanagata N., Minowa T., Takemura T., Koike S. (2009) Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801 [DOI] [PubMed] [Google Scholar]
- 19. Nishimura Y., Shimojima M., Tano Y., Miyamura T., Wakita T., Shimizu H. (2009) Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15, 794–797 [DOI] [PubMed] [Google Scholar]
- 20. Nishimura Y., Wakita T., Shimizu H. (2010) Tyrosine sulfation of the amino terminus of PSGL-1 is critical for enterovirus 71 infection. PLoS Pathog. 6, e1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujita H., Ezaki J., Noguchi Y., Kono A., Himeno M., Kato K. (1991) Isolation and sequencing of a cDNA clone encoding 85kDa sialoglycoprotein in rat liver lysosomal membranes. Biochem. Biophys. Res. Commun. 178, 444–452 [DOI] [PubMed] [Google Scholar]
- 22. Reczek D., Schwake M., Schröder J., Hughes H., Blanz J., Jin X., Brondyk W., Van Patten S., Edmunds T., Saftig P. (2007) LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell 131, 770–783 [DOI] [PubMed] [Google Scholar]
- 23. Yamayoshi S., Koike S. (2011) Identification of a human SCARB2 region that is important for enterovirus 71 binding and infection. J. Virol. 85, 4937–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 25. Shen M. Y., Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goddard T. D., Huang C. C., Ferrin T. E. (2005) Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure 13, 473–482 [DOI] [PubMed] [Google Scholar]
- 27. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 28. Sandoval I. V., Arredondo J. J., Alcalde J., Gonzalez Noriega A., Vandekerckhove J., Jimenez M. A., Rico M. (1994) The residues Leu(Ile)475-Ile(Leu, Val, Ala)476, contained in the extended carboxyl cytoplasmic tail, are critical for targeting of the resident lysosomal membrane protein LIMP II to lysosomes. J. Biol. Chem. 269, 6622–6631 [PubMed] [Google Scholar]
- 29. Arita M., Nagata N., Iwata N., Ami Y., Suzaki Y., Mizuta K., Iwasaki T., Sata T., Wakita T., Shimizu H. (2007) An attenuated strain of enterovirus 71 belonging to genotype a showed a broad spectrum of antigenicity with attenuated neurovirulence in cynomolgus monkeys. J. Virol. 81, 9386–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oberste M. S., Maher K., Kilpatrick D. R., Pallansch M. A. (1999) Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73, 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang P., Mueller S., Morais M. C., Bator C. M., Bowman V. D., Hafenstein S., Wimmer E., Rossmann M. G. (2008) Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. U.S.A. 105, 18284–18289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tuthill T. J., Groppelli E., Hogle J. M., Rowlands D. J. (2010) Picornaviruses. Curr. Top. Microbiol. Immunol. 343, 43–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bubeck D., Filman D. J., Cheng N., Steven A. C., Hogle J. M., Belnap D. M. (2005) The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79, 7745–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang B., Chuang H., Yang K. D. (2009) Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol. J. 6, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin Y. W., Wang S. W., Tung Y. Y., Chen S. H. (2009) Enterovirus 71 infection of human dendritic cells. Exp. Biol. Med. 234, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 36. Yang S. L., Chou Y. T., Wu C. N., Ho M. S. (2011) Annexin II binds to capsid protein VP1 of enterovirus 71 and enhances viral infectivity. J. Virol. 85, 11809–11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eskelinen E. L., Tanaka Y., Saftig P. (2003) At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 13, 137–145 [DOI] [PubMed] [Google Scholar]
- 38. Blanz J., Groth J., Zachos C., Wehling C., Saftig P., Schwake M. (2010) Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand β-glucocerebrosidase. Hum. Mol. Genet. 19, 563–572 [DOI] [PubMed] [Google Scholar]
- 39. Eckhardt E. R., Cai L., Sun B., Webb N. R., van der Westhuyzen D. R. (2004) High density lipoprotein uptake by scavenger receptor SR-BII. J. Biol. Chem. 279, 14372–14381 [DOI] [PubMed] [Google Scholar]
- 40. Simmons N. B., Seymour K. L., Habersetzer J., Gunnell G. F. (2008) Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451, 818–821 [DOI] [PubMed] [Google Scholar]
- 41. Calisher C. H., Childs J. E., Field H. E., Holmes K. V., Schountz T. (2006) Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lau S. K., Woo P. C., Lai K. K., Huang Y., Yip C. C., Shek C. T., Lee P., Lam C. S., Chan K. H., Yuen K. Y. (2011) Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 85, 8819–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chua B. H., Phuektes P., Sanders S. A., Nicholls P. K., McMinn P. C. (2008) The molecular basis of mouse adaptation by human enterovirus 71. J. Gen. Virol. 89, 1622–1632 [DOI] [PubMed] [Google Scholar]
- 44. Arita M., Ami Y., Wakita T., Shimizu H. (2008) Cooperative effect of the attenuation determinants derived from poliovirus Sabin 1 strain is essential for attenuation of enterovirus 71 in the NOD/SCID mouse infection model. J. Virol. 82, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Foo D. G., Alonso S., Phoon M. C., Ramachandran N. P., Chow V. T., Poh C. L. (2007) Identification of neutralizing linear epitopes from the VP1 capsid protein of enterovirus 71 using synthetic peptides. Virus Res. 125, 61–68 [DOI] [PubMed] [Google Scholar]
- 46. Lal S. K., Kumar P., Yeo W. M., Kar-Roy A., Chow V. T. (2006) The VP1 protein of human enterovirus 71 self-associates via an interaction domain spanning amino acids 66–297. J. Med. Virol. 78, 582–590 [DOI] [PubMed] [Google Scholar]
- 47. Lewis J. K., Bothner B., Smith T. J., Siuzdak G. (1998) Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. U.S.A. 95, 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Broo K., Wei J., Marshall D., Brown F., Smith T. J., Johnson J. E., Schneemann A., Siuzdak G. (2001) Viral capsid mobility: a dynamic conduit for inactivation. Proc. Natl. Acad. Sci. U.S.A. 98, 2274–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hogle J. M. (2002) Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol 56, 677–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mendelsohn C. L., Wimmer E., Racaniello V. R. (1989) Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56, 855–865 [DOI] [PubMed] [Google Scholar]
- 51. Greve J. M., Davis G., Meyer A. M., Forte C. P., Yost S. C., Marlor C. W., Kamarck M. E., McClelland A. (1989) The major human rhinovirus receptor is ICAM-1. Cell 56, 839–847 [DOI] [PubMed] [Google Scholar]
- 52. Olson N. H., Kolatkar P. R., Oliveira M. A., Cheng R. H., Greve J. M., McClelland A., Baker T. S., Rossmann M. G. (1993) Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. U.S.A. 90, 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Levy H. C., Bostina M., Filman D. J., Hogle J. M. (2010) Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84, 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Filman D. J., Syed R., Chow M., Macadam A. J., Minor P. D., Hogle J. M. (1989) Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8, 1567–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith T. J., Kremer M. J., Luo M., Vriend G., Arnold E., Kamer G., Rossmann M. G., McKinlay M. A., Diana G. D., Otto M. J. (1986) The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 56. Pérez L., Carrasco L. (1993) Entry of poliovirus into cells does not require a low-pH step. J. Virol. 67, 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tee K. K., Lam T. T., Chan Y. F., Bible J. M., Kamarulzaman A., Tong C. Y., Takebe Y., Pybus O. G. (2010) Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J. Virol. 84, 3339–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.