Background: The active site of mannitol 2-dehydrogenase from Pseudomonas fluorescens is connected with bulk solvent through a narrow protein channel. Glu292 adopts flexible positions down and up the channel.
Results: Evidence linking motions of Glu292 to proton translocation to and from the active site.
Conclusion: Glu292 controls proton transfer through a water chain mechanism.
Significance: Dynamic features of the proton transfer mechanism have been elucidated.
Keywords: Dehydrogenase, Enzyme Kinetics, Enzyme Mechanisms, Isotope Effects, Molecular Dynamics, Proton Transport, Conformational Dynamics, Proton Transfer, Transient Kinetics
Abstract
The active site of mannitol 2-dehydrogenase from Pseudomonas fluorescens (PfM2DH) is connected with bulk solvent through a narrow protein channel that shows structural resemblance to proton channels utilized by redox-driven proton pumps. A key element of the PfM2DH channel is the “mobile” Glu292, which was seen crystallographically to adopt distinct positions up and down the channel. It was suggested that the “down → up” conformational change of Glu292 could play a proton relay function in enzymatic catalysis, through direct proton shuttling by the Glu or because the channel is opened for water molecules forming a chain along which the protons flow. We report evidence from site-directed mutagenesis (Glu292 → Ala) substantiated by data from molecular dynamics simulations that support a role for Glu292 as a gate in a water chain (von Grotthuss-type) mechanism of proton translocation. Occupancy of the up and down position of Glu292 is influenced by the bonding and charge state of the catalytic acid base Lys295, suggesting that channel opening/closing motions of the Glu are synchronized to the reaction progress. Removal of gatekeeper control in the E292A mutant resulted in a selective, up to 120-fold slowing down of microscopic steps immediately preceding catalytic oxidation of mannitol, consistent with the notion that formation of the productive enzyme-NAD+-mannitol complex is promoted by a corresponding position change of Glu292, which at physiological pH is associated with obligatory deprotonation of Lys295 to solvent. These results underscore the important role of conformational dynamics in the proton transfer steps of alcohol dehydrogenase catalysis.
Introduction
The alcohol dehydrogenase (ADH)2 reaction (Equation 1) is a key transformation of biological catalysis. It is exploited by the living cell in a wide variety of metabolic pathways (1–4) and has important practical applications in the synthesis of pharmaceuticals (5–8). The overall enzymatic conversion proceeds with release of one proton for each molecule NAD(P)H formed upon substrate oxidation (Equation 1).
 |
The catalytic chemistry of ADH enzymes involves hydride transfer to NAD(P)+ coupled to a multistep proton transfer functioning in the abstraction of proton from alcohol substrate and delivery of proton to solvent (9, 10). Proton translocation from the ADH active site to bulk water usually occurs through a succession of “proton hops” along a wire of conducting groups, the so-called proton relay, which are connected by hydrogen bonds (9, 11–15). This proton translocation is often regarded as a process in rapid equilibrium. However, kinetic studies of horse liver ADH have shown that the proton transfer can become rate-limiting for the overall enzymatic reaction (16), emphasizing how important it is to consider proton transfer in the ADH mechanism. As in the horse liver enzyme, the overall proton transfer in the different ADHs3 must probably be viewed as a dynamic process in which protein conformational changes at different time and length scales are coupled to the actual protonation/deprotonation events (16, 17). The molecular characterization of dynamic features of the proton transfer is a current theme of fundamental importance in mechanistic enzymology (17–24). Such as in the classical ADHs (Equation 1), coupling of hydride transfer to a multistep proton transfer is a distinctive feature of the catalytic mechanism of various other NAD(P)+-dependent dehydrogenases (24, 25), as well as alcohol oxidases utilizing FAD or FMN as cofactor to promote hydride-transfer oxidation of their substrates (18, 19, 26–29). Elucidation of the involvement of the proton relay system in the steps of the catalytic cycle presents a key scientific problem for each of these enzymes.
Mannitol 2-dehydrogenase from the bacterium Pseudomonas fluorescens (PfM2DH)4 shows a proton shuttle system that is quite noticeable among ADHs and mechanistically related oxidoreductases. Crystal structures of PfM2DH have revealed a narrow channel that is accessible to water and connects the active site with bulk solvent (30). The PfM2DH channel shares various features of molecular anatomy (Fig. 1) with the canonical proton channels of redox-driven proton pumps such as cytochrome c oxidase (31) and bacteriorhodopsin (32–35). It contains a “mobile” carboxylic acid residue (Glu292) accommodated within an otherwise comparatively hydrophobic channel interior. The side chain of Glu292 was seen to adopt distinct conformations down (Fig. 1A) and up (Fig. 1B) the channel in the holoenzyme bound with NAD+ and a ternary complex of PfM2DH with NAD+ and mannitol, respectively. The down → up movement of Glu292 represents the largest (≈5 Å displacement) among a series of small structural rearrangements in and outside of the PfM2DH channel that occur in response to mannitol binding, as shown in Fig. 1 and supplemental Movie S1. Globally, the conformational change of PfM2DH can be characterized as a subtle domain-opening process in which the C-terminal substrate-binding domain rotates away by about ∼2° from the N-terminal NAD+-binding domain, using the loop region of residues 282–287 as the hinge region. At the top of the channel on the PfM2DH surface, two ionizable amino acids (His294, Glu293) are positioned opposite to each other within hydrogen bonding distance (Fig. 1A). The interaction between His294 and Glu293 becomes disrupted as a result of the down → up movement of Glu292 (Fig. 1B). We extrapolate from reported relationships between structure and function of proton channels (36) that the particular arrangement of His294 and Glu293 might serve the role of a local proton reservoir, facilitating uptake and release of protons in each direction through adjustment of trans-channel proton gradient.
FIGURE 1.
Side chain movement of Glu292 in the proton channel of PfM2DH, and proposed proton translocation pathways. Panels A and B show close ups from the enzyme-NAD+ (PDB 1LJ8) and the enzyme-NAD+-mannitol (PDB 1M2W) crystal structures, respectively. Panel C depicts two possible mechanisms of participation of Glu292 in proton translocation, as proton shuttle (I) or gate (II). The water channel is derived from Asp190-Asn195 forming a loop that contains the oxyanion hole residue Asn191 (yellow), Ala227-Val229 from β11 sheet, and Val288-Ile296 forming the α10 helix containing the catalytic base Lys295 (yellow) and Glu292 (magenta). Hydrogen bonding interactions (distance ≤3.1 Å) between water molecules (red spheres) and amino acid residues or backbone amide groups are shown by dashed lines. NAD+ and mannitol are shown in blue and green, respectively.
Two mechanisms of proton transfer via the catalytic base Lys295 (37, 38) were considered for PfM2DH (30). One involves a shuttling motion of Glu292 (Fig. 1, I) comparable with the mode of proton translocation utilized by known proton pumps (34, 35, 39, 40). Of note, assistance from Glu292 to the removal of protons from the active site could provide internal “pull” to oxidation of the bound alcohol substrate. In an alternative scenario, Glu292 acts as a gate that opens as a result of the up-flip of the Glu side chain, thereby establishing a “water wire” along which protons can flow to solvent (Fig. 1C, II). In both mechanisms, proton translocation would be effectively discontinuous, and the essential time switch for the conformational change of Glu292 might be provided by events of the catalytic cycle. In marked contrast to PfM2DH, proton relays of other ADHs are typically built from hydrogen bond networks that seem to operate continuously and do not involve major conformational rearrangements to achieve the proton translocation (11, 16).
In this work, site-directed mutagenesis, kinetics, and molecular dynamics (MD) simulation were used to interrogate the proposed dynamic function of Glu292 during proton transfer by PfM2DH. Replacement of the Glu by Ala was chosen to disrupt shuttling motions (Fig. 1C, I) or to prevent closing of the proton channel (Fig. 1C, II). The requirement to decrease steric bulk at position 292 in the channel explains the use of a small residue as compared with the approximately isosteric Gln. Catalysis to mannitol oxidation could thus be compared for situations in which protein control over proton translocation to solvent was present (wild-type enzyme) or absent (E292A). A dynamic mechanism of proton transfer is suggested in which, once mannitol has bound to the enzyme-NAD+ complex, a down → up conformational change of Glu292 results in positioning of Lys295 such that it connects the reactive C2 hydroxyl of substrate to a chain of water molecules in the now open proton conduit (Fig. 1C, II). The opposite up → down motion of Glu292 in the step post-catalysis results in release of Lys295 from being bonded with the carbonyl group of fructose, thereby promoting the product dissociation.
EXPERIMENTAL PROCEDURES
Materials and chemicals were described elsewhere (37, 38, 41, 42). Wild-type PfM2DH was prepared by known procedures (43).
Site-directed Mutagenesis, Protein Production, and Characterization
Mutation Glu292 → Ala was introduced by inverse PCR (38) using a pair of oligonucleotide primers: 5′-GTG ACA CCC TAT GCA GAG ATG AAG-3′ and 5′-GGA CAT CAC GGT AAA CGC-3′. The mismatched bases are underlined. The mutated gene was expressed in Escherichia coli JM109, and the E292A mutant was isolated according to protocol (43). CD spectra of wild-type PfM2DH and E292A were recorded as described elsewhere (38). Molar enzyme concentrations were determined from absorbance (280 nm) using an extinction coefficient of 55.4 mm−1 cm−1 for wild-type enzyme and mutant (44).
Steady-state Kinetic Analysis
Initial reaction rates were recorded spectrophotometrically at 25 °C, measuring the change in absorbance of NADH at 340 nm (ϵ340 = 6.22 mm−1 cm−1). A full steady-state kinetic analysis was performed at pH 7.1 (100 mm Tris-HCl) and pH 10.0 (100 mm glycine-NaOH). Kinetic data were collected with the concentrations of substrate and coenzyme being varied against one another, and parameters (kcat, KiC, KC, KS) were obtained by nonlinear regression, as described elsewhere (38, 42, 43). kcat is the turnover number, KiC is the dissociation constant of the enzyme-coenzyme complex, KC and KS are Michaelis constants for coenzyme and substrate, respectively. We use subscript O and R on kcat and the transient rate constant kobs (see later) to indicate the direction of mannitol oxidation (kcatO, kobsO) and fructose reduction (kcatR, kobsR), respectively.
The pH dependences of kinetic parameters for E292A were determined in the pH range 7.1–10.5 for mannitol oxidation (kcatO, KC, and KS) and 7.1–10.0 for fructose reduction (kcatR, KS) as described previously in studies of other PfM2DH mutants (41). A two-component buffer (Tris, glycine) having pH-independent ionic strength of 0.1 m was used (45).
Kinetic isotope effects (KIE) resulting from deuteration of one of the reactants (2-[2H]mannitol; S-4-[2H]NADH) or solvent were determined using reported procedures (42, 43). A nomenclature is used where superscript D describes the primary deuterium KIE and superscript D2O describes the solvent KIE on the respective isotope-sensitive parameter (46). DKIEs were obtained in both directions in the reactions at pH 7.1 (100 mm Tris-HCl) and 10.0 (100 mm glycine-NaOH). D2OKIEs were determined for mannitol oxidation under conditions where the examined kinetic parameter was independent of pH(D), that is, pH(D) 10.0.
Analysis of the data from pH dependence and KIE measurements was done by reported procedures (42, 43). Full details are given in supplemental data.
Transient Kinetic Analysis
Stopped-flow kinetic data were acquired at 25 °C, typically by recording the change of NADH absorbance at 340 nm as described elsewhere (41, 42). The instrument used (Applied Photophysics model SX.18 MV) had a dead time of 2.0 ms. All reactions were performed using enzyme (10–20 μm) in a limiting concentration. Procedures applied for determination of transient kinetic constants under conditions of varied pH and solvent (H2O, D2O) are provided in supplemental data.
Measurement of transient proton release (pH 7.1 and 8.0) or uptake (pH 7.1) during the enzymatic reaction was done using the pH indicator phenol red whose change in absorbance was recorded at 556 nm. E292A was gel-filtered twice to a 0.5 mm Tris-HCl buffer (pH 7.1 and 8.0) containing 34 μm phenol red, 25 mm (pH 7.1) or 6 mm (pH 8.0) NAD+, or 200 mm fructose (pH 7.1). NaCl was added to adjust the ionic strength of the solution to that of the reference buffer (100 mm Tris-HCl). The enzyme solution was rapidly mixed with mannitol (1000 mm, pH 7.1; 600 mm, pH 8.0) or NADH (400 μm) dissolved in the same buffer. Proton uptake was measured similarly for the wild-type enzyme (initial concentrations: 200 μm NADH, 250 mm fructose). Proton concentrations were determined as described elsewhere (11). Data fitting is described in supplemental data.
MD Simulations
The MD simulations were performed using a double precision version of the Gromacs MD simulation package (47). Initial coordinates were taken from the crystal structures (binary complex with NAD+, PDB code 1LJ8; ternary complex with mannitol and NAD(H), PDB code 1M2W). PDB codes are subsequently used for identification. The program PROPKA version 3.1 (48) was used to obtain estimates for the acid ionization constants (pKa values) for each residue in PDB 1LJ8 and 1M2W. The pKa of the residues in the active site and the pKa change resulting from formation of the ternary complex were of particular interest and carefully analyzed. Both protein structures were then protonated at pH 7.0 in accordance with the pKa prediction. The Gromacs pdb2gmx program was used for adding hydrogens to the protein. A variation of the Amber99 force field was used for modeling the proteins (49). Using the Antechamber program (50), general amber force field parameters and AM1BCC partial charges were generated for the ligands (51, 52). Modeling of fructose into 1LJ8 used the coordinates for mannitol from 1M2W, except that the C2 oxygen, which was modeled using Chimera (53), had a shorter bond length and slightly different dihedral angles. Note that the bound coenzyme was modeled as NADH in this case. No prior assumptions concerning the conformation of the nicotinamide ring of NADH were made, and the experimental conformation of the bound NAD+ was used. Using the applied force field, only small fluctuations of the conformation of the nicotinamide ring were observed. Further technical details of the MD simulation are given in the supplemental information. Simulations were run for 10–20 ns.
Data analysis was done with respect to backbone r.m.s. deviations for simulated structures as compared with the corresponding initial structure, calculating the r.m.s. deviations for the enzyme as a whole but also for the individual protein domains. The trajectory for the positional change of Glu292 between the up and down conformations was also calculated. Therefore an angle α was defined. This represents the angle between two vectors, one pointing from the γ-C atom of Glu292 to CNAD and another pointing from the γ-C atom of Glu292 to the δ-C atom of Glu292. An angle α of 0 indicates the down position of Glu292, whereas one at 180° marks the up position.
RESULTS
Shifts in the pKa Values of Active Site Groups Associated with Binding of Mannitol to Enzyme-NAD+
We have shown in previous work that the pH dependence of mannitol oxidation by PfM2DH is consistent with a change in pKa of the catalytic base Lys295 from a value of 9.2 in enzyme-NAD+ to about 6.8 in the ternary complex (41). Considering that the protein conformational change connected to formation of 1M2W from 1LJ8 could affect the ionization of different groups in and around the active site, particularly that of Glu292, we applied pKa calculations with PROPKA 3.1. Of note, the probable protonation state of relevant protein residues was considered an important input parameter for the MD simulations to be described below. We have summarized the results in supplemental Table S1. It was interesting that among all ionizable residues present in 1M2W and 1LJ8, only Glu292 and Lys295 displayed significant perturbation in pKa upon changing from enzyme-NAD+ to the ternary complex. The calculations reproduced the experimental pKa change of Lys295 remarkably well, with estimates for the actual pKa values of the lysine in 1LJ8 (pKa = 9.2) and 1M2W (pKa = 7.2) being quite close to the experimentally determined constants. The pKa of Glu292 was predicted to change from a value of 9.6 in 1LJ8 to a lower value of about 8 in 1M2W. Note, however, that the side chain of Glu292 is quite flexible in 1M2W, indicated by a large β-factor of 29–34 Å2. Therefore this precludes direct use of the experimental protein structure for calculation of a well defined pKa for Glu292 in the up position. The implication that Lys295 and Glu292 are both protonated in 1LJ8 appeared somewhat improbable. We think that the unexpected similarity in the calculated pKa values for Lys295 and Glu292 could alternatively be interpreted in terms of a common ionization constant for a molecular group that consists of a water-mediated salt bridge between Lys295 and Glu292, as shown in supplemental Fig. S1. The ionization behavior of such a molecular group cannot be determined in a reliable manner using PROPKA 3.1. However, we will show below that the notion of a singly protonated group involving Lys295/H2O/Glu292 is consistent with the release of protons from the enzyme during mannitol oxidation at pH 8.0.
To glean more information on the ionization of Glu292, we performed two MD simulations of 1M2W in which we assumed Glu292 to be protonated or unprotonated, whereas Lys295 was assumed to be unprotonated in both simulations. The simulation trajectories are depicted in panels VII and IX of supplemental Fig. S2 where the data are presented together with results of a comprehensive MD simulation study that will be discussed below. Representative structure snapshots (n = 50) from the MD simulation trajectories were subjected to pKa calculation, yielding a reasonably defined pKa value of 7.0 (±0.6) for Glu292. Supplemental Table S1 summarizes the results of the analysis performed.
We have previously carried out proton release measurements in the kinetic transient of mannitol oxidation by PfM2DH at pH 8.0 (41). The amount of protons released in the pre-steady state phase exceeded the molar equivalent of enzyme active sites present by a factor of about 1.6. Roughly one-third of the released protons was accounted for by the chemical reaction (see Equation 1) under these conditions, implying that PfM2DH released the molar equivalent of protons due to deprotonation. The experimental stoichiometry of the proton release is therefore inconsistent with a doubly protonated group, Lys295/H2O/Glu292, that would have produced up to 2.6 protons per enzyme active site (see supplemental Table S2). The measured data are in excellent agreement with a scenario in which Lys295/H2O/Glu292 contains a single proton that is released because of the pKa change experienced by the lysine once the Lys295/H2O/Glu292 group has become disrupted by the down → up movement of Glu292. Glu292 in the up position will be ionized at pH 8.0 (see supplemental Table S1). These findings were applied in further MD simulations.
Structural Dynamics of Glu292 Characterized by MD Simulation
We performed MD simulations on different ternary complex structures of wild-type PfM2DH. Each of these structures represented a possible snapshot from the proposed enzymatic reaction coordinate as shown in Scheme 1.5 For each complex, the protonation state of Lys295 (NH2/NH3+) is as indicated in Scheme 1 by the superscript H (e.g. HEAB). Similarly, the protonation of Glu292 (COO−/COOH) was varied in 1LJ8 complexes, whereas in 1M2W complexes, Glu292 was assumed to be ionized. Note, for the reasons given above, doubly protonated 1LJ8 complexes were not considered. In summary, therefore, we simulated the following structures whereby the corresponding simulation run(s) and reaction coordinate complexes are indicated in parentheses: 1LJ8-NADH-fructose (runs I,/II, III; *EPQo/HEPQc), 1LJ8-NAD+-mannitol (runs IV/V and VI; EABo/HEABc) and 1M2W-NAD+-mannitol (runs VII/VIII; EABo/HEABo).6 A key proposal of Scheme 1 is the occurrence of closed (Ec) and open (Eo) enzyme forms that are adopted in dependence of the bonding/ionization state of Lys295. The kinetic isomerization of Ec into Eo constitutes a probable manifestation of the crystallographic domain-opening conformational change of PfM2DH (see supplemental Movies S1 and S2). Formation of Eo is a requirement for proton exchange between the active site and bulk solvent. The transition of Ec into Eo plus the tentative suggestion from Scheme 1 that Glu292 might become flexible in Eo was put to a critical test by MD simulation.
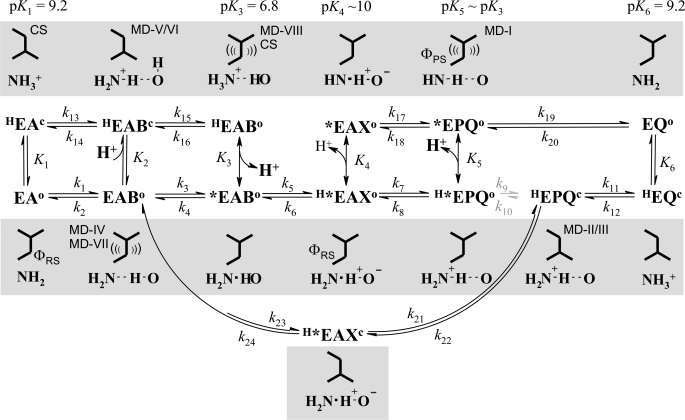
SCHEME 1.
Proposed kinetic reaction coordinate for mannitol oxidation and fructose reduction by wild-type PfM2DH and E292A. The proposed structural dynamics of the Glu292 side chain (up, down) and changes in bonding/protonation state for Lys295 (NH2, NH3+) associated with the reaction progress are shown in the gray boxes. Note: the down and up position of Glu292 is connected with enzyme being in the closed domain (superscript c) and open domain conformation (superscript o), respectively. Complexes observed crystallographically (CS) and in MD simulations (MD run number indicated, see Fig. 2 and supplemental Fig. S2) are shown. Flexibility of Glu292 as revealed by MD simulations is also indicated ((( ))). HE and H*E, protonated enzyme forms, where * indicates the enzyme conformation that has Lys295 connected to the water-wire proton relay; A, NAD+; B, mannitol; X, mannitol having C2-OH (partially) deprotonated; P, fructose; Q, NADH. Deuterium fractionation factors (ΦRS, ΦPS) as relevant for the observed D2OKIEs are indicated (see text in “Discussion”). Protonations at rapid equilibrium are shown as double arrows. Protonations having potential kinetic significance due to the involved position change of Glu292 are shown as reversible reactions. Ionization constants (see supplemental Table S5) are assigned to a molecular enzyme group that contains Lys295. The 2-O(H) of the alcohol/carbonyl group is indicated, with strong hydrogen bonds (−) distinguished from normal hydrogen bonds (--). In mannitol oxidation, the isomerization step k9 (in gray) is probably much slower than proton release from H*EPQ (k5) so that the path via *EPQ is preferred. In fructose reduction, the isomerization of HEPQ (k10) is probably disfavored so that the reaction path from HEPQ to EAB (k21-k24) is used by the wild-type enzyme. In E292A, fructose reduction takes place via k10 from HEPQ to H*EPQ. Proton release/uptake measured in the stopped-flow experiment is shown in bold. See the text for further discussion of the mechanism.
A complete summary of the simulation results is given in supplemental “Results” and Fig. S2. Key findings are presented in Fig. 2. Analysis of r.m.s. deviation trajectories calculated from the simulation data showed enzyme complexes containing negatively charged Glu292 and protonated Lys295, this being a plausible chemical representation of the singly protonated group Lys295/H2O/Glu292, converged readily within ≤5 ns into apparently stable structures that were all highly similar to the corresponding experimental structures. Using the alternative, yet less likely representation of Lys295/H2O/Glu292, where Lys295 is unprotonated and Glu292 is protonated, convergence to a stable r.m.s. deviation level was observed for the 1LJ8 complex having NADH and fructose bound. The corresponding complex with NAD+ and mannitol did not converge, which we noted but did not pursue further. The closed form Ec was preferred in all complexes featuring the singly protonated group Lys295/H2O/Glu292, irrespective of the protonation state of its individual components. Moreover, Glu292 did not change its location down the channel.
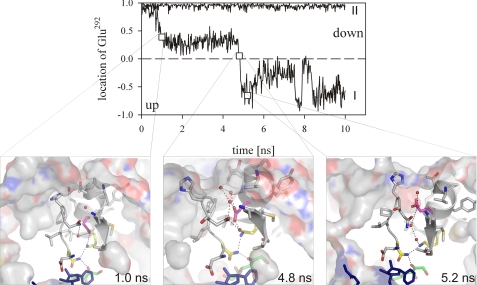
FIGURE 2.
Conformational mobility of Glu292 in 1LJ8-NADH-fructose revealed by MD simulation. Trajectories were obtained for complexes having protonated (I) and unprotonated (II) Lys295. Data on the ordinate show the normalized scalar product of vectors γ-C Glu292/CNAD and γ-C Glu292/δ-C Glu292 forming the angle α. A value of 1 or −1 reflects an angle α of 0° or 180°, meaning that Glu292 adopts the down or up position, respectively. Structure snapshots at the given simulation times are presented below. Residue coloring and representation of hydrogen bonds are the same as described in the legend to Fig. 1.
The overall r.m.s. deviation for enzyme complexes having Lys295 and Glu292 unprotonated did not approach a constant value, suggesting a somewhat unstable conformation. Interestingly, however, in these structures, the r.m.s. deviation trajectories for the individual protein domains were remarkably different from that for the overall r.m.s. deviations, approaching stable and low (≤0.2 nm) values after a comparably short simulation time (≤5 ns). Structural dynamics of the enzyme as a whole is therefore explained from the overall trend of the simulation data to result from an opening movement of the C-terminal relative to the N-terminal domain, in useful agreement with the crystallographically observed reorientation of the two protein domains in 1M2W as compared with 1LJ8 (see supplemental Movie S2).
Enlargement of the interdomain space resulting from domain opening might enhance the flexibility of Glu292, arguably permitting it to change its position in the channel. MD simulations have successfully captured this conformational mobility of Glu292, showing that it is by far greater in unprotonated as compared with protonated complexes of the enzyme (supplemental Fig. S2). The mobility of Glu292 is probably best represented in a superimposition of the trajectories for Glu292 in protonated and unprotonated 1LJ8-NADH-fructose complexes (Fig. 2 and supplemental Fig. S2). Fig. 2 additionally reveals the possibility for Glu292 to switch from the down to the up position in the complex when Lys295 and Glu292 were unprotonated.
1M2W complexes showed a highly flexible Glu292 regardless of the protonation state of Lys295 (supplemental Fig. S2), consistent with the relatively more open domain conformation of 1M2W as compared with 1LJ8. However, among the ensemble of simulated conformations of Glu292 in the two 1M2W complexes, an averaged position up the proton channel was clearly preferred, and there was no switching back of the Glu to a defined down position near the active site in the applied simulation time.
To summarize, MD simulations portray the structural dynamics of Glu292 as a likely accompaniment of the domain-opening conformational change of PfM2DH during formation of the productive ternary complex in mannitol oxidation. The apparent stabilization of the closed enzyme conformation under conditions of protonated Lys295 (1LJ8 complexes; Ec) is explained by charge complementarity in the interdomain space where the lysine is accommodated through interactions with different residues, mainly Asn191 and Asn300 but also water bonded to Glu292 (Fig. 1A). Productive orientation of mannitol in the active site such that the reactive C2 hydroxyl forms a hydrogen bond with NZLys295 will cause substantial perturbation in the lysine site. This perturbation could plausibly serve as trigger for domain opening (HEABc ⇒ HEABo). The finding that preference of Glu292 for adopting a position up and down the water channel is probably linked to a neutral and positively charged NZLys295, respectively, immediately leads to the suggestion that motions of the Glu292 would be directly coupled to steps of the catalytic cycle that involve change in the protonation state of Lys295 (Scheme 1).
Kinetic Characterization of E292A
Results from CD spectroscopy (supplemental Fig. S3) and MD simulation (supplemental Fig. S2, X) show that the overall fold and distribution of secondary structural elements as well as positioning of catalytic reactive groups within the active site were hardly affected by the mutation. Functional consequences in E292A can therefore be interpreted to result from locally disruptive effects of the site-directed substitution.
Like the native enzyme, E292A was best active toward mannitol and fructose at pH values of ∼10 and ∼7, respectively (see Fig. 3). A full steady-state kinetic characterization of E292A was therefore performed under the respective “optimum pH” conditions for oxidation and reduction. Table 1 compares a representative set of kinetic parameters for E292A with those previously reported for wild-type PfM2DH. A complete summary of the kinetic characterization of E292A, including validation of the reported parameters through Haldane relationship analysis, is given in supplemental Table S3.
FIGURE 3.
pH profiles for E292A (closed symbols) and wild-type PfM2DH (open symbols) for mannitol oxidation (A) and fructose reduction (B). pH dependences of log(kcatO/KC), log(kcat/KS), and log kcat are depicted as squares, triangles, and circles, respectively. Lines represent best fits of experimental data and the respective equation (see supplemental “Experimental Procedures”). Corresponding ionization constants are summarized in supplemental Table S5. Note: the pH profile of log(kcatR/KC) for fructose reduction by E292A was not determined. Considering the similarity in all other pH dependences of kinetic parameters for wild-type enzyme and mutant, this profile was not pursued.
TABLE 1.
Kinetic parameters for E292A and wild-type PfM2DH
| Parameter | E292A | Wild-typea |
|---|---|---|
| Mannitol oxidation at pH 10.0 | ||
| kcatO (s−1) | 1.20 ± 0.02 | 40.0 ± 0.5 |
| kobsO (s−1) | 47 ± 2 | 1483 ± 80b |
| KC (μm) | 71 ± 5 | 93 ± 8 |
| KiC (μm) | 22 ± 9 | 300 ± 50 |
| kcatO/KC (m−1 s−1) | 1.7 × 104 | 4.0 × 105 |
| KS (mm) | 0.56 ± 0.03 | 0.40 ± 0.02 |
| kcatO/KS (m−1 s−1) | 2.1 × 103 | 1.0 × 105 |
| Fructose reduction at pH 7.1 | ||
| kcatR (s−1)a | 2.3 ± 0.1 | 61 ± 1.5 |
| kobsR (s−1) | 27 ± 3 | 403 ± 81b |
| KC (μm) | 7 ± 1 | 67 ± 4 |
| KiC (μm) | 17 ± 4 | 80 ± 15 |
| kcatO/KC (m−1 s−1) | 3.3 × 105 | 9.1 × 105 |
| KS (mm) | 0.13 ± 0.01 | 0.24 ± 0.03 |
| kcatR/KS (m−1 s−1) | 1.8 × 104 | 2.5 × 105 |
Substitution of Glu292 by Ala caused a similar (20–40-fold) decrease in kinetic parameters (kcatO, kcatO/KC, kcatO/KS) for mannitol oxidation at pH 10.0. In terms of KiC, binding of NAD+ was about 14-fold tighter by E292A than it was by wild-type enzyme. In fructose reduction at pH 7.1, there was a modest effect of the mutation on kcatR/KC (2.3-fold decrease), whereas more substantial decreases in kcatR (23-fold) and kcatR/KS (12-fold) were observed. In terms of KiC, binding of NADH was about 5-fold stronger in E292A as compared with wild-type enzyme. Observations that in each direction of the reaction, kcat was about as sensitive to the substitution of Glu → Ala as was kcat/KS indicates that the role of Glu292 in the enzymatic mechanism primarily concerns steps of the catalytic cycle. The magnitude of the disruptive effect of the mutation is consistent with an auxiliary function of Glu292 in catalysis. The data also indicate that substitution of Glu292 by Ala is not silent with respect to binding of NAD+ and NADH. However, the kinetic peculiarities of E292A with respect to binding of coenzyme appeared to be not directly related to the mechanistic problems examined and were therefore not further pursued.
To examine the catalytic part of the overall reaction of E292A as a kinetically isolated step, we analyzed mannitol oxidation and fructose reduction by the mutated enzyme in rapid mixing stopped-flow experiments. Irrespective of the pH used (pH 7.1 or 10.0) and the direction of reaction studied, the enzymatic conversion proceeded with a pre-steady state “burst” that was followed by a linear steady-state phase (supplemental Fig. S4). Rate constants (kobs) and amplitudes associated with the kinetic transients were obtained from fits of the data, and results at optimum conditions for reduction (pH 7.1; kobsR) and oxidation (pH 10.0; kobsO) are shown in Table 1. A complete summary of the transient kinetic data are presented in supplemental Table S3. For E292A, the magnitude of the pre-steady state burst decreased as the pH was lowered from 10.0 to 7.1. This decrease is mainly due to changes in the ratio of kobs to kcat in response to variation of pH. Under the conditions used where the concentrations of substrate and coenzyme were saturating, kobsO and kobsR represent the interconversion of enzyme-NAD+-mannitol and enzyme-NADH-fructose in oxidation and reduction direction, respectively.7 Either rate constant of E292A was substantially (11–23-fold, fructose reduction; 30–50-fold, mannitol oxidation) decreased at each pH as compared with the corresponding kobs value for the native enzyme. These results confirm the notion that substitution of Glu292 by Ala partially disrupts the catalytic cycle of PfM2DH.
pH Studies
Despite fundamental disparity in the invoked molecular mechanism, the different proposals for the role of Glu292 in the catalytic reaction of PfM2DH (Fig. 1C) agree in their common implication that site-directed replacement of Glu by Ala could interfere with the proton transfer steps. Beyond that, an ionizable residue is removed in the immediate vicinity of the enzyme active site (Fig. 1A). Therefore, the pH activity dependence of E292A might differ significantly from that of wild-type PfM2DH, and comparison of the two pH dependences could provide vital evidence on the role of Glu292 in the enzymatic mechanism. pH profiles for steady-state kinetic parameters of E292A were measured in each direction of reaction. The results are shown in Fig. 3 where they are collated with the corresponding pH profiles of native PfM2DH (42, 43). The pH profile of log(kcatO/KC) for wild-type enzyme had not previously been reported and was therefore determined in this work. pKa values associated with theses pH profiles are summarized in supplemental Table S5. The results show that the pH activity dependence of E292A was overall very similar to that of the wild-type enzyme. Because some of the pH profiles in Fig. 3 might have a shallow appearance (in the log-log representation), we would like to point out that each pH dependence shown was well fitted with the corresponding pH model and the pKa values thus obtained are statistically well determined.
We also determined the pH dependences of kobs in either direction of reaction and show the results in supplemental Fig. S5. Like in wild-type enzyme, kobsR of E292A was independent of pH. An average value for kobsR of 25 ± 2 s−1 was obtained in the pH range 7.0–10.0. Determination of the pH profile of log kobsO was complicated due to the apparent interference from a buffer effect on E292A. However, the overall trend of the data are interpreted as a decrease in log kobsO at low pH. The pKa associated with this decrease had a value of 7.3 ± 0.1 and can be compared with the pKa of 6.8 observed in the pH profile of log kobsO for the wild-type enzyme (41).
We have previously shown that the essential molecular group in enzyme-NAD+ (kcatO/KS profile) and enzyme-NADH (kcatR/KS profile) that undergoes ionization at a pKa of about 9 contains Lys295 (42, 43). The pKa of this group is shifted down to a value of about 7 in enzyme-NAD+-mannitol (kobsO profile), explained by formation of a hydrogen bond between the reactive hydroxyl of substrate and the ϵ-amino group of Lys295 (41). The set of pH profiles collected herein reveal that Glu292 is not part of this essential group in PfM2DH. This result is difficult to reconcile with a proton transfer that relies on Glu292 to take up the proton abstracted from substrate by Lys295 and shuttle it to solvent (Fig. 1C, I). It is, however, compatible with the alternative “gate” mechanism (Fig. 1C, II) of Glu292. The pH studies, furthermore, show that Glu292 is not accountable for the groups whose ionizations determine the pH-log(kcatO/KC) profile, representing the pH dependence of binding of NAD+ to free enzyme.
Kinetic Isotope Effects
We performed steady-state and transient kinetic experiments to measure DKIEs on the relevant parameters (Dkcat, Dkcat/KS, and Dkobs) for E292A. Analysis of these DKIEs provide important information on the contribution of the isotope-sensitive step of hydride transfer to rate limitation in the overall reaction of the mutant. DKIEs on kcat and kcat/KS were determined above (pH 10.0) and below (pH 7.1) the apparent pKa of Lys295. Results are shown in supplemental Table S6. At the optimum state of protonation of Lys295 for oxidation (pH 10.0) and reduction (pH 7.1), E292A showed lower Dkcat/KS values than wild-type PfM2DH. Hydride transfer is therefore not rate-determining for catalysis by E292A in either direction of the reaction. Contrary to the native enzyme (42), DKIEs for the mutant were not affected by the used variation in pH. The Dkcat/KS for E292A approached a value between unity (=no DKIE) and the equilibrium DKIE (DKeq) in the direction of mannitol oxidation (DKeq = 1.2) and fructose reduction (DKeq = 0.83) (42). These DKIEs imply that there must be relatively slow steps prior to and after the isotope-sensitive step of hydride transfer by E292A, strongly suppressing the observable DKIE in each direction of reaction. We also analyzed the occurrence of a DKIE on kobsO (=1.1 ± 0.1) and kobsR (=0.9 ± 0.1) for E292A at pH 10.0. Both DKIEs were between 1 and DKeq, consistent with the notion that hydride transfer comes close to equilibrium in the reaction catalyzed by E292A.
KIEs due to deuteration of solvent may be useful to capture limitations of the enzymatic rate by proton transfer steps. D2OKIEs on kinetic parameters for mannitol oxidation by E292A were therefore determined and are summarized in supplemental Table S6. The large D2OKIE on kcatO/KS (2.4 ± 0.3) (42) for the wild-type enzyme was eliminated in E292A. There was likewise no D2OKIE on kcatO/KS when reaction of E292A was performed with deuterated mannitol substrate. The D2OKIEs on kcat were similar for both native and mutated enzyme (1.6–1.7) implying that solvent sensitivity of step(s) involving dissociation of NADH from E-NADH were not affected by mutation (42). E292A displayed a D2OKIE of 1.4 ± 0.1, whereas the D2OKIE on the amplitude of the pre-steady state burst was inverse (0.6 ± 0.1). By way of comparison, kobsO for the wild-type enzyme does not show solvent dependence (42). From the hyperbolic dependence of kobsO on the mannitol concentration, we determined a substrate half-saturation constant (Kd,S) of 7 ± 1 mm that describes dissociation of mannitol from E292A-NAD+-mannitol. We showed that this KdS was sensitive to solvent deuteration and determined a D2OKIE of 1.8 ± 0.3. The resulting D2OKIE on kobsO/KdS was 0.8, which is comparable with D2OkcatO/KS.
Transient Kinetic Analysis of Proton Transfer
Stopped-flow experiments were performed to measure proton transfer between enzyme and solvent in the pre-steady state phase of the reaction catalyzed by E292A. Fig. 4 shows representative time courses of proton release/uptake by the enzyme. Transient kinetic parameters associated with the shown time courses are given in supplemental Table S4.
FIGURE 4.
Stopped-flow time courses of proton release/uptake and NAD(H) formation during reactions catalyzed by E292A and wild-type enzyme. Panels A and B show mannitol oxidation by E292A at pH 7.1 and 8.0. Release of H+ (I) and formation of NADH (II) is indicated. Panels C and D and E and F display time courses of NAD+ formation (C and E) and H+ uptake (D and F) at pH 7.1 by wild-type PfM2DH and E292A, respectively. Transient kinetic constants are summarized in supplemental Table S4. Note: the decline of [NAD+]/[E] in the very first phase of the stopped-flow time course shown in panels E and F is apparent. It corresponds to the absorbance signal received within the dead time of the instrument (<2 ms).
In mannitol oxidation by E292A (Fig. 4, A and B), there was a weak transient burst of proton release that was followed by the linear steady-state phase. The steady-state rates of proton release and formation of NADH by E292A were identical, consistent with the enzymatic reaction, mannitol + NAD+ ↔ d-fructose + NADH + H+. The amount of protons produced by E292A in the pre-steady state burst (0.32 at pH 7.1 and 0.84 at pH 8.0) was smaller than the limiting concentration of enzyme used in the experiment. For the wild-type enzyme instead, a much larger molar ratio of 1.7 [H+]/[E] at pH 8.0 was observed (41). The transient rate constant of proton release (kobsO_H+) was decreased in E292A, by at least 115-fold (=1500/13), as compared with the approximate kobsO_H+ (≥1500 s−1) for the wild-type enzyme (41).
In fructose reduction (pH 7.1) by E292A (Fig. 4, C and D) and wild-type enzyme (Fig. 4, E and F), uptake of protons (Fig. 4, D and F) matched the consumption of NADH (Fig. 4, C and E) in the kinetic transient, and kobsR_H+ was about equal to kobsR. kobsR_H+ was lowered about 20-fold as a result of the mutation. The molar ratio of NADH consumed and protons taken up in the kinetic transient was in agreement. Summarizing, these results show partial disruption of the overall proton transfer in E292A as compared with the wild-type enzyme.
DISCUSSION
Lys295 provides Brønsted base catalytic assistance to NAD+-dependent oxidation of mannitol by PfM2DH. Because of the high pK of 9.2 for Lys295 in enzyme-NAD+, reaction at physiological conditions (pH ≈ 7) relies on deprotonation of the ϵ-amino group of the lysine at the ternary complex level (37, 41). Depression of the pK of Lys295 to a value of 6.8 as a result of mannitol binding ensures optimal catalytic participation from the lysine in the direction of substrate oxidation (41). Measurements of proton release in the kinetic transient of oxidation of mannitol by wild-type enzyme and a series of active site mutants of PfM2DH have previously captured the essential “activation” of Lys295 and, furthermore, showed that the proton formed in the chemical reaction is also transferred to solvent prior to dissociation of enzyme-NADH, the rate-limiting step at the steady-state (41). The results reported herein delineate the role of the mobile Glu292 in facilitating these critical proton transfer steps of PfM2DH. Collectively, they provide a strong case for the involvement of protein conformational dynamics in catalytic proton transfer by this ADH. Scheme 1 shows the proposed enzymatic mechanism that is discussed below.
Roles of Glu292 in the Catalytic Cycle of PfM2DH: Evidence from Kinetics
The presence of a (small) pre-steady state burst of product formation in multiple turnover stopped-flow progress curves for mannitol oxidation and fructose reduction by E292A together with the suggestion from DKIE studies that a post-catalytic step at the ternary enzyme complex level is slow in the mutated enzyme leads to the proposition that the overall product release (including a possible isomerization of the ternary enzyme complex prior to the actual dissociation of product) has become a partly rate-limiting step of kcat for each direction of the enzymatic reaction as a result of the Glu292 → Ala substitution.
Results of pH studies indicate that the pH-dependent ionization of Lys295 in enzyme-NAD+ was identical with limits of error in E292A as compared with wild-type enzyme. The data also reveal that binding of mannitol to E292A-NAD+ was associated with a similarly large decrease in pKa for Lys295 as the one observed in the wild-type enzyme. Interestingly, therefore, contrary to native PfM2DH, the mutated enzyme did not show fast proton release when its pre-formed binary complex with NAD+ (in which Lys295 is about half or fully protonated at pH 7.1 or 8.0) was mixed with a saturating concentration of mannitol in the stopped-flow instrument. Deprotonation of the lysine was previously suggested to be part of a pre-catalytic conformational equilibrium, EABo (HEABc) ⇔ *EABo (k3 and k4; k15 and k16), in which the ϵ-amino group of Lys295 and the 2-OH of mannitol are optimally aligned for base catalysis (41, 42). Evidence from transient kinetic analysis of proton transfer from enzyme to solvent indicates that substitution of Glu292 by Ala resulted in slowing down of the “forward isomerization” step of this equilibrium (k3 or k15). An additional decrease in k4 and/or k16 as a result of the mutation is likely and it would explain the slow rate of product dissociation in the direction of fructose reduction, as implied by the KIE data.
The finding that E292A undergoes only partial deprotonation in the pre-steady state reaction phase (pH 8.0: 84%), whereas the wild-type enzyme releases 1.7 times its molar equivalent of protons under the same reaction conditions is primarily due to a low NADH burst (∼0.2 [NADH]/[E]) formed in the transient phase of mannitol oxidation. Another, although minor component of this effect could be that the pre-catalytic equilibrium (k3/k4 and k15/k16) (see supplemental Table S2) changes as a result of the mutation. The experimental pKa (wild-type enzyme, 6.8; E292A, 7.3) is assigned to Lys295 in the isomerized *EABo complex ready to undergo reaction via proton abstraction and hydride transfer.
Solvent KIEs were analyzed in terms of the deuterium fractionation factors of reactant (ΦRS), transition (ΦTS), and product states (ΦPS). In wild-type enzyme, ΦRS and ΦTS have a similar value of ∼0.36 (37). The low ΦRS and ΦTS were explained by development of the partial negative charge on the oxygen atom of the reactive alcohol group in both the reactant state (H*EAXo) and the transition state, resulting from partial proton abstraction by ϵ-NH2 of Lys295. The absence of a D2OKIE on kobsO is understood as cancellation of solvent effects on the reactant state and the transition state because of D2OkobsO = ΦRS/ΦTS (54). The large D2OKIE on kcatO/KS for the wild-type enzyme reflects the contribution from ΦTS because the hydride transfer is rate-limiting for kcatO/KS and the relevant reactant state, enzyme-NAD+ (EAo at pL 10.0), may be assumed to have a ΦRS value of ∼1. In E292A, kcatO/KS is limited by product dissociation and therefore fails in capturing the chemical steps of catalysis. The absence of a D2OKIE on kcatO/KS likely reflects values of ΦRS (EAo) and ΦPS (enzyme-NADH-fructose, *EPQo) that are both close to unity. However, more detailed information on E292A can be gleaned from D2OKIEs on the amplitude of the pre-steady state burst (Π), the dissociation constant for mannitol (KdS), and kobsO. D2OΠ is the ratio of ΦRS and ΦPS (54). With ΦPS ≈ 1, it follows from the measured value of D2OΠ that ΦRS is 0.6. This estimate for ΦRS is validated by analysis of D2OKdS (=1.8), which expresses the solvent sensitivity of the enzyme-NAD+ (EAo) and enzyme-NAD+-mannitol (H*EAXo) reactant states. With a ΦRS for EAo of ≈1, a value of 0.55 is calculated for ΦRS of H*EAX (=1/1.8). Using the value of 1.4 for D2OkobsO, we can further calculate that ΦTS for E292A is 0.4 (=0.55/1.4). The similarity of ΦRS (0.4 for the wild-type and 0.6 for E292A) and ΦTS for wild-type enzyme (0.4) and E292A (0.4) shows that substitution of Glu292 by Ala does not change salient features of the chemical steps of the catalytic reaction coordinate of PfM2DH that include, as shown in Scheme 1, preactivation of the mannitol substrate at the reactant state by partial abstraction of the alcohol proton (H*EAXo, k5) and reaction through an oxyanion-like transition state (k7). Therefore, these findings together with the observation that Glu292 does not contribute to ionizations of enzyme-reactant complexes strongly support the proposal for an indirect participation of Glu292 in catalytic proton transfer of PfM2DH (Fig. 1C, II).
Dynamic Function of Glu292 in the Catalytic Mechanism of PfM2DH
The experimental results combined with evidence from MD simulations suggest a role for Glu292 in the catalytic reaction of the PfM2DH (Scheme 1). Glu292 adopts the position down the channel when initially at physiological pH, Lys295 is protonated in enzyme-NAD+. A water-mediated contact between Lys295 and Glu292 stabilizes the down position and proton conduit to and from the active site is interrupted at this stage. When after substrate binding (HEABc, k13) the reactive alcohol group of mannitol is positioned for hydrogen bonding with Lys295, the interaction between the lysine and Glu292 is broken, and the Glu swings into the now preferred up position (HEABo, k15). The down → up movement of Glu292 enables formation of a chain of four water molecules that establish a functional (von Grotthuss type (55)) proton conduit in the now open protein channel. Moreover, it causes the channel to expand by about 2–3 Å at the exit region, which in turn results in disruption of the hydrogen bond between His194 and Glu293. The consequent loss of the local proton buffering capacity at the surface side of the channel leads to development of a large proton gradient between the active site, where Lys295 now becomes activated for catalysis by deprotonation, and the channel exit. Proton translocation to solvent is strongly favored under these conditions.8 The clear benefit for enzymatic catalysis that is drawn from the overall conformational change associated with the positional switch of Glu292 (k15) is that the reactive C2 hydroxyl of mannitol becomes connected via the ϵ-NH2 group of Lys295 to the water-wire proton relay of the enzyme (*EABo).
The catalytic cycle of PfM2DH proceeds via development of a oxyanion character in substrate (H*EAXo, k5), promoted by partial abstraction of the hydroxyl proton to Lys295 under electrostatic steering of the “oxyanion hole” residues Asn191 and Asn300 (37, 41). From this “activated” reactant state (ΦRS ≈ 0.4, H*EAXo (37)), chemical oxidation by hydride transfer and proton transfer (k7) takes place, leading up to an enzyme-NADH-fructose complex in which Lys295 has become protonated but Glu292 still points toward the exit of the channel (H*EPQo). Because Glu292 switching back and forth will obviously be a much slower process than the actual proton translocation through the water-wire relay,9 it is reasonable to assume that the proton from ϵ-NH3+ of Lys295 in enzyme-NADH-fructose escapes to bulk solution before conformational dynamics of Glu292 has had time to intervene. Additionally, work on other proteins shows that a low pKa < 7 is favored under conditions where the buried lysine is hydrated and linked with bulk solvent by a chain of ordered water molecules (56). Fructose is released from *EPQo (k19) and the original protonation state of Lys295 is restored in enzyme-NADH at the steady state, inducing Glu292 to swing back to the down position, which is accompanied by domain closure (HEQc) and collapsing of the water chain.
Inertia of the up and down motion of Glu292 at the level of enzyme-NADH-fructose is the probable reason for an apparent disjointedness in reaction pathways for mannitol oxidation and fructose reduction, as follows. Binding of fructose to enzyme-NADH gives a ternary complex in which Lys295 is protonated and Glu292 points down the channel (HEPQc). This ternary complex, which has not yet isomerized to bring Glu292 into the up position (H*EPQo, k10), undergoes conversion to enzyme-NAD+-mannitol (k22 and k24), featuring the Glu pointing down toward the active site where Lys295 is unprotonated post-catalysis. Proton uptake by Lys295 is proposed to occur at this point (EABo). This would involve a full cycle of up and down motions of Glu292 (EABo ⇒; HEABc) and could derive the proton from His294 at the exit of water channel. The notion that reaction in fructose reduction is disjoined from reaction in mannitol oxidation is supported by DKIE data showing for wild-type PfM2DH and a series of active-site mutants thereof that enzymatic reactions do not obey the relationship, DKeq = (DkcatR/KS)/(DkcatO/KS), as expected if one path was the exact reverse of the other (41). Only after Glu292 has been replaced in E292A do we observe that the experimental DKIEs in oxidation and reduction become consolidated in DKeq (0.83 ≈ 0.9/1.1). The proposal in Scheme 1 that hydride transfer reduction of fructose does not depend on prior kinetic isomerization at k10 is strongly upheld by results of previous work showing that enzyme-NADH-fructose is almost fully competent for hydride transfer reduction of the substrate irrespective of the protonation state of Lys295 (41, 42). Our MD simulations (supplemental Fig. S2, II) are consistent with this notion, showing that in the closed enzyme conformation with Glu292 pointing down the channel (HEPQc), the reactive carbons of fructose and NADH are positioned stably (10 ns) in approximate distance for hydride transfer (≈3.3 Å). Optimized bonding between the catalytic acid-base and the reactive group of the substrate, which is the eventual result of the pre-catalytic isomerization in each direction, appears to be by far less important for hydride transfer in reduction (ϵ-NH3+ … O = C; k22) than it is in oxidation (ϵ-NH2 … HO-C; k5). One could characterize fructose reduction via steps k22 and k24 as reaction of PfM2DH that is uncoupled from the water-wire proton relay. E292A differs from wild-type enzyme in that the kinetic isomerization steps at enzyme-NAD+-mannitol (k15 and k16; k3 and k4) are slowed down drastically (≥30-fold = 1500/50) and fructose reduction takes place by steps k8 and k6 rather than via step k22. In E292A, protonation and deprotonation of Lys295 post-catalysis occurs at the ternary complex level.
Conformational Dynamics and Proton Transfer: PfM2DH and Other Enzymes
In conclusion, mannitol oxidation by PfM2DH involves a water-wire (von Grotthuss type (55)) mechanism of proton transfer that is linked to the conformational dynamics of Glu292. Molecular motion of the Glu is made possible by a global domain opening conformational change of the enzyme that seems to occur mainly in response to change in the bonding/protonation state of Lys295. This ensures that progress of the enzymatic reaction provides the essential timer switch for opening (Glu292 up) and closing (Glu292 down) of the proton conduit. The role of Glu292 in PfM2DH is distinct from the proposed proton shuttle function of similar mobile Glu/Asp residues in the proton channels of redox-driven proton pumps. However, proton transfer along water wires may also have an important role in these proton pumps, as shown recently for bacteriorhodopsin (57) and cytochrome c oxidase (58). Discontinuous operation of a proton relay as in PfM2DH is an entirely new feature of proton transfer in ADHs. An important implication of our findings is that the scope of conformational dynamics in ADH catalysis not only includes the hydride transfer that was extensively studied by experimental and computational methods (59, 60) but also the proton transfer. Dynamic characteristics of the proton transfer have been carefully examined in carbonic anhydrase (61–65) and p-hydroxybenzoate hydroxylase (27, 28). Unlike PfM2DH, both enzymes use shuttling motions, from a His in carbonic anhydrase and the flavin cofactor in p-hydroxybenzoate hydroxylase, to translocate protons. Furthermore, mobile Glu residues potentially participating in proton translocation between the active site and bulk solvent were also identified in the [NiFe]-hydrogenase from Desulfovibrio fructosovorans (66) or in the γ-class carbonic anhydrase from Methanosarcina thermophila (67).
Supplementary Material
Acknowledgments
We thank Karin Longus, Veronika Milocco, and Valentin Pacher for expert technical assistance. Professor Walter Keller (Institute of Molecular Biosciences, University of Graz, Austria) is acknowledged for assistance during recording CD spectra.
This work was supported by the Austrian Science Fund FWF, Grant P18275-B09 (to B. N.).

This article contains supplemental Figs. S1–S5, Tables S1–S6, Movies S1 and S2, “Experimental Procedures,” and “Results.”
Horse liver ADH represents the group of Zn+-dependent ADHs of the medium-chain dehydrogenase/reductase protein family (3). ADHs from the protein families of short-chain dehydrogenases/reductases (68), long-chain dehydrogenases/reductases (69), and aldo-keto reductases (70) utilize a metal-independent mechanism of catalysis. The proton transfer in these non-Zn2+ ADHs is not well understood.
The enzyme belongs to a family of polyol-specific long-chain dehydrogenases/reductases and utilizes a metal-independent mechanism of catalysis (69).
The classical MD simulation used herein was not suitable for reproducing the “partial deprotonation” type of hydrogen bonding between the mannitol 2-OH and the Lys295 NH2, as in the *EAXo and H*EAXo complexes (Scheme 1), which is thought to be a key catalytic factor in the reaction of PfM2DH (37, 38). This limitation notwithstanding, the MD simulation is shown to capture salient features of enzyme structural dynamics that are probably essential in catalysis.
The requirement of having the residue protonation state fixed in the MD simulation results in two extreme representations of the singly protonated group Lys295/H2O/Glu292, as NH3+-COO− and NH2-COOH. The first representation may be favored chemically but we note that the location of the proton in this group is not known.
Transient rates of mannitol oxidation by E292A were independent of whether the reaction was started with free enzyme or enzyme preincubated with a saturating concentration of NAD+. Therefore, this implies that steps involved in coenzyme binding were fast compared with the steps leading to formation of NADH and protons.
Approximating the protein channel as a cylinder of 0.75 nm diameter and 1.1 nm height, we calculate that the local concentration of a proton released from the active site would be about 1 m. The bulk proton concentration is only 100 nm (pH 7.0).
- ADH
- alcohol dehydrogenase
- PfM2DH
- mannitol 2-dehydrogenase from P. fluorescens
- KIE
- kinetic isotope effect
- MD
- molecular dynamics
- PDB
- protein data base
- r.m.s.
- root mean square.
REFERENCES
- 1. de Smidt O., du Preez J. C., Albertyn J. (2008) The alcohol dehydrogenases of Saccharomyces cerevisiae. A comprehensive review. FEMS Yeast Res. 8, 967–978 [DOI] [PubMed] [Google Scholar]
- 2. Höög J. O., Ostberg L. J. (2011) Mammalian alcohol dehydrogenases, a comparative investigation at gene and protein levels. Chem. Biol. Interact. 191, 2–7 [DOI] [PubMed] [Google Scholar]
- 3. Meijers R., Cedergren-Zeppezauer E. S. (2004) in Handbook of metalloproteins (Messerschmidt A., Cygler M., Bode W., eds) 1st Ed., pp. 5–33, John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 4. Metzler D. E. (2001) in Biochemistry: The Chemical Reactions of Living Cells (Metzler D. E., ed) 2nd Ed., pp. 765–836, Academic Press, San Diego [Google Scholar]
- 5. Roberts S. M. (1989) Use of enzymes as catalysts to promote key transformations in organic synthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 324, 577–587 [Google Scholar]
- 6. Hummel W. (1997) New alcohol dehydrogenases for the synthesis of chiral compounds. Adv. Biochem. Eng. Biotechnol. 58, 145–184 [DOI] [PubMed] [Google Scholar]
- 7. Faber K. (2004) Biotransformations in Organic Chemistry, 6th Ed., Springer-Verlag, Berlin [Google Scholar]
- 8. Hall M., Bommarius A. S. (2011) Enantioenriched compounds via enzyme-catalyzed redox reactions. Chem. Rev. 111, 4088–4110 [DOI] [PubMed] [Google Scholar]
- 9. Plapp B. V. (2006) in Isotope Effects in Chemistry and Biology (Kohen A., Limbach H. H., eds) pp. 811–836, CRC Press, Boca Raton, FL [Google Scholar]
- 10. Pettersson G. (1987) Liver alcohol dehydrogenase. CRC Crit. Rev. Biochem. 21, 349–389 [PubMed] [Google Scholar]
- 11. Klimacek M., Hellmer H., Nidetzky B. (2007) Catalytic mechanism of Zn2+-dependent polyol dehydrogenases: kinetic comparison of sheep liver sorbitol dehydrogenase with wild-type and Glu154 → Cys forms of yeast xylitol dehydrogenase. Biochem. J. 404, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koumanov A., Benach J., Atrian S., Gonzàlez-Duarte R., Karshikoff A., Ladenstein R. (2003) The catalytic mechanism of Drosophila alcohol dehydrogenase. Evidence for a proton relay modulated by the coupled ionization of the active site lysine/tyrosine pair and a NAD+ ribose OH switch. Proteins 51, 289–298 [DOI] [PubMed] [Google Scholar]
- 13. Inoue J., Tomioka N., Itai A., Harayama S. (1998) Proton transfer in benzyl alcohol dehydrogenase during catalysis. Alternate proton-relay routes. Biochemistry 37, 3305–3311 [DOI] [PubMed] [Google Scholar]
- 14. Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 15. Milburn C. C., Lamble H. J., Theodossis A., Bull S. D., Hough D. W., Danson M. J., Taylor G. L. (2006) The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus. J. Biol. Chem. 281, 14796–14804 [DOI] [PubMed] [Google Scholar]
- 16. Kovaleva E. G., Plapp B. V. (2005) Deprotonation of the horse liver alcohol dehydrogenase-NAD+ complex controls formation of the ternary complexes. Biochemistry 44, 12797–127808 [DOI] [PubMed] [Google Scholar]
- 17. Plapp B. V. (2010) Conformational changes and catalysis by alcohol dehydrogenase. Arch. Biochem. Biophys. 493, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leferink N. G., Han C., Antonyuk S. V., Heyes D. J., Rigby S. E., Hough M. A., Eady R. R., Scrutton N. S., Hasnain S. S. (2011) Proton-coupled electron transfer in the catalytic cycle of Alcaligenes xylosoxidans copper-dependent nitrite reductase. Biochemistry 50, 4121–4131 [DOI] [PubMed] [Google Scholar]
- 19. Heyes D. J., Levy C., Sakuma M., Robertson D. L., Scrutton N. S. (2011) A twin-track approach has optimized proton and hydride transfer by dynamically coupled tunneling during the evolution of protochlorophyllide oxidoreductase. J. Biol. Chem. 286, 11849–11854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reece S. Y., Nocera D. G. (2009) Proton-coupled electron transfer in biology. Results from synergistic studies in natural and model systems. Annu. Rev. Biochem. 78, 673–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo J., Bruice T. C. (2001) Dynamic structures of horse liver alcohol dehydrogenase (HLADH). Results of molecular dynamics simulations of HLADH-NAD(+)-PhCH(2)OH, HLADH-NAD(+)-PhCH(2)O(−), and HLADH-NADH-PhCHO. J. Am. Chem. Soc. 123, 11952–11959 [DOI] [PubMed] [Google Scholar]
- 22. Siegbahn P. E., Blomberg M. R. (2010) Quantum chemical studies of proton-coupled electron transfer in metalloenzymes. Chem. Rev. 110, 7040–7061 [DOI] [PubMed] [Google Scholar]
- 23. Hammes-Schiffer S. (2002) Comparison of hydride, hydrogen atom, and proton-coupled electron transfer reactions. Chemphyschem 3, 33–42 [DOI] [PubMed] [Google Scholar]
- 24. Schwartz S. D. (2007) in Hydrogen-Transfer Reactions (Hynes J. T., Klinman J. T., Limbach H. H., Schowen R. L., eds) pp. 1209–1239, WILEY-VCH, Weinheim [Google Scholar]
- 25. Ferrer S., Silla E., Tunon I., Oliva M., Moliner V., Williams I. H. (2005) Dependence of enzyme reaction mechanism on protonation state of titratable residues and QM level description: lactate dehydrogenase. Chem. Commun. 47, 5873–5875 [DOI] [PubMed] [Google Scholar]
- 26. Basran J., Hothi P., Masgrau L., Sutcliffe M. J., Scrutton N. S. (2007) in Hydrogen-Transfer reactions (Hynes J. T., Klinman J. T., Limbach H. H., Schowen R. L., eds) pp. 1341–1359, WILEY-VCH, Weinheim, Germany [Google Scholar]
- 27. Frederick K. K., Ballou D. P., Palfey B. A. (2001) Protein dynamics control proton transfers to the substrate on the His72 → Asn mutant of p-hydroxybenzoate hydroxylase. Biochemistry 40, 3891–3899 [DOI] [PubMed] [Google Scholar]
- 28. Frederick K. K., Palfey B. A. (2005) Kinetics of proton-linked flavin conformational changes in p-hydroxybenzoate hydroxylase. Biochemistry 44, 13304–13314 [DOI] [PubMed] [Google Scholar]
- 29. Furuichi M., Suzuki N., Dhakshnamoorhty B., Minagawa H., Yamagishi R., Watanabe Y., Goto Y., Kaneko H., Yoshida Y., Yagi H., Waga I., Kumar P. K., Mizuno H. (2008) X-ray structures of Aerococcus viridans lactate oxidase and its complex with d-lactate at pH 4.5 show an α-hydroxyacid oxidation mechanism. J. Mol. Biol. 378, 436–446 [DOI] [PubMed] [Google Scholar]
- 30. Kavanagh K. L., Klimacek M., Nidetzky B., Wilson D. K. (2002) Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary complexes. Specificity and catalytic mechanism. J. Biol. Chem. 277, 43433–43442 [DOI] [PubMed] [Google Scholar]
- 31. Namslauer A., Brzezinski P. (2004) Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase. FEBS Lett. 567, 103–110 [DOI] [PubMed] [Google Scholar]
- 32. Lanyi J. K., Schobert B. (2003) Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2′ intermediates of the photocycle. J. Mol. Biol. 328, 439–450 [DOI] [PubMed] [Google Scholar]
- 33. Neutze R., Pebay-Peyroula E., Edman K., Royant A., Navarro J., Landau E. M. (2002) Bacteriorhodopsin, a high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta 1565, 144–167 [DOI] [PubMed] [Google Scholar]
- 34. Wolf S., Freier E., Gerwert K. (2008) How does a membrane protein achieve a vectorial proton transfer via water molecules? Chemphyschem 9, 2772–2778 [DOI] [PubMed] [Google Scholar]
- 35. Wolf S., Freier E., Potschies M., Hofmann E., Gerwert K. (2010) Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules. A proton diode. Angew. Chem. Int. Ed. Engl. 49, 6889–6893 [DOI] [PubMed] [Google Scholar]
- 36. Wraight C. A. (2006) Chance and design, proton transfer in water, channels, and bioenergetic proteins. Biochim. Biophys. Acta 1757, 886–912 [DOI] [PubMed] [Google Scholar]
- 37. Klimacek M., Kavanagh K. L., Wilson D. K., Nidetzky B. (2003) On the role of Brønsted catalysis in Pseudomonas fluorescens mannitol 2-dehydrogenase. Biochem. J. 375, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klimacek M., Nidetzky B. (2002) A catalytic consensus motif for d-mannitol 2-dehydrogenase, a member of a polyol-specific long-chain dehydrogenase family, revealed by kinetic characterization of site-directed mutants of the enzyme from Pseudomonas fluorescens. Biochem. J. 367, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garczarek F., Brown L. S., Lanyi J. K., Gerwert K. (2005) Proton binding within a membrane protein by a protonated water cluster. Proc. Natl. Acad. Sci. U.S.A. 102, 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belevich I., Gorbikova E., Belevich N. P., Rauhamäki V., Wikström M., Verkhovsky M. I. (2010) Initiation of the proton pump of cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 107, 18469–18474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klimacek M., Nidetzky B. (2010) The oxyanion hole of Pseudomonas fluorescens mannitol 2-dehydrogenase. A novel structural motif for electrostatic stabilization in alcohol dehydrogenase active sites. Biochem. J. 425, 455–463 [DOI] [PubMed] [Google Scholar]
- 42. Klimacek M., Nidetzky B. (2002) Examining the relative timing of hydrogen abstraction steps during NAD(+)-dependent oxidation of secondary alcohols catalyzed by long-chain d-mannitol dehydrogenase from Pseudomonas fluorescens using pH and kinetic isotope effects. Biochemistry 41, 10158–10165 [DOI] [PubMed] [Google Scholar]
- 43. Slatner M., Nidetzky B., Kulbe K. D. (1999) Kinetic study of the catalytic mechanism of mannitol dehydrogenase from Pseudomonas fluorescens. Biochemistry 38, 10489–10498 [DOI] [PubMed] [Google Scholar]
- 44. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press Inc., Totowa, NJ [Google Scholar]
- 45. Ellis K. J., Morrison J. F. (1982) Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87, 405–426 [DOI] [PubMed] [Google Scholar]
- 46. Northrop D. B. (1977) in Isotope Effects on Enzyme-catalyzed Reactions (Cleland W. W., O'Leary M. H., Northrop D. B., eds) p. 122, University Park Press, Baltimore, MD [Google Scholar]
- 47. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 48. Rostkowski M., Olsson M. H., Søndergaard C. R., Jensen J. H. (2011) Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bayly C. I., Cieplak P., Cornell W. D., Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges – the RESP Model. J. Phys. Chem. 97, 10269–10280 [Google Scholar]
- 51. DePaul A. J., Thompson E. J., Patel S. S., Haldeman K., Sorin E. J. (2010) Equilibrium conformational dynamics in an RNA tetraloop from massively parallel molecular dynamics. Nucleic Acids Res. 38, 4856–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sorin E. J., Pande V. S. (2005) Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys. J. 88, 2472–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera, a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 54. Quinn D. M., Sutton L. D. (1991) in Enzyme Mechanism from Isotope Effects (Cook P. F., ed) pp. 73–126, CRC Press, Boca Raton, FL [Google Scholar]
- 55. Marx D. (2006) Proton transfer 200 years after von Grotthuss. Insights from ab initio simulations. Chemphyschem 7, 1848–1870 [DOI] [PubMed] [Google Scholar]
- 56. Fitch C. A., Karp D. A., Lee K. K., Stites W. E., Lattman E. E., García-Moreno E. B. (2002) Experimental pK(a) values of buried residues. Analysis with continuum methods and role of water penetration. Biophys. J. 82, 3289–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Freier E., Wolf S., Gerwert K. (2011) Proton transfer via a transient linear water molecule chain in a membrane protein. Proc. Natl. Acad. Sci. U.S.A. 108, 11435–11439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wikström M., Verkhovsky M. I., Hummer G. (2003) Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta 1604, 61–65 [DOI] [PubMed] [Google Scholar]
- 59. Hammes-Schiffer S., Benkovic S. J. (2006) Relating protein motion to catalysis. Annu. Rev. Biochem. 75, 519–541 [DOI] [PubMed] [Google Scholar]
- 60. Nagel Z. D., Klinman J. P. (2006) Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 106, 3095–3118 [DOI] [PubMed] [Google Scholar]
- 61. Toba S., Colombo G., Merz K. M., Jr. (1999) Solvent dynamics and mechanism of proton transfer in human carbonic anhydrase II. J. Am. Chem. Soc. 121, 2290–2302 [Google Scholar]
- 62. Maupin C. M., Voth G. A. (2010) Proton transport in carbonic anhydrase. Insights from molecular simulation. Biochim. Biophys. Acta 1804, 332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fisher S. Z., Kovalevsky A. Y., Domsic J. F., Mustyakimov M., McKenna R., Silverman D. N., Langan P. A. (2010) Neutron structure of human carbonic anhydrase II. Implications for proton transfer. Biochemistry 49, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Silverman D. N., Elder I. (2006) in Isotope Effects in Chemistry and Biology (Kohen A., Limbach H. H., eds) pp. 847–860, CRC Press Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- 65. Braun-Sand S., Strajbl M., Warshel A. (2004) Studies of proton translocations in biological systems. Simulating proton transport in carbonic anhydrase by EVB-based models. Biophys. J. 87, 2221–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dementin S., Burlat B., De Lacey A. L., Pardo A., Adryanczyk-Perrier G., Guigliarelli B., Fernandez V. M., Rousset M. (2004) A glutamate is the essential proton transfer gate during the catalytic cycle of the [NiFe]-hydrogenase. J. Biol. Chem. 279, 10508–10513 [DOI] [PubMed] [Google Scholar]
- 67. Tripp B. C., Ferry J. G. (2000) A structure-function study of a proton transport pathway in the γ-class carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39, 9232–9240 [DOI] [PubMed] [Google Scholar]
- 68. Ladenstein R., Winberg J. O., Benach J. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families. Structure-function relationships in short-chain alcohol dehydrogenases. Cell. Mol. Life Sci. 65, 3918–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klimacek M., Kavanagh K. L., Wilson D. K., Nidetzky B. (2003) Pseudomonas fluorescens mannitol 2-dehydrogenase and the family of polyol-specific long-chain dehydrogenases/reductases. Sequence-based classification and analysis of structure-function relationships. Chem. Biol. Interact. 143, 559–582 [DOI] [PubMed] [Google Scholar]
- 70. Hyndman D., Bauman D. R., Heredia V. V., Penning T. M. (2003) The aldo-keto reductase superfamily homepage. Chem. Biol. Interact. 143, 621–631 [DOI] [PubMed] [Google Scholar]
- 71. Agmon N. (1995) The Grotthuss mechanism. Chem. Phys. Lett. 244, 456–462 [Google Scholar]
- 72. Meiboom S. (1961) Nuclear magnetic resonance study of the proton transfer in water. J. Chem. Phys. 34, 375–388 [Google Scholar]
- 73. Klimacek M., Nidetzky B. (2010) From alcohol dehydrogenase to a “one-way” carbonyl reductase by active-site redesign. A mechanistic study of mannitol 2-dehydrogenase from Pseudomonas fluorescens. J. Biol. Chem. 285, 30644–30653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.