Background: Sifuvirtide (SFT) is an HIV peptide fusion inhibitor under phase II clinical trials.
Results: Crystal structure of SFT complexed with gp41 NHR peptide and the potent activity of SFT against diverse HIV-1 variants were determined.
Conclusion: Crystal structure fully supports earlier peptide design.
Significance: Our data present important information for developing SFT for clinical use and for designing novel HIV fusion inhibitors.
Keywords: Antiviral Agents, Crystal Structure, Fusion Protein, HIV-1, Human Immunodeficiency Virus, Membrane Fusion, Peptides, Virus Entry
Abstract
Sifuvirtide (SFT) is an electrostatically constrained α-helical peptide fusion inhibitor showing potent anti-HIV activity, good safety, and pharmacokinetic profiles, and it is currently under phase II clinical trials in China. In this study, we demonstrate its potent and broad anti-HIV activity by using diverse HIV-1 subtypes and variants, including subtypes A, B, and C that dominate the AIDS epidemic worldwide, and subtypes B′, CRF07_BC, and CRF01_AE recombinants that are currently circulating in China, and those possessing cross-resistance to the first and second generation fusion inhibitors. To elucidate its mechanism of action, we determined the crystal structure of SFT in complex with its target N-terminal heptad repeat region (NHR) peptide (N36), which fully supports our rational inhibitor design and reveals its key motifs and residues responsible for the stability and anti-HIV activity. As anticipated, SFT adopts fully helical conformation stabilized by the multiple engineered salt bridges. The designing of SFT also provide novel inter-helical salt bridges and hydrogen bonds that improve the affinity of SFT to NHR trimer. The extra serine residue and acetyl group stabilize α-helicity of the N-terminal portion of SFT, whereas Thr-119 serves to stabilize the hydrophobic NHR pocket. In addition, our structure demonstrates that the residues critical for drug resistance, located at positions 37, 38, 41, and 43 of NHR, are irreplaceable for maintaining the stable fusogenic six-helix bundle structure. Our data present important information for developing SFT for clinical use and for designing novel HIV fusion inhibitors.
Introduction
The trimeric envelope gp120/gp41 complex of HIV-1 mediates viral entry by binding to cellular receptor CD4 and a co-receptor (CCR5 or CXCR4) and subsequently fusing the viral and cellular membranes (1, 2). In the current fusion model, the receptor binding induces a series of coordinated structural changes in gp120 that trigger gp41 to expose, extend, and insert its N-terminal hydrophobic fusion peptide into cell membranes (1–3). The extended pre-hairpin intermediate conformation persists for at least 15 min and then collapses into a trimer-of-hairpins structure (six-helical bundle, 6-HB)5 that pulls the viral and cellular membranes into close contact for fusion (3–6). In this fusion-active gp41 core structure, three N-terminal heptad repeat regions (NHR) form a central trimeric coiled coil, whereas three C-terminal heptad repeat regions (CHR) around the NHR pack as antiparallel helices into hydrophobic grooves (3–6). A number of synthetic peptides derived from the NHR and CHR of gp41 can efficiently inhibit HIV-1 infection by competitively binding to the exposed NHR or CHR in the gp41 pre-hairpin state, hence blocking 6-HB formation in a dominant-negative manner (7–15). Among them, T20 (Enfuvirtide, Fuzeon) has been approved for clinical use as the first member of a new class of anti-HIV drugs, HIV fusion inhibitors (9, 16, 17). This peptide drug can suppress replication of HIV variants with multidrug resistance to reverse transcriptase and protease inhibitors; however, it also easily induces resistance in both clinical settings and laboratory studies (17–21).
Because of the drug resistance problem, many efforts have been made to develop novel anti-HIV agents, including fusion inhibitors with improved stability and potency. In succession to T20, the second-generation peptide fusion inhibitor T1249 was developed with increased antiviral potency, but its clinical development was halted due to the drug formulation problem (14, 22, 23). Among a series of more potent third generation fusion inhibitors (10, 12–15), sifuvirtide (SFT) is one in advance stages. It has been evaluated by the phase I clinical trials and is currently under phase II clinical studies (13, 24). This peptide inhibitor was originally designed with different sequence and/or location in relationship to CHR peptides C34, T20, and T1249 on the basis of three-dimensional structural information of gp41 (13). Our previous studies demonstrated its potent anti-HIV activity, good safety, and pharmacokinetic profiles (13, 24). In this study, we have performed a leading study to evaluate its antiviral spectrum by using two large panels of HIV-1 Env-pseudotyped viruses, corresponding to subtypes A, B, and C that dominate the AIDS epidemic worldwide, and the subtypes B′, CRF07_BC (B/C), and CRF01_AE (A/E) recombinants that are the major circulating viruses in China. We have also tested the susceptibility of HIV-1 variants that possess cross-resistance to the first and second generation fusion inhibitors (T20 and T1249). To elucidate its mechanism of action, we have solved the crystal structure of SFT in complex with its target NHR sequence by using the peptide N36 as a surrogate. The present results show very strong inhibition of SFT on diverse HIV-1 strains and reveal its key molecular determinants. These data have provided important information for developing SFT for clinical use and for designing novel HIV-1 fusion inhibitors.
EXPERIMENTAL PROCEDURES
Peptides Synthesis
Peptides SFT (acetyl-SWETWEREIENYTRQIYRILEESQEQQDRNERDLLE-NH2), T20 (acetyl-YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF-NH2), and N36 (acetyl-SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARIL-NH2) were synthesized by a standard solid-phase Fmoc (N-(9-fluorenyl)methoxycarbonyl) method. All three peptides were acetylated at the N terminus and amidated at the C terminus. They were purified by reversed-phase HPLC and verified for purity >95% and correct amino acid composition by mass spectrometry. Concentrations of the peptides were determined by UV absorbance and a theoretically calculated molar extinction coefficient (280 nm) of 5500 and 1490 m−1·cm−1 based on the number of tryptophan and tyrosine residues (all the peptides tested contain Trp and/or Tyr), respectively.
HIV-1 Env-expressing Plasmids
A panel of plasmids encoding HIV-1 Envs were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, including subtype A Env clones pSVIII-92RW020.5 and 92UG037.8 from Dr. B. H. Hahn; subtype B HIV-1 reference panel of Env clones SC422661.8, TRO.11, and AC10.0.29 from Drs. D. Montefiori, F. Gao, and M. Li; pRHPA4259.7 and pREJO4541.67 from Drs. B. H. Hahn and J. F. Salazar-Gonzalez; and pCAAN5342.A2 from Drs. B. H. Hahn and D. L. Kothe; subtype C HIV-1 reference Env clones Du172.17 and Du422.1 from Drs. D. Montefiori, F. Gao, S. Abdool Karim, and G. Ramjee; CAP45.2.00.G3 from Drs. L. Morris, K. Mlisana, and D. Montefiori; ZM197M.PB7 from Drs. B. H. Hahn, Y. Li, and J. F. Salazar-Gonzalez; ZM109F.PB4 from Drs. E. Hunter and C. Derdeyn. Six CRF07_BC Env clones (CH064.20, CH070.1, CH091.9, CH110.2, CH119.10, and CH120.6) were generous gifts by Dr. Y. Shao in the Chinese Center for Disease Control and Prevention, Beijing, China. A panel of Env clones derived from subtype B′ (B01, B02, B04, and 43-22), CRF01_AE (SHX335.24, YN192.31, AE01, AE03, GX2010.36, GX11.13, GX2010.36H, and BJ5.11), and CRF07_BC (BC02, BC03, BC05, BC07, BC14, SC19-15, BJ22-5, YN148R-9, XJ50-6, and HB5-3) was kindly provided by Y. Wang from the National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China.
Cell Lines
TZM-bl cells were contributed by J. C. Kappes and X. Wu through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health. This reporter cell line stably expresses high levels of CD4, CCR5, and CXCR4 and contains Tat-responsive reporter genes for firefly luciferase and β-galactosidase under the control of an HIV-1 long terminal repeat promoter (26). 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Both of these adherent cell lines were maintained in Dulbecco's modified Eagle's (DMEM) growth medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (FBS). Cells were harvested using trypsin/EDTA solution (Invitrogen). All cell lines were maintained at 37 °C in humidified air containing 5% CO2.
Inhibition of HIV-1 Single Cycle Infection
HIV-1 pseudoviruses were generated as described previously (27, 28). Briefly, 293T cells (5 × 106 cells in 15 ml of growth medium in a T-75 culture flask) were cotransfected with 10 μg of an Env-expressing plasmid, and 20 μg of a backbone plasmid pSG3Δenv that encodes Env-defective, luciferase-expressing HIV-1 genome using Lipofectamine 2000 reagent (Invitrogen). Pseudovirus-containing culture supernatants were harvested 48 h after transfection, filtered by 0.45-μm pore size, and stored at −80 °C in 1-ml aliquots until use. The 50% tissue culture infectious dose (TCID50) of a single thawed aliquot of each pseudovirus batch was determined in TZM-bl cells. The antiviral activity of SFT or T20 was determined using TZM-bl cells as described previously. Briefly, the peptides were prepared with 10 series of dilutions in a 3-fold stepwise manner, mixed with 100 TCID50 viruses, and incubated for 1 h at room temperature. The mixture was added to TZM-bl cells (104/well) and incubated at 37 °C for 48 h. Luciferase activity was measured using luciferase assay reagents (Promega, Madison, WI) and a luminescence counter (Promega) according to the manufacturer's instructions. The percent inhibition by the peptides and 50% inhibitory concentration (IC50) values were calculated as described previously (27, 28).
Inhibition of HIV-1 NL4-3 Variants
HIV-1 molecular clone NL4-3 carrying L33S or L33V mutation was kindly provided by Dr. Frank Kirchhoff from the Institute of Virology, University of Ulm, Ulm, Germany (29, 30). The mutant viruses were generated by transient transfection of NL4-3 plasmids into 293T cells. The virus stocks were harvested 48 h post-transfection and quantified for TCID50. Inhibition of the peptides (SFT and T20) on the NL4-3 mutants was performed as described for pseudoviruses. In brief, 100 TCID50 viruses were used to infect TZM-bl cells in the absence or presence of serially diluted peptides. Two days post-infection, the cells were harvested and lysed in reporter lysis buffer, and the luciferase activity was measured. The IC50 was calculated as described above using GraphPad Prism software (GraphPad Prism software Inc., San Diego).
Crystallization of SFT-N36 Complex and Structure Determination
Equal amounts of synthetic peptides SFT and N36 were dissolved in denaturing buffer (100 mm NaH2PO4, 10 mm Tris-HCl, pH 8.0, 8 m urea). To refold the peptides, the mixture was dialyzed against buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl at 4 °C overnight. The dialyzed sample was clarified by centrifugation and subsequently loaded on a size-exclusion column (Superdex 75 10/300 GL, GE Healthcare). Elutions corresponding to the molecular weight of a 6-HB were pooled and concentrated prior to the crystallization trials. The SFT-N36 complex was crystallized by mixing an equal volume (0.8 μl) of purified protein (∼10 mg/ml) and reservoir solution (0.1 m Tris-HCl, pH 8.5, 10% w/v PEG4000) in a hanging drop vapor diffusion system at 22 °C. The cryocooling for the crystals was achieved by soaking the crystal 30–60 s in reservoir solution containing 30% glycerol (v/v) followed by flash freezing in liquid nitrogen. Complete data were collected at beamline PX III SLS (Villigen, Switzerland) using x-ray with a wavelength of 1.000 Å. The crystal belonged to the space group of C2, contained a complete 6-HB (three SFT and three N36 peptides) per asymmetric unit, and diffracted the x-ray to the resolution limit of 1.7 Å. The structure of SFT/N36 was solved by molecular replacement (Phaser, CCP4 suite) using a truncated HIV-1 gp41 core structure (Protein Data Bank ID 1ENV) as a searching model. The resolution range of the data used in molecular replacement was 44.390–3.0 Å. Phaser reported a single solution with a final log-likelihood gain value of 2479 and the translation function Z-score value of 25.9, indicating a correct solution. The R-factor of the initial solution was 43.39. The molecular replacement resulted in an interpretable electron density map. The initial electron density map was improved by manual model building (Coot version 0.6). The structures were refined using PHENIX (31). The final atomic models have good refinement statistics and stereochemistry qualities (Table 3). The structure is validated by MolProbity analysis (32). The MolProbity score is 1.89, rating as the 72nd percentile among structures of comparable resolution. The Ramachandran plot finds 99.52% residues in the favored area and 0.48% outliers. Hydrogen bond or salt bridge is defined according to measurements in the final atomic model. To define a hydrogen bond, the distance from donor hydrogen to the electronegative acceptor is ≤3.0 Å, and the angle “donor H-acceptor” is ≥120°. To define a salt bridge, the distance between the positively charged atom of basic residues (Lys or Arg) and negatively charged atom on acidic residues (Glu or Asp) is ≤4 Å.
TABLE 3.
Data collection and refinement statistics
| SFT/N36 | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, and c | a = 88.79 Å |
| b = 49.18 Å | |
| c = 55.82 Å | |
| α, β, and γ | 90.00, 90.65, 90.00° |
| X-ray source | PXIII, SLS |
| Wavelength | 1.000 Å |
| Data range | 44.390 to 1.701 Å |
| Reflections unique | 24,817 |
| Rmergea (last shell, 1.80 to 1.70 Å) | 0.034 (0.25) |
| I/σI (last shell, 1.80 to 1.70 Å) | 18.53 (2.51) |
| Completeness (%) (last shell, 1.80 to 1.70 Å) | 93.1 (63.3) |
| Redundancy (last shell, 1.80 to 1.70 Å) | 3.18 (1.24) |
| Refinement | |
| Resolution range | 34.936 to 1.800 Å |
| Completeness (%) (last shell, 1.88 to 1.80 Å) | 98.81 (92.00) |
| Reflections F >0.81 (cross-validation) | 22,201 (1109) |
| Rworkb/Rfreec (last shell, 1.88 to 1.80 Å) | 0.1818/0.2187 (0.2010/0.2576) |
| Non-hydrogen protein atoms | 2243 |
| Protein | 2126 |
| Water | 93 |
| N-Acetyl group | 24 |
| Wilson B factor | 23.53 |
| B-Factor averages | 31.53 |
| Protein | 31.33 |
| N-Acetyl group | 39.33 |
| Water | 41.08 |
| Root mean square deviation | |
| Bond lengths | 0.018 Å |
| Bond angles | 1.490° |
| MolProbity score | 1.89, 72nd percentile |
| % Favored regions and Outliers in Ramachandran plot | 99.52, 0.48 |
a Rmerge = ΣhklΣj|Ihkl,j − Ihkl|/ΣhklΣjIhkl,j, where Ihkl is the average of symmetry-related observations of a unique reflection.
b Rwork = Σhkl‖Fobs(hkl)|−|Fcalc(hkl)‖/Σhkl|Fobs(hkl)|.
c Rfree = the cross-validation R factor for 5% of reflections against which the model was not refined.
RESULTS
Potent Inhibition of SFT on Diverse HIV-1 Subtypes and Variants
One of the obstacles to treatment of the HIV-1 is its extremely high genetic variability, which results in three major groups with many subtypes and circulating recombinant forms (33, 34). Although SFT has advanced into clinical trials, only limited HIV-1 subtypes and variants have been tested for the susceptibility. Here, we first evaluated SFT with a panel of 14 HIV-1 pseudoviruses derived from subtypes A, B, and C that represent the vast majority of HIV-1 infections worldwide. As shown in Table 1, SFT had potent inhibitory activity against the cellular entry by the diverse viruses. It inhibited subtype A–C viruses with mean IC50 values at 1.81, 10.35, and 3.84 nm, respectively. In comparison, T20 showed much weak inhibition on these three subtypes, with mean IC50 at 13.86, 189.20, and 57.41 nm, respectively.
TABLE 1.
Inhibition of SFT on subtypes A–C primary HIV-1 strains
The inhibitory activity of each peptide was determined in triplicate by a single cycle infectivity assay. The data were derived from the results of three independent experiments and are expressed as means ± S.D.
| Pseudovirus | Subtype | Co-Ra | IC50 |
|
|---|---|---|---|---|
| SFT | T20 | |||
| nm | ||||
| 92RW020 | A | R5 | 1.73 ± 0.63 | 23.72 ± 8.73 |
| 92UG037.8 | A | R5 | 1.88 ± 0.30 | 4.00 ± 0.85 |
| SF162 | B | R5 | 3.87 ± 1.07 | 41.69 ± 6.53 |
| AC10.0.29 | B | R5 | 1.59 ± 0.30 | 8.88 ± 1.24 |
| TRO.11 | B | R5 | 21.18 ± 2.76 | 279.00 ± 126.15 |
| pREJO4541 | B | R5 | 1.05 ± 0.10 | 94.02 ± 11.29 |
| pRHPA4259 | B | R5 | 24.41 ± 0.66 | 298.50 ± 22.91 |
| SC422661.8 | B | R5 | 1.72 ± 0.17 | 13.73 ± 0.36 |
| pCAAN5342 | B | R5 | 18.66 ± 0.64 | 588.55 ± |
| ZM197M.PB | C | R5 | 4.84 ± 0.73 | 71.56 ± 5.32 |
| ZM109F.PB | C | R5 | 4.68 ± 1.35 | 76.38 ± 2.69 |
| Du172.17 | C | R5 | 1.51 ± 0.66 | 10.73 ± 6.53 |
| Du422.1 | C | R5 | 1.66 ± 0.17 | 29.13 ± 4.13 |
| CAP45.2.00. | C | R5 | 6.51 ± 1.19 | 99.26 ± 2.61 |
a Co-R means coreceptor use.
Given SFT has been advanced to the phase II clinical studies in China, it is important to know the susceptibility of the predominating HIV-1 subtypes and variants in this region. Therefore, we constructed a panel of 27 HIV-1 pseudoviruses with their Envs derived from two recombinant forms, CRF01_BC and CRF01_AE, and subtype B′ viruses that are dominating the AIDS epidemic in China (35, 36). The results from the single cycle infection assays are presented in Table 2. Compared with T20, SFT displayed more potent inhibitory activity, with mean IC50 at 2.66 nm for CRF07_BC, 10.40 nm for CRF01_AE, and 3.49 nm for B′. T20 inhibited those of three subtypes at 46.05, 26.85, and 19.34 nm, respectively.
TABLE 2.
Inhibition of SFT on CRF07_BC, CRF01_AE, and B′ HIV-1 variants
The inhibitory activity of each peptide was determined in triplicate by a single cycle infectivity assay. The data were derived from the results of at least three independent experiments and are expressed as means ± S.D.
| Pseudovirus | Subtypea | Co-Rb | IC50 |
|
|---|---|---|---|---|
| SFT | T20 | |||
| nm | ||||
| CH64.20 | B/C | R5 | 1.33 ± 0.09 | 58.91 ± 16.51 |
| CH070.1 | B/C | R5 | 4.23 ± 0.25 | 175.00 ± 18.81 |
| Ch091.9 | B/C | R5 | 1.54 ± 0.32 | 72.97 ± 9.96 |
| Ch110.2 | B/C | R5 | 1.75 ± 0.42 | 21.25 ± 6.94 |
| Ch119.10 | B/C | R5 | 3.76 ± 0.33 | 20.04 ± 5.66 |
| Ch120.6 | B/C | R5 | 8.37 ± 0.98 | 50.32 ± 4.38 |
| SC19-15 | B/C | R5 | 0.59 ± 0.09 | 7.62 ± 0.96 |
| BC02 | B/C | R5 | 0.46 ± 0.11 | 3.32 ± 0.40 |
| BC03 | B/C | R5 | 1.49 ± 0.05 | 3.75 ± 0.01 |
| BC05 | B/C | R5 | 1.28 ± 0.23 | 4.21 ± 0.01 |
| BC07 | B/C | R5 | 1.61 ± 0.33 | 11.17 ± 3.50 |
| BC14 | B/C | R5 | 2.56 ± 0.20 | 68.73 ± 3.68 |
| BJ22-5 | B/C | R5 | 1.32 ± 0.44 | 28.64 ± 0.15 |
| XJ50-6 | B/C | R5 | 1.58 ± 0.42 | 64.83 ± 5.73 |
| YN148r-9 | B/C | R5 | 8.25 ± 0.39 | 125.35 ± 6.58 |
| HB5–3 | B/C | R5 | 2.42 ± 0.36 | 20.65 ± 1.22 |
| SHX335.24 | A/E | R5 | 1.01 ± 0.32 | 39.00 ± 9.62 |
| YN192.31 | A/E | R5 | 5.69 ± 0.62 | 65.16 ± 8.01 |
| AE01 | A/E | R5 | 23.45 ± 4.62 | 25.92 ± 2.63 |
| AE03 | A/E | R5 | 1.84 ± 0.35 | 6.67 ± 0.06 |
| GX11.13 | A/E | R5 | 2.62 ± 0.15 | 6.48 ± 0.16 |
| GX2010.36 | A/E | R5 | 1.77 ± 0.37 | 18.74 ± 4.61 |
| GX2010.36H | A/E | R5 | 36.39 ± 2.36 | 25.94 ± 0.52 |
| B01 | B′ | R5 | 0.66 ± 0.19 | 49.50 ± 2.41 |
| B02 | B′ | R5 | 3.2 ± 0.75 | 8.69 ± 0.95 |
| B04 | B′ | R5 | 6.05 ± 1.56 | 6.60 ± 1.26 |
| 43-22 | B′ | R5 | 4.04 ± 0.43 | 12.55 ± 2.57 |
a HIV-1 subtypes are as follows: B/C, CRF07_BC; A/E, CRF01_AE; B′, and Tai B.
b Co-R means coreceptor use.
Potent Inhibition of SFT on HIV-1 Carrying Mutations That Confer Cross-resistance
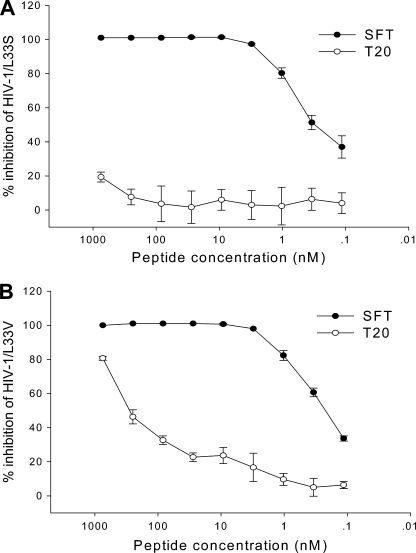
Resistance to T20 usually maps to the 36–45-amino acid region of the peptide-binding site in the NHR domain of the viral gp41, with the 36GIV38 motif being the hot spot for resistance (17–21). Our previous studies demonstrated that SFT possessed efficient potency to inhibit HIV-1 variants that carry single or double T20-resistant mutations (e.g. V38A, V38A/N42T, and N42T/N43K) (13). Here, we investigated whether L33S mutation affects SFT sensitivity, because recent data showed that this point mutation confers cross-resistance to both first generation fusion inhibitor T20 and second generation fusion inhibitor T1249 (14, 29, 30). Prominently, SFT could potently inhibit HIV-1 molecular clone NL4-3 carrying L33S mutation with an IC50 of 0.41 nm, although T20 had no inhibitory activity at a concentration as high as 750 nm (Fig. 1A). Furthermore, we observed whether L33V mutation affects SFT sensitivity, because this naturally occurring mutation in treatment-naive patients has been shown to confer T20 resistance (29). Similarly, SFT had an IC50 value at 0.22 nm, and T20 had an IC50 at 665.40 nm (Fig. 1B). These data highlight that SFT has a potent and broad anti-HIV activity, not only for currently circulating subtypes but also for inhibitor-induced and naturally occurring T20-resistant variants.
FIGURE 1.
Inhibitory activity of SFT on T20-resistant HIV-1 variants. A, inhibition of SFT on replication of HIV-1NL4-3 that carries the L33S mutation at a dose-dependent fashion. B, inhibition of SFT on replication of HIV-1NL4-3 that carries the L33V mutation at a dose-dependent fashion. The experiment was performed in triplicate and repeated twice. The data are presented as means ± S.D.
Crystallization and Structure Determination of SFT-N36 Complex
To elucidate the structural basis of the potent efficacy of SFT, we assembled and crystallized the SFT-N36 complex. We dissolved equal amounts of synthetic N36 and SFT peptides in denaturing buffer. The mixture was dialyzed to allow refolding of the peptides. The 6-HB formed by the SFT/N36 peptides was purified by size-exclusion chromatography. The crystal of the SFT-N36 complex belonged to the space group of C2, contained three pairs of SFT/N36 peptides (one complete 6-HB) per asymmetric unit, and diffracted x-ray to a resolution limit of 1.70 Å. We could build all residues of SFT/N36 peptides, including the N-terminal protection groups of the synthetic peptides in the electron density. The refined model has good refinement statistics and stereochemistry qualities (Table 3).
Structure of 6-HB with SFT/N36
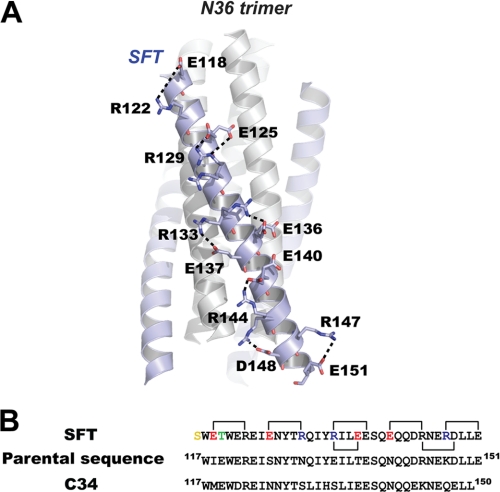
We previously designed SFT based on the structural data of HIV gp41 core domain and computer modeling. We found that SFT could form highly thermostable 6-HB with its NHR counterpart in solution (13). As anticipated, the crystal structure of the SFT-N36 complex resembles a typical 6-HB similar to other HIV gp41 core structures (Fig. 2A and supplemental Fig. S1) (4–6, 37). SFT adopts fully helical conformation when complexed with N36 (Fig. 2A). SFT is composed of 36 amino acids, which is different from its parental sequence in 9 residues and is different from C34 in 16 residues. In the rational design of SFT, we engineered multiple pairs of basic and acidic residues (glutamic acid and arginine) to promote the ion pair formation between i and i + 4 positions on SFT. We could observe all predicted salt bridges in our crystal structure (Fig. 2A). Because these residues are surface-exposed, many of them adopt alternative conformations. For example, Arg-133 adopts two conformations, in which conformation A (35% occupancies) forms a salt bridge with Glu-137 and conformation B (65% occupancies) forms another salt bridge with Glu-136. Arg-144 adopts two conformations, in which conformation A (69% occupancies) forms a salt bridge with Glu-140 and conformation B (31% occupancies) forms another salt bridge with Glu-148 (Fig. 2A). Thus, the salt bridges observed on the SFT helix involve not only the residues at i and i + 4 positions but also other charged residues, which together stabilize the α-helical conformation of the peptide.
FIGURE 2.
Salt bridges formed by charged residues on SFT. A, ribbon model of 6-HB formed by SFT/N36 with the charged residues involving ion pair formation on the SFT helix shown as a stick model with the labels. The salt bridges formed between charged residues are indicated with dashed lines. B, sequence alignment of SFT, its parental sequence (HIV-1 subtype E gp41 CHR(117–151)), and C34. Numbering is based on the sequence of HXB2 gp41. The color codes for SFT sequence are as follows: yellow, N-terminal capping acetylserine; green, mutation to stabilize the hydrophobic pocket; red, residues were changed to negatively charged residues; blue, residues were changed to positively charged residues; black, unchanged residues. Salt bridges observed in the crystal structure of SFT are indicated with the linkers.
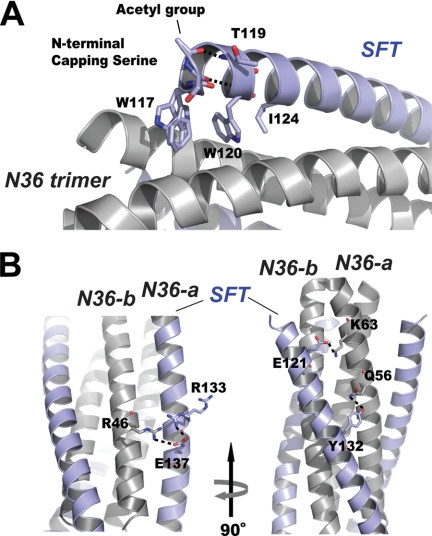
To stabilize the pocket binding domain, we replaced the Glu-119 of the parental sequence by an uncharged residue threonine in the inhibitor design. The crystal structure shows that the side chain of Thr-119 is positioned above the pocket binding domain (Fig. 3A), functioning to stabilize the hydrophobic pocket. We previously introduced a serine residue to the N terminus of SFT to improve the stability of the peptide. As illustrated in Fig. 3A, the acetylserine caps the N terminus of SFT; hence the residue is denoted as capping serine. The carbonyl group of the capping serine accepts a hydrogen bond from NH group of Trp-120, and its acetyl group accepts another hydrogen bond from NH group of Thr-119. Therefore, the capping serine can stabilize the α-helical conformation of SFT.
FIGURE 3.
Interactions between SFT and N36 peptides. A, portion of the ribbon model of 6-HB formed by SFT/N36 with the labels. Residues at the N terminus of SFT are shown in stick model (colored by elements). Trp-117, Trp-120, and Ile-124 were unchanged in SFT design, forming the typical pocket binding domain. The side chain of Thr-119 is located above the pocket binding domain, serving to stabilize the hydrophobic pocket. The carbonyl group of the capping acetylserine at the N terminus of SFT accepts a hydrogen bond (dashed line) from the NH group of Trp-120; the N-terminal acetyl group of the capping serine accepts another hydrogen bond (dashed line) from the NH group of Thr-119. B, portion of ribbon model of the 6-HB formed by SFT/N36 with the labels. Model on the right side is the different view of the model on the left side (rotation 90° around vertical axis to the left). Residues involving salt bridging and hydrogen bonding between SFT and two adjacent N36-a N36-b, are shown as a stick model. The salt bridges and hydrogen bond are indicated with dashed lines.
As the results of SFT design, novel salt bridges and hydrogen bonds were found between SFT and N36 peptides. As shown in Fig. 3B, the side chain of Glu-137 on SFT forms a salt bridge with the side chain of Arg-46 on N36-b. Compared with the HIV gp41 core structure containing C34 sequence (Protein Data Bank code 1ENV), residue Asp-121 is replaced by Glu-121 in SFT. Our structure indicates that the longer glutamic acid side chain of Glu-121 could reach closer to the side chain of Lys-63 on N36-a, resulting a stronger salt bridge. His-132 of C34 is substituted by Tyr-132 in SFT, resulting in a hydrogen bond between Oη atom of the tyrosine and Nϵ2 atom of Gln-56 on N36-a (2.33 Å, 129°).
In summary, the crystal structure of the SFT-N36 complex confirms that the designing of the inhibitor could provide the following advantages. The mutations at N terminus of SFT stabilize the conformation of this portion. The introduction of the charged residues promotes salt bridge formation that favors the helical conformation of the SFT as well as the inter-helical interactions between the inhibitor and the targeting sequence on NHR.
Structural Basis for HIV-1 Resistance to SFT
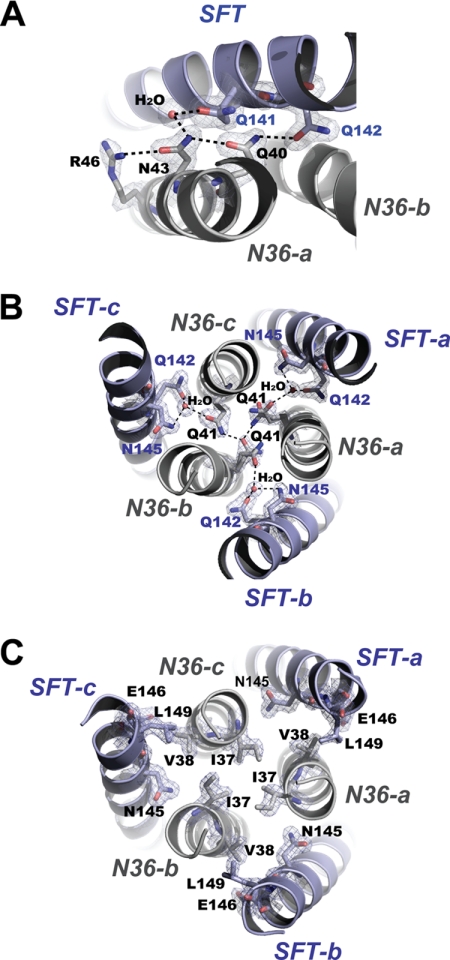
We have recently identified SFT-resistant HIV-1 variants and characterized several novel substitutions on the NHR region of gp41, which are critical in conferring the resistant phenotypes (24). For example, HIV-1 variant with N43K substitution is resistant to both SFT and T20. The mutation can lead to the reduced 6-HB stability and virus infectivity. The structure of the SFT-N36 complex demonstrates that residue Asn-43 is deeply buried inside the 6-HB, where the electron densities are well defined. The residue involves a cluster of highly conserved residues forming an extensive hydrogen bond network that mediates the interactions between N36 and SFT helices (Fig. 4A). The side chain of Asn-43 accepts a hydrogen bond from Arg-46 and donates a hydrogen bond to Gln-40 on the N36 peptide, stabilizing the helical conformation of N36. The side chains of Asn-43 and Gln-141 coordinate an ordered water molecule; Gln-40 donates a hydrogen bond to Gln-142 on SFT, stabilizing the interaction between SFT and N36 helices. Thus, the functionality of an asparagine side chain of Asn-43 is fully required for maintaining the hydrogen network. Substitution of this residue could lead to the disruption of the network, causing the destabilization of the α-helical conformation of NHR and SFT/NHR interactions.
FIGURE 4.
Structural basis of the key residues that are important to SFT resistance. A, Asn-43 is involved in an extensive hydrogen bond network formed by a cluster of residues and an ordered water molecule (stick model, colored by elements), which mediates the interaction between SFT and N36. Final 2Fo − Fc electron density (1.5 s contour) for this site is shown as blue mesh. Dashed lines indicate the hydrogen bonds. B, Gln-41 is the key member of a remarkable glutamine-rich polar layer of the 6-HB. Residues and water molecules within the layer are shown as a stick model with the superimposed final 2Fo − Fc electron density (1.5 s contour) in blue mesh. Dashed lines indicate the hydrogen bonds. C, Ile-37 and Val-38 are the key members of a hydrophobic layer of the 6-HB. Residues within the nonpolar layer are shown as a stick model with the superimposed final 2Fo − Fcelectron density (1.5 s contour) in blue mesh.
Residue Gln-41 belongs to the remarkable glutamine-rich (Gln-rich) polar layer that is critical for the overall stability of the 6-HB structure (4–6). A network of hydrogen bonds connect the residues within the Gln-rich layer (Fig. 3B), in which three Gln-41 residues interact with each other and simultaneously coordinate three water molecules together with Gln-142 and Asn-145 on three SFT. Interestingly, three Gln-41 side chains are not symmetric in 3-fold; two Gln-41 side chains adopt alternative conformations, reflecting a certain level of flexibility of this hydrogen network. The structure indicates that the functionality of the glutamine side chain of Gln-41 is irreplaceable for maintaining the overall structure of the Gln-rich polar layer. Substitutions of Gln-41 may result in the disruption of the layer, leading to not only the destabilization of the trimeric NHR coiled coil but also the interactions between SFT/NHR helices. Evidenced by our previous studies (24), HIV-1 variants carrying Q41K, Q41H, or Q41R mutations are highly resistant to SFT and T20. Substitutions of Gln-41 could lead to the destabilization of 6-HB and reduced virus infectivity.
Residues Ile-37 and Val-38 are members of a nonpolar layer of the 6-HB formed by SFT/N36 helices. There are sufficient electron densities defining the conformations of all residues in the nonpolar layer (Fig. 4C). Three Ile-37 residues are located around the 3-fold symmetry axis of the trimeric N36 coiled coil with their side chains facing each other. Three Val-38 side chains face the hydrophobic region composed of Leu-149, Glu-146, and Asn-145 on SFT. The structure indicates that the hydrophobic interactions among residues in this layer are important to the stability of NHR trimeric core and the interaction between SFT/NHR. Substitutions of Ile-37 and Val-38 by the polar residue threonine or by less hydrophobic residues alanine or methionine can destabilize the trimeric NHR coiled coil and the interaction between SFT/NHR.
DISCUSSION
T20 is the first and only HIV fusion inhibitor approved for salvage therapy of HIV/AIDS patients that failed with the current mixtures of inhibitors directed against viral enzymes (9, 16). The fact that T20 can also induce drug resistance has sparked the development of a new generation of peptide fusion inhibitors that not only preserve the potency against both wild-type and resistant-type viruses but also have improved pharmacokinetic profiles (17–21). Based on this principle, SFT was engineered with several characteristics that facilitate its antiviral activity (13). The charged glutamic acid and lysine residues were introduced into the solvent-accessible site, although the residues responsible for the NHR binding were maintained unchanged. Furthermore, Glu-119 was replaced by threonine with an intention to stabilize the hydrophobic NHR pocket, and a serine residue was added to the N terminus of SFT to increase its stability (13). Like T20, SFT is delivered by subcutaneous injection but has a dramatic and prolonged half-life (13). In this study, we have finely characterized its anti-HIV spectrum by using two large panels of Env-pseudotyped viruses in single cycle infection assays and two HIV-1 variants that carry T20-resistant mutations, and we determined its structural basis by crystallographic studies.
HIV-1 evolves with the great genetic diversity and can be classified into three major groups: M (major), O (outlier), and N (non-M non-O or new) (33, 34). The M group viruses cause the vast majority of HIV-1 pandemics and can be further divided into many subtypes, including A–D, F–H, J, and K, as well as several circulating and unique recombinant forms. Some in vitro and in vivo observations suggest that the various subtypes may respond differently to certain antiretroviral drugs (38–43). The sequence diversity has raised concerns over the peptide fusion inhibitors that target the highly variable HIV-1 Envs. With the first panel of reference viruses, we showed that SFT could efficiently inhibit the subtype A–C viruses that dominate the AIDS epidemics worldwide. Recently, the genetic diversity of HIV-1 subtypes has been found in China. According to the epidemiological data, the recombinant forms CRF07_BC and CRF01_AE, and subtype B′ (Tai B) viruses are responsible for nationwide HIV-1 infections at 50.2, 15.54, and 29.11%, respectively (with a total of ∼95%) (35, 36). It is know that the Env genes of CRF01_BC originate from the subtype C viruses, but some studies indicate that this recombinant form of virus has unique biological characteristics (44–46). For example, it is found that CRF01_BC strains exclusively use CCR5 co-receptor for infection and are highly resistant to the known broadly neutralizing antibodies (44, 45). With the second large panel of HIV-1 pseudoviruses corresponding to the CRF07_BC, CRF01_AE, and B′ subtypes, SFT consistently shows its potency to overcome the genetic diversity. It was noticed that several isolates (e.g. pRHPA4259 and GX2010.36H) were less sensitive to both SFT and T20, but sequence analyses of Envs had not identified the known mutations related to fusion inhibitor-resistance phenotypes (supplemental Fig. S2 and supplemental Table S1).
Previous studies showed that SFT could sustain its inhibitory function on a panel of HIV-1 variants that carry T20-resistant genotypes in the peptide-binding site of the gp41 NHR domain, typically including the 36GIV38 motif and its downstream residues (13). This study further shows that HIV-1 variants with L33S or L33V mutations are highly susceptible on SFT inhibition. The L33S mutant was initially induced by the peptide C34 and showed cross-resistance to T20 and T1249 (29, 47). The L33V mutant was identified as a naturally occurring mutation in treatment-naive patients and showed T20 resistance (29). Taken together, these data indicate that SFT has a high genetic barrier for drug resistance.
Since the finding of T20, several crystallographic studies have been performed and revealed the core structure of HIV-1 gp41 that represents a post-fusion conformation (4–6, 37, 48). On the basis of gp41 core structure, a number of peptide fusion inhibitors have been designed to improve the antiviral and pharmacokinetic properties (14, 49). However, there are limited structural data available for the engineered peptides. Recently, an electrostatically constrained α-helical peptide, SC34EK, was reported with the crystal structure (50). It revealed that the interaction between SC34EK and N36 peptides was well preserved by contacts of hydrophobic interactive residues and that the introduction of the charged EK motifs at the solvent-accessible site of the peptide C34 could provide the salt bridges that stabilize the α-helices and electrostatic interactions (50). In this study, we provide the accurate structural insights into the SFT-N36 complex. The crystal structure fully supports the inhibitor design, offering the detailed molecular basis for the potent antiviral activity (13). SFT is electrostatically constrained by seven salt bridges, conferring the outstanding thermostability. To stabilize the hydrophobic pocket, residue 119 is mutated to threonine in SFT, whereas the corresponding residue in C34 and T1249 is glutamic acid. Although none of the residues is hydrophobic, threonine is more hydrophobic than glutamic acid according to Wimley-White hydrophobicity scales (51). Additionally, glutamic acid side chain is flexible compared with the rigid β-branched threonine side chain that is important for stabilizing the hydrophobic interaction. The structure constraining C34 sequence (Protein Data Bank ID 1ENV) shows that the side chain of Glu-17 points away from the hydrophobic pocket where it could reach the solvent.
We previously identified that HIV-1 mutants with substitutions of Gln-41, Asn-43, or double substitutions Ile-37/Val-38 on gp41 displayed SFT resistance (24). From our structural data, we can deduce the mechanisms for SFT resistance, which is consistent with four inhibitor-resistant mechanisms (reduced contact, steric obstruction, electrostatic repulsion, and electrostatic attraction) proposed by Eggink et al. (14). More importantly, our data suggest the novel resistant mechanisms that are in many cases combined with T20-resistant mechanisms. Gln-41 is located at the center of a Gln-rich polar layer, the conserved structural features in HIV and simian immunodeficiency virus strains, which is crucial for 6-HB stability. Substitutions of Gln-41 could disrupt the hydrogen bond network. Therefore, substitutions Q41K or Q41R cannot only fit the “electrostatic repulsion” mechanism but also fit a novel mechanism of “hydrogen bond disruption.” Similarly, Asn-43 is part of a hydrogen bind network mediating the interaction between N- and C-terminal helices. N43K mutation could disrupt the hydrogen network; therefore, the mutation can fit both “steric obstruction” and hydrogen bond disruption mechanisms. Ile-37 and Val-38 are part of a nonpolar layer of 6-HB. Isoleucine and valine are the most hydrophobic residues (51) with rigid β-branched side chains, which are ideal for stable hydrophobic interactions. Substitutions I37T, V38A, and V38M could lead to the destabilization of hydrophobic interactions; therefore, these mutations can fit the novel resistance mechanism of “hydrophobic contact disruption.” It is worth noticing that residues Ile-37, Val-38, Asn-41, and Gln-43 are involved in the interactions with the unchanged residues in both SFT and T20, supporting the fact that HIV-1 with these mutations displayed cross-resistance against both inhibitors.
In conclusion, our present studies have demonstrated the potent anti-HIV activity of SFT and its structural basis. The data provide important information for developing SFT for clinical use and for designing novel HIV fusion inhibitors.
Supplementary Material
This work was supported by Natural Science Foundation of China Grant 30870123, National Outstanding Youth Award 81025009 from Natural Science Foundation of China, National 973 Program of China Grant 2010CB530100, and National Science and Technology Major Project Grant 2009ZX10004-303.

This article contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 3VIE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 6-HB
- six-helical bundle
- SFT
- sifuvirtide
- NHR
- N-terminal heptad repeat region
- CHR
- C-terminal heptad repeat region
- Env
- envelope.
REFERENCES
- 1. Colman P. M., Lawrence M. C. (2003) The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4, 309–319 [DOI] [PubMed] [Google Scholar]
- 2. Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J. D., Grisé H., Ofek G. A., Taylor K. A., Roux K. H. (2006) Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 [DOI] [PubMed] [Google Scholar]
- 3. Chan D. C., Kim P. S. (1998) HIV entry and its inhibition. Cell 93, 681–684 [DOI] [PubMed] [Google Scholar]
- 4. Chan D. C., Fass D., Berger J. M., Kim P. S. (1997) Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 [DOI] [PubMed] [Google Scholar]
- 5. Tan K., Liu J., Wang J., Shen S., Lu M. (1997) Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 94, 12303–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 [DOI] [PubMed] [Google Scholar]
- 7. Jiang S., Lin K., Strick N., Neurath A. R. (1993) HIV-1 inhibition by a peptide. Nature 365, 113. [DOI] [PubMed] [Google Scholar]
- 8. Wild C. T., Shugars D. C., Greenwell T. K., McDanal C. B., Matthews T. J. (1994) Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U.S.A. 91, 9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kilby J. M., Hopkins S., Venetta T. M., DiMassimo B., Cloud G. A., Lee J. Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M. R., Nowak M. A., Shaw G. M., Saag M. S. (1998) Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 10. Dwyer J. J., Wilson K. L., Davison D. K., Freel S. A., Seedorff J. E., Wring S. A., Tvermoes N. A., Matthews T. J., Greenberg M. L., Delmedico M. K. (2007) Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. U.S.A. 104, 12772–12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He Y., Cheng J., Li J., Qi Z., Lu H., Dong M., Jiang S., Dai Q. (2008) Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure. Implications for designing novel anti-HIV fusion inhibitors. J. Virol. 82, 6349–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., Liu Z., Jiang S., Dai Q. (2008) Potent HIV fusion inhibitors against Enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U.S.A. 105, 16332–16337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Y., Xiao Y., Song H., Liang Q., Ju D., Chen X., Lu H., Jing W., Jiang S., Zhang L. (2008) Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 283, 11126–11134 [DOI] [PubMed] [Google Scholar]
- 14. Eggink D., Langedijk J. P., Bonvin A. M., Deng Y., Lu M., Berkhout B., Sanders R. W. (2009) Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J. Biol. Chem. 284, 26941–26950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baldwin C. E., Sanders R. W., Berkhout B. (2003) Inhibiting HIV-1 entry with fusion inhibitors. Curr. Med. Chem. 10, 1633–1642 [DOI] [PubMed] [Google Scholar]
- 16. Lalezari J. P., Henry K., O'Hearn M., Montaner J. S., Piliero P. J., Trottier B., Walmsley S., Cohen C., Kuritzkes D. R., Eron J. J., Jr., Chung J., DeMasi R., Donatacci L., Drobnes C., Delehanty J., Salgo M. (2003) Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348, 2175–2185 [DOI] [PubMed] [Google Scholar]
- 17. Ashkenazi A., Wexler-Cohen Y., Shai Y. (2011) Multifaceted action of Fuzeon as virus-cell membrane fusion inhibitor. Biochim. Biophys. Acta 1808, 2352–2358 [DOI] [PubMed] [Google Scholar]
- 18. Rimsky L. T., Shugars D. C., Matthews T. J. (1998) Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matthews T., Salgo M., Greenberg M., Chung J., DeMasi R., Bolognesi D. (2004) Enfuvirtide. The first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3, 215–225 [DOI] [PubMed] [Google Scholar]
- 21. Welch B. D., Francis J. N., Redman J. S., Paul S., Weinstock M. T., Reeves J. D., Lie Y. S., Whitby F. G., Eckert D. M., Hill C. P., Root M. J., Kay M. S. (2010) Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J. Virol. 84, 11235–11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin-Carbonero L. (2004) Discontinuation of the clinical development of fusion inhibitor T-1249. AIDS Rev. 6, 61. [PubMed] [Google Scholar]
- 23. Lalezari J. P., Bellos N. C., Sathasivam K., Richmond G. J., Cohen C. J., Myers R. A., Jr., Henry D. H., Raskino C., Melby T., Murchison H., Zhang Y., Spence R., Greenberg M. L., Demasi R. A., Miralles G. D. (2005) T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect. Dis. 191, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 24. Liu Z., Shan M., Li L., Lu L., Meng S., Chen C., He Y., Jiang S., Zhang L. (2011) In vitro selection and characterization of HIV-1 variants with increased resistance to sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 286, 3277–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pancera M., Majeed S., Ban Y. E., Chen L., Huang C. C., Kong L., Kwon Y. D., Stuckey J., Zhou T., Robinson J. E., Schief W. R., Sodroski J., Wyatt R., Kwong P. D. (2010) Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U.S.A. 107, 1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. (2000) Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74, 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He Y., Liu S., Jing W., Lu H., Cai D., Chin D. J., Debnath A. K., Kirchhoff F., Jiang S. (2007) Conserved residue Lys-574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J. Biol. Chem. 282, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 28. He Y., Liu S., Li J., Lu H., Qi Z., Liu Z., Debnath A. K., Jiang S. (2008) Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J. Virol. 82, 11129–11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chinnadurai R., Münch J., Kirchhoff F. (2005) Effect of naturally occurring gp41 HR1 variations on susceptibility of HIV-1 to fusion inhibitors. AIDS 19, 1401–1405 [DOI] [PubMed] [Google Scholar]
- 30. Chinnadurai R., Rajan D., Münch J., Kirchhoff F. (2007) Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 81, 6563–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCutchan F. E. (2000) Understanding the genetic diversity of HIV-1. AIDS 14, S31–S44 [PubMed] [Google Scholar]
- 34. Roux K. H., Taylor K. A. (2007) AIDS virus envelope spike structure. Curr. Opin. Struct. Biol. 17, 244–252 [DOI] [PubMed] [Google Scholar]
- 35. Lu L., Jia M., Ma Y., Yang L., Chen Z., Ho D. D., Jiang Y., Zhang L. (2008) The changing face of HIV in China. Nature 455, 609–611 [DOI] [PubMed] [Google Scholar]
- 36. Liao L., Xing H., Shang H., Li J., Zhong P., Kang L., Cheng H., Si X., Jiang S., Li X., Shao Y. (2010) The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J. Acquir. Immune Defic. Syndr. 53, S10–S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu M., Blacklow S. C., Kim P. S. (1995) A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 38. Palmer S., Alaeus A., Albert J., Cox S. (1998) Drug susceptibility of subtypes A, B, C, D, and E human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retroviruses 14, 157–162 [DOI] [PubMed] [Google Scholar]
- 39. Loemba H., Brenner B., Parniak M. A., Ma'ayan S., Spira B., Moisi D., Oliveira M., Detorio M., Wainberg M. A. (2002) Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against non-nucleoside inhibitors of RT. Antimicrob. Agents Chemother. 46, 2087–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brenner B., Turner D., Oliveira M., Moisi D., Detorio M., Carobene M., Marlink R. G., Schapiro J., Roger M., Wainberg M. A. (2003) A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17, F1–F5 [DOI] [PubMed] [Google Scholar]
- 41. Grossman Z., Istomin V., Averbuch D., Lorber M., Risenberg K., Levi I., Chowers M., Burke M., Bar Yaacov N., Schapiro J. M. (2004) Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS 18, 909–915 [DOI] [PubMed] [Google Scholar]
- 42. Jetté L., Léger R., Thibaudeau K., Benquet C., Robitaille M., Pellerin I., Paradis V., van Wyk P., Pham K., Bridon D. P. (2005) Human growth hormone-releasing factor (hGRF)1–29-albumin bioconjugates activate the GRF receptor on the anterior pituitary in rats. Identification of CJC-1295 as a long-lasting GRF analog. Endocrinology 146, 3052–3058 [DOI] [PubMed] [Google Scholar]
- 43. Ma L., Guo Y., Yuan L., Huang Y., Sun J., Qu S., Yu X., Meng Z., He X., Jiang S., Shao Y. (2009) Phenotypic and genotypic characterization of human immunodeficiency virus type 1 CRF07_BC strains circulating in the Xinjiang Province of China. Retrovirology 6, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chong H., Hong K., Zhang C., Nie J., Song A., Kong W., Wang Y. (2008) Genetic and neutralization properties of HIV-1 env clones from subtype B/BC/AE infections in China. J. Acquir. Immune Defic. Syndr. 47, 535–543 [DOI] [PubMed] [Google Scholar]
- 45. Shang H., Han X., Shi X., Zuo T., Goldin M., Chen D., Han B., Sun W., Wu H., Wang X., Zhang L. (2011) Genetic and neutralization sensitivity of diverse HIV-1 env clones from chronically infected patients in China. J. Biol. Chem. 286, 14531–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang C., Xu S., Wei J., Guo H. (2009) Predicted co-receptor tropism and sequence characteristics of China HIV-1 V3 loops. Implications for the future usage of CCR5 antagonists and AIDS vaccine development. Int. J. Infect. Dis. 13, e212–216 [DOI] [PubMed] [Google Scholar]
- 47. Armand-Ugón M., Gutiérrez A., Clotet B., Esté J. A. (2003) HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59, 137–142 [DOI] [PubMed] [Google Scholar]
- 48. Buzon V., Natrajan G., Schibli D., Campelo F., Kozlov M. M., Weissenhorn W. (2010) Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 6, e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steffen I., Pöhlmann S. (2010) Peptide-based inhibitors of the HIV envelope protein and other class I viral fusion proteins. Curr. Pharm. Des. 16, 1143–1158 [DOI] [PubMed] [Google Scholar]
- 50. Naito T., Izumi K., Kodama E., Sakagami Y., Kajiwara K., Nishikawa H., Watanabe K., Sarafianos S. G., Oishi S., Fujii N., Matsuoka M. (2009) SC29EK, a peptide fusion inhibitor with enhanced α-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Antimicrob. Agents Chemother. 53, 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wimley W. C., White S. H. (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.