Background: The strength of excitatory synapses is determined by the synaptic content of AMPA receptors.
Results: We found that Contactin associated protein 1 (Caspr1) binds to AMPA receptors and regulates their neuronal cell surface and synaptic expression.
Conclusion: Caspr1 is a binding partner for AMPA receptors, which regulates their traffic and synaptic targeting.
Significance: Caspr1 is a new player in plasticity mechanisms in excitatory synapses.
Keywords: Glutamate Receptors Ionotropic (AMPA, NMDA); Neuroscience; Protein-Protein Interactions; Synapses; Synaptic Plasticity; Caspr1; Hippocampal Neurons
Abstract
Glutamate receptors of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) type mediate fast excitatory synaptic transmission in the CNS. Synaptic strength is modulated by AMPA receptor binding partners, which regulate receptor synaptic targeting and functional properties. We identify Contactin-associated protein 1 (Caspr1) as an AMPA receptor interactor. Caspr1 is present in synapses and interacts with AMPA receptors in brain synaptic fractions. Coexpression of Caspr1 with GluA1 increases the amplitude of glutamate-evoked currents. Caspr1 overexpression in hippocampal neurons increases the number and size of synaptic GluA1 clusters, whereas knockdown of Caspr1 decreases the intensity of synaptic GluA1 clusters. Hence, Caspr1 is a regulator of the trafficking of AMPA receptors to synapses.
Introduction
Regulation of the function of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)2-type glutamate receptors is critical for long-lasting changes in synaptic strength, considered to underlie higher brain functions such as learning and memory (1). Protein-protein interactions and posttranslational modification of AMPA receptor subunits regulate receptor function by modulating the localization of receptors at synapses and/or modifying receptor channel properties (2).
Recent studies have identified AMPA receptor auxiliary proteins, which influence various aspects of excitatory synapse function (3). The transmembrane AMPA receptor regulatory proteins (TARPs) regulate the trafficking and channel properties of AMPA receptors (3). Other transmembrane proteins affect AMPA receptor gating, such as the cornichons (4) and cysteine-knot AMPA receptor-modulating protein of 44 kDa (CKAMP44) (5). In a recombinant system, the cornichon protein CNHI-2 abrogates the γ8 TARP-mediated resensitization of AMPA receptors upon prolonged application of glutamate (6), suggesting that different classes of auxiliary subunits can coregulate AMPA receptors. Moreover, proteins such as TARPs and synapse differentiation-induced gene 1 (SynDIG1) (7) affect the cell surface and synaptic expression of AMPA receptors. Fine-tuning of the receptor-gating kinetics and synaptic expression by auxiliary proteins may underlie the different kinetics of gating observed for native AMPA receptors and may depend on the expression pattern of auxiliary subunits, those which have been described and additional ones, yet to be identified.
We searched for novel AMPA receptor binding partners using a pull-down assay in rat cerebellum extracts with the intracellular C terminus of GluA4 as bait. Mass spectrometry analysis of the purified proteins suggested an interaction of GluA4 with the transmembrane protein Contactin associated protein 1 (Caspr1). Biochemical studies confirmed the interaction between Caspr1 and AMPA receptors in the rat brain. Coexpression of Caspr1 with GluA1 increases glutamate-evoked currents. Furthermore, Caspr1 is localized to dendrites and excitatory synapses in hippocampal neurons and regulates the cell surface and synaptic expression of GluA1-containing AMPA receptors.
EXPERIMENTAL PROCEDURES
Antibodies
The anti-GFP polyclonal antibody was purchased from MBL International Corp. (Woburn, USA), the anti-GFP monoclonal antibody was acquired from Roche Diagnostics (Carnaxide, Portugal), and the anti-actin antibody was purchased from Roche Molecular Biochemicals (Indianapolis, IN). The anti-GluA1 polyclonal antibody was from Tocris Bioscience (MO) and the sheep anti-GluA1 N-terminal antibody was a kind gift from Dr. Andrew Irving (University of Dundee, Scotland). The anti-Caspr1 polyclonal antibody was either purchased from Abcam (Cambridge, UK) or a gift from Catherine Faivre-Sarrailh (Université de la Méditerranée Aix-Marseille II, Marseille, France). The anti-myc monoclonal antibody was bought from Cell Signaling Technology, Inc. (Danvers, MA), and the anti-FLAG polyclonal antibody was purchased from Sigma. The anti-GluA2/3, the anti-GluN1 and the anti-PSD95 antibodies were from Millipore (Madrid, Spain), and the anti-MAP2 antibody was from Abcam. The anti-synaptophysin antibody was purchased from Synaptic Systems (Germany).
Constructs for Transfection of Neurons and COS 7 Cells
The following plasmids were kind gifts. The GluA1 and GluA2 constructs were from Dr. Juan Lerma (Instituto de Neurociencias de Alicante, Spain), the N-terminally FLAG-tagged GluA4 plasmid was from Dr. Kari Keinanen (University of Helsinki, Finland), the GluA2L plasmid was from Dr. Pavel Osten (Max Planck Institute for Medical Research, Heidelberg, Germany), the GluN1–1a plasmid was from Dr. Ann Marie Craig (University of British Columbia, Vancouver, Canada), the Caspr1 and CD4-CTCaspr1 constructs were from Dr. Catherine Faivre-Sarrailh, and the TrkB-GFP was from Dr. Volkmar Leβman (Otto-von-Guericke University, Magdeburg, Germany).
For plasmid-based RNA inhibition of Caspr1, the complementary oligonucleotides that target nucleotides 2254–2273 (GAACAGCATTTCTACTGGG) of rat Caspr1 (NM_032061) were annealed and ligated into the HpaI/XhoI sites of the U6 promoter-driven short hairpin RNA expression vector pLentiLox3.7(CMV)EGFP that expresses EGFP under the CMV promoter. The construct for expressing Caspr1* resistant against Caspr1-shRNA was generated by making the following two point mutations, indicated by underlines, in the shRNA-targeting site: GAGCAGCATTTCTATTGGG. All the constructs were verified by DNA sequencing.
Rat Cerebellum Total Extracts
The rat cerebellum was homogenized in 10 volumes of ice-cold 10 mm Tris-HCl buffer (pH 7.4) containing 320 mm sucrose. The homogenate was centrifuged at 700 × g for 10 min at 4 °C. The pellet was homogenized again in the same Tris buffer and centrifuged at 700 × g, for 10 min at 4 °C. Both supernatants were pooled, supplemented with protease inhibitors, and submitted to protein quantification by the BCA method (Pierce). The protein was aliquoted and frozen at −20 °C until needed.
Production of Recombinant Proteins
The cDNA encoding the C terminus of Caspr1 was amplified by RT-PCR from total RNA isolated from rat brain cerebellum using the specific primers 5′-cgcggatcccaaaatcatcgatacaag-3′ and 5′-ccgctcgagtcattcagacctggactc-3′, with the restriction sites for BamHI and XhoI, respectively. The PCR product was subcloned in the pGEX-4T-2 vector, and the construct was sequenced. The plasmid encoding the intracellular carboxy-terminal domain of GluA4 fused to GST has been described previously (8). Recombinant proteins were expressed in BL21 Escherichia coli as described before (8).
Pull-down Assays
Rat cerebellum lysates were diluted with radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 5 mm EGTA, 1% Triton, 0.5% deoxycholate (DOC), and 0.1% SDS at a final pH of 7.5), supplemented with 1 mm DTT and a mixture of protease inhibitors, and sonicated. After centrifugation, the supernatant was incubated with 50 μl of glutathione-Sepharose for 30 min at 4 °C. The supernatant was split in two tubes. One was incubated with 50 μg of GST and the other with 50 μg of the fusion protein of interest at 4 °C for 3 h. 50 μl of radioimmune precipitation assay-equilibrated glutathione-Sepharose was added to both samples and incubated at 4 °C for 30 min. The samples were washed four times with radioimmune precipitation assay buffer, and the proteins were eluted by boiling at 95 °C in 50 μl of sample buffer, separated by SDS-PAGE, and stained with silver nitrate.
MALDI Peptide Mass Fingerprinting and Database Searching
Protein bands were excised manually from the gel and digested automatically using a Proteineer DP protein digestion station (Bruker-Daltonics, Bremen, Germany), according to a previously described protocol (9). For peptide mass fingerprinting (10) spectra acquisition, an aliquot of α-cyano-4-hydroxycinnamic acid in 33% aqueous acetonitrile and 0.1% trifluoroacetic acid was mixed with an aliquot of the digestion solution and the mixture was deposited onto an AnchorChip MALDI probe (Bruker-Daltonics).
MALDI Peptide mass fingerprint spectra were measured on a Bruker Ultraflex TOF/TOF MALDI mass spectrometer (Bruker-Daltonics) (10). Mass measurements were performed in positive ion reflector mode using 140-ns delayed extraction and a nitrogen laser (337 nm). The laser repetition rate was 50 Hz, and the ion acceleration voltage was 25 kV. Mass measurements were performed automatically through fuzzy logic-based software to accumulate 100 single laser shot spectra or manually to accumulate approximately 200 single laser shot spectra. Each spectrum was internally calibrated with the mass signals of two trypsin autolysis ions: (VATVSLPR+H)+ (m/z = 842.510) and (LGEHNIDVLEGNEQFINAAK+H)+ (m/z = 2211.105) to reach a typical mass measurement accuracy of ± 30 ppm. Known trypsin and keratin mass signals as well as potential sodium adducts (+21.982 Da) or signals arising from methionine oxidation (+15.995 Da) were removed from the peak list. The measured tryptic peptide masses were transferred through the MS BioTools program (Bruker-Daltonics) as inputs to search the NCBInr database using Mascot software (Matrix Science, London, UK). This analysis was performed at the Unidad de Proteómica, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid.
Immunoprecipitation Assays
For coimmunoprecipitation assays, lysates of COS 7 cells expressing the proteins of interest or rat brain synaptosomes (1 mg) were solubilized in immunoprecipitation buffer (IPB) (10 mm Tris (pH 7.0), 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 1% Triton X-100, supplemented with protease inhibitors). The samples were sonicated on ice for 30 s, and the insoluble material was removed by centrifugation at 4 °C. Soluble extracts were incubated with 100 μl of a 50% slurry of protein A-Sepharose beads suspended in IPB at 4 °C for 1 h to preabsorb proteins that stick nonspecifically to the protein A-Sepharose beads. The supernatant was incubated either with the antibody of interest or with the same amount of non-immune IgGs at 4 °C, for 3 h and was then incubated with 100 μl of a 50% slurry of protein A-Sepharose beads (2 h at 4 °C). The beads were washed sequentially in IPB + 1% Triton (2×), in IPB + 1% Triton + 500 mm NaCl (3×), and in IPB (2×). The proteins were eluted by boiling in sample buffer, separated by SDS-PAGE, and analyzed by Western blot analysis.
Gel Electrophoresis and Western Blot Analysis
Samples were resolved by SDS-PAGE in 7.5% polyacrylamide gels. For Western blot analysis, proteins were transferred onto a PVDF membrane (Millipore, Madrid, Spain) by electroblotting. The membranes were blocked, incubated with primary and secondary antibodies, and immunostaining was visualized by the enhanced chemifluorescence method on a Storm 860 Gel and Blot Imaging System (GE Healthcare, Carnaxide, Portugal).
Hippocampal and Cortical Cultures
Primary cultures of rat hippocampal and cortical neurons were prepared from the hippocampi or cortices of E18-E19 Wistar rat embryos, after treatment with trypsin (0.06%, 15 min, 37 °C, Invitrogen), in Ca2+- and Mg2+-free Hanks' balanced salt solution (5.36 mm KCl, 0.44 mm KH2PO4, 137 mm NaCl, 4.16 mm NaHCO3, 0.34 mm Na2HPO4.2H2O, 5 mm glucose, 1 mm sodium pyruvate, 10 mm HEPES, 0.001% phenol red). The hippocampal cells were washed with 10% fetal bovine serum prepared in Hanks' balanced salt solution to stop trypsin activity and then washed with Hanks' balanced salt solution to remove serum and avoid glia growth. Cortical cells were washed with Hanks' balanced salt solution six times. Cells were transferred to neurobasal medium (Invitrogen) supplemented with B27 supplement (1:50 dilution, Invitrogen), 25 μm glutamate, 0.5 mm glutamine, and 0.12 mg/ml gentamycin. The cells were mechanically dissociated and then plated in 6-well plates (1 × 105 cells/cm2 for cortical neurons), coated with poly-D-lysine (0.1 mg/ml). For imaging purposes, low-density hippocampal cells were plated at a final density of 3 × 105 cells/dish on poly-D-lysine-coated coverslips in 60 mm culture dishes in neuronal plating medium (minimal essential medium supplemented with 10% horse serum, 0.6% glucose, and 1 mm pyruvic acid). After 2–4 h, coverslips were flipped over an astroglial feeder layer. These neurons grew face-down over the feeder layer but were kept separate from the glia by wax dots on the neuronal side of the coverslips. To prevent the overgrowth of the glia, neuron cultures were treated with 5 μm cytosine arabinoside after 3 days in vitro (DIV). Cultures were maintained in neurobasal medium supplemented with B27 supplement in a humidified incubator of 5% CO2/95% air at 37 °C.
Neuron Transfection
Constructs were recombinantly expressed in primary cultures of hippocampal neurons using the calcium phosphate transfection protocol (adapted from Ref. 11). Briefly, a CaCl2 solution (2.5 m in 10 mm HEPES) was added, dropwise, to plasmid DNA to a final concentration of 250 mm CaCl2. This was then added to an equivalent volume of HEPES-buffered transfection solution (274 mm NaCl, 10 mm KCl, 1.4 mm Na2HPO4, 11 mm dextrose, 42 mm HEPES (pH 7.2)). The mixture was vortexed gently for 2–3 s, and the precipitate was allowed to develop at room temperature for 30 min. The precipitated DNA was added dropwise to the coverslips, and the cultures were incubated for 1–3 h in the presence of kynurenic acid (2 mm). Each coverslip was transferred to a fresh well of the 24-well plate containing 1 ml of culture medium with kynurenic acid (2 mm), slightly acidified with HCl (∼5 mm final concentration), and the plate was incubated at 37 °C in 5% CO2 for 10–15 min. Coverslips were then transferred to a fresh well of the 24-well plate containing conditioned medium and incubated at 37 °C in 5% CO2 to allow expression of the transfected constructs.
Subcellular Fractionation
For purification of rat brain synaptosomes, two rat brains were removed, placed in 5 volumes of ice-cold solution A (5 mm HEPES, 1 mm MgCl2, 0.5 mm CaCl2, 0.31 m sucrose (pH 7.4) supplemented with 0.2 mm PMSF, 1 mm DTT, and a mixture of protease inhibitors) and homogenized with a Potter-Elvehjem homogenizer (8 strokes). The homogenates were centrifuged at 1400 × g for 10 min. The supernatant solution was saved, and the pellet was rehomogenized in 5 volumes (considering the initial weight) of solution A and centrifuged at 710 × g for 10 min. Supernatants were pooled and centrifuged at 13,800 × g for 10 min. The pellet was resuspended in 24 ml/10g of buffer B (6 mm Tris (pH 8.1), 0.32 m sucrose, supplemented with protease inhibitors). This fraction (P2) was used directly for immunoprecipitation studies.
The procedure for purification of synaptic fractions from cultured cortical neurons was adapted from Ref. 12 and modified according to the protocol described in Ref. 13 for high-density cultured neurons. Postsynaptic densities (PSDs) were isolated from 15-DIV high-density cortical rat neurons. Briefly, 38 × 106 cortical cells were collected and homogenized in HEPES-buffered sucrose solution (0.32 m sucrose, 4 mm HEPES (pH 7.4)) containing protease and phosphatase inhibitors (0.2 mm PMSF, 0.1 mm sodium orthovanadate, 50 mm sodium fluoride, 1 μg/ml chymostatin, 1 μg/ml leupeptin, 1 μg/ml antipain, 1 μg/ml pepstatin. Culture homogenate was collected and centrifuged at 900 × g for 15 min to obtain the non-nuclear fraction (S1). The resultant supernatant was centrifuged at 18.000 × g for 15 min to yield the crude synaptosomal pellet (P2). P2 was resuspended in HEPES-buffered sucrose and centrifuged at 18,000 × g for 15 min to yield the washed crude synaptosomal fraction. This fraction was submitted to hypo-osmotic shock by resuspending the pellet in HEPES buffer (4 mm HEPES (pH 7.4) plus protease and phosphatase inhibitors) and incubated 1–2 h with orbital rotation at 4 °C. The lysate was centrifuged at 25,000 × g for 20 min, and the pellet (lysed synaptosomal membrane fraction) was resuspended in HEPES-buffered sucrose (without sodium orthovanadate), placed on top of a discontinuous sucrose gradient (0.8 m, 1 m, 1.2 m) and spun at 150,000 × g for 2 h in a swinging bucket rotor. Synaptic plasma membranes were recovered between the 1.0 m and 1.2 m layers, diluted to 0.32 m sucrose, and centrifuged at 150,000 × g for 30 min. Synaptic plasma membranes were resuspended in HEPES/EDTA buffer (50 mm HEPES, 2 mm EDTA (pH 7.4)) containing protease and phosphatase inhibitors and solubilized in 0.5% Triton X-100 for 15 min with orbital rotation at 4 °C, followed by 20 min of centrifugation at 200,000 × g. The remaining pellet, corresponding to the PSDs, was resuspended in HEPES/EDTA containing 0.5% SDS. All experimental procedures and centrifugations were performed on ice or at 4 °C. For protein quantification, samples were boiled for 5 min at 95 °C. Protein concentrations were determined by the Bio-Rad method, and samples were denatured with 5× concentrated denaturating buffer (125 mm Tris (pH 6.8), 100 mm glycine, 10% SDS, 200 mm DTT, 40% glycerol, 3 mm sodium orthovanadate, and 0.01% bromphenol blue) and separated by SDS-PAGE.
Electrophysiology
HEK293 cells were transfected using FuGENE6 with GFP and GluA1, with or without Caspr1, at a cDNA ratio of 1:1. Two to three days after transfection, cells were bathed in HEPES-buffered solution containing 145 mm NaCl, 2 mm KCl, 2 mm MgCl2, 2 mm CaCl2, 10 mm glucose, and 10 mm HEPES adjusted to 320 mOsm/liter (pH 7.4) with NaOH at room temperature. Whole-cell recordings were performed on green fluorescent cells lifted off the coverslip, placed under the flow of a theta tube, and held at −80 to −40 mV. Recording pipettes (resistance 3–5 MΩ) were filled with a solution containing 130 mm CsCH3SO3, 2 mm NaCl, 2 mm MgCl2, 10 mm EGTA, 10 mm HEPES, 4 mm Na2ATP, adjusted to 310 mOsm/liter (pH 7.2) with CsOH. Currents were evoked by long application of 10 mm glutamate for 100 ms or 1 ms every 20 s by moving the theta tube laterally with a piezoelectric device under computer control.
Immunocytochemistry, Culture Imaging, and Quantitative Fluorescence Analysis
Neurons were fixed for 15 min in 4% sucrose/4%paraformaldehyde in PBS at room temperature and permeabilized with PBS + 0.25% Triton X-100 for 5 min at 4 °C. The neurons were then incubated in 10% BSA in PBS for 30 min at 37 °C to block nonspecific staining and incubated with the indicated primary antibodies diluted in 3% BSA in PBS (2 h, 37 °C). After washing in PBS, cells were incubated with the secondary antibody diluted in 3% BSA in PBS (45 min, 37 °C). The coverslips were mounted using fluorescent mounting medium from DAKO (Glostrup, Denmark). For labeling surface GluA1-containing receptors, live neurons were incubated for 10 min at room temperature with the GluA1 N-terminal antibody diluted in PBS, after which the cells were briefly rinsed in PBS, fixed, and probed as described above. Imaging was performed on a Zeiss Axiovert 200 m microscope using a 63 × 1.4 numerical aperture oil objective.
Images were quantified using image analysis software (ImageJ). For quantitation, sets of cells were cultured and stained simultaneously and imaged using identical settings. The region of interest was randomly selected, and the dendritic length was measured using MAP2 staining. For Caspr1 measurements, the PSD95 and VGLUT signals were thresholded, dilated, and their colocalization was determined. The Caspr1 signal was measured after thresholds were set so that recognizable clusters were included in the analysis, and the Caspr1 signal present in glutamatergic synapses was obtained by measuring the Caspr1 puncta positive for both PSD95 and VGLUT. To calculate the fraction of glutamatergic synapses containing Caspr1, the number of synapses per dendritic length was determined by colocalizing the thresholded PSD95 and VGLUT signals. The number of glutamatergic synapses containing Caspr1 per dendritic length was determined by identifying PSD95- and VGLUT-positive clusters that were also labeled for Caspr1. For quantifying the GluA1 signal, fields for imaging were chosen by the GFP channel for the presence of transfected, GFP-positive neurons. The cell surface GluA1 digital images were subjected to a user-defined intensity threshold to select clusters and measured for cluster intensity, number, and area for the selected region. The synaptic GluA1 clusters were selected by their overlap with thresholded and dilated PSD95 signal. Measurements were performed in a minimum of three independent preparations, and at least nine cells per condition were analyzed for each preparation.
RESULTS
Caspr1 Interacts with AMPA Receptors
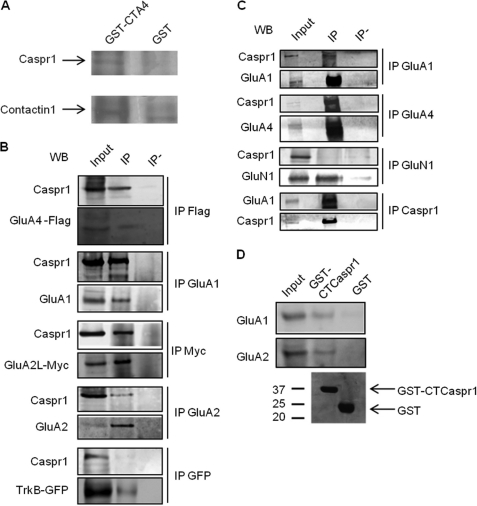
To identify proteins that specifically interact with the C terminus of the long splice isoform of GluA4, pull-down experiments were performed using GST fused to the C terminus of rat GluA4 (GST-CTA4) and adult rat cerebellum lysate because GluA4 is particularly abundant in this brain region (14). MALDI peptide mass fingerprint analysis identified Caspr1 and its binding partner Contactin1 (Fig. 1A and supplemental Figs. 1 and 2) among the proteins that were exclusive to the GST-CTA4 condition. Contactin1 is a glycosyl phosphatidyl inositol-anchored member of the immunoglobulin gene superfamily, which has been implicated previously in synaptic plasticity (15). Caspr1, also known as paranodin, is a 190-kDa type I transmembrane protein highly expressed in the brain. It has an extracellular architecture similar to neurexins and an intracellular region containing a domain that binds to 4.1-band proteins and a proline-rich sequence (16, 17). Mutations in the Caspr1 family member Caspr2 are implicated in autism spectrum disorders (18, 19).
FIGURE 1.
Caspr1 associates with AMPA receptors. A, Caspr 1 was pulled down from rat cerebellum by the GluA4 C terminus. Rat cerebellum lysate was incubated with GST fused to the intracellular C-terminal region of GluA4 (GST-CTA4) or with GST alone. The GST fusion proteins and associated proteins were purified, separated by SDS-PAGE, and silver-stained. The protein bands exclusive to the pull-down assay with the GluA4 C terminus were excised from the gel, and the proteins were identified by MALDI peptide mass fingerprint. B, Caspr1 interacts with AMPA receptor subunits in heterologous cells. AMPA receptor subunits (GluA4-FLAG, GluA1, GluA2L-myc, or GluA2) or TrkB-GFP were cotransfected with Caspr1 in COS 7 cells. AMPA receptor subunits were immunoprecipitated (IP) using specific antibodies, as indicated, and Western blot (WB) analysis revealed that Caspr1 coimmunoprecipitated with AMPA receptor subunits. Immunoprecipitation of TrkB-GFP from cells cotransfected with Caspr1 and TrkB-GFP did not coimmunoprecipitate Caspr1. C, Caspr1 and AMPA receptors associate in brain. Caspr1 coimmunoprecipitated with GluA4 and GluA1, but not with GluN1, from rat brain synaptosomes. Conversely, GluA1 was coimmunoprecipitated with Caspr1 from rat brain synaptosomes. D, Caspr1 associates with AMPA receptors through its C terminus. GluA1 and GluA2 subunits were pulled down from rat cerebellum lysates when the C terminus of Caspr1 was used as bait. Lower panel, Coomassie staining of GST fusion proteins.
We used additional biochemical assays to confirm the interaction between Caspr1 and GluA4 and tested whether the interaction is specific to GluA4. Caspr1 coprecipitated with FLAG-tagged GluA4 from cotransfected heterologous cells (Fig. 1B), whereas precipitation of Caspr1 was not observed when using non-immune immunoglobulins. Notably, Caspr1 also coimmunoprecipitated with cotransfected AMPA receptor subunits GluA1, GluA2, or the longer splice isoform of GluA2, GluA2L, but not with GFP-TrkB (Fig. 1B). To determine whether these interactions also occur in the brain, we used synaptosomes isolated from adult rat brain to immunoprecipitate GluA4, GluA1, or the N-methyl-d-aspartate (NMDA) receptor subunit GluN1 (Fig. 1C). Caspr1 coimmunoprecipitated with both GluA1 and GluA4 isolated from synaptosomes but not with GluN1, and immunoprecipitation of Caspr1 resulted in coimmunoprecipitation of GluA1 (Fig. 1C). These results confirm that Caspr1 interacts with AMPA receptor subunits in the rat brain and that synaptically localized AMPA receptors interact with Caspr1.
Because we identified Caspr1 as a potential GluA4 interactor using the intracellular C terminus of GluA4, we tested whether the interaction between Caspr1 and GluAs occurs between the intracellular regions of these proteins. The intracellular C terminus of Caspr1 fused to GST (GST-CTCaspr1) pulled-down GluA1 and GluA2 from rat cerebellum lysates (Fig. 1D), confirming a role for this region in the interaction with AMPA receptors.
Caspr1 Localizes to Dendrites and Synapses
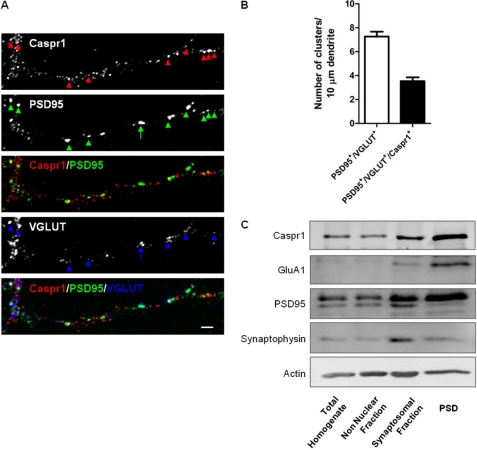
Caspr1 plays important roles in the correct assembly of the paranodes (20), but there is evidence that it is also present in dendrites and enriched in PSD fractions (15). Immunolabeling of cultured hippocampal neurons (15 DIV) for Caspr1 showed that the protein is distributed throughout dendrites and forms clusters that partially colocalize with the glutamatergic synapse markers PSD95, a postsynaptic scaffold, and VGLUT1, a presynaptic vesicular glutamate transporter (Fig. 2A). To evaluate the presence of Caspr1 at excitatory synapses, we identified regions of overlap between the PSD95 and the VGLUT signals and measured the Caspr1 signal at these sites (Fig. 2B). We found that 48.7 ± 4.4% of the clusters positive for both PSD95 and VGLUT contain Caspr1 (Fig. 2B). Furthermore, Caspr1 was present in the PSD95-enriched PSDs isolated from 15 DIV cultured cortical neurons (Fig. 2C). Accordingly, Caspr1 was detected previously in the PSD fraction isolated from adult mouse brain (15). Taken together, this evidence points to a dendritic localization of Caspr1 in neurons and to synaptic localization of a fraction of the protein.
FIGURE 2.
Caspr1 localizes to synapses in hippocampal neurons and to cortical postsynaptic density fractions. A and B, Caspr1 is present in excitatory synapses. Caspr1 is expressed in dendrites in hippocampal neurons (15 DIV) and is significantly colocalized with PSD95 and VGLUT1 (A). Scale bars = 2 μm. The arrowheads point to synapses that contain Caspr1. The arrow indicates a PSD95/VGLUT-positive synapse that lacks Caspr1. B, quantification of the number of dendritic clusters per dendritic length that are positive for both PSD95 and VGLUT (“glutamatergic synapses”) and of the number of glutamatergic synapses that contain Caspr1 (PSD95+/VGLUT+/Caspr1+). C, Caspr1 localizes to postsynaptic density fractions. PSD fractions isolated from 15 DIV rat cortical neurons were analyzed for the presence of Caspr1, GluA1, PSD95, synaptophysin, and actin, as indicated.
Caspr1 Regulates the Synaptic Expression of AMPA Receptors
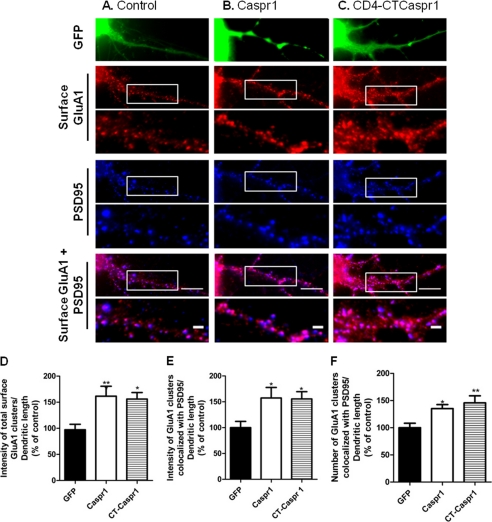
Caspr1 was initially identified as a binding partner for GluA4 in the cerebellum (Fig. 1A), but subsequent analysis found that Caspr1 also binds GluA1 and GluA2 AMPA receptor subunits (Figs. 1, B–D). Because GluA1 is widely expressed in the central nervous system, namely in the hippocampus, where GluA4 has a more limited expression pattern (21), and given the evidence pointing to an important role for the GluA1-containing AMPA receptors in plasticity events (22), we focused on a possible regulation of synaptic GluA1-containing AMPA receptors by Caspr1 in the hippocampus. To explore the effects of the interaction between AMPA receptors and Caspr1 on the cell surface and synaptic expression of GluA1-containing AMPA receptors, we performed quantitative immunofluorescence analysis of the expression of synaptic cell surface GluA1 in hippocampal neurons overexpressing Caspr1. Hippocampal neurons cultured at low density were transfected with EGFP alone or together with Caspr1 and live-stained with an antibody against the N-terminal extracellular region of GluA1. After fixation, neurons were stained with an antibody for PSD95 to visualize excitatory synapses. Compared with neurons transfected with EGFP alone (Fig. 3A), neurons overexpressing Caspr1 (B) showed a significant increase in the fluorescence intensity of total (D) and PSD95-colocalized (E) GluA1 clusters. The density of GluA1 synaptic clusters was also increased in Caspr1-transfected neurons (Fig. 3F). Because the C terminus of AMPA receptor subunits is involved in the interaction with Caspr1, we cotransfected hippocampal neurons with EGFP and a plasmid encoding the C terminus of Caspr1 fused to the extracellular and transmembrane domains of the T-cell surface protein CD4, a transmembrane reporter (CD4-CTCaspr1, Fig. 3C). The CD4-CTCaspr1 construct has been used previously (23), and the encoded protein is targeted to the cell surface. Neurons overexpressing CD4-CTCaspr1 showed a significant increase in the fluorescence intensity of total GluA1 clusters (Fig. 3D) as well as an increase in the fluorescence intensity and density of GluA1 synaptic clusters compared with EGFP-transfected cells (Fig. 3, E and F). These observations suggest a role for Caspr1 in increasing the levels of AMPA receptors at the neuronal surface and at synapses through a mechanism that can be mediated by its C terminus.
FIGURE 3.
Caspr1 overexpression promotes the synaptic expression of GluA1. Hippocampal neurons were transfected at 7 DIV with EGFP (A), Caspr1 and EGFP (B), or CD4-CTCaspr1 and EGFP (C) and live-stained at 15 DIV for cell surface GluA1. After fixation, neurons were stained for PSD95. Synaptic GluA1 is defined as a GluA1 signal that overlaps with PSD95. Transfected neurons, identified by GFP fluorescence, were analyzed for the total GluA1 cell surface fluorescence intensity (D), the GluA1 synaptic cluster fluorescence intensity (E), and number (F) per dendritic length. Results are presented as % of GFP-transfected cells and are averaged from three independent experiments (n ≥ 28 cells). Error bars, mean ± S.E. Significance, analysis of variance followed by Bonferroni's multiple comparison test. *, p < 0.05; **, p < 0.01 relative to GFP-transfected neurons. Scale bars = 10 μm; 2 μm for enlarged images.
Caspr1 Increases Glutamate-evoked Currents Mediated by GluA1
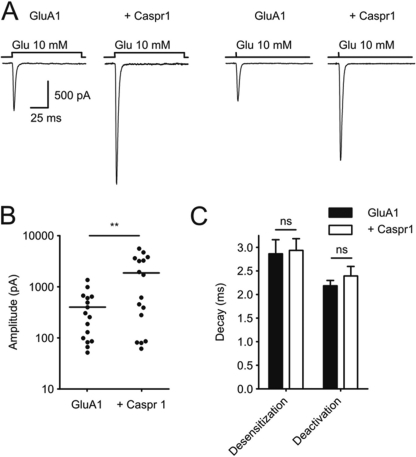
To test the functional consequences of the association of AMPA receptors with Caspr1, we expressed GluA1 with or without Caspr1 in HEK293 cells and recorded whole-cell currents activated by fast application of glutamate at a saturating concentration (10 mm) with either a long (100 ms) pulse revealing desensitization properties or a short (1 ms) pulse indicative of the deactivation kinetics (24). Although the amplitude of AMPA receptor-mediated currents was variable from cell to cell, coexpression of Caspr1 significantly increased the average amplitude of currents evoked by glutamate (527 ± 124 pA, n = 16 versus 2459 ± 622 pA, n = 16 with Caspr1, p < 0.05) without any effect on the desensitization or deactivation kinetics (Fig. 4). These results indicate that Caspr1 increases the expression of recombinant GluA1-containing AMPA receptors at the cell surface without modifying their kinetics.
FIGURE 4.
Caspr1 enhances GluA1 receptor currents. A, effect of sustained (100 ms, left traces) or brief (1 ms, right traces) applications of saturating glutamate concentration (10 mm) on GluA1-mediated currents, with or without Caspr1. B, plot of the amplitude of GluA1 currents (n = 16) and GluA1 coexpressed with Caspr1 (n = 16). Currents are activated by a long (100 ms) application of 10 mm glutamate. **, p < 0.005, Student's t test, difference between GluA1 and GluA1 coexpressed with Caspr1. C, effect of Caspr1 on GluA1 desensitization and deactivation for long or short glutamate (10 mm) applications, respectively. Statistical analysis reveals no significant differences (ns) (Student's t test, p > 0.05). Error bars indicate mean ± S.E.
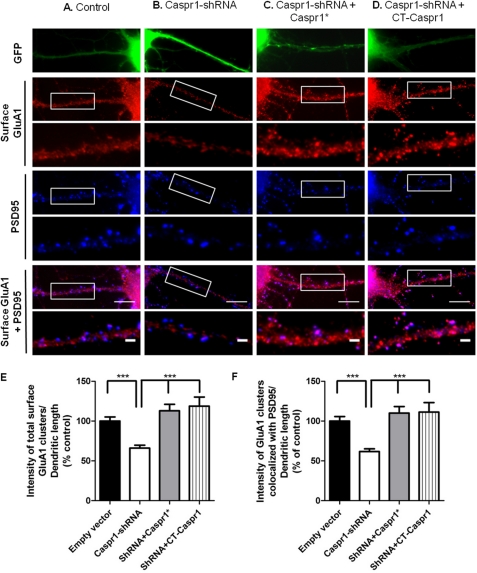
Knockdown of Caspr1 Reduces Synaptic GluA1, an Effect Rescued by the C Terminus of Caspr1
The association of Caspr1 with AMPA receptors suggests that Caspr1 might be required for the expression of AMPA receptors at synapses. To test this possibility, a shRNA sequence against rat Caspr1 mRNA was introduced in the pLentiLox3.7(CMV)EGFP vector. The Caspr1-shRNA construct decreased the expression of synaptic Caspr1 to around 32% of the endogenous expression level in transfected hippocampal neurons identified by the expression of GFP, and compared with neurons transfected with the empty vector (supplemental Fig. 3). Neurons transfected with Caspr1-shRNA showed a 34.0 ± 3.6% decrease in the intensity of cell surface total GluA1 clusters and a 38.4 ± 3.6% decrease of PSD95-colocalized cell surface GluA1 clusters when compared with neurons transfected with the control plasmid (Figs. 5A, B, E, and F). The decrease in the fluorescence intensity of GluA1 clusters was accompanied by a decrease in the area of clusters in neurons transfected with Caspr1-shRNA, whereas the number of GluA1 clusters was not changed (data not shown). To exclude the contribution of off-target effects of the Caspr1-shRNA, a rescue construct was generated with silent mutations in the Caspr1 region targeted by the Caspr1-shRNA (Caspr1*). Neurons were cotransfected with the Caspr1-shRNA plasmid and the Caspr1* construct refractory to Caspr1-shRNA-mediated knockdown and analyzed for cell surface expression of GluA1 (Fig. 5, C, E, and F). Indeed, the expression of the Caspr1* construct rescued the Caspr1-shRNA-mediated decrease on the fluorescence intensity of the cell surface total and synaptic clusters of GluA1. These results indicate that the defects observed for synaptic GluA1 levels with Caspr1-shRNA are specifically due to the loss of Caspr1. The reduction in the intensity of cell surface GluA1 clusters was also fully rescued by expression of the C terminus of Caspr1 (CD4-CTCaspr1, Fig. 5, D–F), indicating that endogenous Caspr1 is required for maintaining the cell surface and synaptic expression of GluA1 through a mechanism dependent on its C-terminal domain.
FIGURE 5.
Loss of Caspr1 decreases the synaptic content of GluA1. Hippocampal neurons in culture were transfected at 7 DIV with pLentiLox3.7 (A), with the Caspr1-shRNA construct (B), or cotransfected with the Caspr1-shRNA and the mutant Caspr1* construct resistant to the Caspr1-shRNA (C), or CD4-CTCaspr1 (D). Neurons were live-stained at 15 DIV for GluA1, fixed, and immunostained for PSD95. Transfected neurons, identified by GFP fluorescence, were analyzed for the fluorescence intensity of the total (E) and PSD95-colocalized (F) GluA1 clusters. Results are presented as % of pLentiLox3.7-transfected cells (three independent experiments, n ≥ 30 cells). Error bars indicate mean ± S.E. Significance, analysis of variance followed by Bonferroni's multiple comparison test. ***, p < 0.001 relative to neurons transfected with pLentiLox3.7. Scale bars = 10 μm; 2 μm for enlarged images.
DISCUSSION
Alterations in the functional properties and in the number of AMPA receptors at synapses are central to the processes of synaptic plasticity that underlie learning and memory formation (1). The number of AMPA receptors at synapses is regulated by receptor interaction with intracellular scaffold proteins (2) and transmembrane proteins such as TARPs, cornichons, SynDIG1, and CKAMP44 (3). TARPs affect the export of AMPA receptors from the endoplasmic reticulum and their synaptic traffic, as well as receptor gating properties (reviewed in Ref. 3). Similarly, the cornichon proteins enhance the surface expression of AMPA receptors and slow down receptor deactivation and desensitization kinetics (4), whereas CKAMP44 does not affect the cell surface expression of AMPA receptors or the amplitude of miniature excitatory postsynaptic currents (mEPSCs) but leads to stronger and faster AMPA receptor desensitization (5). SynDIG1 increases the frequency and amplitude of mEPSCs, and increases GluA1 synapse density and area in hippocampal neurons (7). In this work, we identify Caspr1 as a novel interactor for AMPA receptor subunits that, akin to SynDIG1, increases the synaptic content of GluA1.
Immunocytochemical studies in mature cultures of rat hippocampal neurons confirmed that Caspr1 distributes throughout dendrites and that > 45% of glutamatergic synapses, identified by coincident staining for PSD95 and VGLUT, contain Caspr1 (Fig. 2, A and B). These observations are in agreement with previous immunohistochemical studies in mouse hippocampal slices (15) that showed extensive colocalization of Caspr1 with MAP-2, as well as localization to membrane structures apposed to synaptophysin-positive axon terminals. Analysis of biochemically purified postsynaptic densities from cultured cortical neurons showed an enrichment of Caspr1 in the PSD (Fig. 2C), in conformity with the studies by Murai et al. (15), and with the proteomic analysis of the PSD (25). At the paranodes of myelinated axons, Caspr1 regulates the segregation of Na+ and K+ channels to the nodes of Ranvier and to the juxtaparanodes, respectively (20). The presence of Caspr1 in glutamatergic synapses (Fig. 2) and its interaction with AMPA receptor subunits, both in the brain and in heterologous system (Fig. 1), are compatible with a novel postsynaptic function for Caspr1 in regulating AMPA receptors.
In fact, we demonstrated that coexpression of Caspr1 with GluA1-homomeric AMPA receptors increases the amplitude of glutamate-evoked currents (Fig. 4) and that overexpression of Caspr1 in hippocampal neurons increases the synaptic localization of GluA1-containing AMPA receptors (Fig. 3). On the other hand, endogenous Caspr1 is required for the synaptic localization of AMPA receptors because knockdown of Caspr1 specifically reduces synaptic GluA1 (Fig. 5).
How does Caspr1 regulate the localization of AMPA receptors at the cell surface and at synapses? One possibility is that Caspr1, through its interaction with 4.1-band proteins (17), may reinforce the link between AMPA receptors and 4.1-band proteins, interactors for GluA1 and GluA4 subunits (26, 27). In fact, it has been found that the synaptic targeting of SAP97, another molecule involved in AMPA receptor traffic, depends on its interaction with the protein 4.1 (28). At the paranodal junction, Caspr1 connects through protein 4.1B to cytoskeletal components within the axon (29), including molecules such as ankyrin B, αII spectrin and βII spectrin (30). Interestingly, the interaction of Caspr1 with 4.1B is necessary for the generation of an efficient membrane barrier at the paranodal junction (31). Similarly, the association of Caspr1 with postsynaptic cytoskeleton proteins may allow it to modulate the interaction of AMPA receptors with specialized components of the cytoskeleton at the postsynaptic scaffold. Alternatively, the proline-rich region at the intracellular C terminus of Caspr1 may provide anchoring to other binding partners at the PSD.
In summary, our data support a model in which Caspr1 regulates the content of AMPA receptors at synapses in hippocampal neurons in culture. Caspr1 may constitute a new member of the growing list of AMPA receptor auxiliary proteins.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Irving (University of Dundee, UK) for the antibody against the N-terminal region of GluA1, and Dr. Catherine Faivre-Sarrailh (Université de la Méditerranée Aix-Marseille II, Marseille, France) for the Caspr1 antibody and constructs. We also thank Dr. Juan Lerma (Instituto de Neurociencias de Alicante, Spain) for the GluA1 and GluA2 constructs, Dr. Kari Keinanen (University of Helsinki, Finland) for the N-terminally FLAG-tagged GluA4 plasmid, Dr. Pavel Osten (Max Planck Institute for Medical Research, Heidelberg, Germany) for the GluA2L plasmid, Dr. Ann Marie Craig (University of British Columbia, Vancouver, Canada) for the GluN1–1a plasmid, and Dr. Volkmar Leβman (Otto-von-Guericke University, Magdeburg, Germany) for the TrkB-GFP. We also thank Elisabete Carvalho for technical assistance in the preparation of neuronal cultures.
This work was supported by the Portuguese Foundation for Science and Technology (FCT) Grants POCI/SAU-NEU/58955/2004, PTDC/BIA-BCM/71789/2006, and PTDC/BIA-BCM/113738/2009) and FCT doctoral fellowships SFRH/BD/11826/2003 (to S. D. S.), SFRH/BD/37522/2007 (to J. S. F.), and SFRH/BD/47879/2008 (to L. R.).

This article contains supplemental Figs. 1–3.
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- TARP
- transmembrane AMPA receptor regulatory protein
- IPB
- immunoprecipitation buffer
- E18
- embryonic day 18
- DIV
- days in vitro
- PSD
- postsynaptic density
- VGLUT
- vesicular glutamate transporter.
REFERENCES
- 1. Shepherd J. D., Huganir R. L. (2007) The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643 [DOI] [PubMed] [Google Scholar]
- 2. Santos S. D., Carvalho A. L., Caldeira M. V., Duarte C. B. (2009) Regulation of AMPA receptors and synaptic plasticity. Neuroscience 158, 105–125 [DOI] [PubMed] [Google Scholar]
- 3. Jackson A. C., Nicoll R. A. (2011) The expanding social network of ionotropic glutamate receptors. TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A., Heimrich B., Chisaka O., Jonas P., Schulte U., Fakler B., Klöcker N. (2009) Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 5. von Engelhardt J., Mack V., Sprengel R., Kavenstock N., Li K. W., Stern-Bach Y., Smit A. B., Seeburg P. H., Monyer H. (2010) CKAMP44. A brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science 327, 1518–1522 [DOI] [PubMed] [Google Scholar]
- 6. Kato A. S., Gill M. B., Ho M. T., Yu H., Tu Y., Siuda E. R., Wang H., Qian Y. W., Nisenbaum E. S., Tomita S., Bredt D. S. (2010) Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalashnikova E., Lorca R. A., Kaur I., Barisone G. A., Li B., Ishimaru T., Trimmer J. S., Mohapatra D. P., Díaz E. (2010) SynDIG1. An activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron 65, 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Correia S. S., Duarte C. B., Faro C. J., Pires E. V., Carvalho A. L. (2003) Protein kinase C γ associates directly with the GluR4 α-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor subunit. Effect on receptor phosphorylation. J. Biol. Chem. 278, 6307–6313 [DOI] [PubMed] [Google Scholar]
- 9. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 10. Suckau D., Resemann A., Schuerenberg M., Hufnagel P., Franzen J., Holle A. (2003) A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 376, 952–965 [DOI] [PubMed] [Google Scholar]
- 11. Jiang M., Deng L., Chen G. (2004) High Ca(2+)-phosphate transfection efficiency enables single neuron gene analysis. Gene. Ther. 11, 1303–1311 [DOI] [PubMed] [Google Scholar]
- 12. Peça J., Feliciano C., Ting J. T., Wang W., Wells M. F., Venkatraman T. N., Lascola C. D., Fu Z., Feng G. (2011) Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehlers M. D. (2003) Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6, 231–242 [DOI] [PubMed] [Google Scholar]
- 14. Ripellino J. A., Neve R. L., Howe J. R. (1998) Expression and heteromeric interactions of non-N-methyl-D-aspartate glutamate receptor subunits in the developing and adult cerebellum. Neuroscience 82, 485–497 [DOI] [PubMed] [Google Scholar]
- 15. Murai K. K., Misner D., Ranscht B. (2002) Contactin supports synaptic plasticity associated with hippocampal long-term depression but not potentiation. Curr. Biol. 12, 181–190 [DOI] [PubMed] [Google Scholar]
- 16. Peles E., Nativ M., Lustig M., Grumet M., Schilling J., Martinez R., Plowman G. D., Schlessinger J. (1997) Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 16, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menegoz M., Gaspar P., Le Bert M., Galvez T., Burgaya F., Palfrey C., Ezan P., Arnos F., Girault J. A. (1997) Paranodin, a glycoprotein of neuronal paranodal membranes. Neuron 19, 319–331 [DOI] [PubMed] [Google Scholar]
- 18. Bakkaloglu B., O'Roak B. J., Louvi A., Gupta A. R., Abelson J. F., Morgan T. M., Chawarska K., Klin A., Ercan-Sencicek A. G., Stillman A. A., Tanriover G., Abrahams B. S., Duvall J. A., Robbins E. M., Geschwind D. H., Biederer T., Gunel M., Lifton R. P., State M. W. (2008) Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 82, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peñagarikano O., Abrahams B. S., Herman E. I., Winden K. D., Gdalyahu A., Dong H., Sonnenblick L. I., Gruver R., Almajano J., Bragin A., Golshani P., Trachtenberg J. T., Peles E., Geschwind D. H. (2011) Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bhat M. A., Rios J. C., Lu Y., Garcia-Fresco G. P., Ching W., St Martin M., Li J., Einheber S., Chesler M., Rosenbluth J., Salzer J. L., Bellen H. J. (2001) Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron 30, 369–383 [DOI] [PubMed] [Google Scholar]
- 21. Keinänen K., Wisden W., Sommer B., Werner P., Herb A., Verdoorn T. A., Sakmann B., Seeburg P. H. (1990) A family of AMPA-selective glutamate receptors. Science 249, 556–560 [DOI] [PubMed] [Google Scholar]
- 22. Kessels H. W., Malinow R. (2009) Synaptic AMPA receptor plasticity and behavior. Neuron 61, 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonnon C., Goutebroze L., Denisenko-Nehrbass N., Girault J. A., Faivre-Sarrailh C. (2003) The paranodal complex of F3/contactin and caspr/paranodin traffics to the cell surface via a non-conventional pathway. J. Biol. Chem. 278, 48339–48347 [DOI] [PubMed] [Google Scholar]
- 24. Barberis A., Sachidhanandam S., Mulle C. (2008) GluR6/KA2 kainate receptors mediate slow-deactivating currents. J. Neurosci. 28, 6402–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., Grant S. G. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 [DOI] [PubMed] [Google Scholar]
- 26. Shen L., Liang F., Walensky L. D., Huganir R. L. (2000) Regulation of AMPA receptor GluR1 subunit surface expression by a 4. 1N-linked actin cytoskeletal association. J. Neurosci. 20, 7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coleman S. K., Cai C., Mottershead D. G., Haapalahti J. P., Keinänen K. (2003) Surface expression of GluR-D AMPA receptor is dependent on an interaction between its C-terminal domain and a 4.1 protein. J. Neurosci. 23, 798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rumbaugh G., Sia G. M., Garner C. C., Huganir R. L. (2003) Synapse-associated protein-97 isoform-specific regulation of surface AMPA receptors and synaptic function in cultured neurons. J. Neurosci. 23, 4567–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gollan L., Sabanay H., Poliak S., Berglund E. O., Ranscht B., Peles E. (2002) Retention of a cell adhesion complex at the paranodal junction requires the cytoplasmic region of Caspr. J. Cell Biol. 157, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ogawa Y., Schafer D. P., Horresh I., Bar V., Hales K., Yang Y., Susuki K., Peles E., Stankewich M. C., Rasband M. N. (2006) Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J. Neurosci. 26, 5230–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horresh I., Bar V., Kissil J. L., Peles E. (2010) Organization of myelinated axons by Caspr and Caspr2 requires the cytoskeletal adapter protein 4.1B. J. Neurosci. 30, 2480–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.