Background: TUG controls a specialized membrane trafficking pathway in adipocytes but is widely expressed.
Results: TUG converts p97 ATPase hexamers into monomers and controls Golgi membrane dynamics.
Conclusion: Localized disassembly of p97 hexamers may control a generalized trafficking pathway in eukaryotic cells.
Significance: Understanding how TUG and p97 function provides insight into regulation of both generalized and specialized membrane trafficking.
Keywords: ATPases, GLUT4, Golgi, Membrane Trafficking, Molecular Cell Biology, Protein Assembly, Protein Domains, Protein Motifs, Protein Translocation, Ubiquitin
Abstract
p97/VCP is a hexameric ATPase that is coupled to diverse cellular processes, such as membrane fusion and proteolysis. How p97 activity is regulated is not fully understood. Here we studied the potential role of TUG, a widely expressed protein containing a UBX domain, to control mammalian p97. In HEK293 cells, the vast majority of TUG was bound to p97. Surprisingly, the TUG UBX domain was neither necessary nor sufficient for this interaction. Rather, an extended sequence, comprising three regions of TUG, bound to the p97 N-terminal domain. The TUG C terminus resembled the Arabidopsis protein PUX1. Similar to the previously described action of PUX1 on AtCDC48, TUG caused the conversion of p97 hexamers into monomers. Hexamer disassembly was stoichiometric rather than catalytic and was not greatly affected by the p97 ATP-binding state or by TUG N-terminal regions in vitro. In HeLa cells, TUG localized to the endoplasmic reticulum-to-Golgi intermediate compartment and endoplasmic reticulum exit sites. Although siRNA-mediated TUG depletion had no marked effect on total ubiquitylated proteins or p97 localization, TUG overexpression caused an accumulation of ubiquitylated substrates and targeted both TUG and p97 to the nucleus. A physiologic role of TUG was revealed by siRNA-mediated depletion, which showed that TUG is required for efficient reassembly of the Golgi complex after brefeldin A removal. Together, these data support a model in which TUG controls p97 oligomeric status at a particular location in the early secretory pathway and in which this process regulates membrane trafficking in various cell types.
Introduction
Mammalian p97/VCP and its yeast and plant orthologs, termed CDC48, are hexameric AAA ATPase family members. These ATPases associated with diverse cellular activities couple ATP hydrolysis to mechanical force to generate a variety of effects on various substrate proteins (1). p97 has been implicated in homotypic membrane fusion, ER3-associated degradation, mitotic spindle disassembly, gene expression, and chromatin remodeling (2–5). ATP-driven conformational changes in the p97 hexamer are coupled through adaptor proteins to configure protein complexes and to regulate protein degradation (6–11).
Ubiquitin regulatory X (UBX) domain-containing proteins bind p97/VCP/CDC48, typically through a ubiquitin-like UBX domain near their C termini, and recruit p97 ATPase activity to various cellular targets (3). Outside of the UBX domain itself, these proteins have little similarity, and they have a wide variety of other interacting partners and functions (3, 12–16). The roles of some UBX family members, such as that of p47 to recruit p97 in membrane fusion processes, have been relatively well studied (6, 17–21). The functions of several other UBX proteins are less well understood. Both p97 and many UBX proteins are highly conserved, with orthologs in plants and fungi. These pairings play fundamental roles in cell division, protein homeostasis, and membrane trafficking.
TUG (tether, containing a UBX domain, for GLUT4; also termed ASPL, UBXD9, RCC17) belongs to the family of UBX-domain containing proteins and regulates the trafficking of GLUT4 glucose transporters in 3T3-L1 adipocytes (22–26). GLUT4 is regulated by a cell type-specific mechanism, which is probably an adaptation of a more general trafficking pathway present broadly in eukaryotic cells. TUG is expressed in a wide range of cell types (26–28) and may participate in such a pathway, but how TUG may direct p97 activity is not understood. Here, we approached this question by characterizing the TUG-p97 interaction. We noted that the C-terminal, UBX-containing region of TUG resembles the Arabidopsis protein, PUX1, which was shown to disassemble AtCDC48 hexamers (29, 30). We show that mammalian TUG similarly converts p97 hexamers into monomers, and we characterize requirements for this action. Finally, we provide evidence that TUG resides at the ER-Golgi intermediate compartment (ERGIC) and at ER exit sites (ERES) in HeLa cells and that its activity is essential for Golgi dynamics.
EXPERIMENTAL PROCEDURES
Reagents, Cell Culture, and Molecular Biology
Antisera against TUG were described previously (26) and were affinity-purified using the immobilized peptide immunogen (Pierce). This peptide is uniquely encoded in rodent and human genomes. TUG antisera were used at dilutions of 1:1,000–2,000 on Western blots and 1:200 for immunofluorescence. Other antibodies include those directed to HA epitope (Covance), FLAG epitope (M2, Sigma), ERGIC-53 (Enzo Life Sciences), GM130 (BD Transduction Laboratories), Vti1a (BD Transduction Laboratories), Hsp90 (BD Transduction Laboratories), anti-GST (Millipore), transferrin receptor (Invitrogen), ubiquitin (Covance), and p97/VCP (Abcam and Maine Biotechnology Services). ATP, ADP, and ATPγS were purchased from Sigma and used at a final concentration of 1 mm. MG-132 was purchased from American Peptide and was used at a concentration of 10 μm for 2 h at 37 °C. Iodoacetamide was purchased from Sigma and used at a final concentration of 20 mm in cell lysis buffer.
HeLa cells were grown in DMEM (Invitrogen) containing 10% fetal bovine serum and supplemented with 2 mm l-glutamine. HEK293 cells were grown in DMEM containing 10% bovine growth serum (Hyclone) and supplemented with 2 mm l-glutamine.
To express truncated proteins, PCR was done using oligonucleotides corresponding to the residues indicated for each construct, and products were cloned in bacterial or eukaryotic expression vectors. The N-terminal His6-HA-tagged p97 and truncation mutants were generated by PCR amplification of human p97 clone (31) (Addgene plasmid 23971) and were cloned using BamHI and NotI into the pcDNA3.1 or pET28a vectors. His6-, GST-, or FLAG-tagged full-length and truncation mutants of TUG were generated by PCR amplification of the murine TUG cDNA (26). These tags were at the N terminus, and the coding sequences were cloned using EcoRI and NotI into pcDNA3.1, pBICD2, or pET28a vectors or using BamHI and EcoRI into the pGEX-KG vector, as described previously (25). All constructs were verified by sequencing. Synthetic siRNAs (purchased from Dharmacon) had the following sequences: TUG siRNA A, 5′-CCCUGUGAAUAUGAUCUGAUU-3′; TUG siRNA B, 5′-GCAGGACUCUUUCUGUUCAUU-3′; control (scrambled) siRNA, 5′-CGUACGCGGAAUACUUCGA-3′.
Sequence alignments of TUG and PUX1 (NM_113645.2) were done using Praline (32). The N-terminal regions of TUG were not included. More conserved residues are marked by progressively warmer colors (yellows, oranges, and reds) and less conserved residues are indicated by progressively cooler colors (greens, blues, and purples) in supplemental Fig. 1.
Immunofluorescence Microscopy
Cells were plated on coverslips coated with poly-d-lysine (Sigma) in 6-well dishes (Corning Glass). Two hours prior to fixation, cells were incubated in phenol red-free DMEM (Invitrogen) containing 10% serum. Cells were washed once with phosphate-buffered saline, pH 7.4 (PBS), and then fixed using 4% paraformaldehyde for 30 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 4% normal goat serum (Jackson Immunoresearch) in PBS for 30 min at room temperature. Staining with primary antibodies was done at dilutions of 1:200 in 4% normal goat serum for 1 h at room temperature and was followed by three washes with PBS. Alexa-conjugated goat anti-mouse IgG or anti-rabbit IgG secondary antibodies (Alexa488 or Alexa594, Molecular Probes) were used at dilutions of 1:200 in 4% normal goat serum in PBS for 1 h. Coverslips were then washed three times in PBS and mounted on slides using Prolong Gold Antifade with or without DAPI (Molecular Probes/Invitrogen). Brefeldin A (Cell Signaling Technology) was prepared in DMSO and used at a final concentration of 10 μg/ml. Nocodazole (Sigma) was prepared in DMSO and used at a final concentration of 30 μm. The VSVG-ts045-YFP and Sec13-GFP constructs were kind gifts from the Caplan and Gorelick laboratories, respectively, and were transfected as indicated. For wide field epifluorescence microscopy, images were acquired on a Zeiss Axiovert 100 M microscope using ×40/1.30 or ×63/1.40 oil immersion objectives, and a cooled CCD camera was driven by Zeiss Axiovision software. For confocal microscopy, images were acquired on a Zeiss Axiovert LSM 510 confocal microscope using a ×63/1.20 water immersion objective, and the pinhole was set for an Airy unit of 1.0. To quantify effects on Golgi reassembly, GM130 mean fluorescence intensities were measured in two regions of each cell, first in the perinuclear region, and second in a larger area covering the entirety of the cell. This analysis was done using ImageJ, and 19–24 cells were examined for each condition. For each cell, the ratio of perinuclear to whole cell mean fluorescent intensities was calculated. The mean ratio ± S.E. was then plotted for each condition. Significant differences were assessed using a two-tailed t test.
Transfection and Biochemistry
HEK293 or HeLa cells were transfected with plasmid DNA or siRNA duplexes using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions. Cells were lysed in 0.5% Nonidet P-40, 20 mm HEPES, pH 7.4, 150 mm NaCl containing Complete EDTA-free protease inhibitors (Roche Applied Science). Lysate from one 10-cm plate of cells was used for each immunoprecipitation. Insoluble debris was removed by centrifugation for 10 min at 1,000 × g at 4 °C. Immunoprecipitations were done using 10 μl of antibody or 30 μl of FLAG M2 affinity matrix (Sigma) for 1 ml of lysate for 2 h at 4 °C. Immune complexes were pelleted using Protein G-agarose beads (Pierce) at 500 × g. Immunoprecipitates were washed six times with lysis buffer and eluted in 2× SDS-PAGE sample buffer (Invitrogen) for 10–15 min at 37 °C. Samples were analyzed by SDS-PAGE and immunoblotting. For experiments in which cells were lysed in denaturing conditions, boiling lysis buffer containing 1% SDS, 20 mm Tris pH 8.0, 150 mm NaCl, 0.5 mm EDTA was used. After brief sonication, insoluble debris was pelleted, and samples were analyzed by SDS-PAGE and immunoblotting.
Pull-downs using recombinant proteins were performed in the Nonidet P-40 lysis buffer noted above. 10–50 μg of GST- or His-tagged proteins were incubated in a final volume of 1 ml for 1–2 h, followed by 1 h of incubation with glutathione beads (GE Healthcare) or HisPur Cobalt (Pierce) beads at 4 °C. Beads were washed six times in lysis buffer, eluted in 2× SDS-PAGE sample buffer (Invitrogen), and analyzed by SDS-PAGE and immunoblotting.
For SDS-PAGE and immunoblotting, samples were solubilized in 2× LDS sample buffer or native gel sample buffer (Invitrogen) and separated using 4–12% Novex NuPAGE or 4–20% native polyacrylamide gels (Invitrogen). Proteins were transferred to nitrocellulose using a Bio-Rad semidry transfer apparatus. Membranes were blocked using 5% milk in PBS for 10–30 min. Primary and secondary antibodies were used for 1 h each, and three 10-min washes were done after each incubation using PBS with 0.05% Tween 20. All steps were performed at room temperature. Proteins were detected using chemiluminescence as described previously (25) or using infrared fluorescence on a LI-COR Odyssey imaging system.
Recombinant Protein Expression and Pull-downs
His6- and GST-tagged constructs were expressed in BL21(DE3)pLys GOLD Escherichia coli (Agilent Technologies). Bacterial cultures growing at 37 °C were induced with isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mm once an OD of 0.6 was reached. After 3–4 h, bacteria were lysed in 50 mm Tris, pH 8.0, 300 mm NaCl, including 10 mm imidazole for His-tagged proteins. Bacterial lysates were sonicated, and insoluble debris was pelleted at 16,000 × g for 20 min. Protein from supernatant were purified using HisPur Cobalt Resin (Pierce), Ni2+-NTA-agarose (Qiagen), or glutathione-Sepharose 4B (GE Healthcare) as appropriate. GST-tagged proteins were eluted in 20 mm reduced glutathione, and His6-tagged proteins were eluted in 150 mm imidazole (lysate buffer was used). In both cases, proteins were dialyzed (Slide-A-Lyzer, Pierce) in 150 mm NaCl, 50 mm Tris, pH 7.6, 5% glycerol overnight. Protein concentrations were determined using both an EZQ protein quantitation kit (Invitrogen) and SDS-PAGE with Gelcode Coomassie staining (Pierce) with comparison against known protein standards.
Sucrose Gradient Sedimentation
Samples of 1 ml each were loaded on the top of 0.5–1.2 m sucrose gradients made up in 20 mm HEPES, pH 7.4, 150 mm KCl, 1 mm MgCl2. Analysis of recombinant proteins was done without detergent; however, similar results were obtained when 0.5% Nonidet P-40 was included. Lysates from FLAG-TUG (or mock)-transfected HEK293 cells were prepared 24 h after transfection using 0.5% Nonidet P-40 and were analyzed on gradients containing 0.5% Nonidet P-40. All gradients were prepared by hand by layering 1-ml volumes, in which the sucrose concentrations were successively reduced in each 0.1 m step, to reach a final volume of 8 ml. Samples were centrifuged at 128,000 × g in a SW 41 Ti rotor (Beckman Coulter) for 16–20 h at 4 °C. Fractions of 0.5 ml each were collected by hand, starting at the top of the gradients, and were analyzed by SDS-PAGE and immunoblotting. Standard control proteins, including GST (26 kDa), ovalbumin (45 kDa; Invitrogen), BSA (64 kDa and dimer at 130 kDa; American Bioanalytical), Apoferritin (450 kDa; MP Biomedicals), and thyroglobulin (660 kDa; Calbiochem), were centrifuged in parallel gradients.
For analysis of the oligomeric status of p97, GST-TUG and His6-p97 proteins were expressed in a BL21(DE3)pLys GOLD E. coli isopropyl-β-d-thiogalactopyranoside-inducible system. Varying molar ratios were used, with most experiments done using 3:1 TUG/p97. In experiments using ADP, ATP, or ATPγS, the final concentration of nucleotide was 1 mm. All reactions were incubated on ice for 30 min prior to velocity sedimentation on sucrose gradients.
RESULTS
Interaction of TUG and p97
We identified p97 by mass spectrometry of 3T3-L1 adipocyte proteins that bound to GST-TUG. To confirm the interaction, we coimmunoprecipitated the endogenous TUG and p97 proteins from lysates of HEK293 cells (Fig. 1A). p97 is extremely abundant, and quantitative analyses indicated that only a small fraction (∼5–10%) of total cellular p97 was bound to TUG. By contrast, a large fraction (∼85–90%) of total cellular TUG was bound to p97, as assessed by quantification of the converse immunoprecipitations. The nearly quantitative recovery of TUG with p97 suggests that this interaction has very high affinity and that p97 is a primary binding partner for TUG.
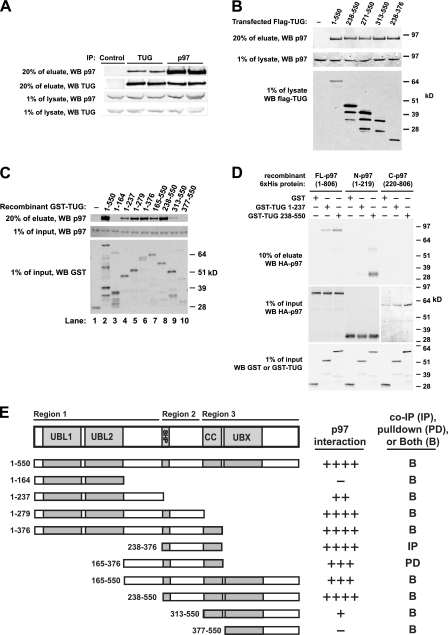
FIGURE 1.
Interaction of TUG and p97. A, anti-TUG or anti-p97 antibodies were used to immunoprecipitate (IP) endogenous TUG and p97 from HEK293 cells lysed using 0.5% Nonidet P-40. Control IgG was used in lane 1, and duplicates of TUG (lanes 2 and 3) and p97 (lanes 4 and 5) immunoprecipitates are shown. Eluates and preimmunoprecipitation lysates were immunoblotted to detect TUG and p97, as indicated. B, HEK293 cells were transfected to express FLAG-tagged TUG proteins containing the indicated residues and lysed using 0.5% Nonidet P-40, and immunoprecipitations were performed using an anti-FLAG affinity matrix. Endogenous p97 was immunoblotted in eluates and pre-immunoprecipitation lysates, as indicated. C, recombinant GST or GST-TUG proteins containing the indicated TUG residues were used to pull down recombinant His6-p97 in 0.5% Nonidet P-40 buffer. Eluates and input proteins were immunoblotted using anti-p97 and anti-GST antibodies as indicated. D, GST or GST-TUG proteins were used to pull down His6-HA-tagged full-length, N-terminal, or C-terminal p97 proteins in 0.5% Nonidet P-40 buffer. Eluates and inputs were immunoblotted using anti-GST and anti-HA antibodies as indicated. Residues present in each p97 and TUG protein are indicated. E, diagram summarizes binding of p97 to TUG proteins containing the indicated residues. For reference, the overall TUG domain structure is shown at top. The relative amount of p97 that associated with each TUG protein is indicated at the right. Binding was tested using both GST pull-downs (PD; as in C and D) and by immunoprecipitation (IP; as in B). Most interactions were tested both ways, as indicated (B), and similar results were obtained using both methods. Each construct was tested at least twice, with most tested three times. WB, Western blot.
TUG contains three potential p97-interacting sites, including an N-terminal ubiquitin-like (UBL) domain (33) referred to here as “UBL1,” an 8-residue SHP box (5, 34), and a UBX domain, as noted above. Within the 550-residue murine TUG protein, these regions reside at positions 10–83, 260–267, and 382–457, respectively, and an additional likely UBL/UBX domain (“UBL2”) is present at residues 92–164. To test if TUG binds p97 through one or more of these regions, we used truncated proteins. Initially, we expressed FLAG-tagged TUG proteins in HEK293 cells, as shown in Fig. 1B, and coimmunoprecipitated endogenous p97. Truncation of the TUG N terminus (lane 3) and SHP box (lanes 4 and 5) did not greatly alter the amount of p97 that was copurified in these experiments. In addition, modest expression of a small construct containing only the SHP box and flanking residues coimmunoprecipitated p97 in amounts similar to those observed using full-length TUG. These results suggested that TUG may interact with p97 through more than one binding region.
We used recombinant proteins to show that TUG-p97 binding is direct and to define the interacting regions. Fig. 1C presents a representative immunoblot, in which various GST-TUG proteins were used to pull down full-length His6-p97. An N-terminal TUG fragment containing residues up to, but not including, the SHP box (residues 1–237) pulled down p97 (lane 4); however, further truncations of this region did not interact. Constructs containing the 8-residue SHP box (residues 1–279, residues 1–376, residues 165–550, and residues 238–550) purified p97 most effectively (lanes 5–8). Moreover, as described below, proteins containing mutations in the SHP box bound to p97 much less well. Interestingly, a C-terminal fragment of TUG containing the UBX domain (residues 377–550) was unable to pull down p97 (lane 10). The TUG UBX domain lacks a key phenylalanine residue that mediates p97 binding by other UBX domains and contains a proline at this position (residue 433), which may account for the reduced binding (20, 35, 36). Inclusion of residues immediately upstream of the UBX domain (residues 313–376), which are predicted to form a coiled-coil, conferred a weak interaction with p97 (residues 313–550; lane 9) but alone did not interact with p97 (not shown). These data indicated that the interaction of TUG with p97 involves multiple regions in TUG and requires sequences N-terminal to the UBX domain, as described further below.
To test which region(s) of p97 interact with TUG, we used full-length, N-terminal, and C-terminal p97 proteins. Full-length TUG was susceptible to degradation during purification of the recombinant protein, and we therefore used N- and C-terminal TUG fragments to determine if binding occurred. As shown in Fig. 1D, GST fusions to both TUG fragments pulled down full-length p97 (residues 1–806). The p97 N terminus (residues 1–219) bound strongly to the TUG C terminus (residues 238–550) and was also copurified to a much lesser degree with the TUG N terminus (residues 1–237). This C-terminal TUG construct contains both the SHP box and the UBX domain. Neither half of TUG bound to the C-terminal portion of p97 (residues 220–806). We conclude that multiple regions of TUG bind to the N-terminal domain of p97.
Fig. 1E summarizes data from several interaction experiments using both GST pull-down and coimmunoprecipitation, which support the idea that three contiguous regions of TUG mediate its interaction with p97. The TUG N terminus, containing UBL1 and UBL2, binds p97 only in the presence of more downstream residues (Region 1). The central region of TUG contains the SHP box, and mediates a second interaction (Region 2). The TUG C terminus mediates a third interaction and requires a predicted coiled-coil motif upstream of the UBX domain (Region 3). Of these, the central region containing the SHP box purifies p97 most efficiently and presumably has the highest affinity for p97. It seems most likely that these regions form an extended interface with the p97 N terminus and confer the nearly quantitative association of TUG with p97 we observed in cells.
TUG Mediates p97 Hexamer Disassembly
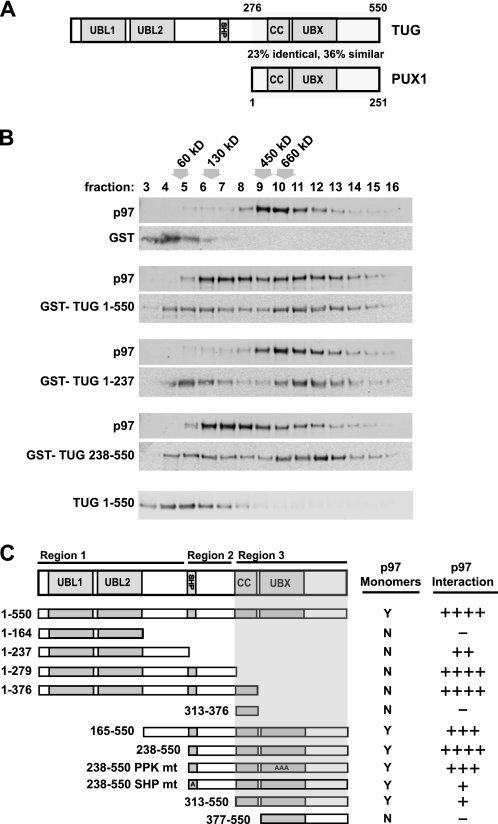
We noted that the C-terminal half of TUG resembles the Arabidopsis thaliana protein, PUX1, as depicted in Fig. 2A. Alignment of the amino acid sequences revealed that TUG and PUX1 are ∼23% identical and ∼36% similar in this region (supplemental Fig. 1). PUX1 is thought to regulate the oligomeric status of the Arabidopsis p97 ortholog, AtCDC48, and it causes disassembly of this hexamer into its monomeric subunits in vitro (29, 30). Thus, we considered whether TUG may function analogously to disassemble mammalian p97 hexamers into monomeric subunits.
FIGURE 2.
TUG generates p97 monomers. A, diagrams of the domain structures of TUG and PUX1; a region of similarity is indicated. The full alignment is shown in supplemental Fig. 1. B, recombinant His6-p97 proteins were incubated with GST alone or with the indicated GST-TUG proteins in equimolar amounts. After 30 min at 4 °C, p97 complexes were separated by sedimentation on sucrose gradients and analyzed by immunoblotting for p97 and TUG, as indicated. Fractions are numbered from the top of the gradient, and the positions of molecular weight standards are indicated above the fraction numbers. The position on the gradient of TUG alone is shown at the bottom. C, the diagrammed TUG proteins were expressed as recombinant GST fusions and were tested for their ability to generate p97 monomers using sedimentation. A 3:1 molar ratio of TUG/p97 proteins was used. All constructs were tested at least twice. The shaded area corresponds to the region of TUG that was necessary for generation of p97 monomers. For reference, the relative ability of each TUG construct to copurify p97 in pull-down and/or coimmunoprecipitation experiments is shown at the right.
Fig. 2B shows that mammalian TUG caused the generation of p97 monomers, as assessed by sedimentation on 0.5–1.2 m sucrose density gradients. In these and subsequent experiments, the samples were loaded at the top of the gradient and were centrifuged for 16 h at 4 °C. Recombinant p97 protein formed the expected hexameric complex (∼530 kDa) in the presence of GST alone and was most abundant in fractions 9–11 (Fig. 2B, top pair of panels). When p97 was incubated with GST-tagged full-length TUG (residues 1–550), p97 sedimented at a much lower molecular weight and appeared in fractions 6–8 (second pair of panels). Similar results were obtained using a truncated form of TUG, containing only C-terminal residues 238–550 (fourth pair of panels). In these samples, the p97 proteins in fractions 6–7 cosedimented near the ∼130 kDa marker and may be p97 monomers or p97-TUG heterodimers. The TUG N terminus (residues 1–237), which is not conserved in PUX1, did not produce p97 monomers (third pair of panels). Rather, this fragment shifted the p97 hexamers to slightly higher molecular weight fractions, suggesting that it associates with but does not disassemble the p97 hexamer. Immunoblots of TUG blots were consistent with this interpretation and are shown below each p97 immunoblot. As a control, TUG alone is found mostly in fractions 4–6. Binding of TUG to p97 can draw TUG up into higher molecular weight fractions. Together with the results in Fig. 1, the data show that TUG 1–237 bound but did not disassemble p97 hexamers, whereas intact TUG 1–550 and TUG 238–550 bound and also disassembled p97 hexamers in vitro.
To further characterize the TUG sequences that mediate p97 hexamer disassembly, we tested several additional truncated forms, using sedimentation on sucrose gradients. As summarized in Fig. 2C, the C-terminal region of TUG was both necessary and sufficient for p97 disassembly. This portion corresponds to PUX1, and all constructs containing it generated p97 monomers with similar efficiency. In contrast, constructs that lacked the UBX domain did not produce monomers but still bound to hexameric p97 and increased its abundance in higher molecular weight fractions.
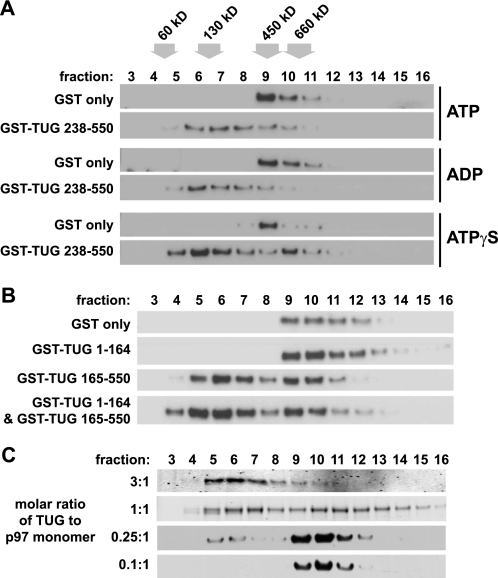
We next tested if the nucleotide-bound state of the p97 ATPase affects its dissociation by TUG. After preincubating p97 hexamers with ATP, ADP, or ATPγS, we added TUG and used sedimentation to assess p97 monomer generation. Neither ATP nor ADP had much effect on the ability of TUG to produce p97 monomers (Fig. 3A, top and middle). The non-hydrolyzable analog, ATPγS, also did not inhibit the ability of TUG to generate p97 monomers (Fig. 3A, bottom). In the ATPγS samples, there was a slight excess of p97 in the hexamer range (fraction 10), as compared with samples incubated with ATP or ADP. This complex may contain TUG bound to the p97 hexamer because it resides one fraction higher than p97 hexamer alone (fraction 9). The main point is that TUG caused the production of p97 monomers regardless of the nucleotide-bound state of p97.
FIGURE 3.
p97 hexamer disassembly in vitro is not affected by nucleotides or by TUG N-terminal domains and requires stoichiometric amounts of TUG. A, recombinant His6-p97 was preincubated with ATP, ADP, or ATPγS at final concentrations of 1 mm for 10 min at 4 °C, and then GST or GST-TUG 238–550 was added, and the incubation was continued for an additional 30 min at 4 °C. The molar ratio of TUG to p97 was 3:1. p97 oligomeric status was analyzed by sedimentation on sucrose density gradients, followed by immunoblotting of the fractions as indicated. The positions of molecular weight markers are shown at the top. The experiment was repeated three times with similar results. B, recombinant His6-p97 was incubated for 30 min at 4 °C with GST alone or with GST TUG 1–164 and/or GST TUG 165–550, as indicated. Molar ratios of 3:1 (TUG/p97) were used. p97 complexes were then separated by sedimentation on sucrose gradients, and the fractions were immunoblotted to detect p97. The experiment was performed twice with similar results. C, GST-TUG 238–550 was incubated with His6-p97 at the indicated molar ratios of TUG/p97 monomer, and then complexes were sedimented on sucrose gradients. Fractions were immunoblotted to detect p97.
We wondered if TUG N-terminal regions might serve a regulatory role to enhance or limit monomer formation by the TUG C-terminal region. In the data presented thus far, the presence or absence of sequences in the N-terminal half of TUG had little effect on p97 monomer formation. To further test if N-terminal regions might regulate p97 disassembly, we performed experiments in which we reconstituted full-length TUG in trans. One such example is shown in Fig. 3B. TUG 1–164 did not cause monomer generation (second panel), whereas TUG 165–550 very effectively led to p97 monomers (third panel). When these constructs were first incubated together and then added to p97, there was little change in the ability of TUG 165–550 to cause monomer formation (fourth panel). This result was also obtained for other constructs tested in trans (e.g. TUG 1–237 and TUG 238–550; not shown). Conversely, monomer formation was not rescued when TUG 1–376 and TUG 377–550 were added together. Together with data summarized in Fig. 2C, these results suggest that the predicted coiled-coil motif must be linked in cis to the UBX domain to produce monomers. We conclude that the N-terminal regions of TUG do not regulate the efficiency of p97 disassembly in vitro.
We tested various molar ratios of TUG to p97 monomer, to determine if similar amounts of TUG and p97 are required for hexamer disassembly. As shown in Fig. 3C, p97 monomer generation was minimal at TUG/p97 molar ratios of 0.1:1 or 0.25:1 but was progressively more marked at ratios of 1:1 and 3:1. These data support the idea that TUG acts stoichiometrically, rather than catalytically, to produce p97 monomers.
Monomerization of Endogenous p97 in Cells
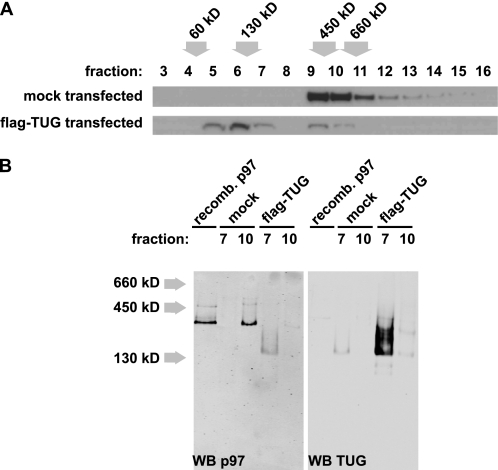
Because the above studies were performed in vitro, we next tested if TUG could generate p97 monomers in cells. Fig. 4A shows that transfection of FLAG-tagged TUG in HEK293 cells caused the nearly quantitative conversion of endogenous p97 hexamers into monomers, as assessed by sedimentation on sucrose gradients. These experiments were also repeated with N- and C-terminal TUG fragments, with results similar to those in Fig. 2C. Thus, endogenous p97 can be acted upon by TUG, despite the possible presence of post-translational modifications and other cofactors present in vivo (37). Monomeric subunits of p97 are thought not to have ATPase activity (29, 38). Therefore, TUG-mediated hexamer disassembly may occur in a controlled or localized manner in cells and may constitute a novel mechanism for regulation of p97 activity in vivo.
FIGURE 4.
Transfected TUG generates and binds p97 monomers. A, HEK293 cells were transfected with FLAG-tagged TUG and lysed using 0.5% Nonidet P-40. Postnuclear supernatants were analyzed by sedimentation on sucrose density gradients, and fractions were immunoblotted to detect p97. The positions of molecular weight standards are indicated. The experiment was repeated with similar results. B, proteins in fractions 7 and 10 from the gradients in A were separated using native PAGE and then immunoblotted as indicated. Recombinant His6-p97 was included in the first lane for comparison, and the position of molecular weight standards is indicated at the left. Western blots (WB) of p97 and TUG were done on the same membrane and were imaged at different wavelengths using a LI-COR Odyssey imaging system. The experiment was repeated with similar results.
p97 proteins isolated from eukaryotic cells, or produced recombinantly in bacteria, are present almost exclusively in hexameric form (29, 30, 38). To test if TUG binds p97 monomers as well as hexamers, we performed sequential separations of cell lysates, as shown in Fig. 4B. We first used sedimentation on sucrose gradients as above. We then used native PAGE to analyze fractions containing predominantly monomeric p97 (fraction 7) or hexameric p97 (fraction 10). We found that endogenous p97 hexamers from HEK293 cells comigrated with recombinant p97 proteins on native PAGE (Fig. 4B, left, lanes 1 and 3). Endogenous TUG ran at an unexpectedly large size on native gels (Fig. 4B, right, lane 2). Transient transfection of TUG caused p97 signal to appear in fraction 7, consistent with the production of p97 monomers. p97 in this fraction migrated as a lower molecular weight smear rather than as a tight band of higher molecular weight (Fig. 4B, left, lane 4). The overexpressed TUG was also present in this fraction and ran as an identical smear on immunoblots of native polyacrylamide gels probed with both antibodies (Fig. 4B, right, lane 4). Together, these data indicate that TUG can bind monomers of endogenous p97 in cells.
Subcellular Localization of TUG in HeLa Cells
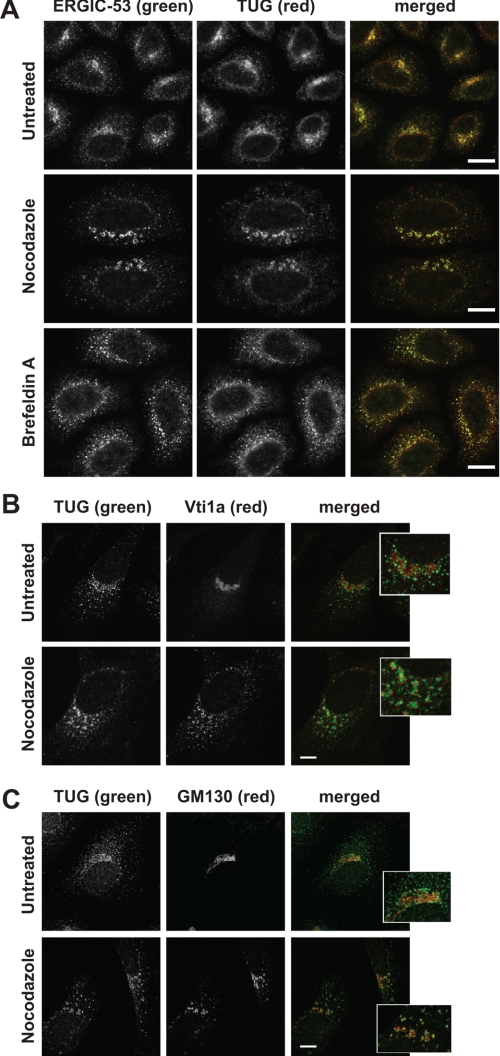
Hexameric p97 is one of the most abundant complexes in cells and is present at multiple locations. We hypothesized that TUG may specifically disassemble p97 hexamers at a particular site. We used immunofluorescence microscopy to determine if TUG had a unique staining pattern, as is observed for other UBX domain-containing proteins (14, 16, 39). Initial studies of HeLa cells showed that TUG was present in both a perinuclear location and in peripheral punctae. This pattern was similar to that observed in 3T3-L1 adipocytes, and the specificity of the antibody was further supported by competition experiments using the immunizing peptide (26) (supplemental Fig. 2). This staining was reminiscent of that of ERES and the ERGIC. Therefore, we costained ERGIC-53, a cargo receptor that is a marker for these compartments, using coimmunofluorescence microscopy (40). Fig. 5A shows that ERGIC-53 localized to peripheral ERES as well as perinuclear ERES/ERGIC. TUG colocalized extensively with ERGIC-53 in both the perinuclear area and in the peripheral punctae, suggesting that it resides specifically at the ERES and ERGIC (Fig. 5A, top panels).
FIGURE 5.
TUG colocalizes with ERGIC-53 by confocal microscopy. A, endogenous TUG and ERGIC-53 were detected in HeLa cells using indirect immunofluorescence and confocal microscopy. Cells were untreated or treated with 30 μm nocodazole or 10 μg/ml brefeldin A for 2 h prior to staining, as indicated. B and C, endogenous TUG and Vti1a (B) and TUG and GM130 (C) were imaged as in A. The insets show enlargements of the perinuclear region of the merged images. Scale bars, 10 μm.
To evaluate this colocalization further, we used nocodazole and brefeldin A (BFA) to alter ERGIC and Golgi morphology. Nocodazole disrupts the microtubule-dependent trafficking between these compartments, and treatment with this drug for 2 h redistributed ERGIC-53 into fragmented, ringlike structures (Fig. 5A, middle panels). TUG was similarly redistributed and remained highly colocalized with ERGIC-53 in nocodazole-treated cells. BFA inhibits the cycling of ARF GTPases, and prolonged treatment with this compound collapses Golgi and ERGIC markers into endosomes and the ER (41, 42). In cells treated with BFA for 2 h, ERGIC-53 was scattered into larger peripheral structures, with reduced perinuclear and slightly increased ER staining (Fig. 5A, bottom panels). TUG was also redistributed and remained highly colocalized with ERGIC-53 in the BFA-treated cells. These data support the notion that TUG resides at ERGIC and ERES membranes in HeLa cells.
To further substantiate the localization of TUG to early and not late compartments of the secretory pathway, we costained HeLa cells for TUG together with Vti1a or GM130. Vti1a is a SNARE protein that localizes primarily to the trans-Golgi network (43, 44). As shown in Fig. 5B, TUG was present near structures staining for Vti1a but did not colocalize with this protein. When cells were treated with nocodazole for 2 h, Vti1a staining became fragmented and peripherally distributed and remained distinct from that of TUG. By contrast, the cis-Golgi marker GM130 (45, 46) was present primarily in the perinuclear region and overlapped only slightly with TUG, consistent with the close juxtaposition of ERGIC and cis-Golgi compartments (Fig. 5C). GM130 also cycles through a subdomain of the ERGIC, and the overlap may occur at this location (47, 48). Most perinuclear TUG staining surrounded structures containing GM130, and the peripheral TUG punctae had no relationship to GM130. Nocodazole treatment dispersed GM130, and TUG maintained its close association and some areas of overlap. These data show that TUG is not highly concentrated on Golgi or trans-Golgi network membranes.
As an additional test of TUG localization in HeLa cells, we used VSVG-ts045-YFP (49, 50). This marker cannot exit the ER when the cells are maintained at 40 °C but moves to the Golgi and to subsequent compartments at 32 °C. In transfected HeLa cells, VSV-G and endogenous TUG colocalized maximally at 4 min after release of the ER exit block (supplemental Fig. 3, A–C). Finally, we also used Sec13, a component of the COPII coat found at ERES, in colocalization studies. A tagged Sec13-GFP protein has been described previously and recapitulates this localization (51). Native TUG colocalized with transfected Sec13-GFP in HeLa cells (supplemental Fig. 3D). Together, the data support the idea that TUG is present at early compartments of the secretory pathway, specifically at ERES and ERGIC, and that it does not localize to late Golgi or trans-Golgi network membranes in HeLa cells.
TUG Overexpression and Knockdown in HeLa Cells
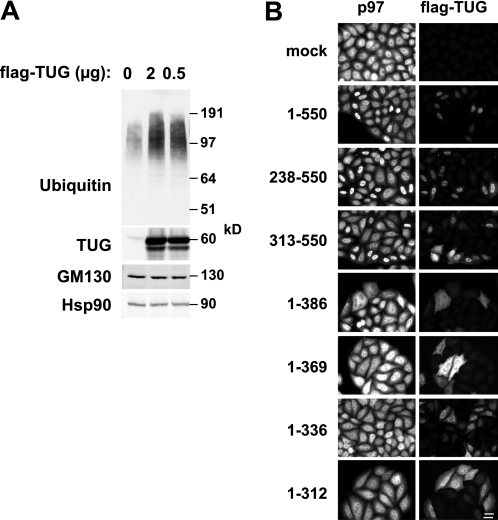
Data shown above revealed that transfected TUG had a marked effect to disassemble p97 hexamers in cells (Fig. 4), which presumably inhibits p97 ATPase activity. To test if this has functional consequences for p97-dependent processes in cells, we examined HeLa cells in which TUG was overexpressed using transient transfection. Fig. 6A shows that in transfected cells, the total abundance of polyubiquitylated proteins was increased. Immunoblots using an antibody to the TUG C terminus showed that TUG was overexpressed by ≥50-fold, compared with the abundance of endogenous TUG protein (Fig. 6A, second panel). No effects on GM130 abundance were observed, and Hsp90 served as a loading control. The data are consistent with the idea that p97 hexamer disassembly results in the accumulation of polyubiquitylated substrates.
FIGURE 6.
TUG overexpression affects ubiquitylated protein abundance and p97 distribution. A, HeLa cells were transfected using the indicated amounts of a plasmid to express full-length, FLAG-tagged TUG. Cells were lysed 48 h after transfection, and immunoblots were done to detect the indicated proteins. An antibody to the TUG C terminus was used to detect endogenous and transfected TUG and to assess the level of overexpression. B, FLAG-tagged intact TUG (residues 1–550) or truncated TUG proteins (containing the indicated residues) were transfected HeLa cells. Cells were fixed, and immunofluorescence microscopy was performed to detect endogenous p97 and FLAG-tagged TUG, as indicated. Scale bar, 20 μm.
Transient transfection of TUG caused the accumulation of both p97 and the exogenous TUG protein in the nucleus, as shown in Fig. 6B. This effect was observed for full-length TUG (residues 1–550) as well as for C-terminal fragments containing residues 238–550 or 313–550. In contrast, N-terminal fragments of TUG were more diffusely localized and did not cause nuclear accumulation of p97 (Fig. 6B, bottom four pairs of images). Some of these N-terminal fragments (e.g. 1–386 and 1–369) appeared to shift p97 distribution slightly out of the nucleus and into the cytosol. Because the overexpressed TUG is present at such high abundance compared with that of the endogenous protein, TUG that resides at its usual location at the ERGIC/ERES might not be well detected by immunofluorescence microscopy. It is uncertain whether the nuclear accumulation of overexpressed TUG represents a physiologic phenomenon. However, the data also raise the possibility that TUG may be targeted to the nucleus under certain conditions. TUG contains a potential nuclear localization signal at residues 350–361. This sequence is not sufficient to target TUG proteins to the nucleus because it is present in TUG fragments (1–386 and 1–369) that do not exhibit nuclear accumulation. Strikingly, the intact and C-terminal TUG proteins that accumulate in the nucleus (containing residues 1–550, 238–550, and 313–550) also disassemble p97 hexamers (Fig. 2C, above). The N-terminal fragments (containing residues 1–386, 1–369, 1–336, and 1–312) are predicted to bind, but not to disassemble, p97 hexamers. Therefore, it is possible that the potential nuclear localization signal is only functional in the context of a heterodimeric TUG-p97 complex and that it then acts to target both TUG and p97 to the nucleus.
The overexpression data support the idea that TUG-mediated p97 disassembly can occur within cells and does not occur only using recombinant proteins or in cell lysates. Previous subcellular fractionation and microscopy data show that most endogenous TUG is present outside the nucleus (26), and data here show that it is localized to the ERES/ERGIC (Fig. 5, above). Therefore, we hypothesize that endogenous TUG may act at this location, possibly by regulating p97 oligomerization and activity.
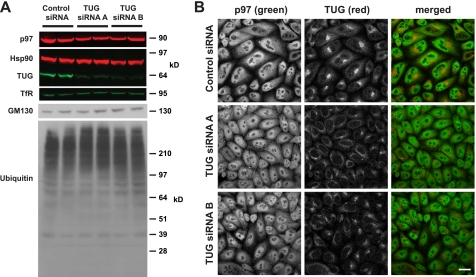
To further investigate the role of TUG in p97-dependent processes, we depleted TUG using transient transfection of siRNA duplexes. Fig. 7A shows that ∼48 h after transfection, ≥75% knockdown (KD) was achieved using each of two independent TUG siRNAs (A and B), compared with a scrambled control siRNA. Hsp90 and transferrin receptor were used as cytoplasmic and membrane loading controls, and we did not observe any cell death or alteration in cell growth. p97 abundance was decreased by ∼5–10% in TUG KD samples, compared with the controls. There was no large effect on GM130 abundance. Although knockdown of some UBX family members can result in an accumulation of ubiquitylated substrates, we did not observe this effect in TUG-depleted cells. We conclude that TUG KD slightly reduced p97 abundance but did not affect the overall abundance of polyubiquitylated proteins.
FIGURE 7.
siRNA-mediated TUG depletion has no large effect on ubiquitylated protein abundance or p97 distribution. A, HeLa were transfected using control (scrambled) siRNA, TUG siRNA A, or TUG siRNA B, as indicated, and were lysed 48 h after transfection. Proteins were separated by SDS-PAGE and immunoblotted as indicated. Hsp90 and transferrin receptor were used as loading controls. B, endogenous TUG and p97 were imaged in HeLa cells using indirect immunofluorescence and confocal microscopy. Cells were treated with the indicated siRNAs 48 h prior to fixation. Scale bar, 20 μm.
We next tested if TUG depletion altered the subcellular distribution of p97. As expected, the TUG immunofluorescent signal was reduced in cells treated with TUG siRNAs, as compared with those treated with a control siRNA (Fig. 7B). p97 was widely distributed throughout the nucleus and cytosol, and the intensity and distribution of p97 staining were not altered by TUG depletion. In addition, we did not observe the vacuolization noted previously after more prolonged p97 knockdown (52). Together, the data show that whereas TUG overexpression affected both ubiquitylated protein abundance and p97 targeting, TUG knockdown had no large effect on either of these end points.
Other UBX proteins, such as p47 and p37, are well known to recruit p97 activity to specific SNARE complexes and are important for the maintenance of Golgi structure (17, 18, 21, 53). Therefore, we examined the requirement of TUG for Golgi structure, using GM130 as a cis-Golgi marker (54). As shown in Fig. 8A, siRNA-mediated TUG depletion slightly dispersed Golgi structures marked by GM130 staining. As noted earlier, TUG knockdown did not affect GM130 abundance (Fig. 7A). Thus, the reduced GM130 peak intensity, observed by immunofluorescence microscopy, was not due to reduced GM130 abundance. Rather, the staining pattern was more diffuse, with reduced perinuclear intensity, in TUG-depleted cells. We observed this effect very consistently. We observed no effect of siRNA-mediated TUG depletion on trafficking of VSV-G to the cell surface (supplemental Fig. 4). The data suggest that there may be subtle alterations in Golgi morphology or dynamics in TUG-depleted cells.
FIGURE 8.
TUG depletion alters GM130 staining intensity and impairs Golgi reformation after brefeldin A washout. A, endogenous TUG and GM130 were imaged using immunofluorescence and confocal microscopy of HeLa cells. Cells were treated with control siRNA, TUG siRNA A, or TUG siRNA B, as indicated, for 48 h prior to fixation. B, HeLa cells were treated with control siRNA or TUG siRNA A for 48 h and then mock-treated or treated with BFA, as indicated. BFA was used at 10 μg/ml at 37 °C for 1 h, and then cells were washed three times and allowed to recover in the absence of BFA for 1 h at 37 °C. Cells were then fixed and stained to detect GM130 as in A. The experiment was repeated with similar results. C, images of cells in B were quantified. For each cell, the ratio of fluorescence intensity in the perinuclear region to that of the entire cell was quantified. 19–24 cells were analyzed per condition, and the mean ratio ± S.E. (error bars) is plotted. Statistical significance was assessed using a two-tailed t test. Scale bars, 20 μm.
To test more directly if TUG depletion alters Golgi dynamics, we examined the reassembly of Golgi membranes after BFA washout. Reassembly of the Golgi after mitosis and during interphase requires p97 activity and homotypic membrane fusion; reassembly after BFA washout may also require p97, yet this is less clear (55, 56). To study the requirement of TUG, we treated HeLa cells with BFA for 1 h to induce Golgi fragmentation. We then removed BFA, allowed cells to recover for 1 h, and imaged GM130 to assess Golgi integrity. As shown in Fig. 8B, cells treated with a control siRNA demonstrated a moderate degree of Golgi reformation, with dispersed GM130 observed in most cells (Fig. 8B, top right panel). As a control, mock-treated cells not exposed to BFA had intact Golgi complexes (Fig. 8B, top left panel). After siRNA-mediated depletion of TUG, Golgi structures were slightly more diffuse in the mock-treated cells (Fig. 8B, bottom left panel), similar to results in Fig. 8A. Strikingly, Golgi reassembly after BFA removal was markedly impaired in cells treated with the TUG siRNA (Fig. 8B, bottom right panel). To quantify these data, we calculated a ratio of perinuclear to whole-cell fluorescence intensities for each of several cells under each condition. As shown in Fig. 8C, this ratio was reduced in control cells after BFA washout, compared with untreated control cells, consistent with incomplete reassembly of Golgi complexes. This was expected because the cells were given only 1 h to recover from BFA treatment; indeed, we chose this time point precisely because recovery was incomplete. In cells treated with TUG siRNA but not BFA, the slight loss of compactness of GM130 staining did not result in a significant decrease in the ratio of perinuclear to whole cell fluorescence intensities. However, after BFA removal, this ratio was significantly reduced in TUG KD cells compared with control cells (compare fourth bar with second bar). This defect in Golgi reassembly remained significant even when untreated TUG KD cells were normalized to untreated control cells, and BFA-treated TUG KD cells were scaled accordingly. We conclude that TUG depletion caused a significant impairment in the recovery of Golgi complexes after BFA washout. These data support the concept that TUG functions in membrane trafficking pathways important for Golgi dynamics.
DISCUSSION
Our data show that TUG regulates the oligomeric structure of the p97 ATPase and that it resides at the ERGIC and ERES. Nearly all TUG is complexed with p97 in cells. Because p97 hexamerization is essential for its ATPase activity (8, 57), the data imply that TUG controls p97 activity on ERES/ERGIC membranes. We examined p97-dependent processes in cells and found that overexpressed TUG can cause the accumulation of ubiquitylated substrates. A physiologic role was revealed by siRNA-mediated depletion, which showed that TUG was required for efficient reassembly of Golgi complexes after brefeldin A removal. Thus, we propose that it may control a specific, p97-dependent step in membrane trafficking.
The data distinguish TUG from other members of the UBX domain-containing protein family, which serve as p97 cofactors. UBX proteins are thought to recruit p97 activity to particular substrate proteins and thus to mediate the disassembly of protein complexes or to extract proteins from membranes. By disassembling p97 hexamers, TUG may inhibit p97 action rather than coupling it to a particular target. This action is fundamentally different from that of other UBX proteins and appears to be conserved through evolution (29, 30).
The mammalian TUG-p97 interaction involves an extended sequence comprising three regions of TUG and the N terminus of p97. This interaction is also distinct from the bipartite binding employed by other UBX proteins (5). No protein containing the three p97-interacting sites, UBL, SHP, and UBX, has been described previously. The SHP box of p47 was recently identified as the most important region for p47-p97 binding, similar to our results with TUG (58). It is remarkable that the TUG UBX domain alone did not bind p97 in vitro because this domain and its interaction with p97 have been considered a defining feature of this family. Another UBX protein, UBXD1, also lacks key residues in the UBX domain and binds p97 through a PUB domain (39, 59). Although TUG does not contain a PUB domain, its predicted coiled-coil region apparently stabilized a UBX-p97 interaction. Very recently, a VCP-interacting motif has been defined (60, 61); this motif is not present in TUG. Structural studies will be required to understand how the TUG regions defined here cooperate to bind and regulate p97.
We find that TUG regulates the oligomeric state of mammalian p97, similar to the Arabidopsis protein PUX1, which regulates oligomerization of AtCDC48 (29, 30). Indeed, a C-terminal TUG fragment corresponding to PUX1 was both necessary and sufficient to disassemble p97 hexamers. Although full-length TUG also disassembled p97 hexamers in vitro, a possibility is that N-terminal regions of TUG confer some regulation on this process in vivo. Such regulation was not apparent using recombinant TUG but could involve post-translational modifications or protein interactions of TUG and/or p97 in cells. The control of p97 oligomeric state would be a unique mechanism to regulate the activity of this ATPase, which, to our knowledge, has not been described for any other member of the AAA ATPase family.
Our data show that TUG binds to both monomeric and hexameric p97. The mechanism for hexamer disassembly is unlikely to be catalytic, because the molar amounts of TUG that were required were similar to or greater than the molar amounts of p97 monomer. It is unclear how p97 hexamer disassembly can occur, given that TUG bound only to the N-terminal domain of p97. The first ATPase domain (D1) of p97 is the region necessary for hexamerization (10). ATP hydrolysis-related conformational changes are coupled to adaptors through N-terminal position changes (6, 9). Possibly, the reverse may also occur, so that TUG binding to the p97 N terminus may impart conformational changes to the D1 domain to mediate disassembly. Because the nucleotide-bound state of the hexamer did not affect monomer generation in vitro, such conformational changes may occur independent of ATP-binding status.
Our data show that TUG is concentrated at the ERES/ERGIC, suggesting that it may disassemble p97 hexamers specifically at this location in cells. We observed marked p97 disassembly when TUG was overexpressed by transient transfection, possibly reflecting more widespread localization as well as overexpression of the transfected TUG protein. Transient transfection of TUG caused an accumulation of total ubiquitylated proteins, as would be expected from such marked p97 disassembly in cells. Although we did not observe an accumulation of ubiquitylated substrates in TUG-depleted HeLa cells, specific ubiquitylated substrates did accumulate in yeast lacking the putative TUG ortholog, Ubx4p (62).
Overexpressed, full-length TUG localized primarily to the nucleus and caused the nuclear accumulation of p97. Endogenous p97 is known to localize to the nucleus (63), as do other UBX cofactors, such as p47 (53). It is interesting to note that putative TUG orthologs in Saccharomyces cerevisiae and Caenorhabditis elegans are also reported to localize to the nucleus in addition to the cytoplasm (see the Saccharomyces Genome Database for YMR067C and WormBase for B0024.10), and the yeast protein, Ubx4p, functions in endoplasmic reticulum-associated degradation as well as to control the degradation of the Rbp1 subunit of RNA polymerase II (62, 64, 65). It may be that TUG, like other p97 cofactors, has functions both within and outside the nucleus. We did observe a correlation of TUG fragments that localized to the nucleus and TUG fragments that are able to disassemble p97 hexamers. Thus, one possibility is that hexamer disassembly is coupled to nuclear import, perhaps suggesting that p97 must be monomeric to pass through nuclear pores. However, the physiologic relevance of the TUG-p97 nuclear accumulation we observed remains uncertain because the vast majority of endogenous TUG resides outside of the nucleus. The idea that TUG may regulate p97 specifically at the ERES/ERGIC, when expressed at physiologic levels, fits well with the established role of p97 to control protein trafficking in the early secretory pathway (5, 21, 53, 56).
Our data implicate TUG in a trafficking pathway involved in Golgi dynamics. Defects in Golgi reassembly are also observed upon disruption of two other p97-interacting proteins, p37 and p47, which couple p97 ATPase activity to SNARE proteins during interphase and at the end of mitosis, respectively (19, 53). Interestingly, the function of p47 is regulated by its sequestration in the nucleus. It may be that TUG modulates p97 activity to control membrane fusion in a similar manner. The Arabidopsis protein, PUX1, was identified for its ability to bind the plant ortholog of syntaxin 5 (29). TUG is not known to bind particular SNARES, and we have been unable to demonstrate an interaction between syntaxin 5 and TUG. However, in addition to UBX proteins, p97 also interacts with coat proteins such as clathrin (66), tethering proteins such as EEA1 (67), cargoes destined for lysosomes (68), and membrane-remodeling factors in the autophagy pathway (69). Thus, TUG regulation of p97 activity could control any of a number of substrates involved in membrane trafficking, which could function in Golgi structure and reassembly.
The studies described here will also shed light on how GLUT4 glucose transporters traffic in adipocytes. As noted above, GLUT4 participates in what is probably a cell type-specific adaptation of a more general trafficking pathway present in non-specialized cells. GLUT4 itself is a cargo that may not be essential for the regulated trafficking of vesicles containing these transporters (70, 71) but may enhance this response (72). GLUT4 regulation has been thought to occur primarily at a postendosomal or post-Golgi location (22). However, ERGIC/ERES and cis-Golgi matrix proteins, such as p115 and Golgin-160, are implicated in GLUT4 targeting to an intracellular, insulin-responsive compartment, similar to TUG (24, 25, 73, 74). Comparable with our results in TUG-depleted cells, VSV-G trafficking was not affected by Golgin-160 knockdown (75) or expression of p115 mutants (76). Together with earlier work (24), our present data raise the possibility that GLUT4 may participate in an unconventional secretion pathway (77). Understanding the cell type specificity of this response and how p97 may participate will require further studies.
In conclusion, our data show that TUG mediates p97 hexamer disassembly, localizes to the ERES/ERGIC, and is required for efficient Golgi reformation after BFA removal. Targeting of TUG and p97 to the nucleus may add an additional layer of control to these processes. These actions are fundamental to the control of subcellular organization and membrane dynamics. Future work to study these effects will require structural characterization of the p97-TUG interaction and identification of other proteins that act together with TUG and p97 to mediate membrane trafficking. The data here provide a basis for these studies and contribute to understanding of how TUG and p97 act coordinately to control cellular functions.
Supplementary Material
Acknowledgments
We thank James Cresswell and Chenfei Yu for assistance; Michael Caplan, Fred Gorelick, and Nico Dantuma for reagents; and Michael Caplan, Fred Gorelick, Derek Toomre, Megan King, and Mingming Hao for helpful discussions. This work used resources of the Yale Diabetes Endocrinology Research Center Cell Biology Core.
This work was supported, in whole or in part, by National Institutes of Health Grants R21 DK70812 and R01 DK075772. This work was also supported by a pilot grant from the O'Brien Kidney Center at Yale (P30 DK079310) and by a W.M. Keck Foundation Distinguished Young Scholar in Medical Research Award (to J. S. B.).

This article contains supplemental Figs. 1–4.
- ER
- endoplasmic reticulum
- BFA
- brefeldin A
- ERES
- endoplasmic reticulum exit site(s)
- ERGIC
- endoplasmic reticulum-Golgi intermediate compartment
- UBL
- ubiquitin-like
- UBX
- ubiquitin regulatory X
- KD
- knockdown
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. White S. R., Lauring B. (2007) AAA+ ATPases. Achieving diversity of function with conserved machinery. Traffic 8, 1657–1667 [DOI] [PubMed] [Google Scholar]
- 2. Brunger A. T., DeLaBarre B. (2003) NSF and p97/VCP. Similar at first, different at last. FEBS Lett. 555, 126–133 [DOI] [PubMed] [Google Scholar]
- 3. Schuberth C., Buchberger A. (2008) UBX domain proteins. Major regulators of the AAA ATPase Cdc48/p97. Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vij N. (2008) AAA ATPase p97/VCP. Cellular functions, disease and therapeutic potential. J. Cell Mol. Med. 12, 2511–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yeung H. O., Kloppsteck P., Niwa H., Isaacson R. L., Matthews S., Zhang X., Freemont P. S. (2008) Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 36, 62–67 [DOI] [PubMed] [Google Scholar]
- 6. Beuron F., Dreveny I., Yuan X., Pye V. E., McKeown C., Briggs L. C., Cliff M. J., Kaneko Y., Wallis R., Isaacson R. L., Ladbury J. E., Matthews S. J., Kondo H., Zhang X., Freemont P. S. (2006) Conformational changes in the AAA ATPase p97-p47 adaptor complex. EMBO J. 25, 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies J. M., Tsuruta H., May A. P., Weis W. I. (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small angle x-ray scattering. Structure 13, 183–195 [DOI] [PubMed] [Google Scholar]
- 8. DeLaBarre B., Brunger A. T. (2005) Nucleotide-dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347, 437–452 [DOI] [PubMed] [Google Scholar]
- 9. Pye V. E., Dreveny I., Briggs L. C., Sands C., Beuron F., Zhang X., Freemont P. S. (2006) Going through the motions. The ATPase cycle of p97. J. Struct. Biol. 156, 12–28 [DOI] [PubMed] [Google Scholar]
- 10. Wang Q., Song C., Yang X., Li C. C. (2003) D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J. Biol. Chem. 278, 32784–32793 [DOI] [PubMed] [Google Scholar]
- 11. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48. A power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 12. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buchberger A., Howard M. J., Proctor M., Bycroft M. (2001) The UBX domain. A widespread ubiquitin-like module. J. Mol. Biol. 307, 17–24 [DOI] [PubMed] [Google Scholar]
- 14. Liang J., Yin C., Doong H., Fang S., Peterhoff C., Nixon R. A., Monteiro M. J. (2006) Characterization of erasin (UBXD2). A new ER protein that promotes ER-associated protein degradation. J. Cell Sci. 119, 4011–4024 [DOI] [PubMed] [Google Scholar]
- 15. Nagahama M., Ohnishi M., Kawate Y., Matsui T., Miyake H., Yuasa K., Tani K., Tagaya M., Tsuji A. (2009) UBXD1 is a VCP-interacting protein that is involved in ER-associated degradation. Biochem. Biophys. Res. Commun. 382, 303–308 [DOI] [PubMed] [Google Scholar]
- 16. Rezvani K., Teng Y., Pan Y., Dani J. A., Lindstrom J., García Gras E. A., McIntosh J. M., De Biasi M. (2009) UBXD4, a UBX-containing protein, regulates the cell surface number and stability of α3-containing nicotinic acetylcholine receptors. J. Neurosci. 29, 6883–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kondo H., Rabouille C., Newman R., Levine T. P., Pappin D., Freemont P., Warren G. (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78 [DOI] [PubMed] [Google Scholar]
- 18. Meyer H. H., Kondo H., Warren G. (1998) The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett. 437, 255–257 [DOI] [PubMed] [Google Scholar]
- 19. Uchiyama K., Jokitalo E., Lindman M., Jackman M., Kano F., Murata M., Zhang X., Kondo H. (2003) The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J. Cell Biol. 161, 1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dreveny I., Kondo H., Uchiyama K., Shaw A., Zhang X., Freemont P. S. (2004) Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 23, 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uchiyama K., Kondo H. (2005) p97/p47-Mediated biogenesis of Golgi and ER. J. Biochem. 137, 115–119 [DOI] [PubMed] [Google Scholar]
- 22. Bogan J. S., Kandror K. V. (2010) Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr. Opin. Cell Biol. 22, 506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rubin B. R., Bogan J. S. (2009) Intracellular retention and insulin-stimulated mobilization of GLUT4 glucose transporters. Vitam. Horm. 80, 155–192 [DOI] [PubMed] [Google Scholar]
- 24. Xu Y., Rubin B. R., Orme C. M., Karpikov A., Yu C., Bogan J. S., Toomre D. K. (2011) Dual mode of insulin action controls GLUT4 vesicle exocytosis. J. Cell Biol. 193, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu C., Cresswell J., Löffler M. G., Bogan J. S. (2007) The glucose transporter 4-regulating protein TUG is essential for highly insulin-responsive glucose uptake in 3T3-L1 adipocytes. J. Biol. Chem. 282, 7710–7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bogan J. S., Hendon N., McKee A. E., Tsao T. S., Lodish H. F. (2003) Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 425, 727–733 [DOI] [PubMed] [Google Scholar]
- 27. Ladanyi M., Lui M. Y., Antonescu C. R., Krause-Boehm A., Meindl A., Argani P., Healey J. H., Ueda T., Yoshikawa H., Meloni-Ehrig A., Sorensen P. H., Mertens F., Mandahl N., van den Berghe H., Sciot R., Dal Cin P., Bridge J. (2001) The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene 20, 48–57 [DOI] [PubMed] [Google Scholar]
- 28. Castorena C. M., Mackrell J. G., Bogan J. S., Kanzaki M., Cartee G. D. (2011) Clustering of GLUT4, TUG, and RUVBL2 protein levels correlate with myosin heavy chain isoform pattern in skeletal muscles, but AS160 and TBC1D1 levels do not. J. Appl. Physiol. 111, 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rancour D. M., Park S., Knight S. D., Bednarek S. Y. (2004) Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of arabidopsis CDC48. J. Biol. Chem. 279, 54264–54274 [DOI] [PubMed] [Google Scholar]
- 30. Park S., Rancour D. M., Bednarek S. Y. (2007) Protein domain-domain interactions and requirements for the negative regulation of Arabidopsis CDC48/p97 by the plant ubiquitin regulatory X (UBX) domain-containing protein, PUX1. J. Biol. Chem. 282, 5217–5224 [DOI] [PubMed] [Google Scholar]
- 31. Tresse E., Salomons F. A., Vesa J., Bott L. C., Kimonis V., Yao T. P., Dantuma N. P., Taylor J. P. (2010) VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simossis V. A., Kleinjung J., Heringa J. (2005) Homology-extended sequence alignment. Nucleic Acids Res. 33, 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tettamanzi M. C., Yu C., Bogan J. S., Hodsdon M. E. (2006) Solution structure and backbone dynamics of an N-terminal ubiquitin-like domain in the GLUT4-regulating protein, TUG. Protein Sci. 15, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sato B. K., Hampton R. Y. (2006) Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast 23, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 35. Shin H. Y., Kang W., Lee S. Y., Yang J. K. (2010) Crystallization and preliminary X-ray crystallographic analysis of the N domain of p97/VCP in complex with the UBX domain of FAF1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 41–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hänzelmann P., Buchberger A., Schindelin H. (2011) Hierarchical binding of cofactors to the AAA ATPase p97. Structure 19, 833–843 [DOI] [PubMed] [Google Scholar]
- 37. Mori-Konya C., Kato N., Maeda R., Yasuda K., Higashimae N., Noguchi M., Koike M., Kimura Y., Ohizumi H., Hori S., Kakizuka A. (2009) p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation. Genes Cells 14, 483–497 [DOI] [PubMed] [Google Scholar]
- 38. Aker J., Hesselink R., Engel R., Karlova R., Borst J. W., Visser A. J., de Vries S. C. (2007) In vivo hexamerization and characterization of the Arabidopsis AAA ATPase CDC48A complex using forster resonance energy transfer-fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Plant Physiol. 145, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Madsen L., Andersen K. M., Prag S., Moos T., Semple C. A., Seeger M., Hartmann-Petersen R. (2008) Ubxd1 is a novel co-factor of the human p97 ATPase. Int. J. Biochem. Cell Biol. 40, 2927–2942 [DOI] [PubMed] [Google Scholar]
- 40. Hauri H. P., Kappeler F., Andersson H., Appenzeller C. (2000) ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113, 587–596 [DOI] [PubMed] [Google Scholar]
- 41. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) Brefeldin A. Insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orci L., Tagaya M., Amherdt M., Perrelet A., Donaldson J. G., Lippincott-Schwartz J., Klausner R. D., Rothman J. E. (1991) Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64, 1183–1195 [DOI] [PubMed] [Google Scholar]
- 43. Kreykenbohm V., Wenzel D., Antonin W., Atlachkine V., von Mollard G. F. (2002) The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81, 273–280 [DOI] [PubMed] [Google Scholar]
- 44. Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., Johannes L. (2002) Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez C., Garcia-Mata R., Hauri H. P., Sztul E. (2001) The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J. Biol. Chem. 276, 2693–2700 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. (1997) The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 89, 445–455 [DOI] [PubMed] [Google Scholar]
- 47. Marra P., Maffucci T., Daniele T., Tullio G. D., Ikehara Y., Chan E. K., Luini A., Beznoussenko G., Mironov A., De Matteis M. A. (2001) The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 48. Marra P., Salvatore L., Mironov A., Jr., Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. (2007) The biogenesis of the Golgi ribbon. The roles of membrane input from the ER and of GM130. Mol. Biol. Cell 18, 1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kreis T. E., Lodish H. F. (1986) Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell 46, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. (1997) ER-to-Golgi transport visualized in living cells. Nature 389, 81–85 [DOI] [PubMed] [Google Scholar]
- 51. Shugrue C. A., Kolen E. R., Peters H., Czernik A., Kaiser C., Matovcik L., Hubbard A. L., Gorelick F. (1999) Identification of the putative mammalian orthologue of Sec31P, a component of the COPII coat. J. Cell Sci. 112, 4547–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wójcik C., Yano M., DeMartino G. N. (2004) RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 117, 281–292 [DOI] [PubMed] [Google Scholar]
- 53. Uchiyama K., Totsukawa G., Puhka M., Kaneko Y., Jokitalo E., Dreveny I., Beuron F., Zhang X., Freemont P., Kondo H. (2006) p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev. Cell 11, 803–816 [DOI] [PubMed] [Google Scholar]
- 54. Nakamura N. (2010) Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J. Pharmacol. Sci. 112, 255–264 [DOI] [PubMed] [Google Scholar]
- 55. Dalal S., Rosser M. F., Cyr D. M., Hanson P. I. (2004) Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell 15, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tang D., Mar K., Warren G., Wang Y. (2008) Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J. Biol. Chem. 283, 6085–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Briggs L. C., Baldwin G. S., Miyata N., Kondo H., Zhang X., Freemont P. S. (2008) Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 283, 13745–13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaneko Y., Tamura K., Totsukawa G., Kondo H. (2010) Isolation of a point-mutated p47 lacking binding affinity to p97ATPase. FEBS Lett. 584, 3873–3877 [DOI] [PubMed] [Google Scholar]
- 59. Kern M., Fernandez-Sáiz V., Schäfer Z., Buchberger A. (2009) UBXD1 binds p97 through two independent binding sites. Biochem. Biophys. Res. Commun. 380, 303–307 [DOI] [PubMed] [Google Scholar]
- 60. Hänzelmann P., Schindelin H. (2011) The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM). Mutually exclusive binding of cofactors to the N-terminal domain of p97. J. Biol. Chem. 286, 38679–38690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stapf C., Cartwright E., Bycroft M., Hofmann K., Buchberger A. (2011) The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J. Biol. Chem. 286, 38670–38678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alberts S. M., Sonntag C., Schäfer A., Wolf D. H. (2009) Ubx4 modulates Cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J. Biol. Chem. 284, 16082–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Decottignies A., Evain A., Ghislain M. (2004) Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast 21, 127–139 [DOI] [PubMed] [Google Scholar]
- 65. Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011) Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pleasure I. T., Black M. M., Keen J. H. (1993) Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365, 459–462 [DOI] [PubMed] [Google Scholar]
- 67. Ramanathan H. N., Ye Y. (2011) The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. doi: 10.1038/cr.2011.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ren J., Pashkova N., Winistorfer S., Piper R. C. (2008) DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J. Biol. Chem. 283, 21599–21611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., Eskelinen E. L., Thumm M. (2010) Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 190, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jordens I., Molle D., Xiong W., Keller S. R., McGraw T. E. (2010) Insulin-regulated aminopeptidase is a key regulator of GLUT4 trafficking by controlling the sorting of GLUT4 from endosomes to specialized insulin-regulated vesicles. Mol. Biol. Cell 21, 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yeh T. Y., Sbodio J. I., Tsun Z. Y., Luo B., Chi N. W. (2007) Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem. J. 402, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kandror K. V., Pilch P. F. (2011) The sugar is sIRVed. Sorting Glut4 and its fellow travelers. Traffic 12, 665–671 [DOI] [PubMed] [Google Scholar]
- 73. Hosaka T., Brooks C. C., Presman E., Kim S. K., Zhang Z., Breen M., Gross D. N., Sztul E., Pilch P. F. (2005) p115 Interacts with the GLUT4 vesicle protein, IRAP, and plays a critical role in insulin-stimulated GLUT4 translocation. Mol. Biol. Cell 16, 2882–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Williams D., Hicks S. W., Machamer C. E., Pessin J. E. (2006) Golgin-160 is required for the Golgi membrane sorting of the insulin-responsive glucose transporter GLUT4 in adipocytes. Mol. Biol. Cell 17, 5346–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hicks S. W., Horn T. A., McCaffery J. M., Zuckerman D. M., Machamer C. E. (2006) Golgin-160 promotes cell surface expression of the β-1 adrenergic receptor. Traffic 7, 1666–1677 [DOI] [PubMed] [Google Scholar]
- 76. Brandon E., Gao Y., Garcia-Mata R., Alvarez C., Sztul E. (2003) Membrane targeting of p115 phosphorylation mutants and their effects on Golgi integrity and secretory traffic. Eur. J. Cell Biol. 82, 411–420 [DOI] [PubMed] [Google Scholar]
- 77. Grieve A. G., Rabouille C. (2011) Golgi and related vesicle proteomics. Simplify to identify. Cold Spring Harb. Perspect. Biol. 3:a005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.