Background: Amylosucrases (AS) hold great potential for glycodiversification.
Results: The first three-dimensional structure of AS from Deinococcus geothermalis solved here revealed an unusual dimer organization. Structures of complex of AS with turanose were also determined.
Conclusion: Dimerization may contribute to thermostability. Turanose versus trehalulose formation is controlled by residues from subsite +1.
Significance: This study improves the comprehension of AS properties and provides new insight for AS design.
Keywords: Biotechnology, Carbohydrate, Enzyme Structure, Glycosidases, Structural Biology, Amylosucrase, Crystal Structure, Glucosylation, Sucrose Isomerization, Thermostability
Abstract
Amylosucrases are sucrose-utilizing α-transglucosidases that naturally catalyze the synthesis of α-glucans, linked exclusively through α1,4-linkages. Side products and in particular sucrose isomers such as turanose and trehalulose are also produced by these enzymes. Here, we report the first structural and biophysical characterization of the most thermostable amylosucrase identified so far, the amylosucrase from Deinoccocus geothermalis (DgAS). The three-dimensional structure revealed a homodimeric quaternary organization, never reported before for other amylosucrases. A sequence signature of dimerization was identified from the analysis of the dimer interface and sequence alignments. By rigidifying the DgAS structure, the quaternary organization is likely to participate in the enhanced thermal stability of the protein. Amylosucrase specificity with respect to sucrose isomer formation (turanose or trehalulose) was also investigated. We report the first structures of the amylosucrases from Deinococcus geothermalis and Neisseria polysaccharea in complex with turanose. In the amylosucrase from N. polysaccharea (NpAS), key residues were found to force the fructosyl moiety to bind in an open state with the O3′ ideally positioned to explain the preferential formation of turanose by NpAS. Such residues are either not present or not similarly placed in DgAS. As a consequence, DgAS binds the furanoid tautomers of fructose through a weak network of interactions to enable turanose formation. Such topology at subsite +1 is likely favoring other possible fructose binding modes in agreement with the higher amount of trehalulose formed by DgAS. Our findings help to understand the inter-relationships between amylosucrase structure, flexibility, function, and stability and provide new insight for amylosucrase design.
Introduction
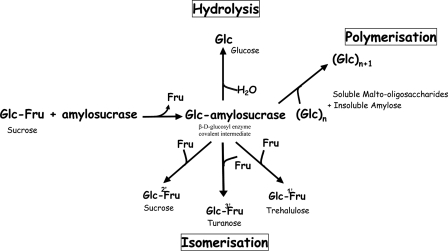
Amylosucrases (AS)4 (E.C. number 2.4.1.4) are glucansucrases belonging to Glycoside-Hydrolase Family 13 of the Carbohydrate-Active EnZymes (CAZy) classification (1–3). The main reaction catalyzed by these enzymes is the formation of an amylose-like glucan with a concomitant release of fructose from sucrose substrate (Fig. 1) (4). Side reactions including sucrose hydrolysis and sucrose isomer synthesis (i.e. turanose, α-d-Glcp(1→3)-β-d-Fru and trehalulose, α-d-Glcp(1→1)-β-d-Fru) also occur (4, 5) although other GH13 enzymes are known to be more specific for these types of reaction (6–8). These molecules are known to be less cariogenic than sucrose and are thus of interest for nutritional applications (9). AS are also able to transfer the glucosyl moieties from sucrose to exogenous acceptors such as maltose, glycogen, maltodextrins, arbutin, and others (10, 11). The ability of AS to convert sucrose (an abundant and inexpensive substrate) into valuable derivatives is a great advantage in comparison to Leloir glucosyltransferases, which use nucleotide-activated sugars as glucosyl donor substrates (12). Moreover, protein engineering techniques also enabled the creation of novel AS with tailored specificity toward unnatural acceptor molecules (13).
FIGURE 1.
Reactions catalyzed by amylosucrases from sole sucrose. Glc, glucose; Fru, fructose; sucrose, α-d-glucopyranosyl-1,2-β-d-fructofuranoside; turanose, α-d-glucopyranosyl-1,3-β-d-fructose; trehalulose, α-d-glucopyranosyl-1,1-β-d-fructose.
AS that were characterized so far are produced by various species from the genus Neisseria (4), Deinococcus (14, 15), and Alteromonas (16). However, Neisseria polysaccharea amylosucrase (NpAS) is the only AS for which several structures, alone or in complex with sucrose substrate or products, are available to date (17–20). The three-dimensional structure of NpAS is organized in five domains, namely A, B, C, N, and B′. B′ and N domains are only found in AS, whereas the three other domains are conserved among GH13 enzymes. Although the potential of NpAS for glycodiversification is large, this enzyme suffers from a low catalytic efficiency and a weak thermostability, limiting its industrial development (15). Directed evolution has been attempted to improve NpAS catalytic efficiency and thermostability (21). Searching for more thermostable and efficient enzymes in the natural diversity is another alternative that has motivated the biochemical characterization of the amylosucrases from Deinococcus geothermalis (DgAS) (15), Deinococcus radiodurans (DrAS) (14), and Alteromonas macleodii (AmAS) (15).
With a specific activity of 44 units mg−1 at the optimal temperature of 50 °C, the recombinant DgAS is the most thermostable AS characterized to date (15). The size distribution of the α-glucan chains produced by DgAS and NpAS differs and the two enzymes do not synthesize equivalent amounts of turanose and trehalulose. Indeed, DgAS produces significantly higher amounts of trehalulose than NpAS (15). Glucosylation of unnatural acceptors such as salicin also revealed that DgAS only produces a monoglucosylated form, whereas NpAS is able to produce a diglucosylated compound (22).
Here, we report the first three-dimensional structure of DgAS in its apo form that reveals an unusual homodimeric arrangement, thereby allowing identification of determinants possibly controlling thermostability. Furthermore, the structures of both DgAS and NpAS in complex with turanose (a fructose acceptor reaction product) were also determined and their analysis highlighted major differences upon fructose binding, which are discussed with regard to the enzyme product specificities.
EXPERIMENTAL PROCEDURES
Expression and Purification
Expression and purification of recombinant glutathione S-transferase-amylosucrase fusion proteins (GST-DgAS, GST-DrAS, and GST-NpAS) were performed as previously described by Emond et al. (15), Pizzut et al. (14), and de Montalk et al. (23), respectively. The GST-AS proteins were purified by affinity chromatography using glutathione-Sepharose 4B support (GE Healthcare). The GST tag was then removed using Prescission protease (GE Healthcare), which left 5 residues (GPLGS) of the cutting site at the N-terminal extremity.
Thermostability of Amylosucrases
The melting point (Tm) of amylosucrases was assayed by differential scanning fluorimetry. A mixture of enzyme (2 μm), Sypro-orange (5 ×) (Invitrogen), and 50 mm Tris, pH 7.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA were incubated using a temperature gradient from 20 to 80 °C with a 0.3 °C increment. The thermal transition was monitored using a RTQ-PCR CFX96 Real-time System (Bio-Rad). Tm was given by the inflection point of the curve relative fluorescence unit = f(T). Circular dichroism spectra were recorded on a JASCO J815 spectropolarimeter equipped with a Peltier cell temperature controller. Tm values were obtained by heating the sample (enzyme 2 μm in 50 mm Tris, pH 7.0, 150 mm NaCl, 70 μm DTT, 70 μm EDTA) at 1 °C/min and recording the ellipticity value at 220 nm from 25 to 80 °C with delays of 30 s. SigmaPlot 10.0 software was used for all graphic analyses, Tm determination, and statistics.
The half-life (t½) at 50 °C of DgAS and NpAS preparations were also determined by incubating AS pure enzymes (300 mg liters−1) in 50 mm Tris, pH 7.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA at 50 °C. At various intervals, aliquots were taken and the enzyme activity was determined as previously described (22).
Size Exclusion Chromatography Multiangle Laser Light Scattering (SEC-MALLS) Experiments
NpAS, DgAS, and DrAS protein samples buffered in 50 mm Tris, pH 7.5, 150 mm NaCl were analyzed on a Shodex KW-803 column (Showa Denko Europe GmbH, Munich, Germany) with multiangle laser light scattering (MALLS). The column was equilibrated in a 0.1-μm filtered 50 mm Tris, pH 7.5, 150 mm NaCl, 0.02% (w/v) NaN3 buffer on a Agilent 1260 Infinity LC chromatographic system (Agilent Technology, Massy, France). Data were collected using a DAWN HELEOS-II 18-angle and Optilab T-rEX refractive index detector (Wyatt Technology Corp., Toulouse France). Sample concentrations were 1.55, 2.1, and 1.85 g liter−1 for NpAS, DgAS, and DrAS solutions, respectively. Protein samples were prepared in the Tris buffer used as the mobile phase to equilibrate the column. 20 μl of each protein sample was loaded on the column and the separation was performed at a flow rate of 0.5 ml min−1 at 22 °C. Results were analyzed using the ASTRA V software (Wyatt Technology Corp.).
Native-PAGE
Native-PAGE was performed with a two-phase gel composed of a 4% stacking gel (4% acrylamide, 125 mm Tris, pH 6.8, 0.1% ammonium persulfate, 0.15% N,N,N′,N′-tetramethylethylenediamine) and a 10% separating gel (10% acrylamide, 375 mm Tris, pH 8.8, 0.1% ammonium persulfate, 0.15% N,N,N′,N′-tetramethylethylenediamine). 15 μl of the protein samples (2 g liter−1) mixed with 2 × loading buffer (40 mm Tris, pH 6.8, 50% glycerol (v/v), 0.04% bromophenol blue (w/v)) were loaded onto the gel. Electrophoresis was performed at room temperature using a mini-PROTEAN system (Bio-Rad) at a voltage of 100 V with an electrophoresis buffer composed of 15 g liter−1 of Tris, pH 8.3, and 72 g liter−1 of glycine. The gels were stained with Coomassie Brilliant Blue. Molecular weight estimation was performed using NativeMark native protein markers (Invitrogen) as standards.
Crystallization
NpAS was crystallized using conditions previously described by Skov et al. (17, 18). DgAS crystallization experiments were carried out at 12 °C using the hanging drop vapor diffusion method. Best crystals were obtained with a 1:1 (v/v) ratio of protein (6 mg ml−1 in 20 mm Tris, pH 8.0) to precipitant solution (1.5 m sodium acetate, 0.1 m sodium cacodylate, pH 7.0). Lens shape crystals appeared after 2 weeks and grew to a maximal size of 140 × 60 × 40 μm3.
Soaking Experiments
Crystals of NpAS were soaked for 20 min in the reservoir solution supplemented by 250 mm turanose (Sigma). DgAS crystals quickly fractured in the presence of such a high concentration of turanose, so they were soaked for a few seconds in the reservoir solution supplemented with only 14 mm turanose.
Data Collection and Structure Determination
X-ray experiments were carried out at 100 K. Prior to flash cooling, native crystals of DgAS were soaked for a few seconds in the reservoir solution supplemented with 20% (v/v) glycerol to avoid ice formation. Conversely, due to the cryoprotection effect of turanose, crystals of AS-turanose complexes were intrinsically cryoprotected. Native DgAS and NpAS-turanose diffraction datasets were collected to a maximum resolution of 1.97- and 1.85-Å, respectively, on beamline ID14-1 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). The DgAS-turanose complex dataset was collected to 2.10 Å on the ESRF beamline ID29. Diffracted intensities were integrated using iMOSFLM (24) and scaled with SCALA (25) from the CCP4 software suite (26, 27) and 5% of the scaled amplitudes were randomly selected and excluded from the refinement procedure. Crystals of DgAS in the apo form and in complex with turanose belong to the C2221 space group with 1 molecule per asymmetric unit giving a Matthews coefficient of 2.3 Å3/Da. Crystals of NpAS-turanose belong to the P21212 space group with 1 molecule per asymmetric unit and a Matthews coefficient of 2.4 Å3/Da. Data collection statistics are given in Table 1. The native structure of DgAS was solved by the molecular replacement method using PHASER (28) and the structure of NpAS (PDB code 1G5A) (18) as a search model. The translation function Z-score was 16.5 and R and Rfree of the refined molecular replacement solution were 0.39 and 0.47, respectively. Structures of the NpAS-turanose or DgAS-turanose complexes were straight refined from their native structures using refmac5 (29).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the outer resolution shell.
| NpAS-turanose | DgAS | DgAS-turanose | |
|---|---|---|---|
| Data collection | |||
| Space group | P21212 | C2221 | C2221 |
| a, b, c (Å) | 96.0, 116.3, 60.5 | 105.3, 110.2, 115.5 | 104.7, 110.4, 115.3 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 29.27-1.85 (1.95-1.85)* | 33.38-1.97 (2.08-1.97) | 63.45-2.10 (2.21-2.10) |
| Rsym | 0.096 (0.332) | 0.086 (0.376) | 0.099 (0.324) |
| I/σI | 6.5 (2.2) | 8.3 (2.0) | 4.9 (2.1) |
| Completeness (%) | 99.6 (100) | 99.6 (99.8) | 100 (98.9) |
| Redundancy | 3.6 (3.6) | 4.0 (3.9) | 6.3 (4.2) |
| No. of molecule/AU | 1 | 1 | 1 |
| Matthews coefficient (Å3/Da) | 2.4 | 2.3 | 2.3 |
| Refinement | |||
| Resolution (Å) | 29.10-1.85 | 33.22-1.97 | 13-2.10 |
| No. of unique reflections | 58,309 (8,452) | 47,355 (6,837) | 39,319 (5,648) |
| Rwork/Rfree (%) | 14.7/18.6 | 14.4/18.7 | 14.6/20.7 |
| Total number of atoms | 5,867 | 5,956 | 5,712 |
| Number of residues in the protein | 632 | 651 | 651 |
| Number of ligand molecules | 2 Turanoses, 1 PEG | 1 Tris, 13 glycerols | 1 Turanose |
| Number of water molecules | 767 | 602 | 394 |
| B-factors (Å2) | |||
| Protein | 11.6 | 17.9 | 20.7 |
| Ligand | 19.9 | 36.9 | 26.4 |
| Water | 21.2 | 27.5 | 26.1 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.022 | 0.024 | 0.023 |
| Bond angles (°) | 1.8 | 1.8 | 1.9 |
Building and Refinement
Structure refinement was performed with refmac5 from the CCP4 GUI (29) and models were manually reconstructed in the SigmaA weighted electron density maps using COOT (30). Water molecules were automatically assigned and ligand molecules were manually fitted in residual maps. Final DgAS structures in the apo form and in complex with turanose contain 651 residues of the 655 theoretical residues with four missing residues at the C-terminal extremity. The final model of NpAS in complex with turanose contains 628 residues of the 632 theoretical residues with four missing residues at the N-terminal extremity. Refinement statistics are given in Table 1.
Coordinates
Coordinates have been deposited at the protein data bank (PDB codes 3UCQ, 3UER, and 3UEQ for DgAS, DgAS-turanose complex, and NpAS-turanose complex, respectively).
In Vitro Synthesis of Sucrose Isomers
Synthesis of sucrose isomers (turanose and trehalulose) was performed with 100 mm sucrose in the presence of 0 or 100 mm fructose at 30 °C or at optimum enzyme temperature (50 °C for DgAS and 37 °C for NpAs). At the end of the reaction (24 h), samples were centrifuged at 12,000 × g for 10 min. The concentration of sucrose isomers was determined by HPAEC-PAD using an analytical CarboPACTM PA100 (4 × 250 mm) column with a CarboPAC PA-100 Guard (4 × 50 mm). Detection was performed using a Dionex ED40 module with a gold working electrode and an Ag/AgCl pH reference electrode (Dionex, Sonnyvale, CA).
Molecular Dynamics (MD) Simulations
All MD simulations were carried out using the AMBER 9 suite of programs and the all-atom ff03 force field (31, 32). The starting models were derived from the high resolution crystal structures of DgAS and NpAS (PDB code 1G5A) (18). Fifteen and 20 Na+ cations were added to neutralize the DgAS and NpAS monomers, respectively. Each protein together with its counterions was embedded in a rectangular parallelepipedal solvent box that left a space of 0.12 nm around the solute. TIP3P water molecules (∼28,000) were added using the LEaP module integrated in the AMBER 9 package (33). Simulation preparation consisted in different phases of minimization, heating, and equilibration under different type of restraints. The simulation was carried out at constant temperature (303 K) and pressure (1 bar) conditions over 60 ns. The temperature and pressure were controlled using a Langevin thermostat (34) and Berendsen barostat (35) with a collision frequency (2 ps−1) and pressure relaxation time (2 ps). Long-range electrostatic forces were handled by using the particle-mesh Ewald method (36). The time step of the simulations was 2.0 fs and the SHAKE algorithm was used to constrain the lengths of all chemical bonds involving hydrogen atoms to their equilibrium values (37).
To avoid artifacts, MD simulations were run three times with different starting velocity distribution. The resulting trajectories were analyzed using the Ptraj module of the AMBER 9 package. The root mean square (r.m.s.) deviation was calculated for protein backbone atoms using least squares fitting. Distances between protein loops were calculated with respect to their center of mass. Atomic positional fluctuations (Δri2) of protein backbone were calculated using the coordinates of the 60-ns trajectories. A mass-weighted average value was then calculated for each residue. These parameters are related to the B-factors through the following relationship.
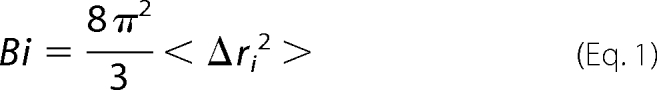 |
Sequence and Structure Alignments
Multiple sequence alignment was performed using ClustalW2 (38) and represented using ESPript (39). Structural alignments were generated using SwissPDB viewer (40) and subsequently served as a basis to align sequences of homologous proteins of unknown three-dimensional structures. Multiple sequence alignment was then manually corrected. The r.m.s. deviations among three-dimensional structures were calculated using Superpose (41) from the CCP4 suite of programs (26, 27).
RESULTS AND DISCUSSION
Overall Structure and Dynamics
Tertiary Structure of DgAS
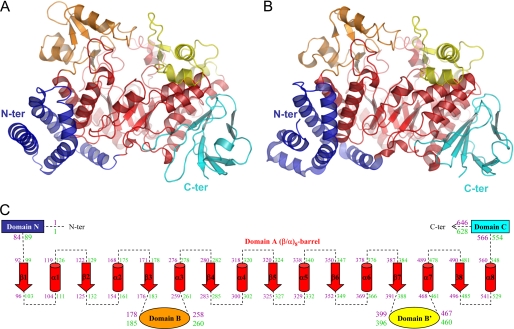
The three-dimensional structure of DgAS is organized into the five distinct domains already described for NpAS (18) (Fig. 2): the all helical N-terminal domain (residues 1 to 83), the central catalytic domain A (residues 84–177, 259–398, and 468–565), two regions protruding from domain A, constituting, respectively, domains B (178–258) and B′ (399–467), and finally the β-stranded C-terminal domain (566–646) adopting a Greek key motif. DgAS and NpAS share 38% sequence identity. Among the five regions, domains A, B, and B′ are the most conserved with a sequence identity of 43, 58, and 52%, respectively. Domains N and C only share 21 and 19% sequence identity, respectively.
FIGURE 2.
Comparison of the three-dimensional architecture of DgAS (PDB code 3UCQ) and NpAS (PDB code 1G5A (18)). Panel A, DgAS. Panel B, NpAS. Panel C, schematic view of DgAS and NpAS primary and secondary structures. The limits of the different domains are given in purple for DgAS and green for NpAS. The same color code is used throughout the different views: catalytic domain A (red), domain N (blue), domain C (cyan), domain B (orange), and domain B′ (yellow).
The overall r.m.s. deviation value obtained after secondary structure superimposition of DgAS with NpAS was 1.3 Å. Lower r.m.s. deviation values, i.e. 1.0, 1.1, and 0.9 Å, were observed for domains A, B, and B′, respectively. More significant structural differences occurred between the less conserved domains N and C leading to larger r.m.s. deviation values of 1.9 and 1.5 Å, respectively.
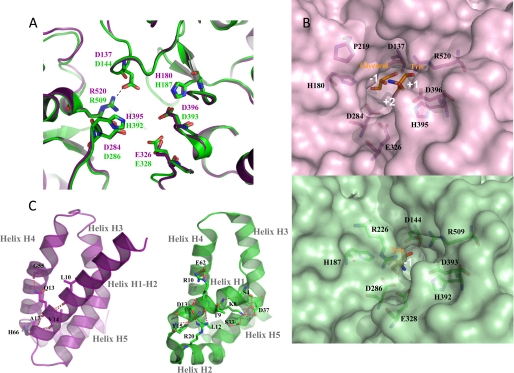
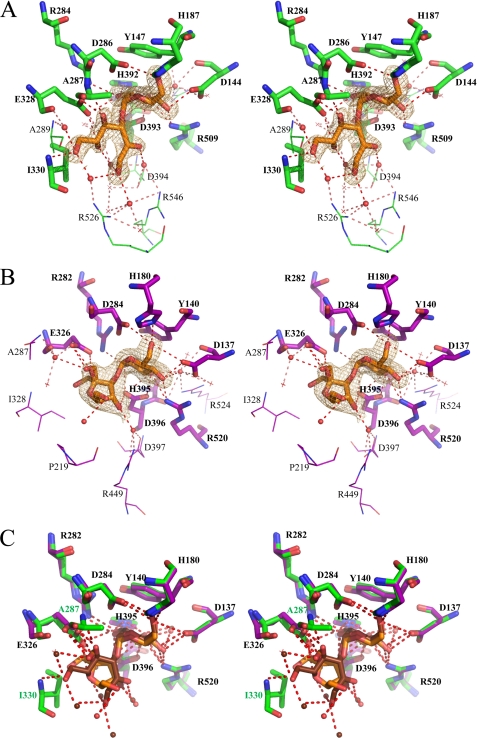
The catalytic domain A of DgAS adopts the typical (β/α)8-barrel of the GH family 13 (2). The active site is highly conserved with respect to that of NpAS (Fig. 3A). Residues Glu326 and Asp284 of DgAS were identified as the general acid/base and the nucleophile, respectively, involved in the formation of the β-glucosyl intermediate occurring in the α-retaining mechanism (corresponding to Glu328 and Asp286 in NpAS (42)). Three additional important residues conserved in the GH13 family and known to assist in catalysis are Asp396, His180, and His395 (corresponding to Asp393, His187, and His392 in NpAS, respectively). A salt bridge formed by residues Arg520 and Asp137 (equivalent to Arg509 and Asp144 in NpAS) blocks the bottom of the catalytic pocket. All the residues defining the catalytic site are conserved between NpAS and DgAS with the exception of Arg226, which is substituted by Pro219 in DgAS. As a result, the active site pocket of DgAS shows an enlarged aperture around subsite +2 compared with NpAS that may facilitate ligand accessibility and binding into the active site (Fig. 3B). Two ligands were found in the active site (Fig. 3A), a glycerol molecule (used as cryoprotectant for x-ray experiments) at subsite −1 and a Tris molecule at subsite +1, which are interacting together through a polar contact involving glycerol-O2 and Tris-O3. Both B and B′ domains are found longer in DgAS than in NpAS. Domain B is composed of 81 amino acid residues in DgAS (versus 76 residues in NpAS), organized in two antiparallel β-strands and two α-helices. As for domain B′, it is composed of four α-helices comprising 69 amino acid residues (versus 65 in NpAS). The domain C of DgAS is composed of 82 amino acid residues folded as an eight-stranded β-sandwich as previously observed in the NpAS structure. The domains C of both enzymes differ in the short α-helix between strands β4 and β5 observed only in DgAS. Additionally, the loop interconnecting strands β2 and β3 is three residues longer in DgAS than in NpAS. The main difference between the DgAS and NpAS helical domains N resides in the N-terminal extremity, which is composed by a long α-helix in DgAS and two small α-helices in NpAS (Fig. 3C). The remaining α-helices of the N-terminal domain (H3, H4, and H5) are structurally conserved in DgAS and NpAS.
FIGURE 3.
Comparison of DgAS (PDB code 3UCQ) and NpAS (PDB code 1G5A (18)) crystallographic structures. Panel A, superimposition of the active sites of DgAS (purple) and NpAS (green). Catalytic residues are shown as sticks. Ligands (Tris and glycerol) were omitted for clarity purpose. Panel B, molecular surface representation of the DgAS (purple) and NpAS (green) catalytic sites. Panel C, representation of the polar interactions involved in the stabilization of the first helix of domain N in DgAS (purple) and NpAS (green).
Insight into Enzyme Plasticity through MD Simulations
To complete our structural comparison of amylosucrases, we undertook the investigation of their dynamic properties. Large scale MD simulations (60 ns) were thus carried out in explicit water for DgAS and NpAS.
As a check for the stability of protein structures during the course of MD simulations, the time evolution of the backbone atoms r.m.s. deviation was calculated after least square fitting (supplemental Fig. S1A). Variations of backbone r.m.s. deviations indicated a distinct conformational behavior of the two enzymes. The r.m.s. deviation of DgAS rapidly increased by 3 Å within the first 12 ns of the simulation and then stabilized for the rest of the simulation. As for NpAS, the r.m.s. deviation slowly increased by 2.2 Å over the first 30 ns before reaching a plateau (supplemental Fig. S1A). Thus, faster (12 versus 30 ns) and larger amplitude (3 versus 2.2 Å) changes were observed for DgAS compared with NpAS. MD simulations further revealed that the r.m.s. deviation of DgAS mainly resulted from a high mobility of the N-terminal part of the protein (residues 1–83). As shown in supplemental Fig. S1B, the r.m.s. deviation of domain N increased by as much as 5.2 Å over the first 12 ns of the simulation for DgAS, whereas it remained unchanged for NpAS. The high flexibility of the N-extremity was also shown by the large calculated B-factor values (supplemental Fig. S1C). Such large conformational fluctuations of DgAS are due to the rearrangement of the first long helix (residues 1–24) into two shorter helices (composed of residues 1–14 and 15–24, respectively). Such conformational changes are not observed for NpAS as its first α-helix is already composed of two short helices in the x-ray structure. These helices are strongly stabilized through a large network of ionic interactions involving charged residues (Arg, Lys, Asp, and Glu) from the three first α-helices belonging to the N-terminal region of NpAS (Fig. 3C). Noteworthy, there are no equivalent charged residues in DgAS. Even if other structural regions also appear flexible, as indicated by the calculated B-factors, differences between both enzymes are, however, less significant. Most flexible regions correspond to loops of the (β/α)8-barrel that confer a pocket topology to the active site. In particular, loops 7 and 3 (encompassed in domains B′ and B, respectively) moved away from each other during the MD simulations (supplemental Fig. S1D) resulting in the opening of the catalytic pocket. With the exception of the N domain in DgAS and surface loops (2, 3, 4, 7, and 8) in both enzymes, the remaining parts of DgAs and NpAS did not exhibit any significant fluctuation (supplemental Fig. S1, E and F).
Oligomerization and Thermostability
The First Dimeric Structure of Amylosucrase
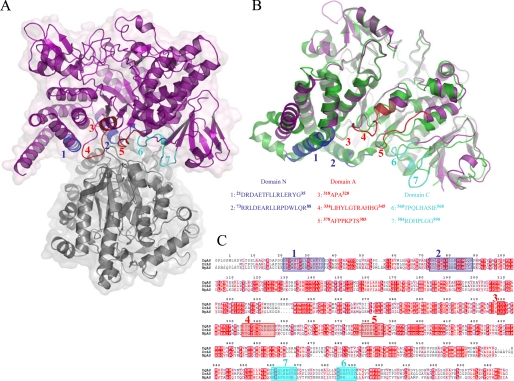
Whereas the asymmetric unit of DgAS is composed of a single peptide chain, exploration of symmetry related molecular interfaces using the PISA server (43) indicated a stable dimeric organization with a buried interface of 2,950 A2 out of a total accessible surface area of 45,330 A2 (Fig. 4A). The buried surface represents 6.1% of the solvent accessible area. The interface is defined by a set of residues located in the catalytic domain (Ala318–Ala320, Leu334–His344, Ala378–Ser385) and also in domains N (Asp21–Gly35, Arg73–Arg88) and C (Thr560–Glu568, Arg584–Gly590) (Fig. 4B). Stabilization between the protomer units of DgAS benefits from strong salt bridge interactions formed between Arg74 and Glu25 and between Arg341 and Asp84, and from a network of direct and water-mediated hydrogen-bonding interactions (supplemental Fig. S2). The residues involved in the interface stabilization of DgAS are not conserved in NpAS. In particular, Arg341, which appears to play a key role in stabilizing the DgAS interface, is one of the five additional residues of the α5–β6 interconnecting loop from the (β/α)8-barrel, which is found to be longer compared with NpAS (Figs. 2C and 4B). Of note, such an insertion region in DgAS is also found to interact through polar interactions with the Arg584–Met591 loop from domain C of the second protomer, which is four residues shorter in NpAS (Fig. 4C).
FIGURE 4.
Quaternary structure organization of DgAS (PDB code 3UCQ). Panel A, visualization of the DgAS dimer. The protomers are symmetry-related following a 2-fold crystallographic symmetry. Panel B, structural superimposition of DgAS (purple) and NpAS (PDB code 1G5A (18)) (green) monomeric units. Structural elements involved in the dimeric interface of DgAS involve 7 regions of domains N, A, and C. The amino acid sequences composing the 7 regions are listed. Panel C, sequence alignment of DgAS, DrAS, and NpAS. The 7 regions are shown on the alignment using the same color code as in panel B.
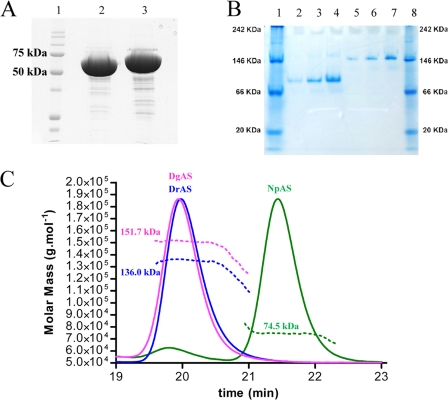
The dimerization interface displays a strong shape complementarity between the protomers. First, a bundle of hydrophobic residues from the N domain involving Leu80 and Leu81 from α-helix H5 of the first protomer fits nicely into the groove formed by the two α-helices, H3 and H5, from the second protomer (supplemental Fig. S2). Residues Leu80, Leu81, and Leu29, belonging to these α-helices and only present in DgAS, are involved in these hydrophobic contacts (supplemental Fig. S2A). In addition, the extremity of the Arg584–Gly590 loop (insertion region situated between strands β3 and β4 of the C-terminal domain in DgAS as described earlier) is found interlocked between two loops, 339–344 (another insertion region in DgAS) and 379–385 (highly conserved in the Deinococcus genus). Interestingly, in such contacting amino acid residues, we observe a bunch of prolines, which are generally known to rigidify the polypeptide backbone and may constrain the polypeptide chain conformation to facilitate dimer formation. Pro587 from the first protomer faces up Pro380, Pro381, and Pro383 from the second one. Although oligomeric states have been previously observed for other members of the GH13 family, such as cyclomaltodextrinases (CDase; EC 3.2.1.54) (PDB code 1H3G (44)), maltogenic amylase (MAase; EC 3.2.1.133 (PDB code 1SMA (45)), and neopullulanase (NPase, EC, 3.2.1.135) (PDB code 1J0H (46)), the structure of DgAS is, to our knowledge, the first homodimeric amylosucrase reported to date. Comparison of electrophoresis under denaturating (SDS-PAGE) and native conditions argue for a dimeric assembly of DgAS in solution (Fig. 5, A and B).
FIGURE 5.
Biochemical analysis of DgAS and NpAS. Panel A, SDS-PAGE of protein markers (lane 1), DgAS (lane 2), and NpAS (lane 3). Panel B, native PAGE of protein markers (lanes 1 and 8), NpAS (lanes 2–4), and DgAS (lanes 5–7); for both enzymes, no effect of dilution is visible (concentration in proteins are 0.5, 1, and 2 g liter−1, respectively). Panel C, SEC-MALLS/RI experiments for NpAS (green line), DgAS (purple line), and DrAS (blue line). Continuous lines represent the light scattering curve, dashed lines define the variation of refractive index. The experimentally measured molecular weight distribution (as horizontal lines) and the average molecular weight are indicated for each elution peaks.
The dimeric conformation of DgAS in solution was confirmed by SEC-MALLS/RI experiments. As shown in Fig. 5C, the elution volume of DgAS differed significantly from that of NpAS, corresponding to a molecule with a molecular mass of 151.7 kDa compatible with a dimeric assembly of DgAS (theoretical molecular mass of 146.3 kDa). As expected, the NpAS mainly eluted as a monomer with a molecular mass of 74.5 kDa (theoretical molecular mass of 71.5 kDa). The monomeric NpAS and the dimeric DgAS remained stable over a few weeks at 4 °C.
To search for a sequence signature of dimeric forms in other amylosucrases, we performed a sequence alignment of DgAS, NpAS and DrAS (Fig. 4C). Among the 7 regions found to be involved in DgAS dimerization from structural analysis (listed in Fig. 4B), regions 4, 6, and 7 are well conserved in DrAS. Notably, the corresponding loops in NpAS are shorter and could prevent dimer formation. To get some evidence of DrAS dimerization, SEC-MALLS analysis of DrAS was performed and it confirmed the homodimeric organization of this enzyme with a molecular mass of 136.0 kDa (theoretical molecular mass of 143.3 kDa) (Fig. 5C). The pattern composed of regions 1–7 could thus be proposed as a signature of dimerization. We screened the identified pattern against sequences of GH13 enzymes (sequence alignment is provided in supplemental Fig. S3). The proposed dimerization pattern was found in Deinococcus desertii DdAS, Deinococcus maripensis DmAS, α-amylases from Meiothermus rubber, Meiothermus silvanus and Truepera radiovictrix sequences, suggesting a similar dimeric organization to DgAS. On the other hand, sequences of amylosucrases from Neisseria and Alteromonas groups did not contain this molecular pattern. Like NpAS, one could infer that they might adopt a monomeric form (23).
DgAS Thermostability
The optimum temperature of DgAS is 50 °C, whereas an optimal temperature of 37 °C was found for NpAS (15). The t½ of freshly purified DgAS and NpAS was 69 h and 15 min at 50 °C, respectively. The higher stability of DgAS was confirmed by Tm measurements, which gave 58.9 ± 0.2 °C for DgAS versus 49.6 ± 0.4 °C for NpAS when measured by differential scanning fluorimetry and 62.0 ± 0.2 °C versus 51.3 ± 0.2 °C when determined by circular dichroism. Close inspection of the primary sequences of DgAS and NpAS gave rise to the following observation: DgAS contains (i) a higher proportion of charged residues (Lys, Arg, His, Asp, and Glu), which represent 28.8% of the total number of residues compared with only 22.9% in NpAS, (ii) a lower number of polar uncharged residues (Gly, Ser, Thr, Asn, Gln, Tyr, and Cys), which represent 24.3% of the total number of residues for DgAS and 33.4% for NpAS, (iii) more hydrophobic residues (Leu, Met, Ile, Val, Trp, Pro, Ala, and Phe) representing 46.9% of the total number of residues compared with 43.7% for NpAS, and (iv) six additional proline residues. All aforementioned features are usually observed in proteins from thermophilic organisms and could contribute to the higher thermostability of DgAS (47, 48). In addition, we have shown using MD simulations on DgAS and NpAS monomers that the N-terminal extremity of DgAS exhibits a higher flexibility. DgAS dimerization might then constrain the movement of helix H1-H2 at the N terminus of the protein and one could thus assume that dimerization is likely to contribute significantly to enzyme stabilization of this enzyme.
Specificity toward Sucrose Isomer Formation
Previous characterization of DgAS and NpAS revealed that both enzymes catalyze turanose and trehalulose synthesis as side products when incubated with sucrose but the ratio of turanose versus trehalulose production was radically different for the two enzymes (15). Other enzymes catalyzing exclusively sucrose isomerization reactions were previously characterized (7). Their primary structures contain a highly conserved motif named the “charged isomerization motif” composed of residues RLDRD, which are involved in the product specificity (49). Additionally, these enzymes contain two conserved phenylalanine forming the so-called “aromatic clamp” motif, which is involved in the tautomerization of fructose occurring during the isomerization reaction (8). Both sites are neither conserved in primary nor in tertiary structures of amylosucrases, which might help to explain the differences observed between product profiles of amylosucrases and sucrose isomerases. To further investigate the specificity for sucrose isomerization of DgAS and NpAS, experiments were carried out in the presence of 100 mm sucrose and exogenous fructose acceptor at either 30 °C or at the optimal temperature of each enzyme, i.e. 50 and 37 °C for DgAS and NpAS, respectively. The results reported in Table 2 show that, for all operational conditions, NpAS preferentially synthesized turanose. In contrast, equivalent amounts of turanose and trehalulose were produced by DgAS. Increasing the initial fructose concentration enabled sucrose isomer production yield to be enhanced. The acceptor reaction onto fructose was forced at the detriment of α-glucan chain formation. At 30 °C, 69 and 64% of the glucosyl units coming from sucrose were transferred onto d-fructose by DgAS and NpAS, respectively. However, the ratio of turanose versus trehalulose remained unchanged compared with that obtained in the absence of exogenous fructose acceptor. Similarly, increasing the temperature had no significant effect on the sucrose isomer ratio. To go further in the understanding of this feature, the crystal structures of DgAS and NpAS in complex with turanose were determined. These are the first complexes of amylosucrases with sucrose isomers.
TABLE 2.
Comparison of turanose and trehalulose formation by DgAS and NpAS expressed as the percentage of the glucosyl units coming from sucrose transferred onto fructose
| Reaction conditions | Enzyme |
|||||||
|---|---|---|---|---|---|---|---|---|
| NpAS |
DgAS |
|||||||
| 30 °C |
37 °C (optimum temperature) |
30 °C |
50 °C (optimum temperature) |
|||||
| Sucrose 100 mm | Sucrose, 100 mm, fructose, 100 mm | Sucrose, 100 mm | Sucrose, 100 mm, fructose, 100 mm | Sucrose, 100 mm | Sucrose, 100 mm, fructose, 100 mm | Sucrose, 100 mm | Sucrose, 100 mm, fructose, 100 mm | |
| % of glucosyl residues in turanose | 20.6 | 51.9 | 19.4 | 44.0 | 12.9 | 34.1 | 12.4 | 33.4 |
| % of glucosyl residues in trehalulose | 4.5 | 11.9 | 4.1 | 9.0 | 12.8 | 35.2 | 10.6 | 26.5 |
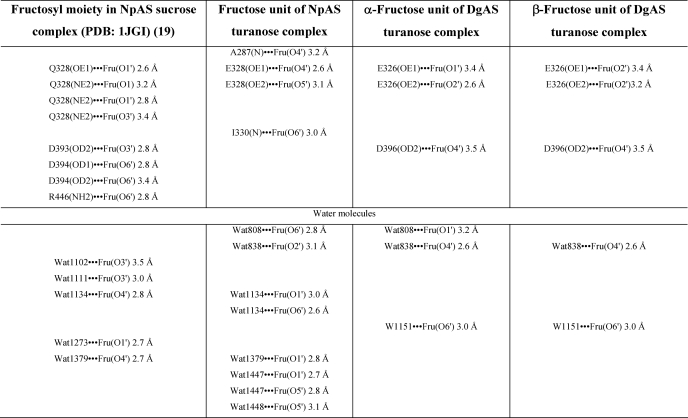
The NpAS-turanose complex was first compared with the structure of the inactive E328Q NpAS mutant in complex with sucrose (PDB code 1JGI) and no difference exceeding the structure coordinate errors was observed between the two structures (19). The structure of the NpAS-turanose complex revealed two turanose-binding sites, which were named TB1 and TB2 for the site located in the catalytic pocket and at the surface of the C domain, respectively. The electron density corresponding to the turanose bound at subsite −1 and +1 of the catalytic pocket is shown in Fig. 6A. The glucosyl moiety occupies catalytic subsite −1 in a 4C1 chair conformation. Its pyranosyl ring can be nicely superimposed onto the sucrose glucosyl ring from the x-ray structure (PDB code 1JGI) and is maintained by the same dense network of hydrogen bonding interactions involving residues Asp144, His187, Glu328, Arg284, Asp286, His392, Asp393, and Arg509 (supplemental Table S1). Glucosyl binding is also reinforced by a stacking interaction with Tyr147 and hydrogen bonds with Arg513 and Asp144 mediated by a water molecule (supplemental Fig. S4). The most striking observation arose from the fructosyl moiety binding conformation. Indeed, at subsite +1, the electron density of the ligand unequivocally revealed that the fructose unit of turanose is bound in the open state. This is quite surprising, considering that in water solution, turanose adopts an equilibria with a predominant proportion of β pyranoid (47%) and β furanoid (37%) tautomers after 20 min at 20 °C as determined by 1H NMR spectroscopy (50). The hydroxyl groups O4′, O5′, and O6′ of the open state fructose form direct hydrogen bonds with residues Ala287, Glu328, and Ile330, respectively. Noteworthy, a dense network of additional hydrogen bonds mediated by water molecules is also observed around fructose (Table 3). Within this network, the water molecule Wat1134 mediates two hydrogen bonds between Arg226 and both O1′ and O6′. In addition O6′ is also within hydrogen bond distance of the main chain NH of Ile330 and water molecule Wat1366. These interactions involving O1′ and O6′ of the fructose unit probably favor fructose binding in the open state by preventing hemiacetal formation. Such a binding mode allows perfect positioning of the O3′ for the nucleophilic attack of the β-glucosyl intermediate and the formation of turanose. It is likely responsible for the predominant turanose formation by NpAS. In the turanose molecule found in TB2, the fructose ring adopts a β-d-furanosyl conformation (supplemental Fig. S5).
FIGURE 6.
Stereoview of turanose (orange) conformation bound to NpAS (PDB code 3UEQ) (green) and DgAS (PDB code 3UER) (purple) with a σA weighted 2Fo − Fc electron density map contoured at 1.0 σ around the ligand. Hydrogen bonding interactions are shown as a red dashed line and water molecules as red spheres. Panel A, TB1 turanose-binding site of NpAS. Panel B, TB1 turanose-binding site of DgAS, the two alternative conformations of the ligand corresponding to the two turanose anomers α and β are represented. Panel C, stereoview of the structural superimposition of TB1 turanose binding sites of NpAS and DgAS using the same color code except for the turanose-NpAS complex where the turanose and water molecules are brown colored for clarity purpose. Residue numbering has been done according to DgAS sequence numbering with the exception of Ile330 and Ala287 (in green) that are only present in the interaction between NpAS and turanose.
TABLE 3.
Interactions of fructose unit from sucrose and turanose with NpAS and DgAS
In DgAS, only one turanose-binding site, equivalent to TB1 of NpAS, was observed (Fig. 6B). The absence of TB2 is likely due to the fact that residues corresponding to Phe559, Asn560, and Asn562 from domain C of NpAS are substituted by three hydrophobic residues: Lue572, Pro573, and Pro575, respectively. Such substitutions prevent stacking and hydrogen bonding interactions with turanose. In the TB1 site of DgAS, the glucosyl moiety of turanose occupies subsite −1 and adopts a 4C1 configuration with a very clear electron density. Interactions between the glucopyranosyl ring and the protein are similar to those described for the NpAS-turanose complex (supplemental Table S1). The electron density of the fructosyl ring is less clear at subsite +1. However, it defines an envelope that fits best with the α-anomer of fructofuranose (Fig. 6B). The β-anomer also probably bound subsite +1 but to a lesser extent. In addition, we cannot exclude the presence of fructose either in open or pyranoid forms. The O3′ glucosylated-α-d-fructofuranose is reported to be one of the less prevalent tautomer of turanose in water solution (1% at 20 °C (50)). Accommodation of this conformation in subsite +1 involves H-bonds with the catalytic residues Glu326 and Asp396 and two water-mediated interactions with O1′, O4′ and O6′. DgAS binds the furanoid tautomers of fructose to form turanose but via a considerably weaker network of interactions than that observed in NpAS. Further comparisons of DgAS and NpAS subsite +1 revealed structural traits that prevent open state conformation of fructose to bind DgAS subsite +1 (Fig. 6C). Indeed, residue Arg226 of NpAS, which interacts via a water molecule with both O1′ and O6′ of the open form of fructose, is replaced by Pro219 in DgAS. Furthermore, residue Ile328 corresponding to Ile330 in NpAS, which was directly involved in H-bond interaction with O6′ of fructose in the open state, cannot play the same role in DgAS due to a slight motion of the loop bearing this residue. The discrimination between the various tautomers of turanose is less drastic in DgAS than in NpAS. DgAS subsite +1 accommodates various fructose tautomers to optimally arrange O1′ or O3′ of the fructose unit for nucleophilic attack of the glucosyl enzyme intermediate therefore yielding equivalent amounts of trehalulose and turanose.
DgAS- and NpAS-turanose complex analysis thus enabled us to explain differences observed in DgAS and NpAS product specificity. This work opens new perspectives for the rational and/or semi-rational redesign of this thermostable amylosucrase to modulate sucrose isomer synthesis with the view of reducing side product formation or controlling sucrose isomer profile for the development of novel syrups enriched in sucrose substitutes.
Supplementary Material
Acknowledgments
We thank the Computing Center of Region Midi-Pyrénées (CALMIP, Toulouse, France) and the Center for Computing Resources (CRI) of INSA-Toulouse for providing computing resources and support. We are grateful to the staff of Synchrotron beamlines ID14-1 and ID29 at the European Synchrotron Radiation Facility (Grenoble, France) for providing assistance in using the beamlines. We are grateful to Javier Perez (Synchrotron SOLEIL) for the fruitful discussions. We also thank Stéphanie Terme (Wyatt technology France) for helpful assistance in the SEC-MALLS experiments.

This article contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 3UCQ, 3UER, and 3UEQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AS
- amylosucrase
- NpAS
- N. polysaccharea amylosucrase
- DgAS
- D. geothermalis amylosucrase
- DrAS
- D. radiodurans amylosucrase
- AmAS
- A. macleodii amylosucrase
- SEC
- size exclusion chromatography
- MALLS
- multiangle laser light scattering
- MD
- molecular dynamics
- r.m.s.
- root mean square
- PDB
- Protein Data Bank.
REFERENCES
- 1. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 3. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Potocki de Montalk G., Remaud-Simeon M., Willemot R. M., Sarçabal P., Planchot V., Monsan P. (2000) Amylosucrase from Neisseria polysaccharea. Novel catalytic properties. FEBS Lett. 471, 219–223 [DOI] [PubMed] [Google Scholar]
- 5. Okada G., Hehre E. J. (1974) New studies on amylosucrase, a bacterial α-d-glucosylase that directly converts sucrose to a glycogen-like α-glucan. J. Biol. Chem. 249, 126–135 [PubMed] [Google Scholar]
- 6. Goulter K. C., Hashimi S. M., Birch R. G. (2012) Microbial sucrose isomerases. Producing organisms, genes, and enzymes. Enzyme Microb. Technol. 50, 57–64 [DOI] [PubMed] [Google Scholar]
- 7. Ravaud S., Robert X., Watzlawick H., Haser R., Mattes R., Aghajari N. (2007) Trehalulose synthase native and carbohydrate-complexed structures provide insights into sucrose isomerization. J. Biol. Chem. 282, 28126–28136 [DOI] [PubMed] [Google Scholar]
- 8. Ravaud S., Robert X., Watzlawick H., Haser R., Mattes R., Aghajari N. (2009) Structural determinants of product specificity of sucrose isomerases. FEBS Lett. 583, 1964–1968 [DOI] [PubMed] [Google Scholar]
- 9. Thompson J., Pikis A. (2012) Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol. Oral Microbiol. 27, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seo D. H., Jung J.-H., Ha S.-J., Song M.-C., Cha J., Yoo S. H., Kim T.-J., Baek N. I., Park C.-S. (2009) Highly selective biotransformation of arbutin to arbutin-α-glucoside using amylosucrase from Deinococcus geothermalis DSM 11300. J. Mol. Catal. B 60, 113–118 [Google Scholar]
- 11. André I., Potocki-Véronèse G., Morel S., Monsan P., Remaud-Siméon M. (2010) Sucrose-utilizing transglucosidases for biocatalysis. Top. Curr. Chem. 294, 25–48 [DOI] [PubMed] [Google Scholar]
- 12. Chang A., Singh S., Phillips G. N., Jr., Thorson J. S. (2011) Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol 22, 800–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Champion E., André I., Moulis C., Boutet J., Descroix K., Morel S., Monsan P., Mulard L. A., Remaud-Siméon M. (2009) Design of α-transglucosidases of controlled specificity for programmed chemoenzymatic synthesis of antigenic oligosaccharides. J. Am. Chem. Soc. 131, 7379–7389 [DOI] [PubMed] [Google Scholar]
- 14. Pizzut-Serin S., Potocki-Véronèse G., van der Veen B. A., Albenne C., Monsan P., Remaud-Simeon M. (2005) Characterization of a novel amylosucrase from Deinococcus radiodurans. FEBS Lett. 579, 1405–1410 [DOI] [PubMed] [Google Scholar]
- 15. Emond S., Mondeil S., Jaziri K., André I., Monsan P., Remaud-Siméon M., Potocki-Véronèse G. (2008) Cloning, purification and characterization of a thermostable amylosucrase from Deinococcus geothermalis. FEMS Microbiol Lett 285, 25–32 [DOI] [PubMed] [Google Scholar]
- 16. Ha S. J., Seo D. H., Jung J. H., Cha J., Kim T. J., Kim Y. W., Park C. S. (2009) Molecular cloning and functional expression of a new amylosucrase from Alteromonas macleodii. Biosci. Biotechnol. Biochem. 73, 1505–1512 [DOI] [PubMed] [Google Scholar]
- 17. Skov L. K., Mirza O., Henriksen A., Potocki de Montalk G., Remaud-Simeon M., Sarcabal P., Willemot R. M., Monsan P., Gajhede M. (2000) Crystallization and preliminary X-ray studies of recombinant amylosucrase from Neisseria polysaccharea. Acta Crystallogr. D 56, 203–205 [DOI] [PubMed] [Google Scholar]
- 18. Skov L. K., Mirza O., Henriksen A., De Montalk G. P., Remaud-Simeon M., Sarçabal P., Willemot R. M., Monsan P., Gajhede M. (2001) Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J. Biol. Chem. 276, 25273–25278 [DOI] [PubMed] [Google Scholar]
- 19. Skov L. K., Mirza O., Sprogøe D., Dar I., Remaud-Simeon M., Albenne C., Monsan P., Gajhede M. (2002) Oligosaccharide and sucrose complexes of amylosucrase. Structural implications for the polymerase activity. J. Biol. Chem. 277, 47741–47747 [DOI] [PubMed] [Google Scholar]
- 20. Jensen M. H., Mirza O., Albenne C., Remaud-Simeon M., Monsan P., Gajhede M., Skov L. K. (2004) Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea. Biochemistry 43, 3104–3110 [DOI] [PubMed] [Google Scholar]
- 21. Emond S., André I., Jaziri K., Potocki-Véronèse G., Mondon P., Bouayadi K., Kharrat H., Monsan P., Remaud-Simeon M. (2008) Combinatorial engineering to enhance thermostability of amylosucrase. Protein Sci. 17, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung J. H., Seo D. H., Ha S. J., Song M. C., Cha J., Yoo S. H., Kim T. J., Baek N. I., Baik M. Y., Park C. S. (2009) Enzymatic synthesis of salicin glycosides through transglycosylation catalyzed by amylosucrases from Deinococcus geothermalis and Neisseria polysaccharea. Carbohydr. Res. 344, 1612–1619 [DOI] [PubMed] [Google Scholar]
- 23. De Montalk G. P., Remaud-Simeon M., Willemot R. M., Planchot V., Monsan P. (1999) Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 181, 375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM, a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 28. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) COOT, model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 32. Lee M. C., Duan Y. (2004) Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins 55, 620–634 [DOI] [PubMed] [Google Scholar]
- 33. Case D. A., Darden T. E., Cheatham I. T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. A. (2006) AMBER 9, University of California, San Francisco, CA [Google Scholar]
- 34. Pastor R. W., Brooks B. R., Szabo A. (1988) An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65, 1409–1419 [Google Scholar]
- 35. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 36. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 37. Ryckaert J.-P., Ciccotti G., Berendsen H. J. C. (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 38. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 39. Gouet P., Robert X., Courcelle E. (2003) ESPript/ENDscript. Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 41. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 42. Sarçabal P., Remaud-Simeon M., Willemot R., Potocki de Montalk G., Svensson B., Monsan P. (2000) Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474, 33–37 [DOI] [PubMed] [Google Scholar]
- 43. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 44. Fritzsche H. B., Schwede T., Schulz G. E. (2003) Covalent and three-dimensional structure of the cyclodextrinase from Flavobacterium sp. No. 92. Eur. J. Biochem. 270, 2332–2341 [DOI] [PubMed] [Google Scholar]
- 45. Kim J. S., Cha S. S., Kim H. J., Kim T. J., Ha N. C., Oh S. T., Cho H. S., Cho M. J., Kim M. J., Lee H. S., Kim J. W., Choi K. Y., Park K. H., Oh B. H. (1999) Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J. Biol. Chem. 274, 26279–26286 [DOI] [PubMed] [Google Scholar]
- 46. Hondoh H., Kuriki T., Matsuura Y. (2003) Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J. Mol. Biol. 326, 177–188 [DOI] [PubMed] [Google Scholar]
- 47. Jaenicke R., Böhm G. (1998) The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8, 738–748 [DOI] [PubMed] [Google Scholar]
- 48. Prakash O., Jaiswal N. (2010) α-Amylase, an ideal representative of thermostable enzymes. Appl. Biochem. Biotechnol. 160, 2401–2414 [DOI] [PubMed] [Google Scholar]
- 49. Zhang D., Li N., Swaminathan K., Zhang L. H. (2003) A motif rich in charged residues determines product specificity in isomaltulose synthase. FEBS Lett. 534, 151–155 [DOI] [PubMed] [Google Scholar]
- 50. Lichtenthaler F. W., Rönninger S. (1990) Studies on ketoses, 4. -d-glucopyranosyl-d-fructoses: distribution of furanoid and pyranoid tautomers in water, dimethyl sulphoxide, and pyridine. J. Chem. Soc. Perkin Trans. 2, 1489–1497 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.