Background: RNA-dependent RNA polymerases of nonsegmented negative-strand RNA viruses (Mononegavirales) harbor multiple catalytic activities.
Results: Domain screening and trans-complementation of Paramyxovirus polymerase fragments with dimerization tags identifies independent folding-competent domains.
Conclusion: Paramyxovirus polymerases harbor at least two independent domains that lack high-affinity interfaces but require molecular compatibility for bioactivity.
Significance: First demonstration of Mononegavirales polymerase trans-complementation sets the stage for structural analyses of folding domains.
Keywords: Protein Domains, RNA Polymerase, RNA Viruses, Viral Polymerase, Viral Replication
Abstract
All enzymatic activities required for genomic replication and transcription of nonsegmented negative strand RNA viruses (or Mononegavirales) are believed to be concentrated in the viral polymerase (L) protein. However, our insight into the organization of these different enzymatic activities into a bioactive tertiary structure remains rudimentary. Fragments of Mononegavirales polymerases analyzed to date cannot restore bioactivity through trans-complementation, unlike the related L proteins of segmented NSVs. We investigated the domain organization of phylogenetically diverse Paramyxovirus L proteins derived from measles virus (MeV), Nipah virus (NiV), and respiratory syncytial virus (RSV). Through a comprehensive in silico and experimental analysis of domain intersections, we defined MeV L position 615 as an interdomain candidate in addition to the previously reported residue 1708. Only position 1708 of MeV and the homologous positions in NiV and RSV L also tolerated the insertion of epitope tags. Splitting of MeV L at residue 1708 created fragments that were unable to physically interact and trans-complement, but strikingly, these activities were reconstituted by the addition of dimerization tags to the fragments. Equivalently split fragments of NiV, RSV, and MeV L oligomerized with comparable efficiency in all homo- and heterotypic combinations, but only the homotypic pairs were able to trans-complement. These results demonstrate that synthesis as a single polypeptide is not required for the Mononegavirales polymerases to adopt a proper tertiary conformation. Paramyxovirus polymerases are composed of at least two truly independent folding domains that lack a traditional interface but require molecular compatibility for bioactivity. The functional probing of the L domain architecture through trans-complementation is anticipated to be applicable to all Mononegavirales polymerases.
Introduction
The Paramyxovirus family comprises major human and animal pathogens including measles virus (MeV),2 mumps virus, Newcastle disease virus (NDV), the recently emerged highly pathogenic Nipah virus (NiV), and respiratory syncytia virus (RSV). Collectively, members of the family are responsible for substantial morbidity and mortality worldwide. Belonging to the Mononegavirales, the Paramyxoviridae store all genetic information in a single RNA genome of negative polarity, which is encapsidated by the viral nucleocapsid (N) protein in a protein-RNA (ribonucleoprotein) complex (1). Cytosolic transcription and replication of the viral genome is mediated by the viral RNA-dependent RNA polymerase (RdRp) complex, which includes the ribonucleoprotein and viral P and ∼2200-residue L proteins. Besides genome replication, the latter is responsible for viral mRNA synthesis, requiring catalysis of RNA capping, methylation, and polyadenylation in addition to phosphodiester bond formation (1).
Different catalytic activities mediated by a single polypeptide are well compatible with a multidomain architecture, in which individually folding structural domains with discrete functions are connected through linker regions with low intrinsic structure, thus ensuring local concentration of enzymatic activities in a flexible three-dimensional scaffold. Consistent with this model, analysis of purified L proteins of segmented and nonsegmented NSV proteins by electron microscopy supported a structural organization in discernible domains (2, 3). Furthermore, a systematic bioinformatics-based analysis of the L protein of Lassa virus, a distantly related segmented NSV of the Arenavirus family, has revealed that the protein can be split at two positions into distinct fragments that are capable of reconstituting RdRp bioactivity through trans-complementation when co-expressed (4). Thus, Lassa virus polymerase is composed of at least three distinct, independently folding-competent structural domains. Comparable trans-complementation studies with Mononegavirales-derived L proteins are limited to date to the L protein of vesicular stomatitis virus (VSV) of the Rhabdoviridae. By contrast to Lassa virus L, a recent report found N- and C-terminal VSV polymerase fragments were unable to trans-complement each other functionally (3), although these VSV L subunits tolerated separation by an enhanced GFP moiety (5). This was interpreted to reflect that proper tertiary folding of the Mononegavirales polymerase may require synthesis of the protein as a single polypeptide (3).
Based on sequence alignments between different NNSV family representatives, six conserved regions (CR I to CR VI) of higher sequence homology have been identified in the L proteins (6, 7). Of these, the N-terminal sections harboring CR I have been implicated in L oligomerization (8–10) and/or L interaction with P (8, 9, 11–14), CR III in RNA polymerization (15, 16), and CR VI in methyltransferase activity (3, 6, 17). A conserved GXXTnHR motif in NNSV L CR V, which was first identified in VSV L, is furthermore thought to mediate unusual capping of the viral mRNAs through transfer of 5′-monophosphate-mRNA onto GDP (18, 19). This polyribonucleotidyltransferase activity differs from that of eukaryotic mRNA capping through guanylyltransferases, which transfer guanosine monophosphate to pp-mRNA to form the cap structure (20). Surprisingly, Paramyxovirus, but not rhabdovirus, L proteins also may contain traditional guanylyltransferase activity. Purified polymerase of rinderpest virus, a member of the Morbillivirus genus within the Paramyxoviridae, reportedly formed covalent guanosine monophosphate L intermediates in vitro (21), and a largely conserved guanylyltransferase consensus motif required for transcriptase activity was identified near the C terminus of the human parainfluenza virus (HPIV) type 2 L protein (22).
Within the Paramyxovirus family, sequence alignments of different Morbillivirus genus polymerases suggested three large regions (LR I to LR III) separated by variable connectors (23, 24). Of these, LR I harbors CR I and II and thus the oligomerization domain, and LR II contains CR III-CR V including the predicted polymerase and polyribonucleotidyltransferase activities, whereas LR III is considered to encompass the methyltransferase and recently proposed guanylyltransferase functions. Similar to recent reports for VSV L (5), L proteins of MeV, the archetype of the Morbillivirus genus, and rinderpest virus furthermore tolerated insertions into the LR II/LR III but not the LR I/LR II junction (23, 25), consistent with at least a two-domain organization. However, a comprehensive analysis of the Paramyxovirus L domain architecture and direct assessment of whether Paramyxovirus L fragments located on either side of the LR II/LR III intersection remain competent for folding when expressed separately remains elusive. In analogy to the findings obtained for VSV L (3), synthesis as an intact polypeptide may constitute a generally conserved prerequisite for Mononegavirales L bioactivity.
To dissect fundamental principles that govern NNSV polymerase folding, we examined in this study three phylogenetically diverse Paramyxovirus L proteins. Commencing with MeV L, we employed in silico structure predictions combined with experimental evaluation to identify individual folding domain candidates. Guided by this screen, we generated split MeV, NiV, and RSV L pairs and explored their ability to reconstitute RdRp activity in biochemical and functional assays. Our results demonstrate that Paramyxovirus polymerases are composed of at least two discrete structural domains that are capable of independent folding. Neither section contains a high affinity protein-protein interface, but close physical proximity and homotypic origin from the same L protein are essential for restoring RdRp bioactivity. These findings illuminate basic principles of NNSV polymerase architecture and provide a tangible path toward characterizing the structural organization of distantly related Mononegavirales polymerases beyond the Paramyxovirus family. They guide the identification of meaningful NNSV substructures that may be more suitable for x-ray crystallographic analysis than full-length L proteins.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Baby hamster kidney (BHK-21) cells stably expressing T7 polymerase (BSR-T7/5) (26) were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and incubated at every third passage in the presence of G-418 (Geneticin) at a concentration of 100 μg/ml. Lipofectamine 2000 (Invitrogen) was used for cell transfections.
Replicon Reporter Systems
Base vectors for all MeV replicon experiments were previously reported plasmids containing the MeV-Edmonston (Edm) strain-derived L, N, or P open reading frames under the control of the T7 promoter (27). To generate an MeV luciferase replicon reporter construct, the terminal untranslated regions of the MeV genome were added to the firefly luciferase open reading frame (Promega) through recombination PCR (all oligonucleotide primers used in this study are listed in supplemental Table 1, entries #1–7) followed by replacement of the chloramphenicol (CAT) reporter cassette in the previously reported MeV-CAT replicon plasmid (27) with the sequence-confirmed recombination product. For NiV, previously described plasmids containing the NiV L, N, or P open reading frames and a NiV-CAT reporter construct were used as starting material (28). Sequence-optimized cDNA copies encoding the RSV L, N, P, or M2–1 genes based on the strain RSV A2 were synthesized in vitro (GeneArt; codon-optimized sequences are shown in supplemental Table 2) and cloned into the pcDNA3.1 expression vector (Invitrogen). The RSV minigenome reporter pRSVlucM5 was constructed reminiscent of a previously described RSV minigenome (29). Four overlapping oligonucleotides were annealed to form a 238-bp DNA fragment containing a terminal BamHI site, the upstream 32 nucleotide (nt) nonstructural protein 1 (NS1) nontranslated region, the 10-nt RSV NS1 gene start signal, a 44-nt RSV leader sequence, a 94-bp hammerhead ribozyme, a 47-bp T7 terminator, and a NotI compatible end (supplemental Table 1; entries #8–11). A 191-bp DNA fragment containing a terminal HindIII site, a 155-nt RSV trailer sequence, the 12-nt RSV L gene end sequence, a 12-nt nontranslated region of RSV L, and an XhoI site was synthesized in vitro (Integrated DNA Technologies; supplemental Table 1, entry #12). These two fragments were ligated along with a BamHI/XhoI fragment of firefly luciferase cDNA (pGEM-luc, Promega) into the NotI and HindIII sites of pcDNA3.1 such that an antisense copy of luciferase flanked by RSV leader and trailer regulatory elements is produced by T7 polymerase transcription.
Mutagenesis and Generation of Expression Plasmids
For linker insertion scanning, the MeV L expression plasmid was subjected to PCR amplification using primers that duplicate the codon at the insertion site and introduce an AfeI restriction site (supplemental Table 1, entries #13–36). Amplicons were subjected to DpnI digest to remove template material followed by digestion with AfeI and ligation. Correct insertion of the linker domain was confirmed by DNA sequencing. Analogous strategies were employed to introduce streptactin SII-triple FLAG tandem tags into MeV L or triple HA tags into MeV, NiV, and RSV L (supplemental Table 1, entries #37–40 for MeV L SII-FLAG tagging; entries # 41–44 for MeV L HA tagging; entries #45–48 for NiV L HA tagging; entries #49–52 for RSV L HA tagging).
To generate the MeV LN-frag expression construct, a stop codon was introduced in-frame downstream of the SII-FLAG tag in the MeV LFLAG expression plasmid (supplemental Table 1, entry #53) using directed mutagenesis after the QuikChange protocol (Stratagene). Correct insertion was confirmed by sequence analysis and immunoblotting. To generate the MeV LC-frag construct, a SpeI site followed by an ATG start codon was introduced in MeV LHA frame upstream of the HA tag (supplemental Table 1, entry #54) through PCR amplification of the C-terminal fragment. A SpeI/SalI fragment of the sequence-confirmed amplicon was then ligated into the SpeI/SalI-digested MeV L expression vector, thus replacing the L gene with the new insert.
To add a GCN4 tag to the MeV LN-frag construct, the BstBI site in the SII-FLAG cassette was first removed through directed mutagenesis (supplemental Table 1, entry #55). Using appropriate primers (supplemental Table 1, entries #56–57), the GCN4 tag was introduced through PCR amplification followed by DpnI digest of the template DNA and recirculation of the amplicon after BstBI digest. An equivalent strategy was employed for GCN4 tagging of the MeV LC-frag construct using primers annealing at the third copy of the HA tag and the ATG start codon, respectively (supplemental Table 1, entries #58–59). To generate SII-FLAG-tagged LN-frag-GCN4 expression constructs of NiV and RSV L, the SII-FLAG-GCN4 cassette of MeV LN-frag-GCN4 was amplified followed by joining in frame to the N termini of the L fragments through recombination PCR (supplemental Table 1, entries #60–63 for NiV; entries #66–69 for RSV). Sequence-confirmed amplicons were transferred into the NiV or RSV L expression plasmids. HA-tagged LC-frag-GCN4 expression constructs of NiV and RSV L were generated through PCR-mediated insertion of the GCN4 sequence (supplemental Table 1, entries #64–65 for NiV LHA; entries #70–71 for RSV LHA). Silent BstBI (NiV L) or BlpI (RSV L) restriction sites introduced into the GCN4-encoding sequences enabled re-circularization of the amplicons after DpnI-mediated removal of the template DNA and digestion with BstBI or BlpI, respectively. All constructs were sequence confirmed before further experimentation.
Minireplicon Reporter Assays of RdRp Activity
BSR-T7/5 cells (2.5 × 105 per well in a 12-well plate format) were transfected with plasmid DNAs encoding the viral RdRp components (unless otherwise specified, DNA amounts were for MeV: MeV-L (1.1 μg), MeV-N (0.4 μg), and MeV-P (0.3 μg); for NiV: NiV-L (0.2 μg), NiV-N (0.63 μg), and NiV-P (0.4 μg); for RSV, RSV-L (0.1 μg), RSV-M2 (0.15 μg), RSV-N (0.17 μg), and RSV-P (0.15 μg)), and 1 μg of the MeV or RSV luciferase replicon reporter or the NiV CAT replicon reporter plasmid. Control wells included identical amounts of reporter and helper plasmids but lacked the plasmids harboring the respective L gene or, for some experiments, L fragment subunits. In all cases vector (pUC19) DNA was added as necessary so that all transfection mixtures contained identical amounts of total DNA. Thirty-eight hours post-transfection, cells were lysed, and luciferase activities in cleared lysates were determined using Bright-Glo luciferase substrate (Promega) and an Envision Multilabel microplate reader (PerkinElmer Life Sciences), or CAT concentrations were assessed using a CAT-ELISA assay system (Roche Applied Science). Statistical significance of results was determined where indicated using Student's t test.
SDS-PAGE and Immunoblotting
BSR-T7/5 cells (5 × 105) were transfected with 3 μg of plasmid DNA encoding MeV, NiV, or RSV L protein or L protein fragments as specified. Thirty-eight hours post-infection cells were washed in phosphate-buffered saline (PBS), lysed for 10 min at 4 °C in lysis buffer (50 mm Tris, pH 8.0, 62.5 mm EDTA, 0.4% deoxycholate, 1% Igepal (Sigma)) containing protease inhibitors (Complete mix (Roche Applied Science)) and 1 mm phenylmethylsulfonyl fluoride (PMSF), and centrifuged at 20,000 × g for 10 min at 4 °C. Cleared lysates were mixed with equal volumes of urea buffer (200 mm Tris-Cl, pH 6.8, 8 m urea, 5% sodium dodecyl sulfate (SDS), 0.1 mm EDTA, 0.03% bromphenol blue, 1.5% dithiothreitol) and incubated for 30 min at 50 °C. Samples were fractionated on SDS-polyacrylamide gels, blotted to polyvinylidene difluoride membranes (Millipore), and subjected to enhanced chemiluminescence detection (Amersham Biosciences) using specific antibodies directed against the FLAG (M2; Sigma) or HA (16B12; Abcam) epitopes, cellular GAPDH (Ambion), or MeV P (Chemicon) as specified. Blots were developed using a ChemiDoc XRS digital imaging system (Bio-Rad), and signals were assessed with the Image Lab software package.
Co-immunoprecipitation
BSR-T7/5 cells (5 × 105/well) were transfected with plasmid DNA encoding MeV, NiV, or RSV L, different LN-frag or LC-frag subunits, or MeV P as specified in the individual experiments. At 38 h post-transfection, cells were washed 5 times with cold PBS and lysed in co-immunoprecipitation buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), protease inhibitors (Roche Applied Science), and 1 mm PMSF). Cleared lysates (20,000 × g; 30 min; 4 °C) were incubated with specific antibodies directed against the FLAG or HA epitopes as specified at 4 °C, followed by precipitation with immobilized protein G (Pierce) at 4 °C. Precipitates were washed 3 times each in buffer A (100 mm Tris, pH 7.6, 500 mm lithium chloride, 0.1% Triton X-100), then buffer B (20 mm HEPES, pH 7.2, 2 mm EGTA, 10 mm magnesium chloride, 0.1% Triton X-100) followed by resuspension in urea buffer. Denatured samples were fractionated on SDS-polyacrylamide gels (4–20% gradient or 10% depending on the antigenic material assessed) followed by immunoblotting and chemiluminescence detection using specific antibodies directed against the FLAG or HA epitopes, cellular GAPDH, or MeV P as described above.
In Silico Assessment of Protein Domain Architecture
For DomCut (30)-based identification of candidate linker insertion sites, Paramyxovirus L protein sequences were aligned using the ClustalW2 (31) and MUSCLE (32) algorithm as alternative solutions in three distinct settings representing 1) different MeV genotypes, 2) a diverse panel of morbilliviruses, or 3) members of all Paramyxovirus genera. For each individual input sequence, interdomain linkers were predicted, and relative DomCut propensity scores then averaged separately based on the different sequence alignments. DomCut values for all residues that lacked an L-Edm homologue were excluded. Input L sequences were derived from MeV-Edm (genotype A) (33), MeV-Gambia (genotype B2) (34), MeV-Gambia (genotype B3.2) (35), MeV-Toulon (genotype C2) (36), MeV-Illinois (genotype D3) (34), MeV-Alaska (genotype H2) (37) for (1), MeV-Edm, MeV-Gambia B2, MeV-Gambia B3.2, MeV-Toulon, MeV-Illinois, MeV-Alaska, rinderpest virus-KabeteO (38), rinderpest virus-RBOK (39), canine distemper virus Onderstepoort (40), canine distemper virus 5804 (41), peste des petits ruminants virus Turkey 2000 (42), dolphin Morbillivirus (43) for (2), or MeV-Edm, canine distemper virus 5804, NIV (44), HPIV type 1 C35 (45), HPIV type 3 LZ22 (46), HPIV type 2 (47), HPIV type 4 SKPIV4 (48), NDV-ISG0210 (GenBankTM JF340367), Tupaia Paramyxovirus (49), RSV A2 (50), human metapneumovirus Sabana (51) for (3). MeDor (52) was employed to independently predict disordered domains in L-Edm using the IUPred (53), GlobPlot2 (54), DisEMBL (55), FoldIndex (56), and RONN (57) algorithms. In addition, MeV L-Edm was submitted to PONDR-FIT (58) and DRIP-PRED (59) for disorder predictions. To assess the consensus of all algorithms quantitatively, GlobPlot2, FoldIndex, and DomCut average values were shifted to positive integers, and positive output scores of all algorithms were normalized for identical hit cut-off values, averaged, transformed to a 0–10 scale, and plotted as a function of L-Edm residues. MeV L-Edm secondary structure prediction was based on the StrBioLib library of the Pred2ary program (60, 61) embedded in the MeDor package.
RESULTS
In Silico Domain Analysis of Paramyxovirus L Protein
Based on the concept that protein domains are the smallest autonomously folding-competent units within a protein structure (62), linker regions connecting individual domains are expected to show little structural order and comparably low sequence conservation. A variety of predictive algorithms have been developed that seek to identify intrinsically disordered proteins or disordered sections within a protein by means of specific sequence signatures of unstructured regions (63, 64). Because combining different predictors that explore discrete biophysical parameters reportedly boosts the accuracy of the prediction (52, 65, 66), we employed a panel of algorithms for a comprehensive in silico analysis of candidate interdomain boundaries within the Paramyxovirus L protein (Fig. 1, A and B).
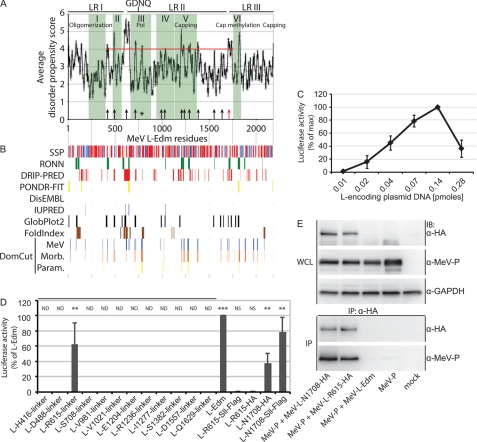
FIGURE 1.
Identification of MeV L folding domains through linker insertion analysis. A, average disorder propensity scores obtained through a combination of eight algorithms (listed in B) are plotted as a function of MeV L. Conserved regions I-VI (green shading; annotations according to predicted activities are detailed in the Introduction), the postulated catalytic center for RNA polymerization (GDNQ), and the area and score cut-off for experimental testing (red line) are highlighted. Black arrows mark residues that were chosen for subsequent linker insertion analysis, the red arrow identifies the position in the LR II/LR III section that was previously demonstrated to tolerate epitope tag insertions (23), and the black asterisk marks a hit candidate that was not tested due to its immediate proximity to the GDNQ catalytic motif. Pol, polymerase domain. B, shown is a graphic representation of individual predictions. To identify candidate disordered domains in MeV L, the MeDor (encompassing RONN, DisEMBL REM465, IUPRED, GLOBPLOT2, and FoldIndex) meta-analysis tool, PONDR-FIT, DRIP-PRED, and DomCut algorithms were used. Secondary structure predictions (SSP) are based on the Pred2ary program embedded in MeDor and show postulated α-helical (red) or β-sheet (blue) regions. Graphic predictions were aligned with the propensity score averages shown in A. C, plasmid titration to determine activity maxima of a firefly luciferase MeV RdRp reporter system is shown. Peak luciferase activities were observed at a molar ratio of 1:0.6 and 0.4 (L-encoding plasmid DNA:N- and P-encoding plasmid DNA). Total amounts of DNA transfected/well were kept constant, and values represent averages of four independent experiments ± S.D. D, linker insertion analysis to map MeV L domain intersections is shown. Twelve 10-residue linkers of the specified sequence were inserted at the indicated positions (corresponding to the black arrows in A). In addition, HA and tandem SII-FLAG epitope tags were inserted after L residues 615 and 1708. Results are expressed relative to luciferase activity observed after transfection of standard L-Edm and represent averages of three independent experiments ± S.D. Statistical analysis probes activity deviation relative to control transfections that received vector DNA in place of the L expression plasmid (**, p < 0.01; ***, p < 0.001; NS, not significant; ND, not determined). E, inactive MeV L615-HA and bioactive L1708-HA efficiently interact with MeV P. Whole cell lysates (WCL) of BSR-T7/5 cells co-expressing MeV P and the different L constructs as indicated were subjected to gel fractionation and immunoblotting (IB) or to reciprocal immunoprecipitation (IP) using specific antibodies directed against the HA epitopes followed by anti-P immunostaining. GAPDH was detected as internal standard. Control samples expressed untagged standard MeV L-Edm or MeV P alone.
The DomCut package predicts candidate linker regions based on a data set of domain/linker segments (30). We targeted three groups of viruses in these studies. Focusing on MeV L as the initial target for experimental assessment, we generated average DomCut interdomain propensity scores for L protein sequences representing a variety of MeV genotypes. We then examined different members of the Morbillivirus genus and finally members of each genus of the Paramyxovirinae and Pneumovirinae subfamilies of the Paramyxovirinae in this way. DomCut output scores then were cross-referenced quantitatively (Fig. 1A) and graphically (Fig. 1B) with the MeDor (MEtaserver of DisOrder (52)), PONDR-FIT meta-predictor (58), and DRIP-PRED (59) predictors of unstructured sections, resulting in a total application of eight algorithms. These studies highlighted 12 candidate regions in MeV L that were located between CR I and CR VI, received combined propensity scores of ≥4, and were identified by at least two of the algorithms employed. Focusing on the L core, potential interdomain regions located in the N-terminal first 408-amino acid section of MeV L, which mediates both L interaction with the viral P protein and polymerase oligomerization (8, 14), and candidates positioned downstream of the LR II/LR III junction (residues 1695–1717 (23, 24)) were not considered for experimental evaluation.
Linker Insertion Analysis of MeV Polymerase Organization
To assess the quality of the in silico predictions, 10-residue hydrophilic linker peptides were engineered into each of the selected regions. In addition, two L variants were generated that contain HA or streptactin-FLAG epitope insertions at position 1708 in the LR II/LR III junction, guided by a previously described epitope-tagged MeV L construct (23, 25). To determine the effect of linker insertions on L bioactivity, we employed a replicon reporter system that contains a firefly luciferase reporter gene flanked by the MeV non-coding genome termini. Co-expression of this construct with MeV N, P, and L proteins generated N-encapsidated RNA of negative polarity that served as the template for luciferase mRNA synthesis by the viral RdRp complex. Variation of the molar ratio of transfected L-encoding plasmid DNA relative to N- and P-encoding plasmids revealed a steep optimum curve with peak luciferase activities measured at 0.14:0.09 and 0.06 pmol (L:N and P) of plasmid DNA transfected (Fig. 1C, signal:background, 320 at peak luciferase activity). Subsequent experiments were performed at this molar ratio.
When examining the different L variants with linker insertions in this assay, we found that the l-Arg-615 linker construct returned significant luciferase activity equivalent to ∼60% that of standard L, whereas all other L variants lacked appreciable bioactivity (Fig. 1D). Replacement of the linker domain at residue 615 with streptactin-FLAG or HA epitope tags, however, substantially reduced L bioactivity to <1% that of standard L. Consistent with previous experiments (23), insertion of these epitope tags at position 1708 in the C-terminal hypervariable region did not abolish L activity (Fig. 1D).
A previous molecular characterization of MeV L regions necessary for interaction with the viral P protein highlighted the N-terminal L residues 1–408 as directly required for P binding (14). To explore whether epitope insertion at L position 615 compromises the interaction with P, we examined the physical interaction of the proteins biochemically. Both bioactive l-Asn-1708-HA and inactive l-Arg-615-HA efficiently and specifically co-immunoprecipitated the P protein (Fig. 1E), suggesting unperturbed hetero-oligomerization of the proteins.
These results support a model in which the overall MeV L architecture comprises at least three sections that tolerate further separation through linker insertion without eliminating bioactivity. Loss of L activity after insertion of FLAG or HA epitope tags at position 615 suggests, however, that this region stands overall under tighter structural scrutiny than the LR II/LR III junction. Although P is still bound efficiently, introduction of a higher content of charged (FLAG) or aromatic/charged (HA) residues is overall not tolerated at position 615.
Independently Expressed L Fragments Assume a Physiological Conformation
To assess whether L domains located up- or downstream of residue 1708 are capable of truly independent folding or require synthesis as a single polypeptide to assume their physiological conformation, we split the L gene at this position and generated separate expression plasmids for the corresponding N- and C-terminal fragments (termed LN-frag and LC-frag). Streptactin-FLAG and HA epitope tags were added to the newly generated N and C termini, respectively, to facilitate the biochemical characterization of the discrete L fragments.
To test the hypothesis that the reciprocal affinity of independently expressed L domains may be low, even if folding-competent, we generated a second set of L expression plasmids in which additional GCN4 affinity tags (67) were added to the N and C termini of the L fragments (detailed in Fig. 2A). These tags were expected to induce intracellular dimerization of the independently synthesized constructs. All four L variants were synthesized and showed the anticipated mobility pattern when expressed individually followed by SDS-PAGE and immunodetection using antibodies directed against the different epitope tags (Fig. 2B).
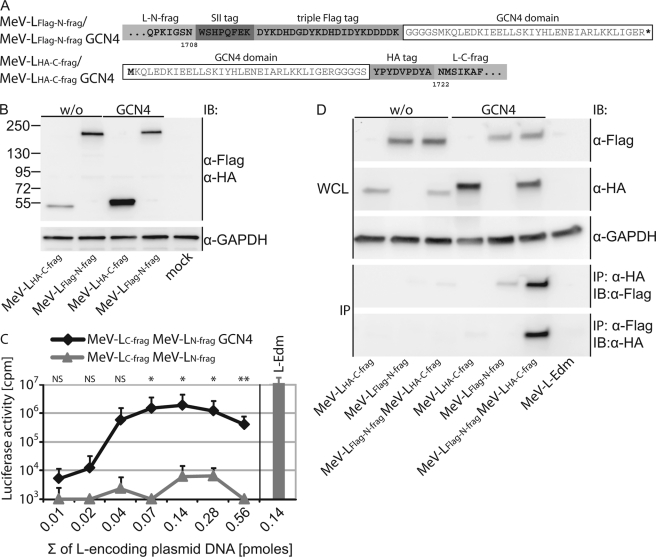
FIGURE 2.
Trans-complementation of bioactivity of MeV L N- and C-terminal fragments. A, shown are schematic and amino acid sequences of MeV-LFLAG-N-frag and MeV-LHA-C-frag constructs. Each L fragment was generated with or without a terminal GCN4 dimerization domain (white boxes). B, expression analysis of the different MeV L fragments is outlined in A. Whole cell lysates of BSR-T7/5 cells transfected with L fragment-encoding plasmids as indicated were gel-fractionated followed by immunoblotting (IB) using specific antibodies directed against the FLAG and HA epitope tags. Numbers represent the migration pattern of a protein standard (in kDa). Cellular GAPDH was analyzed in identical samples as internal standard. C, MeV L fragment trans-complementation in the replicon reporter assay is shown. BSR-T7/5 cells were co-transfected with all replicon components and equimolar amounts of expression plasmids encoding the MeV L N- and C-terminal fragments with or without terminal GCN4 domains. x axis values represent the sum of L fragment-encoding plasmid DNA (in pmol). Controls received standard MeV L-Edm (L-Edm) or vector in place of L fragment-encoding plasmids. Total amounts of DNA transfected/well were kept constant. Values represent relative luciferase activity units and represent the averages of three independent experiments ± S.D. Statistical analysis assesses the significance of activity deviation of GCN4-tagged L fragments from those lacking the dimerization domains (*, p < 0.05; **, p < 0.001; NS, not significant). D, efficient co-immunoprecipitation of MeV L fragments requires the presence of the GCN4 domains. Whole cell lysates (WCL) of BSR-T7/5 cells expressing different combinations of the MeV L fragments with or without additional GCN4 domains were gel-fractionated and immunoblotted (IB) or subjected to reciprocal immunoprecipitation (IP) using specific antibodies directed against the HA or FLAG epitopes followed by anti-FLAG or anti-HA immunostaining, respectively. GAPDH was detected as the internal standard. Control samples expressed untagged standard MeV L-Edm.
Despite efficient fragment expression, co-transfection of plasmids encoding LN-frag and LC-frag without the GCN4 tags at equimolar DNA ratios returned essentially background luciferase activities in the replicon reporter assay. This finding was independent of the combined amount of L fragment-encoding DNA added (Fig. 2C). In striking contrast, the presence of the GCN4 dimerization domains at the L fragments significantly restored L bioactivity, resulting in a luciferase activity optimum curve largely mimicking that described above for standard L (Fig. 2C). Successful trans-complementation was dependent on GCN4-mediated oligomerization of LN-frag GCN4 and LC-frag GCN4, as alternative complementation attempts between LN-frag and LC-frag GCN4 or between LN-frag GCN4 and LC-frag did not restore bioactivity (data not shown).
Reciprocal co-immunoprecipitation of the GCN4-tagged or untagged fragments, either through precipitation with specific antibodies directed against the HA epitope followed by immunodetection with anti-FLAG antibodies or through anti-FLAG precipitation and anti-HA detection, yielded results consistent with the bioactivity data; efficient co-precipitation of the fragments was observed only in the presence of the additional GCN4 dimerization domains (Fig. 2D).
These data demonstrate that the MeV L sections located up- or downstream of residue 1708 have no biochemically detectable affinity for each other, arguing against the presence of a traditional protein-protein interface between the LN-frag and LC-frag subunits. Both fragments are capable, however, of folding into a physiological conformation when expressed separately, as reinstating the physical proximity of the L subunits post-translationally through added GCN4 dimerization domains restored RdRp activity.
Dominant-negative Effect of LN-frag and LC-frag Subunits on Full-length L Bioactivity
To assess the full extent of functional complementation, we generated replicon-based RdRp activity curves for a set of different LN-frag GCN4 and LC-frag GCN4 plasmid DNA ratios ranging from 3.5:1 to 1:3.5. Bioactivity peaked when cells received equimolar amounts of L fragment-encoding plasmid DNA or a slight (1:1.75) excess of LC-frag GCN4, whereas increasing the relative amount of LN-frag GCN4 resulted in a significant reduction of RdRp activity (Fig. 3A). Co-expression of an excess of the LN-frag subunit with standard L confirmed a dominant-negative effect of this fragment on RdRp activity (Fig. 3B). Overexpression of the LC-frag subunit relative to standard L at a 4:1 plasmid DNA ratio did not significantly affect bioactivity of standard L. Surprisingly, we observed a reduction in activity by ∼50% at a 10:1 LC-frag:L ratio, suggesting a dominant negative effect of LC-frag at high relative excess.
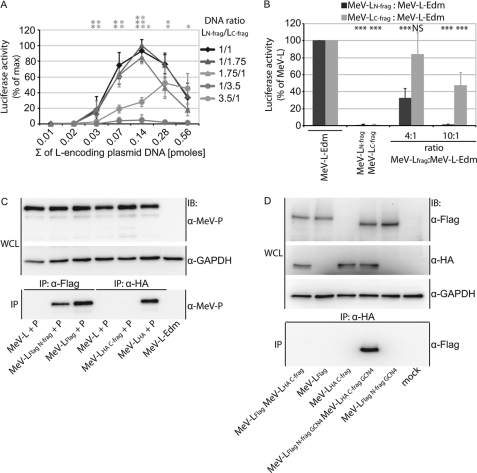
FIGURE 3.
Dominant negative effect of overexpressed L fragments on RdRp activity. A, L trans-complementation efficiency peaks in the presence of a slight excess of the C-terminal L fragment. Trans-complementation MeV replicon activity assays were carried out as outlined for Fig. 2C but after transfection of different molar ratios of the LN-frag and LC-frag expression plasmids as indicated. Values are expressed relative to the L fragment combination that returned the overall highest luciferase activities (molar ratio of LN-frag to LC-frag plasmid DNA 1:1.75, total amount of L fragment-encoding plasmid DNA 0.14 pmol) and represent averages of at least three independent experiments ± S.D. Statistical analysis determines the significance of activity deviation relative to the optimum curve (molar ratio of LN-frag to LC-frag plasmid DNA 1:1.75; *, p < 0.05; **, p < 0.01; ***, p < 0.001). B, co-transfection of BSR-T7/5 cells with replicon helper components, MeV L N- or C-fragment expression constructs, and plasmids encoding full-length MeV L-Edm in either 4:1 or 10:1 molar ratios. Values are expressed relative to MeV L-Edm samples and represent the averages of four independent experiments ± S.D. Statistically significant deviation of luciferase activity was calculated relative to standard MeV L-Edm controls (***, p < 0.001; NS, not significant). In A and B, total amounts of DNA transfected/well were kept constant. C, MeV LN-frag but not LC-frag, interacts with MeV P. Co-immunoprecipitation after transfection of cells with MeV L fragment and MeV P expression plasmids as indicated was carried out as described for Fig. 1E. WCL, whole cell lysates. D, MeV LC-frag does not efficiently co-precipitate standard MeV-L. Immunoprecipitations (IP) were carried out using HA epitope-specific antibodies followed by immunostaining (IB) with anti-FLAG antibodies. As controls, FLAG and HA epitope-containing material and cellular GAPDH were immunostained in cellular lysates (WCL). Mock-transfected cells received empty vector in place of l-encoding plasmid.
Consistent with previous studies locating the P binding domain in the N-terminal 408-residue section of MeV L (14), we found the LN-frag but not the LC-frag subunit to be capable of efficient co-precipitation of the P protein (Fig. 3C). Competition for P binding between LN-frag and standard L thus emerges as a possible basis for the dominant-negative phenotype associated with this fragment in the RdRp activity assays. By analogy, the inhibitory effect of the LC-frag subunit seen at high excess could be due to direct interaction of the fragment with the corresponding C-terminal domain in standard L. To test this hypothesis, we attempted co-immunoprecipitation of LC-frag and full-length L. As seen before (Fig. 3C) for interaction with P, however, no biochemically appreciable interaction between the LC-frag and standard L was detectable in this assay (Fig. 3D), whereas the GCN4-tagged LN-frag and LC-frag subunits added for control efficiently co-precipitated. This result was independent of whether cells were transfected with all plasmids of the minireplicon system (shown in Fig. 3D) or only with plasmids encoding the L variants and P protein (data not shown).
These observations re-emphasize proper folding of the LN-frag subunit, enabling it to compete for interaction with RdRp complex components. Unexpectedly, the data also suggest the possibility of weak interactions between the LC-frag subunit and full-length L or P proteins, which may be responsible for the dominant negative effect associated with the fragment at high molar excess but cannot be detected biochemically.
Epitope Tag Insertions in RSV and NiV L
To explore whether the organization of MeV L into at least two independently folding functional domains constitutes a general feature of Paramyxovirus L proteins, we identified residues homologous to MeV L amino acid 1708 in polymerase proteins of two additional Paramyxoviruses in search of sites suitable for L splitting. For this proof-of-concept approach, NiV and RSV L proteins were chosen based on their diverse phylogenetic proximity to MeV; NiV, like MeV a member of the Paramyxoviridae subfamily albeit of the Henipavirus genus, is closely related to MeV. By contrast, RSV is a member of the Pneumovirus subfamily and thus a far more distantly related representative of the Paramyxoviridae (1). Separate sequence alignments of a panel of Paramyxovirus L proteins using ClustalW2 (31) and MUSCLE (32) as alternative algorithms posited NiV and RSV L proteins to exhibit different patterns relative to MeV polymerase (Fig. 4, A and B). In the case of RSV L in particular, residues predicted to be homologous to MeV L 1708 differed by 54 amino acids, exceeding the total length of the LR II/LR III junction in MeV L.
FIGURE 4.
Identification of HA epitope tag insertion sites in NiV and RSV L. A and B, sequence alignments of L proteins derived from MeV-Edm, NiV (88), HPIV2 (89), MuV-Miyahara (90), HPIV3–14702 (91), NDV-AF2240 (80), and RSV-A2 using the ClustalW2 (A) and MUSCLE (B) algorithms are shown. Regions corresponding to part of the LR II/LR III intersection in MeV L are shown, and numbers reflect amino acid positions. Gray boxes mark the insertion sites of the epitope tags. C, shown is expression analysis of the newly generated NiV and RSV L variants. Whole cell lysates (WCL) of BSR-T7/5 cells transfected with the different L expression plasmids were gel fractionated and immunostained (IB) using specific antibodies directed against the HA epitope or cellular GAPDH. Control cells (mock) received empty vector in place of L-encoding plasmid. D and E, shown is activity testing of the HA-tagged NiV and RSV L variants using specific replicon reporter systems for NiV (D, CAT reporter) and RSV (E, firefly luciferase reporter). Values are expressed relative to activities of standard NiV or RSV L and reflect the averages of four independent experiments ± S.D.
We, therefore, first prescreened the different predictions experimentally through HA epitope insertion in analogy to the MeV LHA-(1708) construct. Gel fractionation of transfected cell lysates and immunoblotting revealed expression of all tagged NiV and RSV L variants (Fig. 4C). However, cytosolic steady state levels varied relative to each other in the case of the NiV L constructs or were substantially reduced in the case of the RSV LHA variants relative to MeV LHA-(1708). When analyzed in homotypic NiV and RSV replicon reporter assays, both NiV L variants nevertheless returned similar RdRp activities, ∼40–60% that observed for standard NiV L, despite the difference in expression levels (Fig. 4D). In contrast, the two RSV L variants showed significant differences in bioactivity ranging from ∼60% that of standard RSV L in the case of RSV LHA-(1749) to only 10% in the case RSV LHA-(1695) (Fig. 4E).
Thus, we identified at least one position for each L protein, at which significant RdRp bioactivity was retained in the reporter assays after HA epitope insertion. In the case of RSV L, ClustalW-based predictions were superior to those obtained through MUSCLE-driven alignment. Based on these results, residues 1763 in NiV L and 1749 in RSV L were chosen for L protein splitting and subsequent trans-complementation experiments.
Homo- and Heterotypic Trans-complementation of MeV, NiV, and RSV L
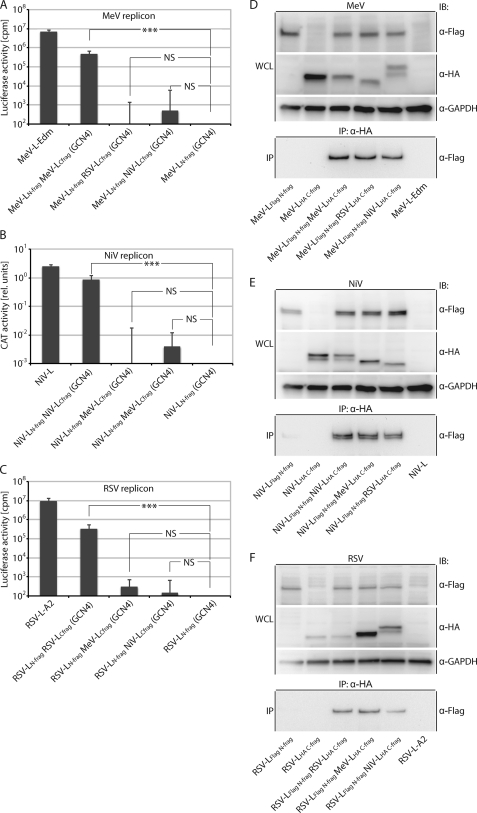
All RSV and NiV L fragments were generated with added FLAG or HA epitope tags and terminal GCN4 dimerization domains in analogy to the MeV LN-frag and LC-frag subunits. When subjected to replicon-based quantification of RdRp activity, we observed that both the NiV and RSV L fragment pairs were capable of significant trans-complementation of bioactivity similar to our initial finding with the MeV L fragments (Fig. 5, A–C). In contrast, co-expressing the different L fragments in all possible heterotypic combinations did not result in the recovery of any significant L bioactivity, indicating the inability of L fragments to interact productively with each other when derived from different Paramyxoviruses (Fig. 5, A–C).
FIGURE 5.
Functional trans-complementation mandates homotypic L fragment combinations. A–C, shown are homo- and hetero-complementation assays after co-transfection of plasmids encoding GCN4 domain-tagged MeV, NiV, or RSV L N- and C-terminal fragments in all combinations in the context of the MeV (A), NiV (B), or RSV (C) replicon reporter systems. Values show relative luciferase or CAT activities and represent the averages of at least three independent experiments ± S.D. Control cells received the respective LN-frag expression plasmid-only instead of the homotypic standard L or co-transfection of LN-frag and LC-frag encoding plasmids. Statistical analysis assesses the significance of deviation of the different L fragment combinations from the controls (***, p < 0.001; NS, not significant). D–F, all homo- and heterotypic L N- and C-fragment combinations analyzed in A–C efficiently co-immunoprecipitate. Whole cell lysates (WCL) of BSR-T7/5 cells expressing the GCN4-tagged homo- and heterotypic L fragment pairs as specified were subjected to immunoprecipitation (IP) with anti-HA antibodies followed by gel fractionation and immunoblotting (IB) using anti-FLAG antibodies. In parallel, lysates were directly analyzed using HA-, FLAG-, or cellular GAPDH-specific antibodies. Controls received expression plasmids encoding standard, untagged L in place of the epitope-tagged L variants.
To test whether the lack of productive interaction was due to inefficient GCN4 domain-mediated L subunit dimerization under heterotypic conditions, we subjected all fragment combinations examined to co-immunoprecipitation experiments relying on the additionally added FLAG and HA epitope tags for precipitation and immunodetection. As expected, all L subunits were synthesized successfully when expressed alone or in combination with the different counterpart fragments (Fig. 5, D and E, whole cell lysate panels). Importantly, all C-terminal L fragments were also capable of efficiently co-precipitating the different N-terminal fragments regardless of their virus origin (Fig. 5, D and E, immunoprecipitation panels), confirming full functionality of the GCN4 domains under both homo- and heterotypic conditions and hence equally productive dimerization of the different L fragment combinations.
These data demonstrate that a molecular organization into at least two independent folding domains constitutes a general feature of Paramyxovirus polymerases, as all homotypic L fragment combinations tested returned significant bioactivity in the replicon assays. Failure of trans-complementation between the heterotypic L pairs despite efficient, GCN4-mediated fragment hetero-oligomerization reveals that molecular compatibility between the subunits is required for RdRp activity. Although a biochemically appreciable high affinity protein-protein interface is lacking between the Paramyxovirus L N- and C-terminal fragments, physical proximity of the subunits alone is necessary but not sufficient for RdRp bioactivity.
DISCUSSION
Reflecting their central position in the life cycle of all RNA viruses, RNA-dependent RNA polymerases are determinants for viral pathogenesis and constitute attractive targets for antiviral therapeutics (68, 69). Crystal structures, either free or complexed with nucleic acid substrates, are available for RdRps derived from a variety of different viral families (70–72). These structures revealed a conserved fundamental organization of the proteins into a geometry resembling a “right-hand” shape in which the “fingers,” “palm,” and “thumb” domains are thought to ensure the correct positioning of substrates and metal ion cofactors (71, 73). Although it has been proposed that this basic shape may be conserved in all nucleotide polymerases (74), our basic understanding of Mononegavirales L protein structure and organization remains in its infancy.
Examining different L proteins derived from representatives of the Paramyxovirinae and Pneumovirinae subfamily, our study illuminates three principles of Paramyxovirus polymerase folding. First, the proteins are composed of a set of at least two truly independent structural units that are competent of proper folding. Second, assuming an enzymatically active tertiary conformation does not require synthesis of the protein as a single polypeptide and, consequently, does not adhere to an integrated folding concept. Third, molecular compatibility between the discrete subunits is essential for functionality, but the subunits lack a high affinity protein-protein interface. Several lines of evidence support these conclusions.
Our bioinformatics analysis has highlighted multiple candidate regions in the Paramyxovirus L protein that may represent domain boundaries. The identification of the MeV L LR II/LR III intersection as a candidate zone by several of the algorithms used supports the validity of the in silico predictions, as successful expansion of this region was reported previously for Morbillivirus L proteins (23, 25). Our subsequent linker insertion scan confirmed these data and, by extending successful epitope insertion to distantly related L proteins, illuminated a conserved Paramyxovirus polymerase organization into at least two distinct sections. In addition, we identified an N-terminal position in MeV L, residue 615 located downstream of the N-terminal L-P and L-L oligomerization domain (8, 14), that likewise tolerates enlargement without eliminating L bioactivity. This finding did not extend, however, to acceptance of FLAG or HA epitope tags, which contain a high density of charged or bulky aromatic amino acids (75, 76) and are thus expected to be structurally more active than the 10-residue linker used in the primary scan. Interestingly, L proteins of the Arenavirus family reportedly comprise at least three independent folding domains (4).
Based on our data, we conclude with confidence that at least two defined structural domains are present in Paramyxovirus polymerases. The N-terminal fragment harbors P binding, polymerase and polyribonucleotidyltransferase activities, whereas the C-terminal section contains methyltransferase and guanylyltransferase motifs and thus contributes predominantly to the generation of proper mRNA cap structures. Although comparative evaluations of different algorithms have demonstrated that a combination of discrete predictors has the highest prospect to maximize the overall accuracy of the prediction (52, 65, 66), our study also highlights the limitations of currently available in silico tools when applied to very large polypeptides such as Mononegavirales L proteins. Therefore, additional domain intersections may well exist in the Paramyxovirus L protein that were not detected by our in silico analysis and experimental evaluation.
We have shown that the addition of GCN4 dimerization domains to independently expressed Paramyxovirus L N- and C-terminal fragments results in significant trans-complementation of RdRp bioactivity. These data demonstrate that post-translational interaction of individually synthesized polypeptides is sufficient for the formation of a catalytically active tertiary structure. Because the replicon reporter system used in our study first generates T7-polymerase-driven RNA molecules of negative polarity, luciferase reporter activity confirms full transcriptase functionality, comprising RNA polymerization, polyadenylation, and capping and methylation of the non-covalently linked N- and C-terminal L fragments. Differently composed NNSV polymerase oligomers are thought to carry out replicase and transcriptase activities. Although replicase activity is mediated by an L-P-N core complex, the transcriptase comprises cellular proteins in addition to L and P (77–79). Although in all cases statistically significant, luciferase activity levels representing reconstituted complexes did not reach the level of those obtained with standard L. This finding does not necessarily reflect, however, that non-covalently linked complexes are inherently less bioactive. A major contributing factor to lower activity may be that GCN4 mediated interactions are not intrinsically geared toward mediating enzymatically productive LN-frag/LC-frag dimerization events but will most likely drive the formation of LN-frag and LC-frag homodimers with equal affinity. Our observation that a 3.5-fold molar excess of either fragment is sufficient to reduce RdRp activity substantially in the replicon trans-complementation assay supports this notion.
Last, the co-precipitation experiments and functional RdRp assays performed in the absence of additional GCN4 tags demonstrate that the L fragments, albeit competent for folding, have no biochemically appreciable inherent affinity for each other. The previously reported lack of VSV L trans-complementation (3) suggests that this may, in fact, constitute a general theme of Mononegavirales L domain interaction, which stands in stark contrast to the strong inherent domain affinities observed for L proteins of segmented negative strand RNA viruses of the Arenavirus family (4). Successful splitting of all three Paramyxovirus polymerase proteins analyzed in our study at homologous positions and the presence of comparable catalytic motifs in methyltransferase in CR VI (6, 17, 81, 82) and guanolyl-transferase near the C terminus (21, 22) in the MeV, NiV, and RSV LC-frag subunits support that the different L C-terminal fragments harbor equivalent enzymatic activities. Clearly, bringing the Paramyxovirus L N- and C-terminal fragments into close physical proximity is a prerequisite for reconstituting RdRp activity. If proximity alone were sufficient for complementation, however, heterotypic trans-complementation between L fragments derived from different Paramyxovirus family members and brought into proximity through GCN4-mediated dimerization should have been successful. Because heterotypic complementation did not restore functionality despite efficient biochemical interaction of the various fragments, our data suggest an architectural model of Mononegavirales L proteins in which independently folding subdomains do not share a traditional protein-protein interface but require low affinity molecular compatibility to achieve functionality.
Mammalian cells are not capable of RNA-dependent RNA polymerization, rendering the viral RdRp complexes attractive targets for antiviral therapies. Novel nucleoside and allosteric inhibitors of hepatitis C virus polymerase, for instance, are at different stages of clinical evaluation and show strong drug potential (83–85). Experimentally tested allosteric blockers of Paramyxovirus polymerase likewise combine high potency with minimal cytotoxicity, opening a desirably large therapeutic window (86, 87). High resolution information about the Mononegavirales polymerase core structure would pave the path for structure-guided de novo drug design efforts, the informed optimization of existing inhibitor scaffolds, and the proactive design of inhibitor protected against rapid viral escape from inhibition. At present, high resolution structures of L have not been solved for any of the NNSV family members. Identifying structurally meaningful fragments that assume a physiological conformation when expressed independently emerges as a valid strategy toward obtaining structural insight. Confirming proper folding of candidate Paramyxovirus L fragments through successful trans-complementation after the addition of GCN4 dimerization tags provides a novel approach toward achieving this goal, which is most likely transferable to the analysis of Mononegavirales L proteins that do not originate from members of the Paramyxovirus family.
Supplementary Material
Acknowledgments
We thank A. L. Hammond for critical reading of the manuscript, P. A. Rota for expression plasmids encoding NIV N, P, and L and the NIV replicon reporter construct, and J. P. Snyder and A. Prussia for helpful discussions concerning implementation of the DomCut algorithm.
This work was supported, in whole or in part, by National Institutes of Health Grants AI071002 and AI085328 (NIAID to R. K. P.).

This article contains supplemental Tables 1 and 2.
- MeV
- measles virus
- NiV
- Nipah virus
- L
- polymerase protein
- N
- nucleocapsid protein
- NNSV
- nonsegmented negative-strand RNA virus
- RSV
- respiratory syncytial virus
- VSV
- vesicular stomatitis virus
- RdRp
- RNA-dependent RNA polymerase, CR, conserved region
- LR
- large region
- HPIV
- human parainfluenza virus
- Edm
- MeV-Edmonston
- CAT
- chloramphenicol
- nt
- nucleotide
- P
- phosphoprotein.
REFERENCES
- 1. Lamb R. A., Parks G. D. (2007) in Fields Virology (Knipe D. M., Howley P. M., eds) pp. 1449–1496, 5 Ed., Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Kranzusch P. J., Schenk A. D., Rahmeh A. A., Radoshitzky S. R., Bavari S., Walz T., Whelan S. P. (2010) Assembly of a functional Machupo virus polymerase complex. Proc. Natl. Acad. Sci. U.S.A. 107, 20069–20074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rahmeh A. A., Schenk A. D., Danek E. I., Kranzusch P. J., Liang B., Walz T., Whelan S. P. (2010) Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 20075–20080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brunotte L., Lelke M., Hass M., Kleinsteuber K., Becker-Ziaja B., Günther S. (2011) Domain structure of Lassa virus L protein. J. Virol. 85, 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruedas J. B., Perrault J. (2009) Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. J. Virol. 83, 12241–12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poch O., Blumberg B. M., Bougueleret L., Tordo N. (1990) J. Gen. Virol. 71, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 7. Sidhu M. S., Menonna J. P., Cook S. D., Dowling P. C., Udem S. A. (1993) Canine distemper virus L gene. Sequence and comparison with related viruses. Virology 193, 50–65 [DOI] [PubMed] [Google Scholar]
- 8. Cevik B., Holmes D. E., Vrotsos E., Feller J. A., Smallwood S., Moyer S. A. (2004) The phosphoprotein (P) and L binding sites reside in the N terminus of the L subunit of the measles virus RNA polymerase. Virology 327, 297–306 [DOI] [PubMed] [Google Scholar]
- 9. Cevik B., Smallwood S., Moyer S. A. (2003) The L-L oligomerization domain resides at the very N terminus of the sendai virus L RNA polymerase protein. Virology 313, 525–536 [DOI] [PubMed] [Google Scholar]
- 10. Smallwood S., Moyer S. A. (2004) The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P, and C proteins. Virology 318, 439–450 [DOI] [PubMed] [Google Scholar]
- 11. Holmes D. E., Moyer S. A. (2002) The phosphoprotein (P) binding site resides in the N terminus of the L polymerase subunit of sendai virus. J. Virol. 76, 3078–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chattopadhyay A., Shaila M. S. (2004) Rinderpest virus RNA polymerase subunits. Mapping of mutual interacting domains on the large protein L and phosphoprotein p. Virus Genes 28, 169–178 [DOI] [PubMed] [Google Scholar]
- 13. Chandrika R., Horikami S. M., Smallwood S., Moyer S. A. (1995) Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213, 352–363 [DOI] [PubMed] [Google Scholar]
- 14. Horikami S. M., Smallwood S., Bankamp B., Moyer S. A. (1994) An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205, 540–545 [DOI] [PubMed] [Google Scholar]
- 15. Malur A. G., Gupta N. K., De Bishnu P., Banerjee A. K. (2002) Analysis of the mutations in the active site of the RNA-dependent RNA polymerase of human parainfluenza virus type 3 (HPIV3) Gene Expr. 10, 93–100 [PMC free article] [PubMed] [Google Scholar]
- 16. Sleat D. E., Banerjee A. K. (1993) Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 67, 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferron F., Longhi S., Henrissat B., Canard B. (2002) Viral RNA polymerases. A predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27, 222–224 [DOI] [PubMed] [Google Scholar]
- 18. Li J., Rahmeh A., Morelli M., Whelan S. P. (2008) A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogino T., Banerjee A. K. (2007) Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97 [DOI] [PubMed] [Google Scholar]
- 20. Shuman S., Lima C. D. (2004) The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 21. Gopinath M., Shaila M. S. (2009) RNA triphosphatase and guanylyl transferase activities are associated with the RNA polymerase protein L of rinderpest virus. J. Gen. Virol. 90, 1748–1756 [DOI] [PubMed] [Google Scholar]
- 22. Nishio M., Tsurudome M., Garcin D., Komada H., Ito M., Le Mercier P., Nosaka T., Kolakofsky D. (2011) Human parainfluenza virus type 2 L protein regions required for interaction with other viral proteins and mRNA capping. J. Virol. 85, 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duprex W. P., Collins F. M., Rima B. K. (2002) Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 76, 7322–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McIlhatton M. A., Curran M. D., Rima B. K. (1997) Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected sequence). J. Gen. Virol. 78, 571–576 [DOI] [PubMed] [Google Scholar]
- 25. Brown D. D., Rima B. K., Allen I. V., Baron M. D., Banyard A. C., Barrett T., Duprex W. P. (2005) Rational attenuation of a Morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. J. Virol. 79, 14330–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buchholz U. J., Finke S., Conzelmann K. K. (1999) Generation of bovine respiratory syncytial virus (BRSV) from cDNA. BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sidhu M. S., Chan J., Kaelin K., Spielhofer P., Radecke F., Schneider H., Masurekar M., Dowling P. C., Billeter M. A., Udem S. A. (1995) Rescue of synthetic measles virus minireplicons. Measles genomic termini direct efficient expression and propagation of a reporter gene. Virology 208, 800–807 [DOI] [PubMed] [Google Scholar]
- 28. Halpin K., Bankamp B., Harcourt B. H., Bellini W. J., Rota P. A. (2004) Nipah virus conforms to the rule of six in a minigenome replication assay. J Gen Virol 85, 701–707 [DOI] [PubMed] [Google Scholar]
- 29. Grosfeld H., Hill M. G., Collins P. L. (1995) RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins. Transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69, 5677–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suyama M., Ohara O. (2003) DomCut. Prediction of interdomain linker regions in amino acid sequences. Bioinformatics 19, 673–674 [DOI] [PubMed] [Google Scholar]
- 31. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 32. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parks C. L., Lerch R. A., Walpita P., Wang H. P., Sidhu M. S., Udem S. A. (2001) Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bankamp B., Bellini W. J., Rota P. A. (1999) Comparison of L proteins of vaccine and wild-type measles viruses. J. Gen. Virol. 80, 1617–1625 [DOI] [PubMed] [Google Scholar]
- 35. Druelle J., Sellin C. I., Waku-Kouomou D., Horvat B., Wild F. T. (2008) Wild type measles virus attenuation independent of type I IFN. Virol. J. 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moulin E., Beal V., Jeantet D., Horvat B., Wild T. F., Waku-Kouomou D. (2011) Molecular characterization of measles virus strains causing subactute sclerosing panencephalitis in France in 1977 and 2007. J. Med. Virol. 83, 1614–1623 [DOI] [PubMed] [Google Scholar]
- 37. Plemper R. K., Doyle J., Sun A., Prussia A., Cheng L. T., Rota P. A., Liotta D. C., Snyder J. P., Compans R. W. (2005) Design of a small molecule entry inhibitor with activity against primary measles virus strains. Antimicrob. Agents Chemother. 49, 3755–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baron M. D., Kamata Y., Barras V., Goatley L., Barrett T. (1996) The genome sequence of the virulent Kabete O strain of rinderpest virus. Comparison with the derived vaccine. J. Gen. Virol. 77, 3041–3046 [DOI] [PubMed] [Google Scholar]
- 39. Baron M. D., Barrett T. (1995) Sequencing and analysis of the nucleocapsid (N) and polymerase (L) genes and the terminal extragenic domains of the vaccine strain of rinderpest virus. J. Gen. Virol. 76, 593–602 [DOI] [PubMed] [Google Scholar]
- 40. Gassen U., Collins F. M., Duprex W. P., Rima B. K. (2000) Establishment of a rescue system for canine distemper virus. J. Virol. 74, 10737–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. von Messling V., Springfeld C., Devaux P., Cattaneo R. (2003) A ferret model of canine distemper virus virulence and immunosuppression. J. Virol. 77, 12579–12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bailey D., Banyard A., Dash P., Ozkul A., Barrett T. (2005) Full genome sequence of peste des petits ruminants virus, a member of the Morbillivirus genus. Virus Res. 110, 119–124 [DOI] [PubMed] [Google Scholar]
- 43. Rima B. K., Collin A. M., Earle J. A. (2005) Completion of the sequence of a cetacean Morbillivirus and comparative analysis of the complete genome sequences of four morbilliviruses. Virus Genes 30, 113–119 [DOI] [PubMed] [Google Scholar]
- 44. Chua K. B., Bellini W. J., Rota P. A., Harcourt B. H., Tamin A., Lam S. K., Ksiazek T. G., Rollin P. E., Zaki S. R., Shieh W., Goldsmith C. S., Gubler D. J., Roehrig J. T., Eaton B., Gould A. R., Olson J., Field H., Daniels P., Ling A. E., Peters C. J., Anderson L. J., Mahy B. W. (2000) Nipah virus. A recently emergent deadly Paramyxovirus. Science 288, 1432–1435 [DOI] [PubMed] [Google Scholar]
- 45. Takimoto T., Bousse T., Portner A. (2000) Molecular cloning and expression of human parainfluenza virus type 1 L gene. Virus Res. 70, 45–53 [DOI] [PubMed] [Google Scholar]
- 46. Yang H. T., Jiang Q., Zhou X., Bai M. Q., Si H. L., Wang X. J., Lu Y., Zhao H., He H. B., He C. Q. (2011) Identification of a natural human serotype 3 parainfluenza virus. Virol. J. 8, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nishio M., Tsurudome M., Ito M., Garcin D., Kolakofsky D., Ito Y. (2005) Identification of Paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J. Virol. 79, 8591–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yea C., Cheung R., Collins C., Adachi D., Nishikawa J., Tellier R. (2009) The complete sequence of a human parainfluenza virus 4 genome. Viruses 1, 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tidona C. A., Kurz H. W., Gelderblom H. R., Darai G. (1999) Isolation and molecular characterization of a novel cytopathogenic Paramyxovirus from tree shrews. Virology 258, 425–434 [DOI] [PubMed] [Google Scholar]
- 50. Whitehead S. S., Juhasz K., Firestone C. Y., Collins P. L., Murphy B. R. (1998) Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J. Virol. 72, 4467–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palacios G., Lowenstine L. J., Cranfield M. R., Gilardi K. V., Spelman L., Lukasik-Braum M., Kinani J. F., Mudakikwa A., Nyirakaragire E., Bussetti A. V., Savji N., Hutchison S., Egholm M., Lipkin W. I. (2011) Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerg. Infect. Dis. 17, 711–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lieutaud P., Canard B., Longhi S. (2008) BMC Genomics 9, S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dosztányi Z., Csizmok V., Tompa P., Simon I. (2005) IUPred. Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 54. Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) GlobPlot. Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Protein disorder prediction. Implications for structural proteomics. Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 56. Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L. (2005) FoldIndex. A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438 [DOI] [PubMed] [Google Scholar]
- 57. Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) RONN. The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 58. Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N. (2010) PONDR-FIT. A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deleted in proof.
- 60. Chandonia J. M., Karplus M. (1999) New methods for accurate prediction of protein secondary structure. Proteins 35, 293–306 [PubMed] [Google Scholar]
- 61. Chandonia J. M. (2007) StrBioLib. A Java library for development of custom computational structural biology applications. Bioinformatics 23, 2018–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schaeffer R. D., Daggett V. (2011) Protein folds and protein folding. Protein Eng. Des. Sel. 24, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oldfield C. J., Cheng Y., Cortese M. S., Brown C. J., Uversky V. N., Dunker A. K. (2005) Comparing and combining predictors of mostly disordered proteins. Biochemistry 44, 1989–2000 [DOI] [PubMed] [Google Scholar]
- 64. Uversky V. N., Radivojac P., Iakoucheva L. M., Obradovic Z., Dunker A. K. (2007) Prediction of intrinsic disorder and its use in functional proteomics. Methods Mol. Biol. 408, 69–92 [DOI] [PubMed] [Google Scholar]
- 65. Ferron F., Longhi S., Canard B., Karlin D. (2006) A practical overview of protein disorder prediction methods. Proteins 65, 1–14 [DOI] [PubMed] [Google Scholar]
- 66. Ferron F., Rancurel C., Longhi S., Cambillau C., Henrissat B., Canard B. (2005) VaZyMolO. A tool to define and classify modularity in viral proteins. J. Gen. Virol. 86, 743–749 [DOI] [PubMed] [Google Scholar]
- 67. Harbury P. B., Zhang T., Kim P. S., Alber T. (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 [DOI] [PubMed] [Google Scholar]
- 68. De Clercq E. (2004) Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2, 704–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ferrer-Orta C., Agudo R., Domingo E., Verdaguer N. (2009) Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase. Curr. Opin. Struct. Biol. 19, 752–758 [DOI] [PubMed] [Google Scholar]
- 70. Ng K. K., Arnold J. J., Cameron C. E. (2008) Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320, 137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ferrer-Orta C., Arias A., Escarmís C., Verdaguer N. (2006) A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16, 27–34 [DOI] [PubMed] [Google Scholar]
- 72. Tao Y. J., Ye Q. (2010) RNA virus replication complexes. PLoS Pathog. 6, e1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brautigam C. A., Steitz T. A. (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 [DOI] [PubMed] [Google Scholar]
- 74. Cheetham G. M., Steitz T. A. (2000) Insights into transcription. Structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10, 117–123 [DOI] [PubMed] [Google Scholar]
- 75. Hopp T. P., Gallis B., Prickett K. S. (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology 6, 1204–1210 [Google Scholar]
- 76. Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. (1988) Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8, 2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Horikami S. M., Curran J., Kolakofsky D., Moyer S. A. (1992) Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66, 4901–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Qanungo K. R., Shaji D., Mathur M., Banerjee A. K. (2004) Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. U. S. A. 101, 5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tawar R. G., Duquerroy S., Vonrhein C., Varela P. F., Damier-Piolle L., Castagné N., MacLellan K., Bedouelle H., Bricogne G., Bhella D., Eléouët J. F., Rey F. A. (2009) Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283 [DOI] [PubMed] [Google Scholar]
- 80. Kusumaningtyas E., Tan W. S., Zamrod Z., Eshaghi M., Yusoff K. (2004) Existence of two forms of L protein of Newcastle disease virus isolates due to a compensatory mutation in Domain V. Arch. Virol. 149, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 81. Grdzelishvili V. Z., Smallwood S., Tower D., Hall R. L., Hunt D. M., Moyer S. A. (2005) A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 79, 7327–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ogino T., Kobayashi M., Iwama M., Mizumoto K. (2005) Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 280, 4429–4435 [DOI] [PubMed] [Google Scholar]
- 83. Deore R. R., Chern J. W. (2010) NS5B RNA dependent RNA polymerase inhibitors. The promising approach to treat hepatitis C virus infections. Curr. Med. Chem. 17, 3806–3826 [DOI] [PubMed] [Google Scholar]
- 84. Koch U., Narjes F. (2007) Recent progress in the development of inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. Curr. Top. Med. Chem. 7, 1302–1329 [DOI] [PubMed] [Google Scholar]
- 85. Vermehren J., Sarrazin C. (2011) New HCV therapies on the horizon. Clin. Microbiol. Infect. 17, 122–134 [DOI] [PubMed] [Google Scholar]
- 86. White L. K., Yoon J. J., Lee J. K., Sun A., Du Y., Fu H., Snyder J. P., Plemper R. K. (2007) Nonnucleoside inhibitor of measles virus RNA-dependent RNA polymerase complex activity. Antimicrob. Agents Chemother. 51, 2293–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yoon J. J., Krumm S. A., Ndungu J. M., Hoffman V., Bankamp B., Rota P. A., Sun A., Snyder J. P., Plemper R. K. (2009) Target analysis of the experimental measles therapeutic AS-136A. Antimicrob. Agents Chemother. 53, 3860–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Harcourt B. H., Tamin A., Halpin K., Ksiazek T. G., Rollin P. E., Bellini W. J., Rota P. A. (2001) Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287, 192–201 [DOI] [PubMed] [Google Scholar]
- 89. Ohgimoto S., Bando H., Kawano M., Okamoto K., Kondo K., Tsurudome M., Nishio M., Ito Y. (1990) Sequence analysis of P gene of human parainfluenza type 2 virus. P and cysteine-rich proteins are translated by two mRNAs that differ by two nontemplated G residues. Virology 177, 116–123 [DOI] [PubMed] [Google Scholar]
- 90. Okazaki K., Tanabayashi K., Takeuchi K., Hishiyama M., Okazaki K., Yamada A. (1992) Molecular cloning and sequence analysis of the mumps virus gene encoding the L protein and the trailer sequence. Virology 188, 926–930 [DOI] [PubMed] [Google Scholar]
- 91. Roth J. P., Li J. K., Smee D. F., Morrey J. D., Barnard D. L. (2009) A recombinant, infectious human parainfluenza virus type 3 expressing the enhanced green fluorescent protein for use in high-throughput antiviral assays. Antiviral Res. 82, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.