Background: Notch signaling has been implicated in the development and function of the immune system, whereas its role in TLRs-triggered innate inflammatory responses remains unclear.
Results: Notch signaling suppresses TLR-triggered inflammatory responses by inhibiting ERK1/2-mediated NF-κB activity in macrophages.
Conclusion: Cross-regulation exists between Notch and TLR signaling.
Significance: Understanding Notch signaling in the context of the TLR-triggered macrophage inflammatory response may provide insights into the mechanisms for inflammation regulation.
Keywords: ERK, Inflammation, Macrophages, NF-κB, Notch Pathway, Toll-like receptors (TLRs)
Abstract
Multiple signaling pathways are involved in the tight regulation of Toll-like receptor (TLR) signaling, which is important for the tailoring of inflammatory response to pathogens in macrophages. It is widely accepted that TLR signaling can activate Notch pathway; however, whether full activation of Notch signaling can feedback modulate TLR signaling pathway so as to control inflammation response remains unclear. Here, we demonstrated that stimulation with TLR ligands up-regulated Notch1 and Notch2 expression in macrophages. The expression of Notch target genes including Hes1 and Hes5 was also induced in macrophages by LPS, suggesting that TLR4 signaling enhances the activation of Notch pathway. Importantly, overexpression of constituted active form of Notch1 (NICD1) and Notch2 (NICD2) suppressed production of TLR4-triggered proinflammatory cytokines such as TNF-α and IL-6 but promoted production of antiinflammatory cytokine IL-10, which is dependent on the PEST domain of NICD. In addition, NICD1 and NICD2 suppressed TLR-triggered ERK phosphorylation, which is indispensable for Notch-mediated inhibition of TLR4-triggered proinflammatory cytokine production. Furthermore, activation of Notch signaling inhibited NF-κB transcription activity by MyD88/TRAF6 and TRIF pathways, which was dependent on ERK activity. Therefore, our results showed that Notch signaling negatively regulates TLR-triggered inflammation responses, revealing a new mechanism for negative regulation of TLR signaling via Notch pathway.
Introduction
Toll-like receptors (TLRs),4 the key pattern-recognition receptors expressed on antigen-presenting cells, play critical roles in the host defense against invading microbial pathogens (1). After the recognition of pathogen-associated molecule patterns, Toll-like receptors initiate shared and distinct signaling pathways depending on adapter MyD88 and/or TRIF, leading to the activation of transcription factors NF-κB, AP-1, IRF3, and/or IRF7, which consequentially induce the production of proinflammatory cytokines and type I interferon (2). Less efficient activation of Toll-like receptors may not evoke potent antiinfection immunity; however, excessive activation of the TLR-triggered response may also induce inflammatory disorders such as endotoxin shock or inflammatory autoimmune diseases (3). Therefore, the tight regulation of TLR signaling pathways is important for the control of the inflammatory response. Up until now, various intracellular signaling molecules have been shown to be involved in the regulation of the TLR pathway to maintain the immunological balance (4). In addition, some other signaling pathways, such as integrin CD11b (5) and immunoreceptor tyrosine-based activation-associated receptors (6), have cross-talk with TLR signaling pathways, resulting in fine tuning of TLR-triggered innate inflammatory responses. This raises one important question, that other new signaling pathways, especially signals via cell membrane receptors, should be further clarified for their involvement in cross-talks with TLR signaling and tight regulation of inflammatory response.
Notch signaling is a highly conserved pathway determining cell fate including cell growth, differentiation, and survival (7–9). Up to date, four Notch receptor family members (Notch1–4) and five Notch ligands Delta-like-1 (DLL-1), DLL-3, DLL-4, Jagged-1 and Jagged-2 have been identified in mammals cells (10, 11). The Notch receptors are large type I transmembrane receptors that undergo proteolytic processing by a furin-like convertase during transit to the cell surface. Binding of a ligand triggers sequential receptor cleavage by a disintegrin and metalloproteinase domain (ADAM)-type metalloproteinases and γ-secretase, resulting in the release of a nuclear translocation of Notch intracellular domain (NICD). The NICD translocates to the nucleus, where it interacts with the transcription factor RBP-J at the jκ site (also named CSL), leading to the formation of a transcriptional activator complex and induction of the transcription of Notch downstream target genes such as basic helix-loop-helix family (Hes1 and Hes5) and hairy and enhancer of split-related (HESR) family (Hey1 and Hey2). However, in the absence of NICD, CSL protein complexes with co-repressor proteins to repress gene transcription (12).
Recent evidence suggests that there is cross-talk between Notch and TLR signaling pathways (13, 14). TLR activation up-regulates the expression of Notch ligands, receptors, and RBP-J, favoring Notch signaling pathway. For example, LPS activates Notch signaling through a JNK-dependent pathway that subsequently regulates the inflammatory response (15). However, LPS was also reported to suppresses Notch signaling via nitric oxide in macrophages (16). A recent study demonstrated that Notch and TLR signaling pathways cooperate to activate the transcription of Notch target gene Hes1 and Hey and to increase the production of TLR-triggered cytokines such as TNF-α, IL-6, and IL-12 (17). Several studies also indicated that Notch signaling plays an important role in inflammatory disorders (18, 19). So it seems that there is reciprocal regulation between Notch and TLR pathway. However, the detailed relationship and underlying mechanisms between Notch and TLR signaling remain far from being understood.
In this study, we showed that the TLR4 signal up-regulated the expression of Notch and its target gene in macrophages. Overexpression of NICD1 and NICD2 decreased TLR4-triggered proinflammatory cytokines but increased antiinflammatory cytokine production. Furthermore, we found that NICD1 and NICD2 suppressed TLR-triggered ERK phosphorylation, which is indispensable for Notch-mediated inhibition of TLR4-triggered inflammatory cytokine production. Besides, we found that Notch inhibited NF-κB transcription activity by MyD88/TRAF6 and TRIF pathways, which was dependent on ERK inactivation by Notch signaling. Therefore, we show for the first time that Notch signaling negatively regulates the TLR-triggered inflammatory response.
MATERIALS AND METHODS
Reagents and Antibodies
RAW264.7 and HEK293T cells were obtained from ATCC (Manassas, VA). LPS (0111:B4) was from Sigma-Aldrich. Phosphorothioate-modified CpG oligodeoxynucleotide (ODN) was synthesized by Shenggong, and its sequence is 5′-TCC ATG ACG TTC CTG ATG CT-3′. PD98059 and γ-secretase inhibitor X (GSI) were purchased from Calbiochem. Primary antibodies against Hes1and Hes5 were from Santa Cruz Biotechnology, anticleaved Notch1, cleaved Notch2, phosphor-ERK1/2, JNK1/2, and p38 antibodies were from Cell Signaling Technology, and anti-β-actin antibody was from Sigma.
Plasmids and Vectors
NICD1, NICD2, and pCMV plasmids were a kind gift from Dr. Xiaolin Tu (Indiana University School of Medicine, Indianapolis, IN). N1-6MT plasmid was a kind gift from Prof. Raphael Kopan (Washington University School of Medicine, St. Louis, MO). pCS2 plasmid was a kind gift from Dr. Junyu Zhang (Fudan University, Shanghai, China). MyD88, TRAF6, and TRIF constructs and NF-κB luciferase reporter plasmids were described previously by us (5, 20, 21). All plasmids were confirmed by DNA sequencing.
Cell Culture and Transfection
Mouse macrophage cell line RAW264.7 and human HEK293T cell line were cultured and transfected with JetPEI (Polyplus Transfection, Illkirch, France) as described (20). Thioglycolate-elicited mouse peritoneal macrophages were isolated from C57BL/6 mice (6–8 weeks) obtained from Joint Ventures Sipper BK Experimental Animal Co. (Shanghai, China) and cultured as described previously (5, 20, 21) and nucleoinfected with the Mouse Macrophage Nucleofect kit using Nucleofector II Biosystems (Amaxa, Gaithersburg, MD) (20, 22).
RNA Interference
The Notch1-specific siRNA (siNotch1-#1084, 5′-CGGCGUGAAUACCUACAAUTT-3′; siNotch1-#3635, 5′-GAGGGAGAUAAACAUUACUTT-3′), and scrambled control siRNA (siNon, 5′-UUCUCCGAACGUGUCACGUTT-3) were synthesized by GenePharma (Shanghai, China) (23). ERK1- and ERK2-specific siRNA (siERK1, sc-29308, and siERK2, sc-35336) were purchased from Santa Cruz Biotechnology. siRNA duplexes were delivered into RAW264.7 cells or mouse peritoneal macrophages at a final concentration of 10 nm using INTERFERin (Illkirch), according to the standard protocol.
Western Blot Analysis
Extraction protein from cells was measured by the BCA protein assay reagent kit (Pierce). 50 μg of protein was resolved by 10% SDS-PAGE and immunoblotted with the indicated antibodies with appropriate HRP-conjugated antibodies as secondary antibodies (Cell Signaling Technology) (5, 23).
ELISA
IL-6, TNF-α, and IL-10 levels in the culture supernatants were quantified with enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems) according to the manufacturer's instructions (5, 20, 21).
Real-time Quantitative PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) following the manufacturer's instructions. Real-time quantitative RT-PCR analysis was performed as described previously (20, 23) using Light Cycler (Roche Diagnostics) and the SYBR RT-PCR kit (Takara, Kyoto, Japan).The primers used are showed in supplemental Table 1.
Luciferase Reporter Assay
RAW264.7 cells or HEK293T cells were co-transfected with the mixture of indicated luciferase reporter plasmid and NF-κB reporter plasmid and the indicated amounts of NICD1, NICD2, N1-6MT, MyD88, TRAF6, or TRIF construct for 24 h, with PRL-TK-Renilla as the luciferase internal control reporter gene. Total amounts of plasmid DNA were equalized with empty control vector. The cells were left treated or untreated with LPS. Luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. Data are normalized for transfection efficiency by dividing firefly luciferase activity with that of Renilla luciferase as we described previously (22).
Statistical Analysis
Statistical significance was determined by Student's t test, with a p value <0.05 considered to be statistically significant.
RESULTS
TLR Ligation Up-regulates Notch Expression and Activates Notch Signaling in Macrophages
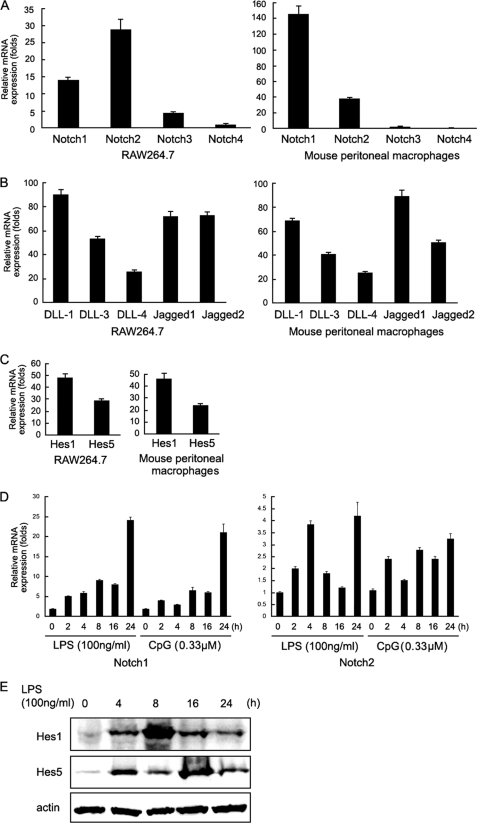
As shown in Fig. 1A, Notch1 and Notch2 were expressed in RAW264.7 cells and peritoneal macrophages, whereas the expression of Notch3 and Notch4 was weak. Therefore we focused on the roles of Notch1 and Notch2 in the TLR response. Notch ligands including DLL-1, DLL-3, DLL-4, Jagged-1, and Jagged-2 were also expressed in RAW264.7 cells and peritoneal macrophages (Fig. 1B). In addition, we further examined the activation of Notch signaling pathway by detecting Notch target gene, Hes1 and Hes5 expression. As shown in Fig. 1C, Hes1 and Hes5 were expressed in RAW264.7 cells and peritoneal macrophages, indicating that Notch signaling was active in macrophages without any stimulation.
FIGURE 1.
TLR ligands up-regulate Notch expression and activate Notch signaling on macrophages. A–C, RAW264.7 cells and mouse peritoneal macrophages were subjected to real-time quantitative PCR analysis for mRNA expression of Notch receptors (A) and Notch ligands (B), and Notch target genes (C). Mouse β-actin was amplified as a control. D, mouse peritoneal macrophages were stimulated with LPS (100 ng/ml) or CpG-ODN (0.33 μm) for the indicated time, and Notch1 and Notch2 mRNA expression were detected by real-time quantitative PCR. Data are shown as mean ± S.D. (error bars; n = 4) of one representative experiment. Similar results were obtained in three independent experiments. E, mouse peritoneal macrophages were stimulated with LPS (100 ng/ml) for the indicated time, and then Hes1 and Hes5 protein expression was detected by immunoblotting. Data are representative of three separate experiments.
Previous studies demonstrated that TLR signaling could modulate Notch signaling in macrophages (13–15). So, we first detected Notch expression under stimulation with LPS (100 ng/ml) and CpG ODN (0.33 μm). As shown in Fig. 1D, stimulation of macrophages with LPS and CpG ODN up-regulated the expression of Notch1 and Notch2. By detecting the expression of Hes1 and Hes5, we found that LPS could induce Notch activation in peritoneal macrophages (Fig. 1E), which was in agreement with a previous report (13). Taken together, these data indicate that Notch activation could be further enhanced by TLR ligands in macrophages, raising a possibility that Notch signaling may be involved in the regulation of TLR-triggered inflammatory responses.
Notch Suppresses Proinflammatory Cytokine Production but Promotes Antiinflammatory Cytokine Production in TLR-triggered Macrophages
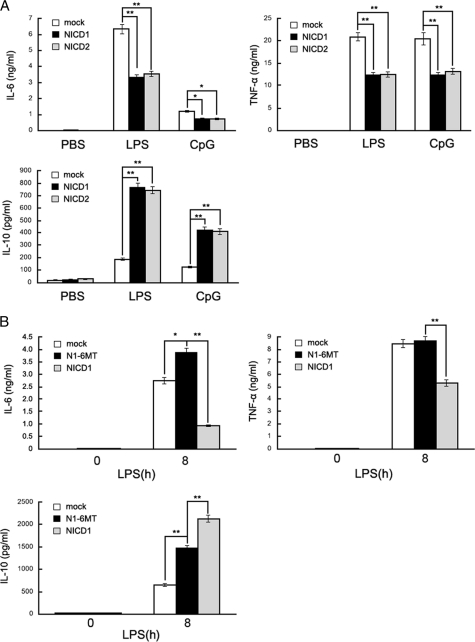
TLR4 can initiate a complex signaling pathway leading to inflammatory cytokine production in macrophages, resulting in inflammation response (24). So we evaluated whether Notch signaling is involved in TLR-triggered inflammatory response in macrophages. Constitutive expression of NICD in target cells could produce an “activated” Notch phenotype. We transfected the intracellular domain of Notch1 or Notch2 (referred to as NICD1 and NICD2, respectively) into peritoneal macrophages. The activation of Notch was confirmed by detecting cleaved Notch using Western blotting in peritoneal macrophages when transfected with NICD1 and NICD2 (supplemental Fig. S1A). As shown in Fig. 2A, overexpression of NICD1 decreased LPS- and CpG ODN-induced secretion of proinflammatory cytokines such as IL-6 and TNF-α while increased the production of antiinflammatory cytokine IL-10. A similar result was also observed in NICD2-overexpressing macrophages in response to the stimuli with LPS and CpG ODN. We further found that the replacement of the 3′ PEST domain of NICD1 with a 6Myc tag (N1–6MT) reversed the NICD1-mediated down-regulation of TNF-α and IL-6 production and the up-regulation of IL-10 production (Fig. 2B). Therefore, the PEST domain, whose phosphorylation triggers Notch degradation (25, 26), plays key roles in mediating regulatory effects of Notch on TLR-mediated inflammatory response.
FIGURE 2.
Effects of Notch activation on LPS-triggered IL-6, TNF-α, and IL-10 production. A, mouse primary peritoneal macrophages (5 × 105) were transiently transfected with NICD1 or NICD2 or mock vector for 48 h and then were stimulated with 100 ng/ml LPS or 0. 33 μm CpG-ODN for 8 h. IL-6, TNF-α, and IL-10 in the supernatants were measured by ELISA. B, mouse primary peritoneal macrophages were transiently transfected with NICD1, N1-6MT, or mock vector for 48 h, and then were stimulated with 100 ng/ml LPS for 8 h. IL-6, TNF-α, and IL-10 in the supernatants were measured by ELISA. Data are shown as mean ± S.D. (error bars; n = 3) of one representative experiment. Similar results were obtained in three independent experiments. *, p < 0.05; **, p < 0.01.
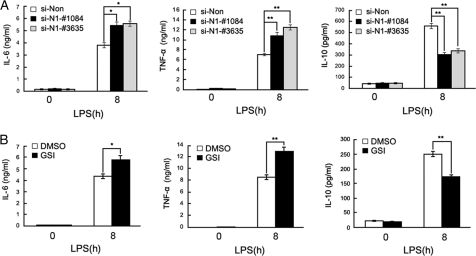
We next silenced Notch1 expression in mouse peritoneal macrophages with specific siRNA (si-N1-#1084 and #3635). The silencing efficacy was confirmed by Western blotting (supplemental Fig. S1B). We found that silencing of Notch1 expression increased LPS-induced TNF-α and IL-6 production but decreased IL-10 production (Fig. 3A). Furthermore, blockade of Notch signaling by a GSI, γ-secretase inhibitor X, significantly increased TNF-α and IL-6 production but decreased IL-10 production (Fig. 3B). These results further confirmed the role of Notch signaling in the negative regulation of TLR-triggered inflammatory response.
FIGURE 3.
Effects of Notch blockade on the TLR4-triggered production of IL-6, TNF-α, and IL-10 in macrophages. A, mouse primary peritoneal macrophages were transfected with siNotch1 (si-N1-#1084, si-N1-#N1–3635) or control siRNA (si-Non) for 48 h and then stimulated with LPS (100 ng/ml) for 8 h. The supernatant IL-6, TNF-α, and IL-10 were measured by ELISA. B, macrophages were pretreated with 2 μm GSI or dimethyl sulfoxide (DMSO) for 24 h and stimulated with LPS (100 ng/ml) for 8 h. The supernatant IL-6, TNF-α, and IL-10 were measured by ELISA. Data are shown as mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01.
Notch Suppresses TLR-triggered Inflammatory Response through ERK1/2 Inactivation
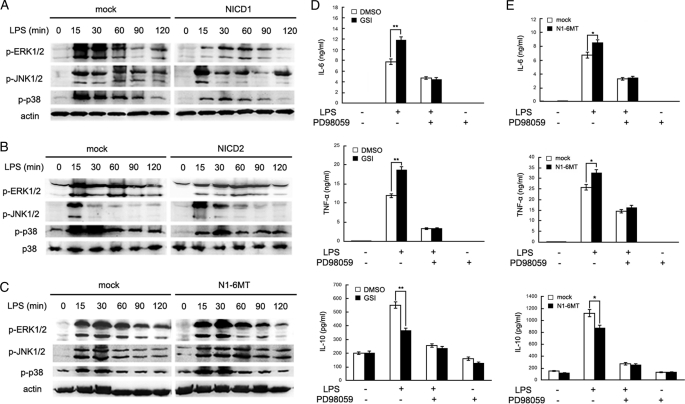
We then investigated whether Notch signaling participates in regulation of TLR-mediated signaling pathway in macrophages. Multiple signaling pathways were involved in the TLR-mediated inflammatory response, including MAPKs and NF-κB (27–29). We first detected the effect of Notch on the MAPK pathway in peritoneal macrophages. As shown in Fig. 4A, whereas LPS stimulation alone enhanced the phosphorylation of ERK1/2, JNK1/2, and p38, overexpression of NICD1 significantly inhibited LPS-induced ERK1/2 phosphorylation compared with mock. A similar result was observed in NICD2-transfected macrophages in response to LPS stimulation (Fig. 4B). On the other hand, overexpression of N1-6MT reversed Notch-mediated inhibition of ERK1/2 activation (Fig. 4C). Blockade of Notch signaling by GSI also promoted LPS-induced ERK activation (supplemental Fig. S2).
FIGURE 4.
Notch signal inhibits TLR-triggered inflammatory cytokine production by ERK1/2 inactivation. A–C, mouse primary peritoneal macrophages were transfected with NICD1 (A), NICD2 (B), N1-6MT (C), or mock vector. After 48 h, cells were stimulated with 100 ng/ml LPS for the indicated time. Phospho-ERK, JNK, and p38 were detected by immunoblotting. Data are representative of three separate experiments. Similar results were obtained in three independent experiments. D, primary peritoneal macrophages were pretreated with 2 μm GSI or dimethyl sulfoxide (DMSO) for 24 h and stimulated with 100 ng/ml LPS and 10 nm PD98059 together or alone for 8 h. The supernatant IL-6, TNF-α, and IL-10 were measured by ELISA. E, mouse primary peritoneal macrophages were transiently transfected with N1-6MT or mock vector. After 48 h, cells were stimulated with 100 ng/ml LPS and 10 nm PD98059 together or alone for 8 h. IL-6, TNF-α, and IL-10 in the supernatants were measured by ELISA. Data are shown as mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01.
We pretreated peritoneal macrophages with a specific MEK inhibitor, PD98059, for 30 min before LPS stimulation and observed that the effect of Notch on TLR-triggered inflammatory cytokine production. Blockade of ERK activation by PD98059 completely reversed the increase in GSI or N1-6MT overexpression-mediated IL-6 and TNF-α production, and the decrease in GSI or N1–6MT overexpression-mediated IL-10 production (Fig. 4, D and E). These results suggest that ERK1/2 inactivation is required for Notch-mediated inhibition of TLR-triggered inflammatory cytokine production.
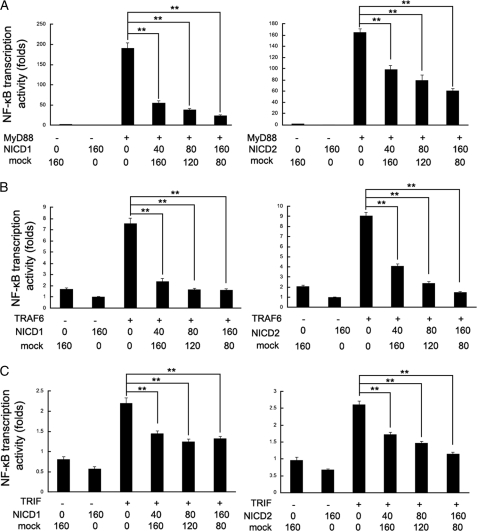
Notch Signaling Inhibits TLR-triggered NF-κB Transcription Activity by MyD88/TRAF6 and TRIF Pathways
NF-κB is one of the most important regulators of proinflammatory gene expression including TNF-α and IL-6. We found that TLR-triggered NF-κB transcription was significantly suppressed by overexpression of NICD1 and NICD2 (Fig. 5A). It is well known that TLR signaling pathway is mediated by both the MyD88-dependent pathway and the TRIF (MyD88-independent) pathway. As shown in Fig. 5A, co-transfection of NICD1 or NICD2 with MyD88 inhibited NF-κB transcription activity in a dose-dependent manner in HEK293T cells; and transfection of NICD1 or NICD2 together with TRAF6 has a similar effect (Fig. 5B). However, co-transfection of N1–6MT with MyD88, TRAF6, or TRIF reversed Notch-mediated inhibition of NF-κB transcription activity (supplemental Fig. S3). In addition, co-transfection of NICD1 or NICD2 with TRIF also dose-dependently inhibited NF-κB transcription activity (Fig. 5C). These results suggest that Notch signaling inhibits TLR-induced NF-κB transcription activity, which is dependent on both MyD88/TRAF6 and TRIF pathways.
FIGURE 5.
Notch signaling inhibits TLR-triggered NF-κB transcription activity via MyD88/TRAF6 and TRIF pathway. HEK293T cells were transfected with 40 ng of NF-κB reporter gene plasmid, 10 ng of pTK-Renilla-luciferase, and 40 ng of MyD88 (A), TRAF6 (B), or TRIF (C) expressing plasmid alone or with 160 ng of NICD1 or NICD2 expressing plasmid. After 24 h, NF-κB reporter activities in lysates were measured by luciferase assay. Empty control vector was added to each sample to ensure transfection of the equal amount of DNA. Luciferase activity is normalized to Renilla luciferase activity and is presented relative to basal luciferase activity. Data are the mean ± S.D. (error bars) of five samples in one experiment representative of similar results obtained in three independent experiments. **, p < 0.01.
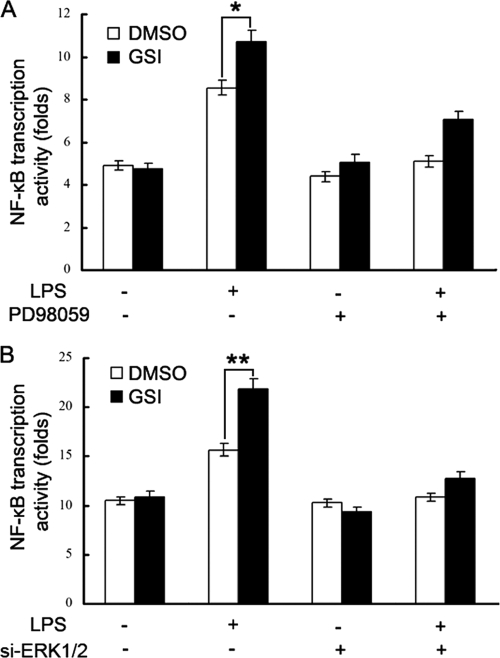
Notch-mediated ERK Inactivation Is Responsible for Inhibition of NF-κB Transcription Activity
Because both the MAPK cascade and the NF-κB signaling pathway participate in the TLR-triggered inflammatory response (30, 31), we investigated the relationship between ERK1/2 activation and NF-κB activity in the Notch-mediated inhibitory effect on TLR inflammatory response. We used PD98059, the specific MEK inhibitor, to treat RAW264.7 cells and then observed the effect of Notch inhibition on LPS-triggered NF-κB transcription activity. As shown in Fig. 6A, inhibition of Notch signaling by GSI increased LPS-triggered NF-κB transcription activity, whereas PD98059 reversed the GSI-mediated effect on NF-κB transcription activity. To confirm further the role of ERK1/2 in the Notch-mediated inhibitory effect of NF-κB, we silenced ERK1/2 expression in RAW246.7 cells with specific siRNA for ERK1 and ERK2 (siERK1 and siERK2), and then observed the effect of the inhibition of Notch by GSI on LPS-triggered NF-κB transcription activity. As shown in Fig. 6B, GSI-mediated promotion of NF-κB activation was also reversed by down-regulation of ERK1/2 expression. These results suggest that Notch-mediated ERK inactivation is responsible for the inhibition of NF-κB transcription activity by Notch.
FIGURE 6.
Notch-mediated ERK1/2 inactivation is responsible for the inhibition of NF-κB transcription activity. A, RAW264.7 cells were transfected with 200 ng of NF-κB luciferase reporter plasmids, 20 ng of pTK-Renilla luciferase reporter plasmids. After 6 h, cells were pretreated with 20 nm PD98059 for 30 min and then pretreated with 2 μm GSI or dimethyl sulfoxide (DMSO). After 24 h, the cells were stimulated with 100 ng/ml LPS for 6 h. Luciferase activity was measured. Data are shown as mean ± S.D. (error bars; n = 5) of one representative result from three independent experiments with similar results. *, p < 0.05; **, p < 0.01. B, RAW264.7 cells were transfected with 5 nm siERK1 and siERK2 or control siRNA (si-Non) for 24 h. The cells were transfected with 200 ng of NF-κB luciferase reporter plasmids, 20 ng of pTK-Renilla luciferase reporter plasmids. After 6 h, cells were pretreated by 2 μm GSI or dimethyl sulfoxide. After 24 h, the cells were stimulated with 100 ng/ml LPS for 6 h. Luciferase activity was measured. Data are shown as mean ± S.D. (error bars; n = 5) of one representative result from three independent experiments with similar results. *, p < 0.05; **, p < 0.01.
DISCUSSION
The present study affirms the hypothesis that the Notch pathway plays an important role in TLR response in macrophages. Evidence supporting this idea includes the expression of multiple Notch receptors and ligands in macrophages; markedly increased Notch1 and Notch2 expression in macrophages stimulated with LPS or CpG ODN, an event that likely involves TLR4/9 and NF-κB; the LPS-induced expression of Notch target genes; the suppression of TLR4-triggered secretion of proinflammatory cytokines by Notch activation; and the regulation of TLR-induced MAPK and NF-κB pathways in macrophages overexpressing NICD1/2.
In the immune system, the role of Notch signaling in the function of professional antigen-presenting cells, especially macrophages, has recently been a hot topic. Many in vitro studies have implicated Notch signaling in macrophage TLR responses. Here, we showed that Notch signaling suppresses TLR-triggered production of proinflammatory cytokines (IL-6 and TNF-α) by inhibiting ERK1/2-mediated NF-κB activity in macrophages. Recently, several studies have reported inconsistent outcomes on the cross-talk of Notch and TLR pathways to regulate TLR responses in macrophages. Tsao et al. (15) showed that disruption of Notch signaling by DAPT (a GSI) attenuated the LPS-induced increase in the levels of released IL-1β and IL-6 but did not alter the level of TNF-α. Using another GSI (IL-CHO) to block Notch signaling, decreased LPS and IFN-γ-induced IL-6 and iNOS mRNA and increased IL-10 mRNA at late stages of activation (12–24 h) were observed in murine bone marrow-derived macrophages (13). Discrepant results were also reported concerning the activation of NF-κB pathway by DLL4-induced Notch signaling (32) or unaltered NF-κB activity by Notch activation (33). These data, together with ours, highlight the profound functional outcome of TLR and Notch interaction. Several issues may contribute to this complexity of interpreting these experiments. First, these studies employed different approaches to manipulate Notch activation or inhibition. For GSIs (DAPT, IL-CHO, or JLK6) (13, 15), GSI activities also regulate other signaling pathways (34); so using different GSIs may alter outcomes in a Notch-independent fashion. In addition, different ligands (DLL4 (32) or Jagged1 (15)) were used for Notch ligation. The amount of ligand used likely does not correspond to physiologically attainable levels and may be quite different among the studies. Finally, few studies have used defined models of macrophages. Macrophages from different species (human or murine) and of various derivations (cell lines (15), bone marrow-derived (13), peripheral blood mononuclear cell-derived (15), or peritoneal macrophages (Fig. 2–4)) were used. Because each study employed distinct model and stimulation conditions, the relevance of the conclusions for the macrophage TLR response remains to be demonstrated. Given these seemingly discrepant results, we speculated that the functional outcome of TLR and Notch interaction is complex, and like other physiological processes, the outcome of one signal pathway (Notch) on another (TLR) is a question of balance (17, 35), in the aforementioned cases among the potentials to activate or repress TLR-driven responses by different Notch components and target genes.
Accumulating evidence supports the existence of important but incompletely understood cross-talk between Notch and other signaling pathways like MAPK, Akt, and NF-κB that regulate cell growth and inflammation (36, 37). The present study demonstrates that Notch activation in macrophages inhibits the activation of the MAPK and NF-κB pathways (Figs. 4 and 6). Notch signaling also suppressed the TLR4-triggered transcription activity of reporter genes such as TNF-α (data not shown) that may contribute to a proinflammatory macrophage phenotype. Furthermore, whereas stimulation through the TLR4/9 pathway (e.g. by LPS or CpG ODN) induces Notch1/2 expression in macrophages (Fig. 1), Notch signaling suppresses the TLR-triggered inflammatory response (Fig. 2), which is reminiscent of a Notch target genes Hes1/Hey1-mediated feedback inhibition of cytokine production that fine tunes the TLR response (17, 35). This adds to the number of signaling pathways including the nucleotide binding domain and leucine-rich repeat-containing gene family, C-type lectins, and glucocorticoids that either positively or negatively affect the TLR responses (4). It will be important to determine whether these reciprocal regulation pathways identified here are restricted to particular cell types or are broadly operational.
Notch signaling has been reported to regulate T cells directly to produce cytokines including IL-2, IFN-γ, IL-4, IL-6, IL-10, and IL-12 (39–43). Because macrophages also produce IFN-γ, IL-6, and IL-10, it will be interesting to explore the role of Notch signaling in regulating cytokine production in macrophages. The effects of NICD and GSI on LPS-induced cytokine production are reminiscent of those of SOCS3 (44). SOCS3 plays an important role in regulating expression of TNF-α, IL-6, and iNOS in macrophages, and its expression is induced by IL-10 (44, 45). SOCS3 negatively regulates expression of IL-6 and iNOS at mRNA and protein levels and TNF-α expression at the protein level (44). It has recently been shown that Notch signaling mediates Bacillus Calmette-Guerin-induced SOCS3 up-regulation (46). Given that LPS-induced IL-10 production was increased in NICD1/2-overexpressing macrophages and decreased in the presence of GSI, and that SOCS3 is a downstream molecule of Notch signaling (46), it will be speculative that SOCS3 be involved in LPS-induced IL-10 production by Notch in macrophages. Interestingly, Notch has been shown to induce IL-10 production in T helper cells in vitro and in vivo (38, 40, 43). Therefore, the relationships among Notch signaling, IL-10, and SOCS3 need to be further investigated.
Taken together, our findings in this study demonstrate that Notch signaling negatively regulates TLR-triggered inflammatory cytokine and signaling pathway in macrophages. Thus, further understanding of the Notch signaling in the context of TLR-triggered macrophage biology will likely provide insights into the mechanisms of inflammation and new approach for rational therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Prof. R. Kopan for N1-6MT plasmid; Dr. X. Tu for NICD1, NICD2, and pCMV plasmids; and Dr. J. Zhang for pCS2 plasmid. In addition, we thank Y. Li and M. Jin for excellent technical assistance.
This work was supported by National Key Basic Research Program of China Grants 2010CB911903 and 2012CB910202, National Natural Science Foundation of China Grants 81123006, 30801009, and 31070789, and Shanghai Committee of Science and Technology Grant 10DZ1910300.

This article contains supplemental Table 1 and Figs. S1–S3.
- TLR
- Toll-like receptor
- DLL
- Delta-like
- GSI
- γ-secretase inhibitor
- NICD
- Notch intracellular domain
- ODN
- oligodeoxynucleotide
- TRAF
- TNF receptor-associated factor
- TRIF
- TIR domain-containing adapter-inducing interferon-β.
REFERENCES
- 1. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 2. O'Neill L. A., Bowie A. G. (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signaling. Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 3. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Negative regulation of Toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 4. O'Neill L. A. (2008) When signaling pathways collide: positive and negative regulation of Toll-like receptor signal transduction. Immunity 29, 12–20 [DOI] [PubMed] [Google Scholar]
- 5. Han C., Jin J., Xu S., Liu H., Li N., Cao X. (2010) Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11, 734–742 [DOI] [PubMed] [Google Scholar]
- 6. Ivashkiv L. B. (2009) Cross-regulation of signaling by ITAM-associated receptors. Nat. Immunol. 10, 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan J. S., Kousis P. C., Suliman S., Visan I., Guidos C. J. (2010) Functions of Notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–365 [DOI] [PubMed] [Google Scholar]
- 8. Aster J. C., Blacklow S. C., Pear W. S. (2011) Notch signaling in T cell lymphoblastic leukemia/lymphoma and other hematological malignancies. J. Pathol. 223, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heitzler P. (2010) Biodiversity and noncanonical Notch signaling. Curr. Top. Dev. Biol. 92, 457–481 [DOI] [PubMed] [Google Scholar]
- 10. Artavanis-Tsakonas S., Muskavitch M. A. (2010) Notch: the past, the present, and the future. Curr. Top. Dev. Biol. 92, 1–29 [DOI] [PubMed] [Google Scholar]
- 11. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 12. Bray S., Bernard F. (2010) Notch targets and their regulation. Curr. Top. Dev. Biol. 92, 253–275 [DOI] [PubMed] [Google Scholar]
- 13. Palaga T., Buranaruk C., Rengpipat S., Fauq A. H., Golde T. E., Kaufmann S. H., Osborne B. A. (2008) Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 38, 174–183 [DOI] [PubMed] [Google Scholar]
- 14. Foldi J., Chung A. Y., Xu H., Zhu J., Outtz H. H., Kitajewski J., Li Y., Hu X., Ivashkiv L. B. (2010) Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J. Immunol. 185, 5023–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsao P. N., Wei S. C., Huang M. T., Lee M. C., Chou H. C., Chen C. Y., Hsieh W. S. (2011) Lipopolysaccharide-induced Notch signaling activation through JNK-dependent pathway regulates inflammatory response. J. Biomed. Sci. 18, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim M. Y., Park J. H., Mo J. S., Ann E. J., Han S. O., Baek S. H., Kim K. J., Im S. Y., Park J. W., Choi E. J., Park H. S. (2008) Down-regulation by lipopolysaccharide of Notch signaling, via nitric oxide. J. Cell Sci. 121, 1466–1476 [DOI] [PubMed] [Google Scholar]
- 17. Hu X., Chung A. Y., Wu I., Foldi J., Chen J., Ji J. D., Tateya T., Kang Y. J., Han J., Gessler M., Kageyama R., Ivashkiv L. B. (2008) Integrated regulation of Toll-like receptor responses by Notch and interferon-γ pathways. Immunity 29, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okamoto M., Takeda K., Joetham A., Ohnishi H., Matsuda H., Swasey C. H., Swanson B. J., Yasutomo K., Dakhama A., Gelfand E. W. (2008) Essential role of Notch signaling in effector memory CD8+ T cell-mediated airway hyperresponsiveness and inflammation. J. Exp. Med. 205, 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niranjan T., Bielesz B., Gruenwald A., Ponda M. P., Kopp J. B., Thomas D. B., Susztak K. (2008) The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat. Med. 14, 290–298 [DOI] [PubMed] [Google Scholar]
- 20. Liu X., Yao M., Li N., Wang C., Zheng Y., Cao X. (2008) CaMKII promotes TLR-triggered proinflammatory cytokine and type I interferon production by directly binding and activating TAK1 and IRF3 in macrophages. Blood 112, 4961–4970 [DOI] [PubMed] [Google Scholar]
- 21. Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., Cao X. (2010) The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nat. Immunol. 11, 487–494 [DOI] [PubMed] [Google Scholar]
- 22. Liu X., Zhan Z., Li D., Xu L., Ma F., Zhang P., Yao H., Cao X. (2011) Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat. Immunol. 12, 416–424 [DOI] [PubMed] [Google Scholar]
- 23. Wang C., Qi R., Li N., Wang Z., An H., Zhang Q., Yu Y., Cao X. (2009) Notch1 signaling sensitizes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human hepatocellular carcinoma cells by inhibiting Akt/Hdm2-mediated p53 degradation and up-regulating p53-dependent DR5 expression. J. Biol. Chem. 284, 16183–16190 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Takeda K., Kaisho T., Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 25. Rechsteiner M. (1988) Regulation of enzyme levels by proteolysis: the role of pest regions. Adv. Enzyme Regul. 27, 135–151 [DOI] [PubMed] [Google Scholar]
- 26. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong C., Davis R. J., Flavell R. A. (2002) MAP kinases in the immune response. Annu. Rev. Immunol. 20, 55–72 [DOI] [PubMed] [Google Scholar]
- 28. Kawai T., Akira S. (2007) Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 29. Cao W., Bao C., Padalko E., Lowenstein C. J. (2008) Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling. J. Exp. Med. 205, 1491–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lawrence T., Gilroy D. W., Colville-Nash P. R., Willoughby D. A. (2001) Possible new role for NF-κB in the resolution of inflammation. Nat. Med. 7, 1291–1297 [DOI] [PubMed] [Google Scholar]
- 31. Mathur R. K., Awasthi A., Wadhone P., Ramanamurthy B., Saha B. (2004) Reciprocal CD40 signals through p38MAPK and ERK-1/2 induce counteracting immune responses. Nat. Med. 10, 540–544 [DOI] [PubMed] [Google Scholar]
- 32. Fung E., Tang S. M., Canner J. P., Morishige K., Arboleda-Velasquez J. F., Cardoso A. A., Carlesso N., Aster J. C., Aikawa M. (2007) Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation 115, 2948–2956 [DOI] [PubMed] [Google Scholar]
- 33. Monsalve E., Pérez M. A., Rubio A., Ruiz-Hidalgo M. J., Baladrón V., García-Ramírez J. J., Gómez J. C., Laborda J., Díaz-Guerra M. J. (2006) Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J. Immunol. 176, 5362–5373 [DOI] [PubMed] [Google Scholar]
- 34. Hass M. R., Sato C., Kopan R., Zhao G. (2009) Presenilin: RIP and beyond. Semin. Cell Dev. Biol. 20, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hertzog P. (2008) A notch in the toll belt. Immunity 29, 663–665 [DOI] [PubMed] [Google Scholar]
- 36. Bellavia D., Campese A. F., Alesse E., Vacca A., Felli M. P., Balestri A., Stoppacciaro A., Tiveron C., Tatangelo L., Giovarelli M., Gaetano C., Ruco L., Hoffman E. S., Hayday A. C., Lendahl U., Frati L., Gulino A., Screpanti I. (2000) Constitutive activation of NF-κB and T cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19, 3337–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haruki N., Kawaguchi K. S., Eichenberger S., Massion P. P., Olson S., Gonzalez A., Carbone D. P., Dang T. P. (2005) Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 65, 3555–3561 [DOI] [PubMed] [Google Scholar]
- 38. Cua D. J., Stohlman S. A. (1997) In vivo effects of T helper cell type 2 cytokines on macrophage antigen-presenting cell induction of T helper subsets. J. Immunol. 159, 5834–5840 [PubMed] [Google Scholar]
- 39. Zhang Y., Sandy A. R., Wang J., Radojcic V., Shan G. T., Tran I. T., Friedman A., Kato K., He S., Cui S., Hexner E., Frank D. M., Emerson S. G., Pear W. S., Maillard I. (2011) Notch signaling is a critical regulator of allogeneic CD4+ T cell responses mediating graft-versus-host disease. Blood 117, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kassner N., Krueger M., Yagita H., Dzionek A., Hutloff A., Kroczek R., Scheffold A., Rutz S. (2010) Cutting edge: plasmacytoid dendritic cells induce IL-10 production in T cells via the Delta-like-4/Notch axis. J. Immunol. 184, 550–554 [DOI] [PubMed] [Google Scholar]
- 41. Okamoto M., Matsuda H., Joetham A., Lucas J. J., Domenico J., Yasutomo K., Takeda K., Gelfand E. W. (2009) Jagged1 on dendritic cells and Notch on CD4+ T cells initiate lung allergic responsiveness by inducing IL-4 production. J. Immunol. 183, 2995–3003 [DOI] [PubMed] [Google Scholar]
- 42. Ong C. T., Sedy J. R., Murphy K. M., Kopan R. (2008) Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS One 3, e2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rutz S., Janke M., Kassner N., Hohnstein T., Krueger M., Scheffold A. (2008) Notch regulates IL-10 production by T helper 1 cells. Proc. Natl. Acad. Sci. U.S.A. 105, 3497–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berlato C., Cassatella M. A., Kinjyo I., Gatto L., Yoshimura A., Bazzoni F. (2002) Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide-induced macrophage activation. J. Immunol. 168, 6404–6411 [DOI] [PubMed] [Google Scholar]
- 45. Qasimi P., Ming-Lum A., Ghanipour A., Ong C. J., Cox M. E., Ihle J., Cacalano N., Yoshimura A., Mui A. L. (2006) Divergent mechanisms utilized by SOCS3 to mediate interleukin-10 inhibition of tumor necrosis factor α and nitric oxide production by macrophages. J. Biol. Chem. 281, 6316–6324 [DOI] [PubMed] [Google Scholar]
- 46. Narayana Y., Balaji K. N. (2008) Notch1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J. Biol. Chem. 283, 12501–12511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.