Background: HypA and HypB are metallochaperones for the activities of [NiFe]-hydrogenase and urease in Helicobacter pylori.
Results: Key residues are identified for the GTP-dependent dimerization of HypB. The HypA-HypB interfaces are also identified.
Conclusion: Self-dimerization is critical for the regulation of GTPase activity. HypA-HypB interaction facilitates further downstream Ni2+ delivery.
Significance: The study is important to understanding [NiFe]-hydrogenase and urease maturation.
Keywords: Chaperone Chaperonin, Helicobacter pylori, Metalloenzymes, Protein Metal Ion Interaction, Protein-Protein Interactions, Metal Delivery
Abstract
The maturation of [NiFe]-hydrogenase is highly dependent on a battery of chaperone proteins. Among these, HypA and HypB were proposed to exert nickel delivery functions in the metallocenter assembly process, although the detailed mechanism remains unclear. Herein, we have overexpressed and purified wild-type HypB as well as two mutants, K168A and M186L/F190V, from Helicobacter pylori. We demonstrated that all proteins bind Ni2+ at a stoichiometry of one Ni2+ per monomer of the proteins with dissociation constants at micromolar levels. Ni2+ elevated GTPase activity of WT HypB, which is attributable to a lower affinity of the protein toward GDP as well as Ni2+-induced dimerization. The disruption of GTP-dependent dimerization has led to GTPase activities of both mutants in apo-forms almost completely abolished, compared with the wild-type protein. The GTPase activity is partially restored for HypB(M186L/F190V) mutant but not for HypB(K168A) mutant upon Ni2+ binding. HypB forms a complex with its partner protein HypA with a low affinity (Kd of 52.2 ± 8.8 μM). Such interactions were also observed in vivo both in the absence and presence of nickel using a GFP-fragment reassembly technique. The putative protein-protein interfaces on H. pylori HypA and HypB proteins were identified by NMR chemical shift perturbation and mutagenesis studies, respectively. Intriguingly, the unique N terminus of H. pylori HypB was identified to participate in the interaction with H. pylori HypA. These structural and functional studies provide insight into the molecular mechanism of Ni2+ delivery during maturation of [NiFe]-hydrogenase.
Introduction
Helicobacter pylori is currently the only discovered bacterial species that can survive under the highly acidic conditions of the human stomach. It is reported that more than 50% of the world population harbor this pathogen in their upper gastrointestinal tract (1). This pathogenic bacterium was found to be associated with a series of human diseases, such as gastritis, peptic ulcer, and even stomach cancer (2, 3). The colonization and survival of H. pylori in human stomach mucosa are heavily dependent on the production of two nickel-containing enzymes, [NiFe]-hydrogenase and urease. The former can be used to provide energy by oxidizing molecular hydrogen (H2) produced by intestinal bacteria (4), whereas the latter hydrolyzes urea into carbamate and ammonia, maintaining the neutral pH of bacterial cytoplasm (5).
Similar to other metalloenzymes, the maturation of [NiFe]-hydrogenase and urease are mediated by a battery of chaperone proteins (6–11). Particularly, the assembly of the Ni2+-containing metallocenter requires the cooperation of a series of metallochaperones to ensure proper metal incorporation (12). Nickel ions, when present in excess, are lethal; thus, the level of the metal ions must be tightly regulated in H. pylori (13–17). The Ni2+ incorporation process mediated by metallochaperones is the pivotal step for maturation of the enzyme.
Previous studies have demonstrated that two metallochaperones, HypA and HypB, are critical for the maturation of [NiFe]-hydrogenase. The enzyme activity of [NiFe]-hydrogenase is severely impaired in hypA or hypB gene knock-out bacterial strains but can be partially restored by the addition of high concentration of nickel ions in the culture medium (18). These studies indicated that HypA and HypB were probably responsible for the Ni2+ delivery in the [NiFe]-hydrogenase metallocenter assembly process. Intriguingly, a recent study demonstrated that disruption of hypA and hypB genes in H. pylori resulted in reduction of the activity of both urease and [NiFe]-hydrogenase, implying a possible “cross-talk” between the maturation pathways of urease and [NiFe]-hydrogenase in H. pylori (19).
The solution structure of HypA from H. pylori was reported recently and exhibits a unique elongated shape with two distinct metal-binding sites. The functional Ni2+ site is located at the N terminus with a typical square planar geometry facilitating further Ni2+ transfer. Zn2+ coordinates to four conserved cysteines tetrahedrally in the zinc site and plays a pivotal role to stabilize the entire protein structure (20). H. pylori HypB was identified as a small GTPase with a highly conserved metal-binding site at its G-domain (21). Although both Ni2+ and Zn2+ could coordinate to this site, only Ni2+ and not Zn2+ induces the dimerization of H. pylori HypB, indicative of the potentially physiological role of Ni2+ (22). It has been demonstrated that the H. pylori HypA-HypB heterodimeric complex could be detected by chemical cross-linking in vitro (21). However, there is still a lack of comprehensive study of H. pylori HypB protein and its interaction with H. pylori HypA.
In the present study, HypB from the H. pylori 26695 bacterial strain was expressed and characterized. A highly conserved Lys residue (Lys-168) and a hydrophobic residue pair (Met-186 and Phe-190) (Fig. 1) were identified to be critical for the GTPase activity of H. pylori HypB. Ni2+ binds to the conserved site of the G-domain of H. pylori HypB proteins (both WT and mutants), functioning as distinctive regulators to modulate the GTP-dependent dimerization of H. pylori HypB. Importantly, the protein-protein interfaces on H. pylori HypA and HypB were identified for the first time by combined use of biochemical and biophysical techniques. A possible effect exerted by HypA and HypB is proposed for Ni2+ delivery in the maturation of [NiFe]-hydrogenase.
FIGURE 1.
Sequence alignment of HypB proteins. The alignment was performed with HypB proteins from bacteria H. pylori 26695 (H. pylori), E. coli strain K12 (E. coli), M. jannaschii DSM 2661 (M. jannaschii), Bradyrhizobium japonium (B. japonium), and Clostridium botulinum strain ATCC 3502 (C. botulinum). The N-terminal residues involved in HypA binding are marked with black arrows. The highly conserved metal binding residues are marked with black asterisks. The invariant lysine residue and the two residues on the hydrophobic core are marked with black triangles.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonucleases and T4 polynucleotide kinase were purchased from New England Biolabs. Phusion high fidelity DNA polymerase was obtained from Finnzymes. Pfu DNA polymerase was obtained from Promega. Primers for PCRs synthesized by Invitrogen are listed in supplemental Table S1. Chromatography columns and the FPLC system were from GE Healthcare. All chemical reagents were purchased from Sigma, unless otherwise specified. All solutions were prepared with Milli-Q water (18.2 megaohms).
HypA and HypB Expression Plasmids and Mutants
The generation of pGBHIS-hypA and pET28a-hypB expression plasmids was described previously (20). The hypB gene was amplified by PCR from genomic DNA of H. pylori 26695. The amplified hypB gene fragment contains NdeI and EcoRI restriction sites at the 5′-end and 3′-end, respectively. The amplified DNA fragment and expression plasmid pET28a were digested with the corresponding restriction enzymes and ligated together with T4 ligase, generating the expression plasmid pET28a-hypB. The construct for expression of HypBΔ24N (residues 25–242) was amplified by PCR using pET28a-hypB vector (full-length) as a template and cloned into pET28a vector. Constructs of HypB(M186L/F190V), HypB(K168A), HypB(K18A), HypB(V20A), HypB(K21A), and HypB(I22A) were generated using Phusion high fidelity DNA polymerase according to the provided protocol. The linear constructs were phosphorylated using T4 polynucleotide kinase and digested with DpnI before ligation. The reaction mixture was subsequently transformed into XL-1 Blue Escherichia coli. All of the constructed plasmids were sequenced (Invitrogen) to verify the cloned sequences.
Protein Expression and Purification
The expression and purification of HypA and 15N-HypA were carried out as described previously (20). HypB was expressed in the E. coli BL21(DE3) strain. When OD at 600 nm reached 0.6 in LB medium, 0.2 mm IPTG2 was added to induce protein expression, and cells were harvested by centrifugation after 16 h of incubation at 25 °C. The cells were lysed by sonication in buffer A (20 mm Tris-HCl containing 500 mm NaCl, pH 7.4) in the presence of 0.1 mm PMSF. Cell supernatant was loaded into a HisTrap column (GE Healthcare) pre-equilibrated with buffer A supplemented with 50 mm imidazole. Bound proteins were eluted using buffer A containing 300 mm imidazole. His-tagged HypB elution fraction was dialyzed against protease buffer (20 mm Tris-HCl, pH 7.4, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT) overnight, followed by the addition of 100 NIH units of thrombin (Sigma) and 30 units of alkaline phosphatase (calf Intestinal; New England Biolabs) to remove the His tag and bound nucleotides. After incubation at room temperature for 3 h, EDTA was added into the cleaved protein to a final concentration of 20 mm and further incubated at 4 °C overnight. The protein was subsequently pooled and applied to a Superdex-75 size exclusion column equilibrated in buffer A in the presence of 1 mm DTT. The yield of purified protein is around 40 mg/liter of LB medium. The metal contents of the purified HypB were determined by inductively coupled plasma MS. DTT was then removed by a HiTrap desalting column (GE Healthcare), and the protein was frozen at −80 °C in 0.5-ml aliquots for further use. All HypB mutants were expressed and purified similarly. The concentrations of all protein samples were determined by a BCA assay.
Electronic Absorption Spectroscopy
All electronic absorption spectra were collected on a Cary 50 UV-visible spectrophotometer with 1-cm quartz cuvette at ambient temperature. Spectra were recorded from 600 to 240 nm with a scan rate of 360 nm/min. For Ni2+ titration experiments, 50 μm apo-HypB solutions or its mutants HypB(K168A) and HypB(M186L/F190V) were prepared in a titration buffer (20 mm Tris-HCl, pH 7.4, containing 100 mm NaCl, 200 μm TCEP). Aliquots of Ni2+ stock solution (10 mm in the titration buffer) were added stepwise into protein solutions. Similarly, GTP was titrated into Ni2+-bound HypB or its mutants except that the titration buffer was supplemented with 1 mm MgSO4, which is required for GTP binding. The UV titration curves were fitted to the Ryan-Weber nonlinear equation (23),
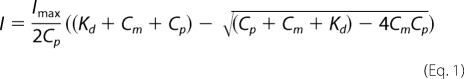 |
where I represents UV absorbance intensity; Imax is the maximal UV absorbance when all of the ligands are bound; Cp and Cm are the total concentrations of proteins and ligands, respectively; and Kd is the dissociation constant.
Size Exclusion Chromatography
Gel filtration analysis of oligomeric states of the proteins was performed on an ÄKTA FPLC system using a Tricorn 30/100 Superdex 75 column at 4 °C eluted with a flow rate of 0.5 ml/min. The column was calibrated with a low molecular weight gel filtration calibration kit (GE Healthcare). For Ni2+-dependent dimerization experiments, a 200 μm concentration of either the apo-forms of HypB proteins or their Ni2+-bound forms, which were prepared by preincubation of 2 mol eq of Ni2+ prior to analysis, were freshly prepared and subjected to chromatography. The proteins were eluted with a gel filtration buffer (20 mm HEPES buffer, 100 mm NaCl, 50 μm TCEP, pH 7.0) supplemented with 20 μm Ni2+. GTP-dependent dimerization experiments were carried out similarly except that 5 mm MgSO4 and 500 μm GTP instead of Ni2+ were included in the gel filtration buffer.
Isothermal Titration Calorimetry (ITC)
Metal-bound HypB was prepared as described in the supplemental material. Freshly purified apo-, Zn2+-, and Ni2+-HypB were prepared in ITC buffer (20 mm HEPES buffer, 100 mm NaCl, 1 mm MgSO4, pH 7.0). GTP (or GTPγS) and GDP with concentrations of 600 μm were used as stock solutions and titrated into 50 μm protein samples. Titration of ligands into the buffer was recorded as background. To determine the binding affinity of HypA and HypB, about 1.1 mm HypA in 20 mm HEPES containing 100 mm NaCl, 2 mm TCEP, pH 7.0, was titrated into 0.1 mm apo-HypB in the same buffer. All experiments were carried out on an ITC200 isothermal titration calorimeter (MicroCal) at 25 °C. Data were fitted by the Origin 7 software package.
GTPase Activity Assay
The GTPase activity assay for WT HypB and its mutants was performed as described previously (22). The reaction mixture consisting of 200 μm GTP, 1 mm MgSO4, ∼10 μm proteins in a reaction buffer (20 mm HEPES, 100 mm NaCl, and 1% glycerol, pH 7.0) was incubated at 37 °C. Aliquots of the mixture were taken out in a time course for determination of released free phosphate by the Malachite Green phosphate assay kit (Cayman).
GFP-Fragment Reassembly
GFP-fragment reassembly was used to verify the interaction between HypA and HypB inside E. coli cytoplasm (24). Details of plasmid construction are described in the supplemental material, and the primers used are listed in supplemental Table S2.
The constructed plasmids pET32a-NGFP, pBAD33-CGFP, pET32a-NGFP-linker-HypA, and pBAD33-HypB-linker-CGFP were subsequently co-transformed into the BL21(DE3) strain with different combinations. The transformed bacteria were cultured in LB medium at 37 °C until A600 reached 0.6, and 20 μm IPTG and 0.2% arabinose were added to induce the protein expression. The bacteria were further cultured overnight at 20 °C. The bacteria were then diluted with PBS buffer and examined by an Axiovert 200 M fluorescent microscope (Carl Zeiss) using a 100× oil immersion objective lens. The effects of Ni2+ on intracellular fluorescence were investigated similarly except that the co-transformed BL21(DE3) was cultured in M9 minimal medium with or without a supplement of 10 mm Ni2+. About 100 μm IPTG and 0.015% arabinose were used to induce protein expression, and intracellular fluorescence was examined after culture for 48 h at 25 °C.
NMR Spectroscopy
The binding interface on HypA was mapped based on NMR titration experiments. All NMR titration experiments were performed at 25 °C on a Bruker Avance 600 spectrometer operating at a 1H frequency of 600.13 MHz, equipped with a TCI CryoProbeTM. Unlabeled monomeric apo-HypB (∼1 mm) was gradually added into 0.3 mm 15N-labeled HypA in 20 mm Tris-HCl buffer, pH 7.4, containing 100 mm NaCl. Two-dimensional 1H-15N HSQC spectra were recorded before and after each addition of HypB. The chemical shift perturbation was quantified in a manner similar to that described previously (25). The intensity of each resonance was plotted against the molar percentage of added HypB, and the resulting slope was used to estimate the extent of line broadening. The chemical shift perturbations (Δ) for each residue were quantified based on Δ = ((Δδ2NH + Δδ2N/25)/2)½. The dissociation constant (Kd) of HypA-HypB complex was obtained from least squares fitting of the chemical shift changes of residue His-17 of 15N-HypA (the largest perturbed residue) as a function of total HypB concentration according to Equation 2 (26),
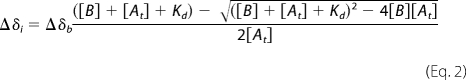 |
where Δδi represents the observed change of chemical shift, [B] is the HypB concentration at each titration point, [At] is the total HypA concentration, and Kd is the dissociation constant.
Chemical Cross-linking
Purified HypA (40 μm), HypB, or its mutant proteins (20 μm) were preincubated in 20 mm HEPES buffer containing 100 mm NaCl at pH 7.4. Cross-linking chemical reagent bis(sulfosuccinimidyl) suberate (BS3) was added into the protein solution to a final concentration of 0.2 mm. The reaction mixtures were further incubated at room temperature for 1 h. The cross-linking reaction was terminated by the addition of 1 m Tris solution to a final concentration of 50 mm. Reaction samples were then subjected to 13.5% SDS-PAGE and stained with Coomassie Blue.
RESULTS
Protein Analysis
All of the purified HypB proteins have more than 95% purity as demonstrated on the SDS-PAGE (Fig. 2). The metal contents of the purified HypB proteins were determined by inductively coupled plasma MS, and the molar ratios of metals to the protein are less than 0.01:1 for Ni2+, Zn2+, and Mg2+, indicating that HypB was purified as an apo-form.
FIGURE 2.
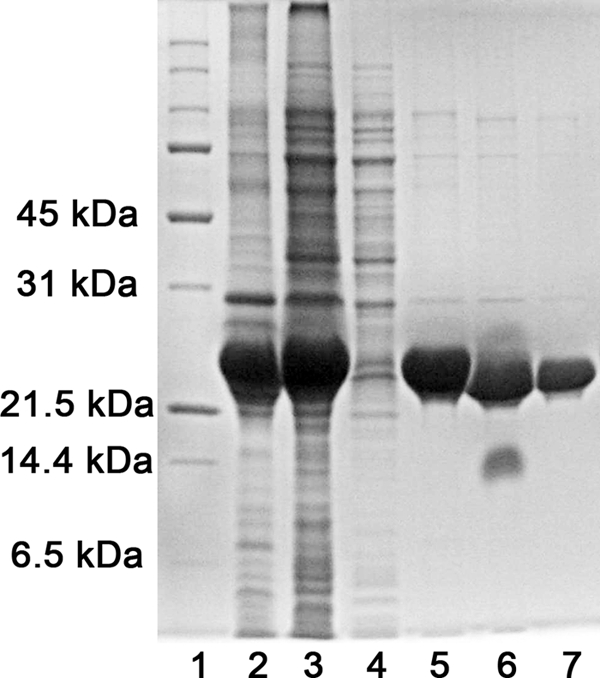
SDS-PAGE analysis of the purified wild-type HypB. Lanes 2–7 correspond to pellet and supernatants after cell lysis, flow-through of Ni2+ column, elution fraction of Ni2+ column, proteins after thrombin cleavage, and purified WT HypB. Lane 1 is the protein marker.
Interaction of Ni2+/GTP with HypB
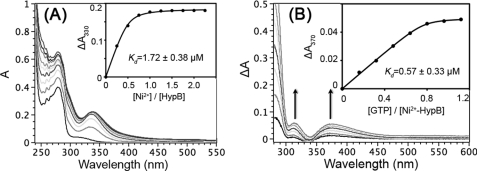
The interactions between Ni2+ and apo-forms of WT HypB or two mutants were monitored by UV-visible spectroscopy. Upon the addition of Ni2+ into apoprotein solutions, two new absorption bands appeared at ∼280 and 340 nm, which represent typical ligand-to-metal charge transfer (LMCT) between Cys-Sγ and Ni2+ (27). The intensities of these bands increased with the further addition of Ni2+ and leveled off at a molar ratio of Ni2+ to the protein of 1.0, indicative of binding of Ni2+ to HypB with a stoichiometry of 1.0 (i.e. one Ni2+ per HypB monomer) (Fig. 3A and supplemental Fig. S1).
FIGURE 3.
Binding of Ni2+ and GTP to H. pylori HypB. A, UV-visible spectra of 50 μm H. pylori HypB in the absence and presence of different molar equivalents of Ni2+ (as 10 mm NiSO4) in 20 mm Tris-HCl buffer, pH 7.4, containing 100 mm NaCl, 200 μm TCEP. Two absorption bands centered at 330 and 280 nm appeared, and their intensities increased until around 1 mol eq of Ni2+ was added (inset). Kd was determined to be 1.72 ± 0.38 μm. The inset shows the titration curve for Ni2+ binding to H. pylori HypB monitored at 330 nm. B, difference UV-visible spectra of Ni2+-HypB with increasing amounts of GTP. The addition of GTP to Ni2+-HypB yielded two absorption bands centered at ∼310 and 370 nm, indicative of conformational changes of the protein at the Ni2+ binding site. The increase in intensities of the bands leveled off at a molar ratio of GTP to Ni2+-HypB of about 1:1 (inset). The dissociation constant (Kd) was determined to be 0.57 ± 0.33 μm. The titration curves were fitted to a Ryan-Weber nonlinear equation to determine the Kd.
GTP binding and hydrolysis were proposed to be critical in the Ni2+ delivery process (28). Titration of GTP to the solutions of Ni2+-HypB (WT or mutants) resulted in a strong absorption band at ∼260 nm, which originated from the self-absorption of GTP molecules. Intriguingly, two relatively weak absorption bands were also observed in the difference UV spectra at around ∼310 and 370 nm, close to the Ni2+ LMCT band (∼340 nm) (Fig. 3B and supplemental Fig. S1), whereas no such absorption peaks were observed when GTP was titrated into Ni2+ solution (supplemental Fig. S3). The absorbance increased with the addition of GTP and saturated at a molar ratio of GTP to Ni2+-HypB of 1.0, indicative of the binding of one GTP molecule per HypB monomer (29). The perturbation of the Ni2+ LMCT band upon nucleotide binding was also observed previously in a Ni2+-binding ATPase (27). Such phenomena are probably due to conformational changes at the Ni2+-binding site induced by binding of GTP or ATP to proteins.
The Ryan-Weber nonlinear equation was applied to fit both the Ni2+ and GTP titration curves. The dissociation constants (Kd) were calculated to be 1.72 ± 0.38 and 0.57 ± 0.33 μm for the binding of Ni2+ to HypB and GTP to Ni2+-HypB, respectively (Fig. 3).
Conserved Lys Residue and Hydrophobic Residue Pair Are Important for GTP-dependent Dimerization and GTPase Activity of HypB
Nucleotide-dependent dimerization was proposed to be a common feature to activate G proteins (30). Previously, it has also been demonstrated that GTP binding induced the dimerization of HypB proteins (22, 29).
Analysis of the structure of HypB from Methanocaldococcus jannaschii (Protein Data Bank entry 2HF9) revealed two unique structural elements that might contribute to the GTP-dependent dimerization. A highly conserved Lys residue, Lys-153 (corresponding to Lys-168 in H. pylori), formed a hydrogen bond with bound GTP, bridging two HypB monomers together. In addition, a hydrophobic residue pair (i.e. Leu-171 and Val-175, corresponding to Met-186 and Phe-190 in H. pylori), located at an α-helical region, may play an important role in stabilizing the dimeric structures by hydrophobic interactions between Val and Leu from each monomer (supplemental Fig. S3). To investigate whether the corresponding structural elements also contribute to the GTP-dependent dimerization of H. pylori HypB, two HypB mutants (i.e. K168A and M186L/F190V) were constructed, and their GTP-dependent dimerization properties and GTPase activities were examined.
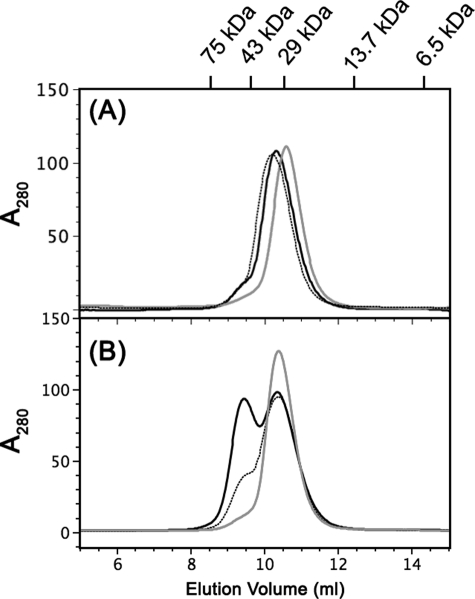
In the absence of GTP, WT HypB, HypB(K168A), and HypB(M186L/F190V) mutants were eluted as a single peak corresponding to the monomeric form of the protein (Fig. 4A). When GTP was included in the gel filtration buffer, ∼50% of WT HypB was eluted at a molecular mass of 52 kDa corresponding to dimeric form. In contrast, HypB(K168A) was eluted as a monomeric form. The majority (∼80%) of HypB(M186L/F190V) was eluted as a monomer (10.4 ml) with a small portion eluted at an elution volume of 9.0 ml, corresponding to a dimeric form (Fig. 4B). These results indicated that mutations at the conserved Lys residues and the two hydrophobic residues completely or partially abolished GTP-dependent dimerization of H. pylori HypB.
FIGURE 4.
The effects of GTP binding on the oligomeric states of the HypB and its mutants. A, gel filtration analysis of HypB and its mutants in the absence of GTP; WT HypB (black solid line) and two mutants, HypB(K168A) (gray solid line) and HypB(M186L/F190V) (dotted line), were eluted as monomers. B, approximately 50% of WT HypB formed a dimer after preincubation with GTP (black solid line), whereas HypB(K168A) (gray solid line) and the majority of HypB(M186L/F190V) (dotted line) mutants were eluted as monomers in the presence of GTP.
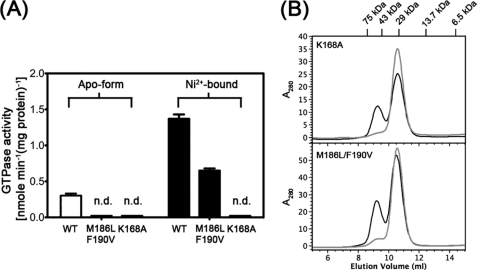
The GTPase activities of two mutants were determined by the Malachite Green method and compared with the WT HypB. The rates of GTP hydrolysis of all protein samples were linear over 80 min (supplemental Fig. S4). In contrast to the WT HypB, which has a low GTP hydrolysis rate of 0.30 ± 0.03 nmol·min−1(mg of protein)−1, both apo-HypB(K168A) and apo-HypB(M186L/F190V) exhibited no detectable GTPase activities (Fig. 5A). Upon Ni2+ binding, the GTPase activity increased dramatically for the wild-type protein (1.37 ± 0.03 nmol min−1(mg of protein)−1) but partially restored for HypB(M186L/F190V) (0.65 ± 0.03 nmol min−1(mg of protein)−1). In contrast, no detectable GTPase activity was found for HypB(K168A) even in the Ni2+-bound form. Similar to the WT HypB for which Ni2+ binding induces dimerization of the protein (22), both HypB(K168A) and HypB(M186L/F190V) mutants could also form dimer upon Ni2+ binding (Fig. 5B).
FIGURE 5.
The effects of Ni2+ binding on the GTPase activities and oligomeric states of HypB mutants. A, GTPase activities of HypB and its mutants in apo- and Ni2+-bound forms. Note that binding of Ni2+ enhanced the GTPase activities for both WT HypB and HypB(M186L/F190V) but not for HypB(K168A). HypB(K168A) mutant in apo- and Ni2+-bound forms showed no detectable GTPase activities (n.d.). B, both mutants HypB(K168A) and HypB(M186L/F190V) were eluted as monomers in the absence of Ni2+ (gray solid line) but partially formed dimers when preincubated with 2 eq of Ni2+ (black solid line). Around 0.1 mm apo-form of HypB mutants in the absence or presence of 2 mol eq of Ni2+ were applied to gel filtration, and proteins were eluted with a standard buffer (20 mm HEPES, 100 mm NaCl, pH 7.0).
Metal Ions Serve as Regulators for Nucleotide Binding to HypB
Previous studies have demonstrated that Ni2+ instead of Zn2+ could promote the dimerization of H. pylori HypB; binding of Ni2+ enhanced GTPase activity, whereas Zn2+ inhibited it (22).
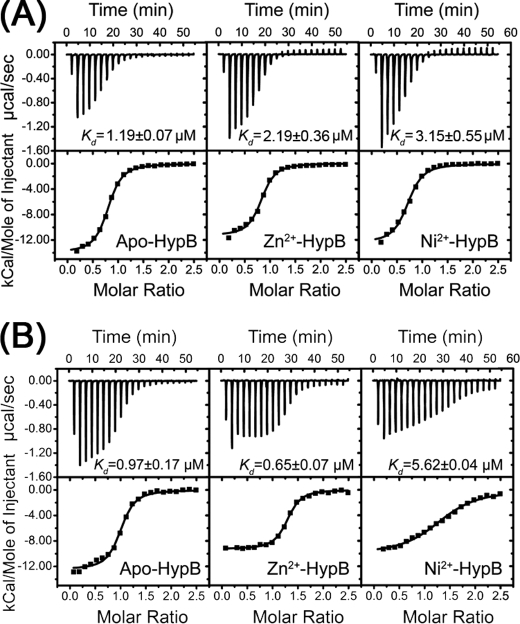
It is known that GTPase activity is also controlled by the affinity to guanine nucleotide and the nucleotide exchange rate (31). The nucleotide-binding properties of H. pylori HypB in its apo- and metal-bound forms were investigated by ITC. The dissociation constants of GTPγS, a non-hydrolyzable GTP analog, to H. pylori HypB were determined to be 1.19 ± 0.07 μm for apo-HypB, 2.19 ± 0.36 μm for Zn2+-HypB, and 3.15 ± 0.55 μm for Ni2+-HypB (Fig. 6A). Binding of GTP to H. pylori HypB yielded similar results to GTPγS (supplemental Fig. S5), implying that metal binding did not significantly affect the binding affinities of GTP to H. pylori HypB. Unexpectedly, the binding of GDP to H. pylori HypB in the presence of Zn2+ and Ni2+ revealed different properties. GDP binds apo- and Zn2+-HypB at least 5 times more tightly than Ni2+-HypB, with dissociation constants of 0.97 ± 0.17, 0.65 ± 0.07, and 5.62 ± 0.64 μm, respectively (Fig. 6B), suggesting that Ni2+-triggered dimerization affected the GDP binding to the protein.
FIGURE 6.
GTPγS (A) and GDP (B) binding to HypB by isothermal titration calorimetry. Metal binding did not affect GTPγS binding, whereas Ni2+ binding led to lower GDP binding affinity to HypB.
HypB Binding Interface Mapping on HypA by NMR Chemical Shift Perturbation
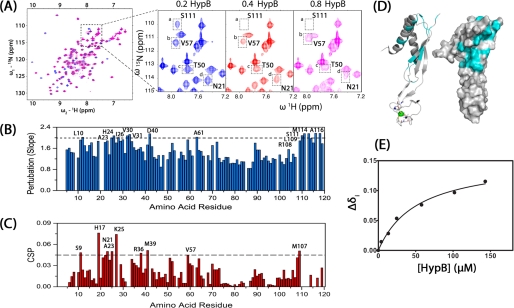
Previous cross-linking experiments demonstrated that monomeric HypA and HypB formed a heterodimeric complex (21). To experimentally map the binding interfaces on HypA, NMR spectroscopy was used to examine the residues that experience chemical shift perturbations upon HypB binding. 15N-HypA was titrated with unlabeled monomeric HypB, and a series of two-dimensional 1H-15N HSQC spectra were recorded (Fig. 7A). Certain resonances in the spectra underwent either significant line broadening or chemical shift changes upon the addition of HypB, indicative of an intermediate or fast exchange on the NMR time scale (32). To quantify the extent of line broadening, the intensities of 115 peaks (of 125 assigned peaks) were plotted against the concentration of HypB, and the slopes obtained for each resonance were plotted and shown in Fig. 7B. The remaining 10 resonances are either overlapped with each other or decreased too fast to be monitored. The most perturbed peaks involved Leu-10, Ala-23, His-24, Ile-26, Val-30, Val-31, Asp-40, Ala-61, Arg-108, Leu-109, Ser-111, Met-114, and Ala-116 when setting the cut-off value to 2.0. The resonance chemical shift perturbation (Δ) can be quantified based on the equation, Δ = ((Δδ2NH +Δδ2N/25)/2)½, and is summarized in Fig. 7C. The resonances experiencing significant chemical shift perturbation included Ser-9, His-17, Asn-21, Ala-23, Lys-25, Arg-36, Met-39, Val-57, and Met-107, which were mapped to the structure of HypA (Fig. 7D). These residues were mainly located at the cleft between helix 1 and strands 1 and 6 (β1/β6).
FIGURE 7.
Identification of protein-protein interfaces on H. pylori HypA. A, two-dimensional 1H,15N HSQC spectra of HypA upon the addition of 0.2, 0.4, and 0.8 mol eq of H. pylori HypB. Residues experiencing changes in intensities (e.g. Ser-111) and chemical shifts (e.g. Asn-21, Thr-50, and Val-57) are highlighted. B, perturbation of resonance intensities of HypA by HypB. The absolute values of the slopes of peak intensities versus HypB concentration for each residue were calculated as described under “Experimental Procedures.” The residues with slope values larger than 2.0 are labeled. C, chemical shift perturbations of HypA upon HypB binding. The residues with chemical shift perturbations larger than 0.45 are highlighted. D, schematic diagram (left) and surface (right) models of HypA structure. Those residues that are perturbed by HypB binding are highlighted in cyan. E, the changes of chemical shift of His-17 versus the concentration of HypB is plotted and used to calculate the HypA-HypB binding constant (Kd of 52.2 ± 8.8 μm).
The dissociation constant for HypA and HypB was determined to be 52.2 ± 8.8 μm by fitting the changes of chemical shifts (e.g. His-17) as a function of HypB concentration (Fig. 7E), indicative of a relatively weak interaction. The binding constant was also measured by ITC, a technique commonly used for quantification of macromolecule-ligand interactions (33). The dissociation constant (Kd of 57.9 ± 8.5 μm) is comparable with the affinity by NMR (supplemental Fig. S6).
Identification of HypB Binding Site to HypA
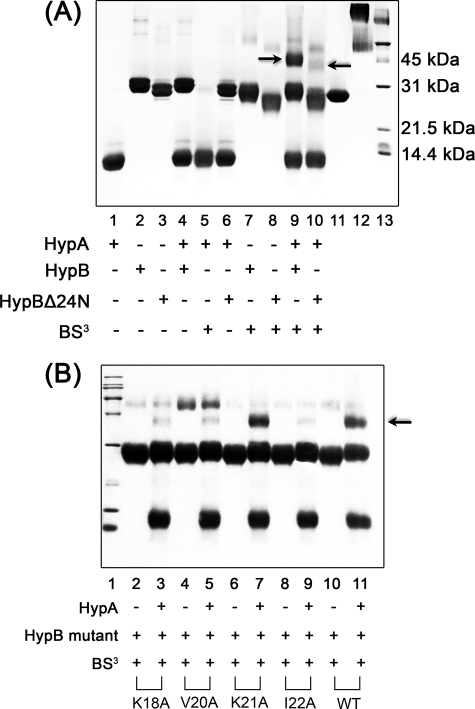
The interaction between HypA and HypB is believed to be important in Ni2+ delivery (21). Unexpectedly, no interaction between HypA and HypB could be detected by GST pull-down experiments (data not shown). The chemical cross-linking method was therefore applied to detect the protein-protein interaction. To investigate whether the N-terminal region of H. pylori HypB is involved in the interaction as previously proposed based on the crystal structure of HypB from M. jannaschii (29), a mutant with N-terminal 24 residues truncated (HypBΔ24N) was expressed and purified. The interactions between HypA and HypB (or HypBΔ24N) were examined by the chemical cross-linking method (Fig. 8A). Incubation of BS3, a water-soluble chemical cross-linking agent, with HypA, HypB, and HypBΔ24N resulted in weak bands corresponding to the molecular weight of a homodimer for each protein (∼25 kDa for HypA, ∼50 kDa for HypB and HypBΔ24N). Incubation of BS3 with a mixture of HypA and HypB resulted in a strong band with molecular mass around 45 kDa. This band was subjected to MALDI-TOF mass spectrum analysis, which revealed the presence of both HypA and HypB proteins (supplemental Table S3), indicative of formation of a heterodimer between HypA and HypB. In contrast, no such a band was observed between HypBΔ24N and HypA, indicating that the N-terminal 24 residues are required for the formation of a HypA-HypB complex.
FIGURE 8.
Identification of the protein-protein interfaces on HypB. A, purified HypA (∼40 μm) and/or full-length HypB (20 μm; lane 9) or HypBΔ24N (lane 10) were incubated in the presence or absence of 0.2 mm BS3. A strong heterodimer band was observed for full-length HypB (indicated by an arrow) but not for HypBΔ24N, indicative of the important role of the N-terminal 24 residues for the formation of HypA-HypB complex. Around 20 μm GST protein was used as a positive control (lanes 11 and 12). B, HypB(K18A), HypB(V20A), HypB(K21A), HypB(I22A), and WT HypB were incubated with or without HypA in the presence of 0.2 mm BS3. The heterodimer band is indicated by a black arrow. Both lane 13 in Fig. 8A and lane 1 in Fig. 8B are protein markers.
Sequence alignments (Fig. 1) of the N-terminal region of HypB from different species revealed three relatively conserved residues (i.e. Lys-18, Val-20, and Ile-22) in the H. pylori HypB sequence. To validate whether these residues involved in HypA-HypB interaction, single-residue mutants of these three conserved residues (K18A, V20A, I22A) and an unconserved residue (K21A) were expressed and purified. The interactions between these HypB mutants and HypA were examined similarly. As shown in Fig. 7B, incubation of HypB(K18A), HypB(V20A), and HypB(I22A) mutants with HypA yielded much weaker heterodimer complex bands compared with the WT HypB, indicative of the involvement of these three conserved residues in HypA-HypB interaction. In contrast, the HypB(K21A) mutant exhibited a strong heterodimer complex band with HypA, which is comparable with that of WT HypB.
Intracellular Interaction of HypA and HypB
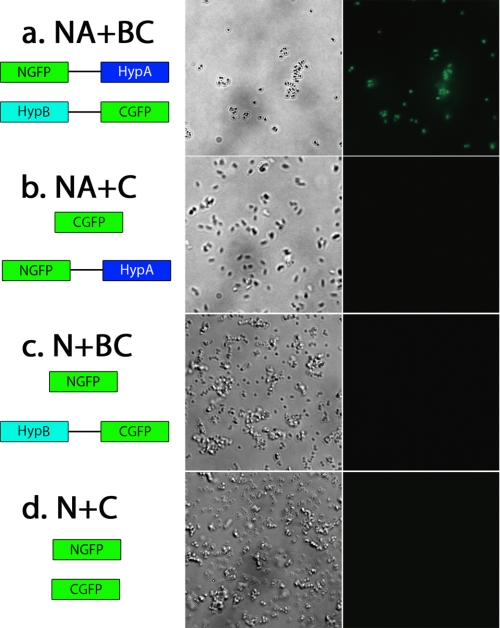
Although in vitro experiments showed the formation of HypA-HypB complex (21), there was a lack of in vivo evidence. GFP-fragment reassembly, a powerful tool to visualize the intracellular interaction between proteins (34), was therefore used. The constructed plasmids were co-transformed into BL21(DE3) bacteria strain with different combinations. The fused proteins were co-expressed, and the phase-contrast fluorescence images of the bacteria are shown in Fig. 9. Only the bacteria containing the pET32a-NGFP-linker-HypA and pBAD33-HypB-linker-CGFP plasmids (NA + BC) exhibited the strongly emitted green fluorescence. In contrast, no fluorescence was detected for the other three control combinations (i.e. E. coli cells that co-expressed CGFP and NGFP-HypA, NGFP and HypB-CGFP, and NGFP and CGFP) (Fig. 9, b–d). The results clearly demonstrated that the interaction between HypA and HypB occurred even in a much more competitive cellular cytoplasm condition. To further investigate the effects of Ni2+ on the intracellular fluorescence, the co-transformed BL21(DE3) bacteria were cultured in M9 minimal medium in the presence or absence of 10 mm Ni2+. The plasmid combination NA + BC exhibited intracellular fluorescence under both conditions (supplemental Fig. S7), indicating that the intracellular interaction between HypA and HypB is independent of the availability of Ni2+. This is consistent with in vitro data indicating that a HypA-HypB complex could be observed in the presence or in the absence of Ni2+ using a cross-linking assay (21).
FIGURE 9.
Visualization of HypA and HypB interaction by GFP reassembly method in E. coli BL21(DE3) cells. Different combinations of fusion proteins (a–d, left) were co-transformed into E. coli cells and co-expressed by the addition of 20 μm IPTG and 0.2% arabinose. Phase-contrast (a–d, middle) and fluorescence images (a–d, right) of the cells were recorded. E. coli cells that co-expressed NGFP-HypA and HypB-CGFP proteins (a) yielded green fluorescence, indicative of interaction between HypA and HypB.
DISCUSSION
As nickel chaperones, HypA and HypB proteins are known to play an important role in the maturation of both [NiFe]-hydrogenase and urease in H. pylori (19). HypB belongs to the G3E family of GTPase (35) and contains a highly conserved metal-binding site on its G-domain. HypA from both H. pylori and Thermococcus kodakaraensis shows that the protein consists of a zinc domain and a nickel-binding domain (20, 36). Zinc coordinates to four conserved cysteines tetrahedrally in the loop region of the domain, whereas nickel binds in the N terminus of the protein. However, there appears to be no detailed study on HypA-HypB interaction.
The homodimeric structure of M. jannaschii HypB revealed that the GTP binding site is composed of residues from both monomers. An invariant lysine, Lys-153 (corresponding to Lys-168 in H. pylori HypB), from monomer B directly contacts the γ-phosphate group of bound GTP molecule in monomer A, forming a salt bridge to stabilize the dimeric structure (29). We showed that substitution of this lysine by alanine in H. pylori HypB(K168A) completely abrogated the GTP-dependent dimerization of the protein, which was probably due to destruction of the salt bridge between the bound GTP γ-phosphate and the lysine side chain. Similarly, substitution of the residues on a hydrophobic core on the dimeric interfaces (M186L and F190V) also abolished the GTP-dependent dimerization of H. pylori HypB. Consequently, no GTPase activities could be detected for both mutants (i.e. apo-HypB(K168A) and apo-HypB(M186L/F190V)).
Upon Ni2+ binding, HypB(M186L/F190V) mutant formed dimers (Fig. 5), resulting in the formation of the intact GTP hydrolysis active site at the interface of the dimer of the protein. As a result, the GTPase activity of Ni2+-HypB(M186L/F190V) significantly increased compared with its apo-form. Our data clearly indicated that monomeric HypB is in a low activity state. Ni2+ acts as a “metallic bridge” that holds two monomers into an active dimer, in which two GTP hydrolysis active sites are formed by the invariant Lys-168 residue from both monomers and thus fully activated.
In contrast, no GTPase activity of Ni2+-HypB(K168A) was observed, although the mutant protein underwent Ni2+-dependent dimerization (Fig. 5B). Because Lys-168 may directly contact with the bound GTP molecule, serving as an important component at the GTP hydrolysis site, mutation of Lys-168 to an alanine led to disruption of the GTP-dependent dimerization of HypB as well as the active site for GTP hydrolysis. Clearly, Lys-168 plays a dual role, whereas Met-186 and Phe-190 only play a role in the dimerization of HypB via hydrophobic contact at the dimeric interface.
Surprisingly, we found that Ni2+-H. pylori HypB exhibited a much lower affinity toward GDP compared with either apo- or Zn-HypB (Kd of 5.6 versus <1 μm, respectively; Fig. 6, A and B). Such lower affinity of GDP may facilitate the GDP/GTP exchange, thus elevating the GTPase activity. It would be of interest to further investigate whether the lower affinity of GDP to Ni2+-H. pylori HypB is due to a conformational change of the protein upon Ni2+ binding.
The interaction of HypA and HypB has been found to be crucial for Ni2+ delivery to the metalloenzymes. In agreement with a previous report (21), we found a heterodimer of HypA and HypB by the cross-linking method. The binding affinity is ∼55 μm as measured by NMR and ITC, indicative of weak interaction. This may explain why a GST pull-down assay failed to detect the interaction between HypA and HypB. Similarly, an interactomic study by tandem affinity purification also failed to demonstrate the interaction of HypA and HypB (37).
NMR is known to be a particularly powerful technique in characterization of weak macromolecule interactions (38). Some protein-protein interactions that cannot be detected by the pull-down assay are readily monitored by NMR (39). Our 1H,15N HSQC experiments clearly showed that HypA interacts with HypB, as demonstrated by the chemical shift changes of specific resonances on HypA (Fig. 7). Furthermore, we found that the binding interface on HypA is at the cleft between helix 1 (α1) and β1/β6. It is noteworthy that binding of both Ni2+ and HypB caused similar chemical shift perturbations on residues from HypA (supplemental Fig. S8), implying a potential competition between HypB and Ni2+. It is likely that binding of HypB could modulate Ni2+ release from Ni2+-HypA, and our preliminary data indeed implied potential transfer of Ni2+ from Ni2+-HypA to HypB.3
The interface of the complex on HypB was examined by mutagenesis and chemical cross-linking. The N terminus of HypB was found to participate in the HypA-HypB interaction, although this region is flexible and absent from the reported M. jannaschii HypB structure (29). Particularly, Lys-18, Val-20, and Ile-22 are involved in the interaction. Our alanine scanning mutagenesis studies on a HypB homologue of Archaeoglobus fulgidus also showed that the N terminus of A. fulgidus HypB participated in the interaction with A. fulgidus HypA,3 indicative of a conserved function of the N-terminal region. In contrast, UreG, a zinc-binding G-protein that participates in the maturation of urease and shares a high sequence similarity with HypB, lacks of the N terminus (40) (supplemental Fig. S9). This indicates that the N-terminal residues of HypB are unique and could be specifically recognized by HypA.
To validate the HypA-HypB interaction in vivo, we then used a GFP reassembly method. As shown in Fig. 9 and supplemental Fig. S7, the green fluorescence of E. coli cells containing the pET32a-NGFP-linker-HypA and pBAD33-HypB-linker-CGFP plasmids (NA + BC), clearly verified the formation of the HypA-HypB complex in vivo (Fig. 9), implying the biologically relevant function of such an interaction. Interestingly, the interaction between HypA and HypB is not dependent on the availability of Ni2+ because intracellular fluorescence could be observed in M9 medium with or without a supplement of Ni2+, in agreement with the NMR data.
The GTPase activity of H. pylori HypB can be enhanced by the binding of Ni2+, the same metal ion that the protein delivers. Similar features have also been found for E. coli ArsA, a soluble ATPase for arsenite and antimonite efflux (41). Intriguingly, a metallochaperone ArsD was identified to transfer trivalent metalloids to ArsA through weak or transient interaction for subsequent metalloid efflux (42–44). Based on the functional similarity between ArsA and HypB, we anticipate that HypA functions similarly to ArsD (i.e. delivering Ni2+ to HypB for subsequent Ni2+ transfer) (Fig. 10). Considering that both Zn2+ and Ni2+ bind H. pylori HypB, it is reasonable to speculate that H. pylori HypB utilizes an indirect approach, such as through specific protein-protein interactions, to achieve selective binding to Ni2+ in vivo, Protein-mediated Ni2+ delivery pathway is therefore postulated. First, Ni2+ is sequestered by HypA via its N-terminal specific Ni2+ binding site (20) and is subsequently transferred to HypB through a specific HypA-HypB interaction, initiating the dimerization of HypB. The active dimeric HypB will then deliver Ni2+ to its downstream receptor (e.g. large subunit of [NiFe]-hydrogenase). Additional chaperone proteins, such as SlyD, which interacts with HypB (45), may also participate in this process. Therefore, further work is warranted to clarify this.
FIGURE 10.
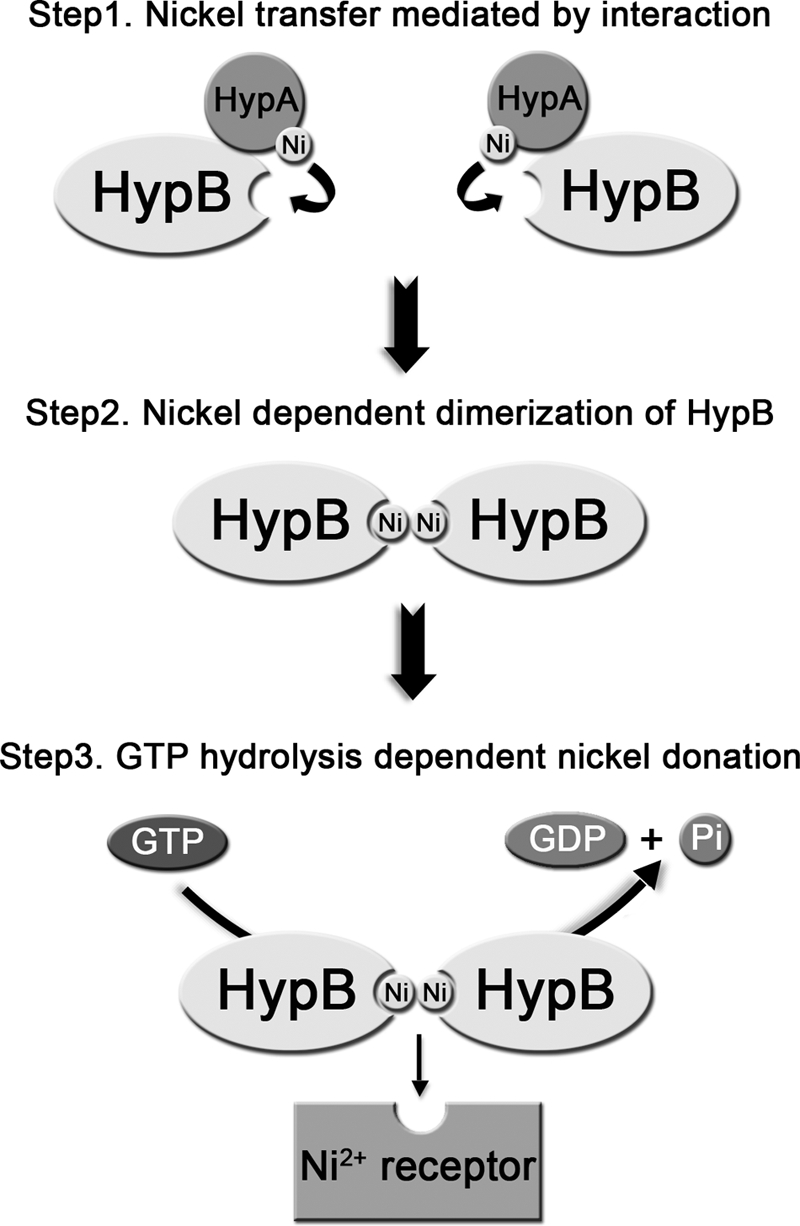
Proposed mechanism of nickel transfer. Nickel is transferred from HypA to the conserved metal-binding site of HypB through a specific protein-protein interaction. Upon acceptance of nickel ions, HypB undergoes nickel-dependent dimerization, and the GTPase activity of HypB is enhanced. The conformational change of HypB coupled with GTP hydrolysis triggers the nickel donation from HypB to its downstream receptor (e.g. the large subunit of [NiFe]-hydrogenase).
Supplementary Material
Acknowledgments
pBAD33 plasmid was obtained from the National BioResource Project (Japan). We thank Prof. Sunny I. Chan (Caltech and Academia Sinica) for helpful comments.
This work was supported by Research Grants Council of Hong Kong Grants HKU1/07C, HKU7042/07P, HKU7049/09P, N_HKU752/09, and CUHK4610/06, the Croucher Foundation, and the University of Hong Kong.

This article contains supplemental Tables S1–S3 and Figs. S1–S9.
W. Xia, H. Li, X. Yang, K.-B. Wong, and H. Sun, unpublished data.
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- BS3
- bis(sulfosuccinimidyl) suberate
- HSQC
- heteronuclear single quantum coherence
- ITC
- isothermal titration calorimetry
- LMCT
- ligand-to-metal charge transfer
- TCEP
- tris(2-carboxyethyl)phosphine
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Covacci A., Telford J. L., Del Giudice G., Parsonnet J., Rappuoli R. (1999) Helicobacter pylori virulence and genetic geography. Science 284, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 2. Marshall B. J., Warren J. R. (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1, 1311–1315 [DOI] [PubMed] [Google Scholar]
- 3. Parsonnet J., Vandersteen D., Goates J., Sibley R. K., Pritikin J., Chang Y. (1991) Helicobacter pylori infection in intestinal- and diffuse-type gastric adenocarcinomas. J. Natl. Cancer Inst. 83, 640–643 [DOI] [PubMed] [Google Scholar]
- 4. Olson J. W., Maier R. J. (2002) Molecular hydrogen as an energy source for Helicobacter pylori. Science 298, 1788–1790 [DOI] [PubMed] [Google Scholar]
- 5. Bauerfeind P., Garner R., Dunn B. E., Mobley H. L. (1997) Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forzi L., Sawers R. G. (2007) Maturation of [NiFe]-hydrogenases in Escherichia coli. BioMetals 20, 565–578 [DOI] [PubMed] [Google Scholar]
- 7. Maier R. J., Benoit S. L., Seshadri S. (2007) Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. BioMetals 20, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng T., Li H., Xia W., Sun H. (2011) Multifaceted SlyD from Helicobacter pylori. Implication in [NiFe]-hydrogenase maturation. J. Biol. Inorg. Chem. doi: 10.1007/s00775-011-0855y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaluarachchi H., Zhang J. W., Zamble D. B. (2011) Escherichia coli SlyD, more than a Ni(II) reservoir. Biochemistry 50, 10761–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cun S., Sun H. (2010) A zinc-binding site by negative selection induces metallodrug susceptibility in an essential chaperonin. Proc. Natl. Acad. Sci. U.S.A. 107, 4943–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cun S., Li H., Ge R., Lin M. C., Sun H. (2008) A histidine-rich and cysteine-rich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori. Implication for nickel homeostasis and bismuth susceptibility. J. Biol. Chem. 283, 15142–15151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leach M. R., Zamble D. B. (2007) Metallocenter assembly of the hydrogenase enzymes. Curr. Opin. Chem. Biol. 11, 159–165 [DOI] [PubMed] [Google Scholar]
- 13. Danielli A., Scarlato V. (2010) Regulatory circuits in Helicobacter pylori. Network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 34, 738–752 [DOI] [PubMed] [Google Scholar]
- 14. Ge R., Watt R. M., Sun X., Tanner J. A., He Q. Y., Huang J. D., Sun H. (2006) Expression and characterization of a histidine-rich protein, Hpn. Potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 393, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ge R., Zhang Y., Sun X., Watt R. M., He Q. Y., Huang J. D., Wilcox D. E., Sun H. (2006) Thermodynamic and kinetic aspects of metal binding to the histidine-rich protein, Hpn. J. Am. Chem. Soc. 128, 11330–11331 [DOI] [PubMed] [Google Scholar]
- 16. Zeng Y. B., Yang N., Sun H. (2011) Metal-binding properties of an Hpn-like histidine-rich protein. Chem. Eur. J. 17, 5852–5860 [DOI] [PubMed] [Google Scholar]
- 17. Macomber L., Hausinger R. P. (2011) Mechanisms of nickel toxicity in microorganisms. Metallomics 3, 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blokesch M., Paschos A., Theodoratou E., Bauer A., Hube M., Huth S., Böck A. (2002) Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30, 674–680 [DOI] [PubMed] [Google Scholar]
- 19. Olson J. W., Mehta N. S., Maier R. J. (2001) Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39, 176–182 [DOI] [PubMed] [Google Scholar]
- 20. Xia W., Li H., Sze K. H., Sun H. (2009) Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J. Am. Chem. Soc. 131, 10031–10040 [DOI] [PubMed] [Google Scholar]
- 21. Mehta N., Olson J. W., Maier R. J. (2003) Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185, 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sydor A. M., Liu J., Zamble D. B. (2011) Effects of metal on the biochemical properties of Helicobacter pylori HypB, a maturation factor of [NiFe]-hydrogenase and urease. J. Bacteriol. 193, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai Y. C., Wu F. C., Liu C. Q., Li W., Guo J. Y., Fu P. Q., Xing B. S., Zheng J. (2008) Ultraviolet absorbance titration for determining stability constants of humic substances with Cu(II) and Hg(II). Anal. Chim. Acta 616, 115–121 [DOI] [PubMed] [Google Scholar]
- 24. Wilson C. G., Magliery T. J., Regan L. (2004) Detecting protein-protein interactions with GFP-fragment reassembly. Nat. Methods 1, 255–262 [DOI] [PubMed] [Google Scholar]
- 25. Cunha L., Kuti M., Bishop D. F., Mezei M., Zeng L., Zhou M. M., Desnick R. J. (2008) Proteins 71, 855–873 [DOI] [PubMed] [Google Scholar]
- 26. Tugarinov V., Kay L. E. (2003) Quantitative NMR studies of high molecular weight proteins. Application to domain orientation and ligand binding in the 723-residue enzyme malate synthase G. J. Mol. Biol. 327, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 27. Jeoung J. H., Giese T., Grünwald M., Dobbek H. (2009) CooC1 from Carboxydothermus hydrogenoformans is a nickel-binding ATPase. Biochemistry 48, 11505–11513 [DOI] [PubMed] [Google Scholar]
- 28. Maier T., Lottspeich F., Böck A. (1995) GTP hydrolysis by HypB is essential for nickel insertion into hydrogenases of Escherichia coli. Eur. J. Biochem. 230, 133–138 [PubMed] [Google Scholar]
- 29. Gasper R., Scrima A., Wittinghofer A. (2006) Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 281, 27492–27502 [DOI] [PubMed] [Google Scholar]
- 30. Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. (2009) It takes two to tango. Regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 31. Kunzelmann S., Praefcke G. J., Herrmann C. (2005) Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1). Methods Enzymol. 404, 512–527 [DOI] [PubMed] [Google Scholar]
- 32. Roberts G. C. K. (1993) NMR of Macromolecules: A Practical Approach, pp. 153–182, IRL Press, New York [Google Scholar]
- 33. Freyer M. W., Lewis E. A. (2008) Isothermal titration calorimetry. Experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 84, 79–113 [DOI] [PubMed] [Google Scholar]
- 34. Morell M., Espargaró A., Avilés F. X., Ventura S. (2007) Detection of transient protein-protein interactions by bimolecular fluorescence complementation. The Abl-SH3 case. Proteomics 7, 1023–1036 [DOI] [PubMed] [Google Scholar]
- 35. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 36. Watanabe S., Arai T., Matsumi R., Atomi H., Imanaka T., Miki K. (2009) Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe]-hydrogenase maturation. J. Mol. Biol. 394, 448–459 [DOI] [PubMed] [Google Scholar]
- 37. Stingl K., Schauer K., Ecobichon C., Labigne A., Lenormand P., Rousselle J. C., Namane A., de Reuse H. (2008) In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol. Cell. Proteomics 7, 2429–2441 [DOI] [PubMed] [Google Scholar]
- 38. Vinogradova O., Qin J. (2011) NMR as a unique tool in assessment and complex determination of weak protein-protein interactions. Top. Curr. Chem. doi: 10.1007/128_2011_216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma C., Li W., Xu Y., Rizo J. (2011) Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol. 18, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ciurli S., Zambelli B., Turano P., Musiani F., Neyroz P. (2009) Proteins 74, 222–239 [DOI] [PubMed] [Google Scholar]
- 41. Rosen B. P. (2002) Biochemistry of arsenic detoxification. FEBS Lett. 529, 86–92 [DOI] [PubMed] [Google Scholar]
- 42. Ajees A. A., Yang J., Rosen B. P. (2011) The ArsD As(III) metallochaperone. BioMetals 24, 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J., Rawat S., Stemmler T. L., Rosen B. P. (2010) Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry 49, 3658–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin Y. F., Walmsley A. R., Rosen B. P. (2006) An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. U.S.A. 103, 15617–15622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J. W., Butland G., Greenblatt J. F., Emili A., Zamble D. B. (2005) A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J. Biol. Chem. 280, 4360–4366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.