Background: Dysregulation of 5-HT1A receptors is associated with depression, but the transcriptional mechanisms are unclear.
Results: Deaf-1 binds the 5-HT1A promoter, and loss of Deaf-1 results in altered expression of 5-HT1A receptors and reduced serotonin levels.
Conclusion: Deaf-1 is a key regulator of 5-HT1A expression in vivo that is affected by a promoter polymorphism prevalent in human depression.
Significance: Understanding transcriptional regulators of serotonin could lead to improved antidepressant strategies.
Keywords: 7-Helix Receptor, Depression, Promoters, Receptor Regulation, Serotonin, Transcription Factors
Abstract
Altered regulation of the serotonin-1A (5-HT1A) receptor gene is implicated in major depression and mood disorders. The functional human 5-HT1A C(−1019)G promoter polymorphism (rs6295), which prevents the binding of Deaf-1/NUDR leading to dysregulation of the receptor, has been associated with major depression. In cell models Deaf-1 displays dual activity, repressing 5-HT1A autoreceptor expression in serotonergic raphe cells while enhancing postsynaptic 5-HT1A heteroreceptor expression in nonserotonergic neurons. A functional Deaf-1 binding site on the mouse 5-HT1A promoter was recognized by Deaf-1 in vitro and in vivo and mediated dual activity of Deaf-1 on 5-HT1A gene transcription. To address regulation by Deaf-1 in vivo, Deaf-1 knock-out mice bred to a C57BL/6 background were compared with wild-type siblings for changes in 5-HT1A RNA and protein by quantitative RT-PCR, in situ hybridization, and immunofluorescence. In the dorsal raphe, Deaf-1 knock-out mice displayed increased 5-HT1A mRNA, protein, and 5-HT1A-positive cell counts but reduced 5-HT levels, whereas other serotonergic markers, such as tryptophan hydroxylase (TPH)- or 5-HT-positive cells and TPH2 RNA levels, were unchanged. By contrast, 5-HT1A mRNA and 5-HT1A-positive cells were reduced in the frontal cortex of Deaf-1-null mice, with no significant change in hippocampal 5-HT1A RNA, protein, or cell counts. The region-specific alterations of brain 5-HT1A gene expression and reduced raphe 5-HT content in Deaf-1−/− mice indicate the importance of Deaf-1 in regulation of 5-HT1A gene expression and provide insight into the role of the 5-HT1A G(−1019) allele in reducing serotonergic neurotransmission by derepression of 5-HT1A autoreceptors.
Introduction
Reduced activity of the brain serotonin system has been implicated in major depression and suicide (1–3), and antidepressant treatments including serotonin reuptake inhibitors (SSRIs)5 increase serotonergic neurotransmission, either directly or indirectly (4, 5). The serotonin-1A (5-HT1A) somatodendritic autoreceptor inhibits the firing of raphe serotonin neurons to negatively regulate the serotonin system (4). Postsynaptic 5-HT1A receptors are strongly expressed in limbic and cortical brain regions (6, 7) and mediate serotonin action to reduce fear and anxiety. 5-HT1A-null mice display increased anxiety behaviors (8–10) and lack response to SSRI treatment (11), whereas transgenic overexpression of the 5-HT1A receptor decreases anxiety (12). Rescue of 5-HT1A receptor expression in the early postnatal forebrain rescues the anxiety phenotype (13). Hence, the 5-HT1A receptor functions as a critical regulator of the serotonin system and is implicated in mood disorders.
The 5-HT1A receptor gene is regulated by a proximal promoter and an upstream repressor region that determines basal expression in neuronal cells (14, 15). In the human 5-HT1A repressor region, we previously identified a functional C(−1019)G polymorphism associated with major depression and suicide, as well as anxiety-related phenotypes, which has been replicated in independent studies (16, 17). The G-allele prevents the binding of the transcription factor deformed epidermal autoregulatory factor-1 (Deaf-1) (18). In serotonergic cells, Deaf-1 represses 5-HT1A autoreceptor expression, whereas in nonserotonergic neuronal cells Deaf-1 enhances 5-HT1A promoter activity (19). However, the dual activity of Deaf-1 on endogenous 5-HT1A expression in vivo has not been demonstrated. Based on promoter studies, the G-allele should increase 5-HT1A autoreceptor levels to reduce 5-HT neuron firing and decrease postsynaptic 5-HT1A receptors to synergistically reduce serotonin neurotransmission. In depressed subjects, the homozygous 5-HT1A G/G(−1019) genotype was associated with increased 5-HT1A autoreceptor binding potential (20–22), consistent with increased 5-HT1A autoreceptors in postmortem studies of depressed suicides (23, 24). Thus, the 5-HT1A G(−1019) allele may alter 5-HT1A receptor expression in vivo because of its inability to bind Deaf-1.
The role of Deaf-1 in the regulation of 5-HT1A receptor expression in vivo was addressed using the Deaf-1−/− mouse (25). In Deaf-1-null mice compared with littermate wild-type controls, 5-HT1A mRNA and protein levels were increased in the dorsal raphe region, whereas serotonin cell number and tryptophan hydroxylase 2 (TPH2) mRNA were unchanged. By contrast, postsynaptic 5-HT1A receptors were down-regulated in the frontal cortex of Deaf-1 mutant mice. Thus, consistent with results in cell lines, we find a region-specific dysregulation of 5-HT1A receptor expression in Deaf-1−/− mice. The Deaf-1 knock-out mice indicate the importance of Deaf-1 in regulation of the 5-HT1A receptor gene in vivo.
MATERIALS AND METHODS
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was conducted as described previously (19). Briefly, sense and antisense oligonucleotides of the human (26C) and mouse (mouse#1, 5′-GGAGAGTCTCTGAGGGTTTTCCTCGTGCCTGATA) Deaf-1 elements synthesized with GG 5′-overhangs were hybridized and labeled with [32P]dCTP using Klenow fragment DNA polymerase (15). Purified bacterially expressed human Deaf-1 protein (5 or 7.5 μg/reaction) was preincubated with or without competitor DNA in a 20-μl reaction containing labeled probe in DNA-binding buffer (20 mm HEPES, 0.2 mm EDTA, 0.2 mm EGTA, 100 mm KCl, 5% glycerol, and 2 mm DTT, pH 7.9), 1 μg of poly(dI-dC), at 22 °C for 20 min and resolved on a nondenaturing 5% acrylamide/Tris-glycine gel at 4 °C. Unlabeled doubled-stranded polyoma virus enhancer activator 3 (PEA3) (5′-GGGATCCAGGAAGTGA) and E2F (5′-ATTTAAGTTTCGCGCCTTTTC) oligonucleotides were used as nonspecific competitor.
Plasmids, Transfections, and Luciferase Assays
The mouse 5-HT1A promoter (position −1 to −984 or −942 relative to the ATG) was amplified by PCR using the following primers: forward, 5′-CTCGAGCCCATCCTTCAGAGTCTCTGAGGG or 5′-GCCACACTGTGAACCTGGCTC; reverse, 5′-AAGCTTGCCTGCCTGCACTCCCG) and SN48 cells genomic DNA as a template and verified by sequence analysis. The forward primer included an XhoI site and the reverse primer a HindIII site for cloning into the pGL3B luciferase reporter vector (Promega). The 3×-mDeaf-1RE (three copies of mouse Deaf-1 response element) construct was generated by cloning complementary 78-bp primers containing three copies of the mouse Deaf-1 palindrome 1 (AGTCTCTGAGGGTTTTCCTCGTGCCT) in the NheI/XhoI site of pGL3P. For luciferase assays, HEK293 cells or Ltk− cells were plated at a density of 2 × 105 and 1.5 × 105 cells/ml respectively, in 6-well plates. The following day cells were transfected with 1 μg of reporter plasmid, 1 μg of pCMV-β-galactosidase, and 1 μg of either pcDNA3 or pcDNA3 His-Deaf-1 using calcium phosphate (HEK293) or Lipofectamine 2000 (Ltk−) with a lipid:DNA ratio of 2:1. Cells were incubated 24–36 h following transfection and lysed in reporter lysis buffer (Promega). β-Galactosidase activity was used to normalize luciferase activity to correct for transient transfection efficiency.
Chromatin Immunoprecipitation (ChIP)
SN48 cells were fixed with 1% formaldehyde for 10 min at 22 °C. Fixation was stopped following addition of 0.125 m glycine for 5 min, and cells were then washed 3 times with PBS. Cells were scraped in 10 mm Tris, pH 8.0, 85 mm KCl, and 0.5% Nonidet P-40 and harvested by centrifugation. Cells were then lysed in 50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100 supplemented with protease inhibitors and sonicated with 16 pulses of 30 s. Extracts were then incubated with preimmune serum or Deaf-1 rabbit antibody (1/100 dilution) overnight at 4 °C. Extracts were then incubated for 2 h with protein A-Sepharose beads at 4 °C, washed once with 1 ml of lysis buffer, 4 times with 1 ml of wash buffer (100 mm Tris-HCl, pH 7.5, 500 mm LiCl, 1% Nonidet P-40, and 1% deoxycholate) and once with 1 ml of TE. DNA was eluted in 0.1 m NaHCO3 with 1% SDS and samples reverse cross-linked overnight at 65 °C. DNA was purified following phenol/chloroform extraction and precipitated with 2 volumes of EtOH and 20 μg of linear acrylamide for 2 h at −20 °C. PCR amplifications were performed using 5 μl of each elution with the following primers (forward/reverse): 5-HT1A promoter, ATGAAAAGGTCACAGGCACC/TCACAGTGTGGCTATCAGGC; 5-HT1A open reading frame, CCGCAAGACGGTCAAGAAGG/GGCGTCGTCCTCACCCTG; GAPDH promoter, GTCCAAAGAGAGGGAGGAGG/AGGGCTGCAGTCCGTATTTA.
Animals and Genotyping
All animal studies were conducted in accordance with the University of Ottawa Animal Care Committee guidelines using protocol NSI-57. Animals were maintained on a 12-h light/dark cycle with lights on at 6:00 a.m., at 21 °C with food and water ad libitum. Deaf-1 knock-out mice were received from Dr. Jane Visvader (25) and rederived by embryo transfer (Taconic, Hudson, NY) on a mixed C57BL/6-BALB/c-129 background. Pilot experiments revealed no significant differences in 5-HT1A mRNA levels between mixed background Deaf-1-null (−/−) and wild-type (+/+) mice, but differences were observed compared with pure C57BL/6 mice. Hence, the mice were back-crossed for five generations onto the C57BL/6 background. All experiments were performed on adult (8–10 weeks old) knock-out mice with age-matched littermate wild-type controls from the C56BL/6 background, unless otherwise specified.
For genotyping, tail DNA was isolated using DNA lysis buffer (100 mm Tris, pH 8.0, 5 mm EDTA, 0.2% SDS, 200 mm NaCl, 10 μg/ml proteinase K). PCR was done using the primers: forward, 5′-GTGCTCTCATCCTCATAGCCTC; reverse1, 3′-GAAACAAGAAAACACATCGGGAGAATCA; reverse2, 3′-AAGAATAGAACCACCACCCAGCT, with 267-bp wild-type and 421-bp knock-out products. Each mouse used in this study was genotyped at weaning and verified at sacrifice.
Two-dimensional SDS-PAGE
Prefrontal cortex was dissected, rinsed in ice-cold 10 mm BES pH 7.4, 0.3 m sucrose, homogenized in isoelectric focusing buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% DTT, 0.001% bromphenol blue) and incubated for 90 min at 22 °C. Samples were centrifuged at 10,000 × g for 5 min, the supernatant was precipitated in 90% acetone and centrifuged (2000 × g for 5 min), and the pellet was resuspended in isoelectric focusing buffer supplemented with biolytes. IPG strips (pH 3–10; Bio-Rad) were rehydrated overnight with 300 μl of isoelectric focusing buffer containing 1 mg of total protein. Isoelectric focusing was done using the following conditions: desalting for 20 min at 500 V; 2.5 h ramp to 10,000 V; 40,000 VH focusing at 10,000 V. IPG strips were then treated for 15 min with a reducing solution (375 mm Tris-HCl, pH 8.8, 20% glycerol, 2% SDS, 65 mm DTT) followed by a 15-min incubation with an alkylation solution (375 mm Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, 215 mm iodoacetamide) and placed on top of a 12% SDS-polyacrylamide gel using a 0.5% agarose overlay. Gels were run for 5 h at 30 mA and transferred to PVDF membranes. Membranes were blocked in 5% milk and incubated overnight at 4 °C with rabbit anti-Deaf-1 antibody (1:1000; custom raised against full-length Deaf-1 peptide; Cedarlane).
Immunofluorescence
Mice were anesthetized by lethal intraperitoneal injection (0.01 ml/g of body weight) of sodium pentobarbital (Somnitol; MTC Pharmaceuticals, Cambridge, ON, Canada) and perfused by cardiac infusion of 20 ml of saline, then 20 ml of 4% paraformaldehyde. Whole brains were isolated, postfixed overnight in 4% paraformaldehyde, washed with 10% sucrose changed twice daily for five consecutive days, and frozen. Coronal brain slices (12-μm) were prepared using the following coordinates (26): rostral raphe (Bregma −4.2 to −4.96 mm), hippocampus (Bregma 1.46 to −2.30 mm), and prefrontal cortex (Bregma 2.96 to −2.80 mm). For 5-HT1A immunostaining and dual immunofluorescence, sections were incubated in 1× target retrieval solution (DakoCytomation; Mississauga, ON, Canada) at 95 °C for 30 min and washed three times in PBS at 22 °C. Sections were blocked for 1 h in blocking solution (Stockholm's PBS with 1% BSA, 10% goat serum, 0.1% Triton X-100) and incubated overnight with the following antibodies: 1:50 rabbit serum anti-5-HT1A (custom made primary antibody designed to the i2 loop of the 5-HT1A receptor sequence; Cedarlane, Hornby, ON, Canada); 1:100 rat anti-5-HT antibody (Chemicon International, Temecula, CA); 1:100 sheep anti-TPH antibody (Chemicon). As control, preimmune rabbit serum was used at a dilution of 1:5000 (Cedarlane). Sections were washed three times in PBS followed by a 1-h incubation at 22 °C with secondary antibodies in blocking solution: 1:1000 Alexa Fluor 488 donkey anti-rabbit IgG; Alexa Fluor 594 goat anti-rat (1:1000), or anti-sheep (1:200) IgG (Molecular Probes). Images were acquired on an Axiovert S100 Zeiss microscope using Northern Eclipse imaging software (EMPIX, Toronto, ON, Canada). Blinded counts of positive-labeled cells were performed in triplicate using a constant area and minimum-threshold of 15% brightness (compared with background) in Adobe Photoshop CS3. Intensity of staining was quantified based on percent brightness of each cell and normalized to average of background brightness, for each section analyzed. A constant pixel area was selected in the brightest region of the cell body. Each staining was replicated three times per brain region, with at least three animals used per group.
In Situ Hybridization
Plasmid containing a 417-bp sequence of the mouse 5-HT1A receptor cDNA for sense and antisense 5-HT1A riboprobes was a kind gift from Dr. Alexandre Bonnin (27). The plasmid was linearized and transcribed to incorporate digoxygenin-UTP (Roche Diagnostics) to generate sense or antisense riboprobes using SP6 or T7 polymerases, respectively. In situ hybridization was done as described (28). Tissue sections were incubated overnight with riboprobes (1:1000) in hybridization buffer (1× SSC, deionized formamide, 10% dextran sulfate, 1 mg/ml rRNA, 1× Denhardt's solution). Slides were washed three times for 30 min each, followed by two washes, 30 min each, with 1× MABT (0.5 m maleic acid, 0.75 m NaCl, 0.5% Tween 20). Slides were then blocked for 1 h at 22 °C in blocking reagent (Roche Diagnostics) followed by overnight incubation with 1:1000 anti-DIG antibody (Roche Diagnostics). Slides were washed five times for 20 min with MABT, equilibrated with prestaining buffer (100 mm NaCl, 50 mm MgCl2, 100 mm Tris, pH 9.5, 0.1% Tween 20) three times for 10 min at 22 °C and developed using staining solution ((100 mm NaCl, 50 mm MgCl2, 100 mm Tris, pH 9.5, 0.1% Tween 20, 10% polyvinyl alcohol) containing 3.5 μl/ml 5-bromo-4-chloro-3-indolyl-phosphate (Roche Diagnostics) and 4.5 μl/ml 4-nitro blue tetrazolium chloride (Roche Diagnostics) for 2–24 h at 22 °C and terminated by rinsing in PBS. Sections were examined using a Zeiss Axiophot II light microscope at ×10 magnification. Images were acquired using Northern Eclipse software and a Axiovert S100 Zeiss microscope (EMPIX).
RNA Extraction/Reverse Transcription
Adult mice were euthanized by CO2 asphyxiation followed by decapitation and removal and dissection of brains. For preparation of total RNA, brain samples were triturated and extracted in 600 μl of TRIzol (Invitrogen), centrifuged in chloroform, and the supernatant was precipitated and resuspended in 30 μl of diethylpyrocarbonate-treated water to 0.5–2 μg/μl. Samples were treated with 1 μl of DNase/5 μg of total RNA for 30 min at 37 °C using the Turbo DNA-free kit (Ambion, Foster City, CA), terminated in DNase inactivation buffer, and quantified at OD260. Reverse transcription was carried out using 0.05 μg/μl DNase-treated RNA, with 1:10 random decamer oligonucleotides (Ambion) and 0.4 mm dNTP mix. Samples were incubated for 3 min at 70 °C and annealed on ice. First-strand buffer (Ambion) was then added along with 25 units of M-MLV reverse transcriptase (Ambion) and 10 units of RNase inhibitor (Promega), and reactions were incubated for 60 min at 42 °C and inactivated for 10 min at 95 °C. Parallel reactions lacking M-MLV enzyme served as no reverse transcription controls. Newly synthesized cDNA was stored at −20 °C.
Quantitative Real-time RT-PCR
Quantitative real-time RT-PCR was performed as described (29) using TaqMan Gene Expression Assay kits (Applied Biosystems). Product numbers were as follows: TPH2, Mm00557717_m1; Deaf-1, Mm00516805_m1; 5-HT1AR, Mm00434106_s1; and GAPDH, 4352339E. TPH2, Deaf-1, and 5-HT1A probes are FAM-labeled probes, whereas GAPDH is a VIC-labeled probe. GAPDH amplification was used as internal control and was included in every multiplex reaction. Each reaction contained 5 μl of TaqMan Universal PCR Mastermix (Applied Biosystems), 0.5 μl each of FAM- and VIC-labeled probes, 3 μl of nuclease-free water, and 1 μl of sample. Standard curves (r2 ≥ 0.996) from serial dilution of the most concentrated cDNA sample spanned the range of test sample duplicates and at least one standard was included in duplicate in each experiment for quantification. Control samples without template DNA (no template controls) or reverse-transcriptase (no RT) controls were run for each gene analyzed. The cycling program used was: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 60 s on the Rotor Gene 3000 cycler (Corbett Research, Sydney, Australia). Data were analyzed using Rotor Gene v6.0 software using the ΔΔCt method (30). Briefly, relative amounts of each target gene to GAPDH were obtained as 10(CtT–CtG), where CtT is the threshold cycle for the target gene and CtG is the threshold cycle for GAPDH in the same tube. To verify the deleted exon sequence in Deaf-1 knock-out mice, SYBR Green technology (Molecular Probes) was applied. The following probes were designed to the deleted sequence of the Deaf-1 gene: forward, 5′-CTGTCGGAACATCAGTGGCA; reverse, 5′-CACAGGCAACTCGCTGTCATAC. The amplification cycles were 92 °C for 10 min, 92 °C for 30s, 58 °C for 15s, 72 °C for 20s (40 cycles), terminated at 72 °C for 10 min. Ct values were quantified using SYBR Green incorporation following amplification with QtaqTM DNA polymerase mix (Clontech). Averages were normalized to GAPDH.
High Performance Liquid Chromatography (HPLC) Analysis
Frozen cryostat sections of the raphe region from mixed background Deaf-1−/− or wild-type littermates were pooled (five/animal), placed in 0.3 m monochloroacetic acid containing 10% methanol and internal standards, and stored at −80 °C for HPLC analysis of levels of 5-HT and its metabolite 5-HIAA. Tissue samples were sonicated in 300 μl of homogenizing solution (14.17 g of monochloroacetic acid, 18.6 mg of EDTA, 5.0 ml of methanol, and 500 ml of H2O). Following centrifugation, the supernatants were used for HPLC analysis using an Agilent pump, guard column, radial compression column (5 m, C18 reverse phase, 8 mm × 10 cm), and coulometric electrochemical detector (ESA model 5100A). Supernatant (40 μl/sample) was passed through the system at a flow rate of 1.5 ml/min (1400–1600 p.s.i.). The mobile phase contained 90 mm NaH2PO4, 1.7 mm sodium 1-octase sulfonate, 50 mm EDTA, 50 mm citric acid, 5 mm KCl, and 10% acetonitrile and was filtered (0.22-mm filter paper) and degassed. The area and height of the peaks were determined using an Agilent integrator. The protein content of each sample was determined using bicinchoninic acid with a protein analysis kit (Pierce) and a Fluorostar colorimeter (BMG, Durham, NC). The lower limit of detection for the monoamines and metabolites was ∼1.0 pg.
RESULTS
Deaf-1 Binding and Dual Activity at Mouse 5-HT1A Promoter
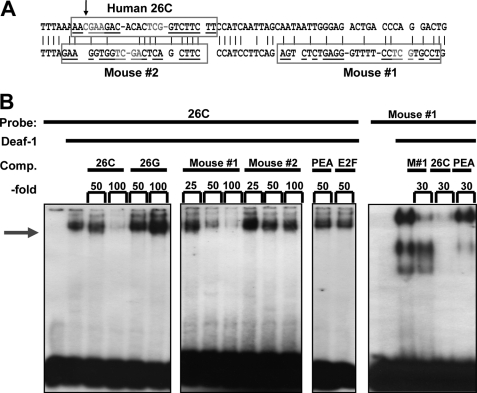
Previous studies have characterized the allele-specific recognition by Deaf-1 at the C-allele of a 26-bp palindrome sequence at −1019 bp of the human 5-HT1A promoter sequence (18, 19), but the corresponding site in the mouse 5-HT1A promoter sequence has not been identified. Based the location and palindrome structure of the human 5-HT1A Deaf-1 site, two imperfect palindrome mouse promoter sequences (mouse#1 and #2) were identified containing the minimal Deaf-1 consensus TCG sequence (31) (Fig. 1A). EMSA was done using recombinant purified Deaf-1 protein incubated with labeled 26-bp human 5-HT1A C-allele Deaf-1 site (26C), in the absence or presence of excess unlabeled double-stranded primers (Fig. 1B). Deaf-1 binding was competed with equal potency by the human 26C and mouse#1 primers, but not mouse#2 primers. As a control, the human 26-bp G-allele palindrome (26G) that does not bind Deaf-1, or unrelated DNA elements for PEA3 or E2F, failed to compete Deaf-1 binding. Conversely, Deaf-1 bound to the labeled mouse#1 DNA element and was competed by unlabeled mouse or human Deaf-1 elements but not the PEA3 element (left panel). Thus, the mouse 5-HT1A promoter contains a Deaf-1-binding element located 947 bp upstream of the translation start site.
FIGURE 1.
Identification of a Deaf-1 element in the mouse 5-HT1A promoter. A, 5-HT1A promoter alignment. Alignment (5′–3′) of human (upper, −1027/−959 bp) and mouse (lower, −971/−946 bp) 5-HT1A promoter regions shows human Deaf-1 site (26C, −1021/−998 bp), mouse palindrome 1 (mouse#1, −971/−947 bp), and mouse palindrome 2 (mouse#2, −1003/−983 bp) in boxes. Consensus minimal Deaf-1 sites (TCG) are highlighted. Palindrome sequences are underlined, palindrome stems are separated by dashes, and palindrome gaps are indicated by a space; an arrow indicates the location of C(−1019)G human 5-HT1A polymorphism. B, EMSA using 5-HT1A Deaf-1 elements. EMSA was done using purified recombinant human Deaf-1 protein (5 or 7.5 μg/lane; left) incubated with labeled human Deaf-1 element (26C; right) or putative mouse Deaf-1 element (mouse#1; left), in the absence or presence of the indicated -fold excess of unlabeled competitor double-stranded primers. Deaf-1 binding (arrow) to 26C was competed by increasing concentrations of mouse#1 but not mouse#2 palindrome element, whereas binding to mouse#1 was competed by unlabeled mouse#1 (M#1) or 26C oligonucleotides. The C- or G-allele (26C or 26G) of human 5-HT1A 26-bp palindrome elements was used as positive or negative control, respectively, and PEA3 or E2F elements were included as nonspecific competitors.
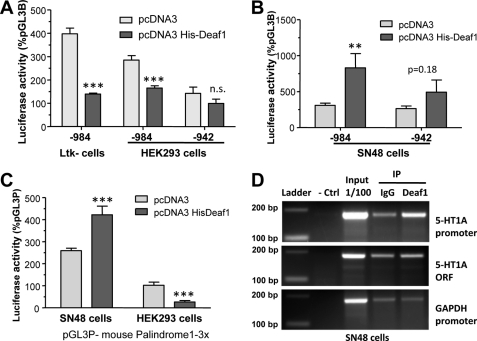
To determine whether Deaf-1 regulates the transcriptional activity of the mouse HTR1A promoter, luciferase transcriptional reporter assays were done using the mouse 5-HT1A promoter construct that includes (−974) or lacks (−942) the mouse#1 site (Fig. 2A). In the presence of Deaf-1, luciferase activity of the mouse 5-HT1A reporter construct was decreased by 60% in mouse Ltk− cells and by 40% in human HEK293 cells, whereas repression was not observed upon deletion of the mouse#1 element. In SN48 cells, a model of mouse septal neurons that expresses 5-HT1A heteroreceptors, Deaf-1 significantly increased the activity of the −984 5-HT1A promoter fragment by 3.5-fold, but had no significant effect in the −942 fragment lacking the Deaf-1 element (Fig. 2B). Taken together, these results highlight the dual activity of Deaf-1 at the mouse 5-HT1A promoter, consistent with findings previously reported for the human promoter (18). We examined Deaf-1 activity using a reporter construct containing three copies of mouse#1 (3×-Deaf-1RE) to address whether this element is sufficient to confer cell specific regulation by Deaf-1 (Fig. 2C). In the presence of Deaf-1 this construct was repressed in HEK293 cells, whereas it was activated in SN48 cells. These results indicate that Deaf-1 acts at its response element to mediate dual regulation of the mouse 5-HT1A promoter, as observed for the human 5-HT1A promoter (18). To specifically demonstrate that Deaf-1 is present at the 5-HT1A promoter in vivo, we performed ChIP assays. In SN48 cells, the mouse 5-HT1A promoter region containing the palindrome was enriched upon immunoprecipitation with Deaf-1 antibody compared with preimmune serum, whereas there was no enrichment of the 5-HT1A coding sequence or the GAPDH promoter (Fig. 2D). These results implicate Deaf-1 in regulating the 5-HT1A promoter in vivo.
FIGURE 2.
Deaf-1 recruitment and dual activity at the mouse 5-HT1A promoter. A and B, dual activity of Deaf-1 at the mouse 5-HT1A promoter. The mouse HTR1A promoter extending from −984 bp (containing mouse#1) or −942 bp (lacking mouse#1) to −1 bp was cloned into pGL3B and co-transfected with vector control (pcDNA3) or pcDNA3 His-Deaf-1 in mouse Ltk− or human HEK293 cells (A) or in mouse SN48 cells (B). Luciferase activity was normalized to β-galactosidase activity, and data are presented as percentage of pGL3B vector activity, as mean ± S.E. (error bars) of three independent experiments. ***, p < 0.001; **, p < 0.01, unpaired t test. C, dual activity of Deaf-1 at mouse#1 5-HT1A element. A pGL3P construct containing three copies of the mouse#1 Deaf-1 response element (3×-Deaf-1RE) was co-transfected with pcDNA3 or pcDNA3 His-Deaf-1 in SN48 or HEK293 cells. Luciferase activity was normalized to β-galactosidase activity, and data are presented as percentage of pGL3P activity, as mean ± S.E. of three independent experiments. ***, p < 0.001, unpaired t test. D, Deaf-1 recruited to the 5-HT1A promoter in vivo. A ChIP assay was performed on chromatin extracted from SN48 cells, using either a Deaf-1 antibody or IgG (preimmune serum) for immunoprecipitation (IP) and amplifying the mouse 5-HT1A promoter, 5-HT1A coding sequence (ORF), or GAPDH promoter regions. DNA size markers and amplification of 1/100 fraction of SN48 extract (input) are shown at right. The 5-HT1A promoter, but not 5-HT1A ORF or GAPDH promoter, was specifically enriched upon immunoprecipitation using the Deaf-1 antibody. Data are representative of three independent assays.
Characterization of Deaf-1-deficient Mice
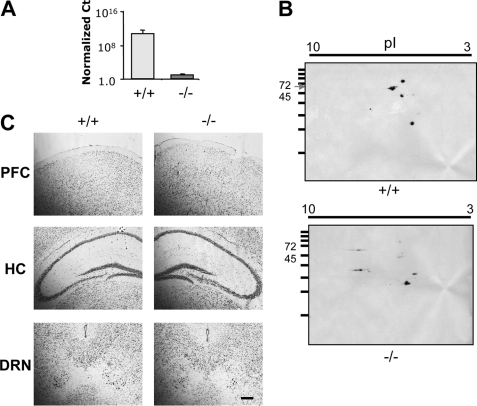
The importance of Deaf-1 in regulation of the 5-HT1A receptor in vivo was addressed using Deaf-1 knock-out mice (25), which were back-crossed from a mixed background onto the C57BL/6 background (see “Materials and Methods”). After genotyping, the presence of the knock-out mutation in genomic DNA and mRNA was verified by DNA sequence analysis. In Deaf-1−/− mice, the loss Deaf-1 coding RNA was shown by RT-PCR analysis (Fig. 3A), and the loss of Deaf-1 protein was resolved by two-dimensional gel electrophoresis/Western blot analysis (Fig. 3B). Morphological analysis of the brains of Deaf-1−/− mice using cresyl violet staining showed no gross alteration (Fig. 3C).
FIGURE 3.
Loss of Deaf-1 mRNA and protein expression but normal brain morphology in Deaf-1−/− mice. A, Deaf-1 RNA. Real-time PCR amplification of Deaf-1 mRNA from the frontal cortex of Deaf-1 wild-type (+/+) and knock-out (−/−) mice using primers located within the deleted exon 4 cDNA sequence is indicated. Ct values were normalized to GAPDH. n = 3 mice/group. B, Deaf-1 protein. Two-dimensional SDS-PAGE shows Deaf-1 protein expression in frontal cortex tissue of wild-type (+/+) and knock-out (−/−) mice. Molecular mass standards (kDa) are shown on the ordinate, and pI range on the abscissa. Deaf-1-deficient mice lacked the 68-kDa Deaf-1 protein species. C, brain morphology of Deaf-1-deficient mice. Representative sections of prefrontal cortex (PFC), hippocampus (HC), and DRN from wild-type (+/+) and knock-out (−/−) mice were stained with cresyl violet. Representative images are shown at magnification of ×10. Scale bar, 100 μm. n = 3 mice/group.
Altered 5-HT1A mRNA Levels in Deaf-1-deficient Mice
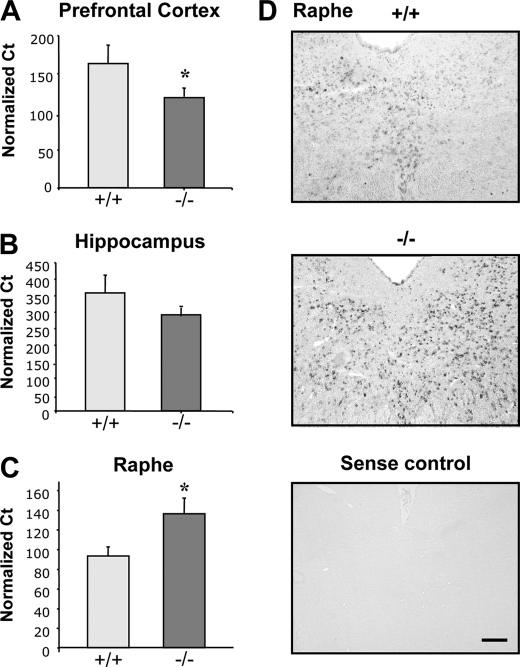
To address whether loss of Deaf-1 affects 5-HT1A receptor expression, brain tissues from Deaf-1-null (−/−) and wild-type (+/+) littermates were examined for levels of 5-HT1A receptor RNA using quantitative RT-PCR (29). Interestingly, we observed a significant 38% decrease in 5-HT1A mRNA levels in the frontal cortex when compared with wild-type littermate controls (Fig. 4A) and a trend toward decreased 5-HT1A mRNA levels (23%) in the hippocampus (Fig. 4B). Oppositely, there was a significant 46% increase in 5-HT1A mRNA in midbrain tissue from the rostral raphe region (Fig. 4C). Although the midbrain tissue dissection used was enriched for serotonin neurons (29), in situ hybridization was performed to determine the precise localization of 5-HT1A RNA in the dorsal raphe nuclei (DRN) (Fig. 4D). Compared with wild-type littermates, 5-HT1A RNA was markedly up-regulated in the DRN of Deaf-1-null mice, consistent with the up-regulation of 5-HT1A autoreceptor expression in serotonin neurons of the DRN. By contrast, no change in 5-HT1A mRNA was observed in the median raphe nuclei of these mice (data not shown), implying that up-regulation of 5-HT1A receptor mRNA in Deaf-1−/− mice is specific to the DRN. The DRN is composed primarily of rhombomere-1-derived 5-HT neurons that are distinct from MRN 5-HT neurons and are implicated in anxiety behavior (32, 33).
FIGURE 4.
Brain region-specific alterations in 5-HT1A mRNA expression in Deaf-1 knock-out mice. RNA isolated from prefrontal cortex (A), hippocampus (B), and rostral raphe region (C) was analyzed by Multiplex RT-PCR for 5-HT1A mRNA and GAPDH RNA and quantified as 5-HT1A RNA normalized to GAPDH RNA using the 10(CtT–CtG) formula. Data are presented as mean ± S.E. (error bars), n = 4 mice/group; *, p < 0.05 by two-tailed unpaired t test. In Deaf-1 knock-out (−/−) compared with wild-type mice (+/+), 5-HT1A RNA was reduced by 38% in prefrontal cortex, 23% in hippocampus, and increased by 46% in raphe region. D, increased 5-HT1A mRNA in DRN of Deaf-1−/− mice. In situ hybridization showing representative sections of the DRN hybridized with 5-HT1A antisense RNA probe in wild-type (+/+) versus Deaf-1 knock-out (−/−) mice. Increased 5-HT1A-positive cell counts were observed in Deaf-1−/− mice. Sense probe was used as negative control. Representative images are shown at magnification of ×20. Scale bar, 100 μm.
5-HT1A Immunostaining in Deaf-1 Mutant Mice
To determine whether altered 5-HT1A mRNA corresponded with changes in 5-HT1A protein levels, immunofluorescent staining for 5-HT1A receptors was performed in the DRN, prefrontal cortex, and dentate gyrus region of the hippocampus (Fig. 5A). No staining was observed in sections not incubated with primary antibody (control) or in sections of cerebellum, which lacks 5-HT1A receptors, whereas cells in the adjacent caudal raphe nuclei were stained in the same section. To demonstrate antibody specificity further, HEK293 cells that were transfected with the mouse 5-HT1A receptor cDNA displayed strong staining compared with cells transfected with empty vector (supplemental Fig. 1).
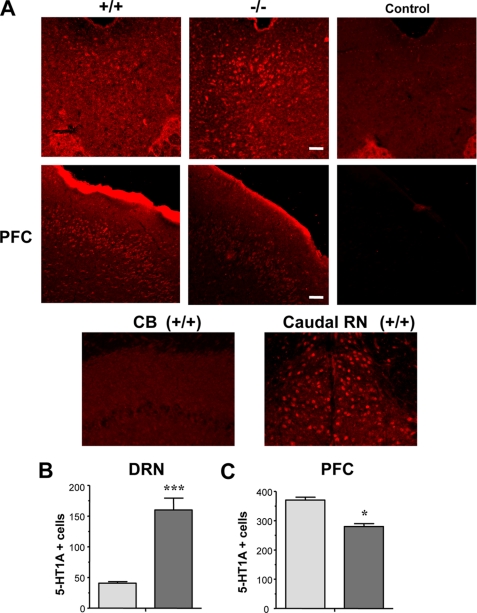
FIGURE 5.
Regional changes in 5-HT1A protein expression in Deaf-1 knock-out mice. A, 5-HT1A receptor immunoreactivity in wild-type (+/+) and Deaf-1 knock-out (−/−) mice in DRN and prefrontal cortex (PFC), cerebellum (CB, negative control), and adjacent caudal raphe nuclei (Caudal RN). Secondary antibody alone was used as a negative control (Control). Representative images are shown at magnification of ×10 (DRN and PFC) and ×20 (CB and Caudal RN). Scale bars, 100 μm. B and C, quantitative analysis of 5-HT1A receptor-positive cells in DRN (B) and prefrontal cortex (C) of wild type (+/+) and Deaf-1 knock-out (−/−) mice. Counts are presented as number of 5-HT1A + cells ± S.E. (error bars), n ≥ 3 mice. *, p < 0.05 by two-tailed unpaired t test; ***, p < 0.01 by two-tailed t test.
In Deaf-1-null mice, a 3-fold increase in 5-HT1A receptor-labeled cells was observed in the DRN (Fig. 5B), which correlated with the observed up-regulation of 5-HT1A mRNA (Fig. 3). The increase in 5-HT1A-labeled cells in Deaf-1−/− DRN was greater than the change in 5-HT1A mRNA levels (38%) and may reflect the limit of detection using the 5-HT1A antibody for cell counting (see below). The number of cells labeled with anti-glial fibrillary acidic protein antibody in the DRN region was comparable between knock-out and wild-type mice (data not shown), implying no alteration in activated astroglia. Oppositely, in the prefrontal cortex, a significant 25% decrease in 5-HT1A-labeled cells was observed (Fig. 5C), consistent with reduced levels of 5-HT1A mRNA in this region. No significant change in the number of 5-HT1A-labeled cells was observed in the hippocampal dentate gyrus (data not shown). Thus, Deaf-1 deficiency leads to brain region-specific changes in 5-HT1A receptor protein levels that mirrored changes in 5-HT1A mRNA.
Deaf-1 Mutant Mice Display Altered 5-HT1A Autoreceptor and 5-HT Levels, but Unchanged TPH2 RNA Levels
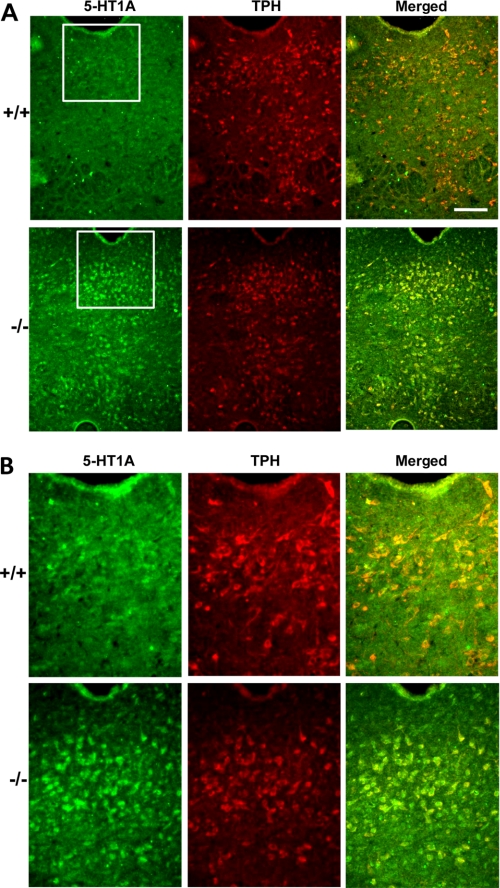
To examine whether raphe 5-HT1A receptors represent autoreceptors, dual immunofluorescence labeling was done with antibodies to 5-HT1A receptor and TPH, the rate-limiting enzyme for serotonin synthesis, to identify 5-HT neurons (Fig. 6, A and B). In wild-type and knock-out DRN, >80% of cells were co-labeled with 5-HT1A and TPH, indicating that the 5-HT1A receptors in this region are largely 5-HT1A autoreceptors expressed on serotonergic neurons. Similarly, 5-HT1A receptors and 5-HT were also largely co-localized in DRN (supplemental Fig. 2). The number of TPH-positive cells in DRN of both genotypes did not differ, suggesting no change in the number of serotonin neurons. Interestingly, in knock-out animals only 18% of TPH+ cells were 5-HT1A−. This was in sharp contrast with wild-type animals, where 68% of TPH+ cells were 5-HT1A−. This difference can be explained by the much lower number of 5-HT1A+ cells detected in the DRN of wild-type animals due to the sensitivity threshold of the 5-HT1A antibody.
FIGURE 6.
Co-localization of 5-HT1A and TPH in the DRN of Deaf-1 knock-out animals. A, dual immunofluorescence analysis of the DRN of Deaf-1+/+ and Deaf-1−/− mice at magnifications of ×10 (A) and ×20 (insets). Representative images of sections co-stained with rabbit anti-5-HT1A antibody (green) and sheep anti-TPH antibody (red) and the merged image are shown. The proportion of labeled cells in wild-type animals was (mean ± S.E., n = 3): 5-HT1A+/TPH+, 82 ± 11%; 5-HT1A+/TPH−, 18 ± 11%; TPH+/5-HT1A−, 68 ± 6%. The proportion of labeled cells in knock-out animals was (mean ± S.E., n = 3): 5-HT1A+/TPH+, 84 ± 3%; 5-HT1A+/TPH−, 17 ± 2%; TPH+/5-HT1A−, 18 ± 4%. Scale bar, 100 μm.
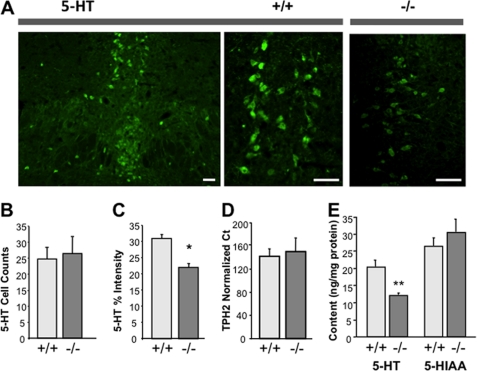
To determine whether the increase in autoreceptor levels in Deaf-1-deficient animals altered serotonergic function, immunofluorescence was done using anti-5-HT antibody to detect serotonin in the DRN (Fig. 7A). As observed for TPH+ cells, there was no change in the number of 5-HT-positive cells (Fig. 7B) or the number of TPH-positive cells (Fig. 6, A and B) in Deaf-1-null versus wild-type raphe, but the intensity of serotonin staining was reduced by 30% in Deaf-1-null tissue (Fig. 7C). A similar reduction (40%) was observed upon measurement of the raphe 5-HT content by HPLC, which validates the quantification of 5-HT by immunofluorescence, whereas there was no change in 5-HIAA content (Fig. 7E). These data indicate that Deaf-1 deficiency is associated with up-regulation of 5-HT1A autoreceptors and reduced raphe 5-HT levels, consistent with increased inhibition of serotonergic tone by 5-HT1A autoreceptors. The lack of change in 5-HIAA levels suggests that release and metabolism of 5-HT were compensated in these mice under basal conditions, despite lower 5-HT levels.
FIGURE 7.
Reduced serotonin in 5-HT1A-positive cells of Deaf-1 knock-out mice. A, representative images of immunofluorescent labeling of 5-HT-positive cells in DRN of wild-type (+/+) at magnifications of ×10 and ×20 compared with Deaf-1 knock-out (−/−) DRN at magnification of ×20. Scale bars, 50 μm. B, quantification of 5-HT-positive cells in wild-type and Deaf-1 knock-out DRN. Cell counts are presented as mean percentage of total cells ± S.E. (error bars) from combined rostral and caudal DRN. C, reduced 5-HT intensity/cell in Deaf-1−/− DRN. 5-HT intensity is presented as percent brightness above background. n ≥ 3 mice/group. *, p < 0.05 by two-tailed unpaired t test. D, TPH2 mRNA expression in raphe region. Multiplex real-time PCR shows TPH2 RNA levels in the dorsal raphe region dissected from wild-type (+/+) and Deaf-1 knock-out (−/−) mice. Data are normalized to GAPDH using the 10(CtT–CtG) method and are presented as mean ± S.E., n = 3 mice/group. E, 5-HT and 5-HIAA content in raphe region. Sections of dorsal raphe tissue from three to five Deaf-1+/+ or Deaf-1−/− mice were pooled and the total content of 5-HT and metabolite 5-HIAA quantified by HPLC analysis. **, p < 0.01 by two-tailed unpaired t test.
To examine whether Deaf-1 deficiency has a global action on serotonergic neurons, we examined the level of TPH2 mRNA in raphe tissue (Fig. 7D). TPH2 is rate-limiting for brain 5-HT synthesis and is not expected to be regulated by Deaf-1. Levels of TPH2 mRNA were unchanged between Deaf-1 mutant and wild-type mice. Taken together, these results indicate that the absence Deaf-1 did not affect the number of serotonin neurons or the expression of TPH2 despite reduced levels of 5-HT. This suggests that Deaf-1 deficiency did not affect serotonergic differentiation but specifically increased 5-HT1A autoreceptor expression, which was associated with reduced serotonin levels at the cell body of neurons in the DRN.
DISCUSSION
Altered 5-HT1A Receptor Expression in Deaf-1 Knock-out Mice
The identification of Deaf-1 as an allele-specific regulator of the human 5-HT1A receptor gene and the association of the 5-HT1A G(−1019) allele with depression and suicide suggest that Deaf-1 may have an important role in the regulation of 5-HT1A receptor expression in vivo. Our previous results demonstrated that Deaf-1 has differential activity depending on the cell model, with repressor activity in serotonergic raphe RN46A cells but enhancer activity in several nonserotonergic neuronal cell lines (18, 19), implying that Deaf-1 regulates 5-HT1A autoreceptors differently than 5-HT1A heteroreceptors expressed on nonserotonergic neurons. By analogy with the human 5-HT1A promoter, we identified a palindromic site located at −947 bp in the mouse 5-HT1A promoter that contains the minimal TCG Deaf-1 consensus sequence (31) and is specifically recognized by Deaf-1 protein. Although the level of nucleotide identity between human and mouse genes in this region is limited (Fig. 1A), the presence of a Deaf-1 element in the same region suggests its importance in regulating 5-HT1A gene transcription. Similar to the human 5-HT1A promoter Deaf-1 exhibited dual activity at the mouse 5-HT1A promoter that is mediated directly by the Deaf-1 element (Fig. 2). Furthermore, we show for the first time using the ChIP assay that Deaf-1 binds directly to the 5-HT1A promoter in vivo, indicating that Deaf-1 regulates 5-HT1A expression directly via this element in neuronal cells. The evidence from these in vitro and in vivo studies make a strong case for a direct effect of Deaf-1 on 5-HT1A expression in vivo.
To address the effect of Deaf-1 on 5-HT1A receptor gene regulation in vivo, we have examined the levels of 5-HT1A mRNA and protein in Deaf-1 knock-out mice on the C57BL/6 genetic background. The C57BL/6 strain displays higher relative levels of forebrain 5-HT1A receptor RNA (data not shown) and responds to chronic SSRI treatment with increased hippocampal neurogenesis and reduced anxiety-like behavior. These responses are abolished in 5-HT1A knock-out mice (11) but are not observed in the BALB/c strain (34). The C57BL/6 strain also has the higher activity Arg477 variant of TPH2 (35) and the low activity GK-5-HTT variant (36), suggesting a more active serotonin system. Thus, the Deaf-1−/− mouse provides a useful model to study 5-HT1A receptor regulation in vivo.
Deaf-1 binds directly to the mouse 5-HT1A promoter, and mediates dual activity through the Deaf-1 element on the promoter (Figs. 1 and 2). The cell specificity of this activity corresponds to that observed in Deaf-1-null mice, where 5-HT1A mRNA was increased in 5-HT neurons, consistent with the role of Deaf-1 as a repressor of 5-HT1A autoreceptor transcription in serotonergic neurons. Oppositely, in nonserotonergic neurons such as SN48 cells, Deaf-1 activated 5-HT1A transcription, and deficiency of Deaf-1 in vivo reduced 5-HT1A receptor RNA levels in the cortex, a nonserotonergic brain region. These results indicate that altered 5-HT1A regulation in Deaf-1−/− mice is primarily due to loss of Deaf-1-dependent regulation of the 5-HT1A gene, although loss of Deaf-1 may also indirectly regulate 5-HT1A receptor expression in Deaf-1−/− mice. Importantly, the expression of other genes (e.g. TPH2) in 5-HT neurons was not affected in Deaf-1 knock-outs, arguing against a nonspecific global effect, which would be expected to produce a general inhibition or induction of 5-HT1A and other genes. For example, a developmental abnormality that leads to impaired serotonergic differentiation such as Pet-1 knock-out, globally reduces 5-HT1A and other serotonin markers like TPH2 (37, 38), which we did not observe in the Deaf-1−/− mice. Thus, the effects of Deaf-1 deficiency that we observed are both cell- and gene-specific and indicate a direct effect.
Increased presynaptic 5-HT1A autoreceptor expression in Deaf-1 knock-out mice would tend to reduce raphe firing rate and decrease serotonin release. Consistent with this, we observed reduced 5-HT levels in dorsal raphe neurons from Deaf-1−/− mice, suggesting an inhibition of serotonin synthesis despite the lack of change in TPH2 mRNA. Oppositely, in 5-HT1A−/− mice, the lack of 5-HT1A autoreceptors results in increased raphe basal firing and fluoxetine-induced 5-HT release (39–41). Similarly, specific suppression of raphe 5-HT1A autoreceptor expression also increases raphe firing activity and 5-HT release (42–44). Interestingly, the number of serotonin-positive cells was not altered in adult Deaf-1 knock-out mice, suggesting that Deaf-1 deficiency does not significantly alter survival or differentiation of serotonin neurons. However, dysregulation of 5-HT1A autoreceptor levels and serotonin neurotransmission induced by the lack of Deaf-1 during development could impact the behavioral phenotype of adult knock-out mice, as previously observed for 5-HT1A-null mice (13).
By contrast to the up-regulation of 5-HT1A autoreceptors, 5-HT1A mRNA and 5-HT1A-positive cells were reduced in the frontal cortex of Deaf-1 knock-out mice, suggesting that Deaf-1 acts as an enhancer in the cortex, consistent with a dual regulation of pre- and postsynaptic 5-HT1A receptors by Deaf-1 in vivo. Although Deaf-1 enhanced 5-HT1A receptor transcription in nonserotonergic neurons, Hes1, which represses at the 26-bp 5-HT1A palindrome sequence, retained repressor activity in these cells (19, 45). In contrast to Deaf-1-deficient mice, Hes1−/− mice display widespread up-regulation of 5-HT1A receptor RNA and protein in embryonic midbrain and hindbrain regions (45). Thus, the loss of Deaf-1 regulation results in a dual action to decrease serotonergic neurotransmission: induction of 5-HT1A autoreceptors to decrease 5-HT neuronal activity and decreased cortical 5-HT1A receptors to reduce serotonin responsiveness.
Deaf-1 and Human 5-HT1A C(−1019)G Polymorphism
Reduced activity of the serotonin system is implicated in major depressive disorder (3), and antidepressant therapies act directly or indirectly to enhance serotonergic neurotransmission (4). Upon chronic treatment with SSRIs, 5-HT1A autoreceptors become desensitized whereas postsynaptic 5-HT1A receptors remain responsive to enhanced serotonergic neurotransmission, correlating with clinical improvement (4, 46–49). Genetic suppression of 5-HT1A autoreceptor expression by only 30% in mice reduces depression and stress reactivity phenotypes and permits an accelerated response to chronic SSRI treatment, suggesting that relatively small changes in autoreceptor expression can have a major impact on behavioral phenotype (42). The 5-HT1A C(−1019)G polymorphism is associated with depression, with the G allele preventing Deaf-1 regulation of 5-HT1A transcription, which could reduce serotonergic neurotransmission (16). Based on our results, the human 5-HT1A G/G(−1019) genotype would be predicted to result in opposite changes in 5-HT1A receptor RNA and protein levels in pre- and postsynaptic brain regions. Recent studies support an up-regulation of 5-HT1A autoreceptors in antidepressant-free depressed patients with the G/G genotype (21, 22, 50), but not in healthy subjects (51). This suggests that compensatory mechanisms may be recruited in healthy subjects to normalize 5-HT1A autoreceptor expression whereas they are incompletely effective in depressed subjects. However, the effect of the G/G genotype on postsynaptic 5-HT1A receptors is less clear. The G/G genotype in depressed subjects was associated with no change or increased 5-HT1A receptors in some regions such as the hippocampus (21, 22, 50), which could reflect a compensatory up-regulation due to lower serotonergic neurotransmission in these subjects (52) or a decrease in forebrain Deaf-1 levels, leading to reduced transcription of the 5-HT1A heteroreceptors (53). Several studies report reduced 5-HT1A RNA or binding in the prefrontal cortex of subjects with major depression (24, 53–56), consistent with reduced transcription of the 5-HT1A receptor. Similarly, in female cynomolgus monkeys, depressed status was associated with reduced 5-HT1A receptor levels in several forebrain regions (especially cingulate cortex, with lesser change in hippocampus and amygdala) with no change in the raphe region (57). In addition to the presence of the G(−1019) allele, reduction in Deaf-1 expression has also been associated with reduced 5-HT1A receptor expression in the prefrontal cortex of female depressed subjects (53). Thus, both the C(−1019)G polymorphism and Deaf-1 levels may differentially influence 5-HT1A receptor levels in the brains of healthy versus depressed subjects, conferring distinct phenotypes and treatment responses. However, association studies are limited by frequency of the risk allele in the population size, heterogeneity of the genetic background, as well as environmental influences, and are not always replicated.
Hence, better animal models are needed to address the etiology and treatment of depression. Deaf-1-deficient mice provide an animal model of perturbed 5-HT1A receptor regulation that corresponds to the expected effects of G/G(−1019) genotype in humans. In addition to its association with depression, anxiety, and suicide, the C(−1019)G polymorphism has been associated with reduced response to antidepressant treatment (14, 58–64). In mice and rats, 5-HT1A receptors are implicated in the activity of chronic SSRI treatment to induce hippocampal neurogenesis and to reduce anxiety and depression-like behaviors (11, 65). It is possible that the combination of reduced cortical 5-HT1A receptors and increased 5-HT1A autoreceptors in the Deaf-1-null mice may attenuate SSRI action. However, unlike the Deaf-1-null mice, the human G allele would affect not only Deaf-1 repression but also that of Hes1 and Hes5 (45). Nevertheless, Deaf-1-deficient mice provide a useful model for investigating the role of Deaf-1 binding at the C(−1019)G polymorphism and its impact on 5-HT1A receptor expression, behavior, and antidepressant response (16).
Supplementary Material
Acknowledgments
We thank Dr. Kelly McClellan for assistance with in situ hybridization and Matthew Mount for assistance with perfusions.
This work was supported by Canadian Institutes of Health Research Grants MOP-36437 and MOP-115098 (to P. R. A.) and a grant from the National Health and Medical Research Council, Australia (to J. E. V.). The Heart and Stroke Foundation Centre for Stroke Recovery supported some equipment used for these studies.

This article contains supplemental Figs. 1 and 2.
- SSRI
- serotonin-selective reuptake inhibitor
- BES
- 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid
- Deaf-1
- deformed epidermal autoregulatory factor-1
- DRN
- dorsal raphe nuclei
- 5-HIAA
- 5-hydroxyindole acetic acid
- 5-HT
- 5-hydroxytryptamine (serotonin)
- PEA
- polyoma virus enhancer
- TPH
- tryptophan hydroxylase.
REFERENCES
- 1. Charney D. S., Krystal J. H., Delgado P. L., Heninger G. R. (1990) Serotonin-specific drugs for anxiety and depressive disorders. Annu. Rev. Med. 41, 437–446 [DOI] [PubMed] [Google Scholar]
- 2. Mann J. J. (1999) Role of the serotonergic system in the pathogenesis of major depression and suicidal behavior. Neuropsychopharmacology 21, 99S–105S [DOI] [PubMed] [Google Scholar]
- 3. Jans L. A., Riedel W. J., Markus C. R., Blokland A. (2007) Serotonergic vulnerability and depression: assumptions, experimental evidence and implications. Mol. Psychiatry 12, 522–543 [DOI] [PubMed] [Google Scholar]
- 4. Piñeyro G., Blier P. (1999) Autoregulation of serotonin neurons: role in antidepressant drug action. Pharmacol. Rev. 51, 533–591 [PubMed] [Google Scholar]
- 5. Blier P. (2003) The pharmacology of putative early-onset antidepressant strategies. Eur. Neuropsychopharmacol. 13, 57–66 [DOI] [PubMed] [Google Scholar]
- 6. Chalmers D. T., Watson S. J. (1991) Comparative anatomical distribution of 5-HT1A receptor mRNA and 5-HT1A binding in rat brain: a combined in situ hybridization/in vitro receptor autoradiographic study. Brain Res. 561, 51–60 [DOI] [PubMed] [Google Scholar]
- 7. Pompeiano M., Palacios J. M., Mengod G. (1992) Distribution and cellular localization of mRNA coding for 5-HT1A receptor in the rat brain: correlation with receptor binding. J. Neurosci. 12, 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramboz S., Oosting R., Amara D. A., Kung H. F., Blier P., Mendelsohn M., Mann J. J., Brunner D., Hen R. (1998) Serotonin receptor 1A knock-out: an animal model of anxiety-related disorder. Proc. Natl. Acad. Sci. U.S.A. 95, 14476–14481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parks C. L., Robinson P. S., Sibille E., Shenk T., Toth M. (1998) Increased anxiety of mice lacking the serotonin 1A receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 10734–10739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heisler L. K., Chu H. M., Brennan T. J., Danao J. A., Bajwa P., Parsons L. H., Tecott L. H. (1998) Elevated anxiety and antidepressant-like responses in serotonin 5-HT1A receptor mutant mice. Proc. Natl. Acad. Sci. U.S.A. 95, 15049–15054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santarelli L., Saxe M., Gross C., Surget A., Battaglia F., Dulawa S., Weisstaub N., Lee J., Duman R., Arancio O., Belzung C., Hen R. (2003) Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301, 805–809 [DOI] [PubMed] [Google Scholar]
- 12. Kusserow H., Davies B., Hörtnagl H., Voigt I., Stroh T., Bert B., Deng D. R., Fink H., Veh R. W., Theuring F. (2004) Reduced anxiety-related behavior in transgenic mice overexpressing serotonin 1A receptors. Brain Res. Mol. Brain Res. 129, 104–116 [DOI] [PubMed] [Google Scholar]
- 13. Gross C., Zhuang X., Stark K., Ramboz S., Oosting R., Kirby L., Santarelli L., Beck S., Hen R. (2002) Serotonin 1A receptor acts during development to establish normal anxiety-like behavior in the adult. Nature 416, 396–400 [DOI] [PubMed] [Google Scholar]
- 14. Lemonde S., Rogaeva A., Albert P. R. (2004) Cell type-dependent recruitment of trichostatin A-sensitive repression of the human 5-HT1A receptor gene. J. Neurochem 88, 857–868 [DOI] [PubMed] [Google Scholar]
- 15. Ou X. M., Jafar-Nejad H., Storring J. M., Meng J. H., Lemonde S., Albert P. R. (2000) Novel dual repressor elements for neuronal cell-specific transcription of the rat 5-HT1A receptor gene. J. Biol. Chem. 275, 8161–8168 [DOI] [PubMed] [Google Scholar]
- 16. Le François B., Czesak M., Steubl D., Albert P. R. (2008) Transcriptional regulation at a HTR1A polymorphism associated with mental illness. Neuropharmacology 55, 977–985 [DOI] [PubMed] [Google Scholar]
- 17. Kishi T., Tsunoka T., Ikeda M., Kawashima K., Okochi T., Kitajima T., Kinoshita Y., Okumura T., Yamanouchi Y., Inada T., Ozaki N., Iwata N. (2009) Serotonin 1A receptor gene and major depressive disorder: an association study and meta-analysis. J. Hum. Genet. 54, 629–633 [DOI] [PubMed] [Google Scholar]
- 18. Lemonde S., Turecki G., Bakish D., Du L., Hrdina P. D., Bown C. D., Sequeira A., Kushwaha N., Morris S. J., Basak A., Ou X. M., Albert P. R. (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J. Neurosci. 23, 8788–8799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czesak M., Lemonde S., Peterson E. A., Rogaeva A., Albert P. R. (2006) Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J. Neurosci. 26, 1864–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parsey R. V., Oquendo M. A., Ogden R. T., Olvet D. M., Simpson N., Huang Y. Y., Van Heertum R. L., Arango V., Mann J. J. (2006) Altered serotonin 1A binding in major depression: a [carbonyl-C-11]WAY100635 positron emission tomography study. Biol. Psychiatry 59, 106–113 [DOI] [PubMed] [Google Scholar]
- 21. Sullivan G. M., Ogden R. T., Oquendo M. A., Kumar J. S., Simpson N., Huang Y. Y., Mann J. J., Parsey R. V. (2009) Positron emission tomography quantification of serotonin-1A receptor binding in medication-free bipolar depression. Biol. Psychiatry 66, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parsey R. V., Ogden R. T., Miller J. M., Tin A., Hesselgrave N., Goldstein E., Mikhno A., Milak M., Zanderigo F., Sullivan G. M., Oquendo M. A., Mann J. J. (2010) Higher serotonin 1A binding in a second major depression cohort: modeling and reference region considerations. Biol. Psychiatry 68, 170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boldrini M., Underwood M. D., Mann J. J., Arango V. (2008) Serotonin-1A autoreceptor binding in the dorsal raphe nucleus of depressed suicides. J. Psychiatr. Res. 42, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stockmeier C. A., Shapiro L. A., Dilley G. E., Kolli T. N., Friedman L., Rajkowska G. (1998) Increase in serotonin-1A autoreceptors in the midbrain of suicide victims with major depression-postmortem evidence for decreased serotonin activity. J. Neurosci. 18, 7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahm K., Sum E. Y., Fujiwara Y., Lindeman G. J., Visvader J. E., Orkin S. H. (2004) Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol. Cell. Biol. 24, 2074–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paxinos G., Franklin K. (2001) The Mouse Brain, 2nd Ed., Academic Press, New York [Google Scholar]
- 27. Bonnin A., Peng W., Hewlett W., Levitt P. (2006) Expression mapping of 5-HT1 serotonin receptor subtypes during fetal and early postnatal mouse forebrain development. Neuroscience 141, 781–794 [DOI] [PubMed] [Google Scholar]
- 28. Ferguson K. L., McClellan K. A., Vanderluit J. L., McIntosh W. C., Schuurmans C., Polleux F., Slack R. S. (2005) A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 24, 4381–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Czesak M., Burns A. M., Lenicov F. R., Albert P. R. (2007) Characterization of rat rostral raphe primary cultures: multiplex quantification of serotonergic markers. J. Neurosci. Methods 164, 59–67 [DOI] [PubMed] [Google Scholar]
- 30. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 31. Huggenvik J. I., Michelson R. J., Collard M. W., Ziemba A. J., Gurley P., Mowen K. A. (1998) Characterization of a nuclear deformed epidermal autoregulatory factor-1 (DEAF-1)-related (NUDR) transcriptional regulator protein. Mol. Endocrinol. 12, 1619–1639 [DOI] [PubMed] [Google Scholar]
- 32. Jensen P., Farago A. F., Awatramani R. B., Scott M. M., Deneris E. S., Dymecki S. M. (2008) Redefining the serotonergic system by genetic lineage. Nat. Neurosci. 11, 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim J. Y., Duan X., Liu C. Y., Jang M. H., Guo J. U., Pow-anpongkul N., Kang E., Song H., Ming G. L. (2009) DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holick K. A., Lee D. C., Hen R., Dulawa S. C. (2008) Behavioral effects of chronic fluoxetine in BALB/cJ mice do not require adult hippocampal neurogenesis or the serotonin 1A receptor. Neuropsychopharmacology 33, 406–417 [DOI] [PubMed] [Google Scholar]
- 35. Zhang X., Beaulieu J. M., Sotnikova T. D., Gainetdinov R. R., Caron M. G. (2004) Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science 305, 217. [DOI] [PubMed] [Google Scholar]
- 36. Carneiro A. M., Airey D. C., Thompson B., Zhu C. B., Lu L., Chesler E. J., Erikson K. M., Blakely R. D. (2009) Functional coding variation in recombinant inbred mouse lines reveals multiple serotonin transporter-associated phenotypes. Proc. Natl. Acad. Sci. U.S.A. 106, 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jacobsen K. X., Czesak M., Deria M., Le François B., Albert P. R. (2011) Region-specific regulation of 5-HT1A receptor expression by Pet-1-dependent mechanisms in vivo. J. Neurochem. 116, 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C., Maejima T., Wyler S. C., Casadesus G., Herlitze S., Deneris E. S. (2010) Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 13, 1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He M., Sibille E., Benjamin D., Toth M., Shippenberg T. (2001) Differential effects of 5-HT1A receptor deletion upon basal and fluoxetine-evoked 5-HT concentrations as revealed by in vivo microdialysis. Brain Res. 902, 11–17 [DOI] [PubMed] [Google Scholar]
- 40. Richer M., Hen R., Blier P. (2002) Modification of serotonin neuron properties in mice lacking 5-HT1A receptors. Eur. J. Pharmacol. 435, 195–203 [DOI] [PubMed] [Google Scholar]
- 41. Bortolozzi A., Amargós-Bosch M., Toth M., Artigas F., Adell A. (2004) In vivo efflux of serotonin in the dorsal raphe nucleus of 5-HT1A receptor knock-out mice. J. Neurochem. 88, 1373–1379 [DOI] [PubMed] [Google Scholar]
- 42. Richardson-Jones J. W., Craige C. P., Guiard B. P., Stephen A., Metzger K. L., Kung H. F., Gardier A. M., Dranovsky A., David D. J., Beck S. G., Hen R., Leonardo E. D. (2010) 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron 65, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richardson-Jones J. W., Craige C. P., Nguyen T. H., Kung H. F., Gardier A. M., Dranovsky A., David D. J., Guiard B. P., Beck S. G., Hen R., Leonardo E. D. (2011) Serotonin-1A autoreceptors are necessary and sufficient for the normal formation of circuits underlying innate anxiety. J. Neurosci. 31, 6008–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bortolozzi A., Castane A., Semakova J., Santana N., Alvarado G., Cortes R., Ferres-Coy A., Fernandez G., Carmona M. C., Toth M., Perales J. C., Montefeltro A., Artigas F. (August 2, 2011) Mol. Psychiatry 10.1038/mp.2011.92 [DOI] [PubMed] [Google Scholar]
- 45. Jacobsen K. X., Vanderluit J. L., Slack R. S., Albert P. R. (2008) HES1 regulates 5-HT1A receptor gene transcription at a functional polymorphism: essential role in developmental expression. Mol. Cell. Neurosci. 38, 349–358 [DOI] [PubMed] [Google Scholar]
- 46. Rueter L. E., De Montigny C., Blier P. (1998) Electrophysiological characterization of the effect of long term duloxetine administration on the rat serotonergic and noradrenergic systems. J. Pharmacol. Exp. Ther. 285, 404–412 [PubMed] [Google Scholar]
- 47. Le Poul E., Boni C., Hanoun N., Laporte A. M., Laaris N., Chauveau J., Hamon M., Lanfumey L. (2000) Differential adaptation of brain 5-HT1A and 5-HT1B receptors and 5-HT transporter in rats treated chronically with fluoxetine. Neuropharmacology 39, 110–122 [DOI] [PubMed] [Google Scholar]
- 48. Hensler J. G. (2002) Differential regulation of 5-HT1A receptor-G protein interactions in brain following chronic antidepressant administration. Neuropsychopharmacology 26, 565–573 [DOI] [PubMed] [Google Scholar]
- 49. Castro M. E., Diaz A., del Olmo E., Pazos A. (2003) Chronic fluoxetine induces opposite changes in G protein coupling at pre- and postsynaptic 5-HT1A receptors in rat brain. Neuropharmacology 44, 93–101 [DOI] [PubMed] [Google Scholar]
- 50. Parsey R. V., Olvet D. M., Oquendo M. A., Huang Y. Y., Ogden R. T., Mann J. J. (2006) Higher 5-HT1A receptor binding potential during a major depressive episode predicts poor treatment response: preliminary data from a naturalistic study. Neuropsychopharmacology 31, 1745–1749 [DOI] [PubMed] [Google Scholar]
- 51. David S. P., Murthy N. V., Rabiner E. A., Munafó M. R., Johnstone E. C., Jacob R., Walton R. T., Grasby P. M. (2005) A functional genetic variation of the serotonin (5-HT) transporter affects 5-HT1A receptor binding in humans. J. Neurosci. 25, 2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abbas S. Y., Nogueira M. I., Azmitia E. C. (2007) Antagonist-induced increase in 5-HT1A-receptor expression in adult rat hippocampus and cortex. Synapse 61, 531–539 [DOI] [PubMed] [Google Scholar]
- 53. Szewczyk B., Albert P. R., Burns A. M., Czesak M., Overholser J. C., Jurjus G. J., Meltzer H. Y., Konick L. C., Dieter L., Herbst N., May W., Rajkowska G., Stockmeier C. A., Austin M. C. (2009) Gender-specific decrease in NUDR and 5-HT1A receptor proteins in the prefrontal cortex of subjects with major depressive disorder. Int. J. Neuropsychopharmacol. 12, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. López J. F., Chalmers D. T., Little K. Y., Watson S. J. (1998) A.E. Bennett Research Award. Regulation of serotonin 1A, glucocorticoid, and mineralocorticoid receptor in rat and human hippocampus: implications for the neurobiology of depression. Biol. Psychiatry 43, 547–573 [DOI] [PubMed] [Google Scholar]
- 55. Drevets W. C., Thase M. E., Moses-Kolko E. L., Price J., Frank E., Kupfer D. J., Mathis C. (2007) Serotonin-1A receptor imaging in recurrent depression: replication and literature review. Nucl. Med. Biol. 34, 865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sargent P. A., Kjaer K. H., Bench C. J., Rabiner E. A., Messa C., Meyer J., Gunn R. N., Grasby P. M., Cowen P. J. (2000) Brain serotonin 1A receptor binding measured by positron emission tomography with [11C]WAY-100635: effects of depression and antidepressant treatment. Arch. Gen. Psychiatry 57, 174–180 [DOI] [PubMed] [Google Scholar]
- 57. Shively C. A., Friedman D. P., Gage H. D., Bounds M. C., Brown-Proctor C., Blair J. B., Henderson J. A., Smith M. A., Buchheimer N. (2006) Behavioral depression and positron emission tomography-determined serotonin 1A receptor binding potential in cynomolgus monkeys. Arch. Gen. Psychiatry 63, 396–403 [DOI] [PubMed] [Google Scholar]
- 58. Arias B., Catalán R., Gastó C., Gutiérrez B., Fañanás L. (2005) Evidence for a combined genetic effect of the 5-HT(1A) receptor and serotonin transporter genes in the clinical outcome of major depressive patients treated with citalopram. J. Psychopharmacol. 19, 166–172 [DOI] [PubMed] [Google Scholar]
- 59. Baune B. T., Hohoff C., Roehrs T., Deckert J., Arolt V., Domschke K. (2008) Serotonin receptor 1A-1019C/G variant: impact on antidepressant pharmacoresponse in melancholic depression? Neurosci. Lett. 436, 111–115 [DOI] [PubMed] [Google Scholar]
- 60. Hong C. J., Chen T. J., Yu Y. W., Tsai S. J. (2006) Response to fluoxetine and serotonin 1A receptor (C−1019G) polymorphism in Taiwan Chinese major depressive disorder. Pharmacogenomics J. 6, 27–33 [DOI] [PubMed] [Google Scholar]
- 61. Serretti A., Artioli P., Lorenzi C., Pirovano A., Tubazio V., Zanardi R. (2004) The C(−1019)G polymorphism of the 5-HT1A gene promoter and antidepressant response in mood disorders: preliminary findings. Int. J. Neuropsychopharmacol. 7, 453–460 [DOI] [PubMed] [Google Scholar]
- 62. Wang L., Fang C., Zhang A., Du J., Yu L., Ma J., Feng G., Xing Q., He L. (2008) The –1019 C/G polymorphism of the 5-HT(1)A receptor gene is associated with negative symptom response to risperidone treatment in schizophrenia patients. J. Psychopharmacol. 22, 904–909 [DOI] [PubMed] [Google Scholar]
- 63. Yu Y. W., Tsai S. J., Liou Y. J., Hong C. J., Chen T. J. (2006) Association study of two serotonin 1A receptor gene polymorphisms and fluoxetine treatment response in Chinese major depressive disorders. Eur. Neuropsychopharmacol. 16, 498–503 [DOI] [PubMed] [Google Scholar]
- 64. Yevtushenko O. O., Oros M. M., Reynolds G. P. (2010) Early response to selective serotonin reuptake inhibitors in panic disorder is associated with a functional 5-HT1A receptor gene polymorphism. J. Affect. Disord. 123, 308–311 [DOI] [PubMed] [Google Scholar]
- 65. Greene J., Banasr M., Lee B., Warner-Schmidt J., Duman R. S. (2009) Vascular endothelial growth factor signaling is required for the behavioral actions of antidepressant treatment: pharmacological and cellular characterization. Neuropsychopharmacology 34, 2459–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.