Background: Molecular mechanisms underlying target of rapamycin complex 2 (TORC2) signaling are poorly understood.
Results: The TORC2 component Avo1 directly interacts with a downstream substrate Ypk2.
Conclusion: Avo1-Ypk2 interaction is essential for TORC2-Ypk2 coupling and activation of the downstream effector pathway(s) regulating cell integrity and actin organization.
Significance: Physical associations between TORC2 components and specific downstream effectors provide the molecular basis for selective downstream signaling.
Keywords: Actin, Protein Kinases, Protein-Protein Interactions, Signal Transduction, Signaling, TOR Complex (TORC), Yeast, Yeast Genetics, Cell Wall Integrity
Abstract
The conserved Ser/Thr kinase target of rapamycin (TOR) serves as a central regulator in controlling cell growth-related functions. There exist two distinct TOR complexes, TORC1 and TORC2, each coupling to specific downstream effectors and signaling pathways. In Saccharomyces cerevisiae, TORC2 is involved in regulating actin organization and maintaining cell wall integrity. Ypk2 (yeast protein kinase 2), a member of the cAMP-dependent, cGMP-dependent, and PKC (AGC) kinase family, is a TORC2 substrate known to participate in actin and cell wall regulation. Employing avo3ts mutants with defects in TORC2 functions that are suppressible by active Ypk2, we investigated the molecular interactions involved in mediating TORC2 signaling to Ypk2. GST pulldown assays in yeast lysates demonstrated physical interactions between Ypk2 and components of TORC2. In vitro binding assays revealed that Avo1 directly binds to Ypk2. In avo3ts mutants, the TORC2-Ypk2 interaction was reduced and could be restored by AVO1 overexpression, highlighting the important role of Avo1 in coupling TORC2 to Ypk2. The interaction was mapped to an internal region (amino acids 600–840) of Avo1 and a C-terminal region of Ypk2. Ypk2334–677, a truncated form of Ypk2 containing the Avo1-interacting region, was able to interfere with Avo1-Ypk2 interaction in vitro. Overexpressing Ypk2334–677 in yeast cells resulted in a perturbation of TORC2 functions, causing defective cell wall integrity, aberrant actin organization, and diminished TORC2-dependent Ypk2 phosphorylation evidenced by the loss of an electrophoretic mobility shift. Together, our data support the conclusion that the direct Avo1-Ypk2 interaction is crucial for TORC2 signaling to the downstream Ypk2 pathway.
Introduction
The target of rapamycin (TOR)2 protein, a conserved Ser/Thr protein kinase in diverse species, serves as a pivotal regulator of cell growth in response to nutrients, growth factors, the energy status of the cell, and environmental stress (1, 2). Dysfunction of the TOR signaling pathway can result in human diseases, including obesity, diabetes, and cancer (2, 3). TOR exerts its functions mainly from two distinct multi-protein complexes, i.e. TOR complex 1 (TORC1) and TORC2, of which the components and functions are also well conserved (4–10). In Saccharomyces cerevisiae, there exist two TOR proteins (Tor1 and Tor2) (11), and either one can complex with Kog1 and Lst8 to form the rapamycin-sensitive TORC1, serving to modulate diverse growth/proliferation-related cellular processes such as ribosome biogenesis and translation, transcription, cell cycle regulation, and autophagy (12–15). Tor2 but not Tor1 forms TORC2 together with the TORC-common component Lst8, the two essential proteins Avo1 and Avo3, and the two nonessential proteins Avo2 and Bit61 (5, 16). The function of TORC2 in the budding yeast is rapamycin-insensitive and has been linked to the regulation of actin organization, cell wall integrity, and sphingolipid metabolism (5, 17, 18). The highly conserved components of TORCs are reported to participate in regulating TOR functions. For example, Lst8 is important for the kinase activity of TORCs, whereas the TORC2-specific proteins Avo1 and Avo3 maintain the complex integrity (19, 20). However, the roles of specific TORC components in regulating the downstream signaling remain largely unknown.
The distinctive functions of TORCs may be attributed to the fact that each complex has its own specific substrates and downstream signaling effectors. Well characterized mammalian TORC1 substrates include eIF4E-binding protein 1 (4E-BP1) and S6 kinase (S6K); S6K is a member of the AGC (cAMP-dependent, cGMP-dependent, and PKC) protein kinase family (21). Sch9, a S6K homolog in yeast, is also directly phosphorylated and activated by TORC1 (22). To date, most of the known TORC2 substrates are AGC kinases, including the mammalian Akt, SGK, PKC, and their counterparts in other organisms (23–26). Although the substrate specificity of TORCs is presumably determined by the subunit composition of each complex, the underlying molecular interactions await further elucidation.
In S. cerevisiae, one of the identified TORC2 substrates is Ypk2 (yeast protein kinase 2), an AGC kinase highly homologous to mammalian SGK and Akt (27, 28). TORC2 phosphorylates Ypk2 at Ser641 in the turn motif and Thr659 in the hydrophobic motif; both sites are located in a region C-terminal to the kinase domain of Ypk2 (27). It is believed that the TORC2-mediated phosphorylation of the C-terminal tail relieves Ypk2 from the autoinhibition by its own N-terminal regulatory region, allowing phosphorylation of the activation loop by PDK1-like kinases Pkh1 and Pkh2 and the full activation of Ypk2 (29). Other TORC2 substrates found in yeast are Slm1 and Slm2; these two very similar proteins can bind to phosphatidylinositol 4,5-bisphosphate and are known to be involved in the regulation of actin organization (30). However, Ypk2 appears to mediate most of the TORC2 functions, including the regulation of actin rearrangement and ceramide biosynthesis (17, 27). The evidence for Ypk2 being the major TORC2 downstream effector is the observation that constitutively active Ypk2 mutants suppress phenotypes of tor2 mutants (27).
In this study, to further understand the functions of individual TORC2 components in mediating downstream signaling, we investigated the molecular interactions involved in the TORC2-mediated Ypk2 regulation. We found Avo3 essential for the TORC2-Ypk2 interaction in yeast cells, likely because it serves to maintain the integrity of TORC2 (20). We identified Avo1 as a direct binding partner of Ypk2. The interaction regions were mapped to an internal region (aa 600–840) of Avo1 and the N-terminal 99 aa, as well as a C-terminal region (aa 467–677) of Ypk2. In the wild-type background, overexpression of Ypk2334–677, a truncated mutant able to interfere with the interaction between wild-type Ypk2 and Avo1, resulted in phenotypes reminiscent of TORC2 mutants. Taken together, our data suggest that the direct interaction between Avo1 and Ypk2 mediates the coupling of TORC2 to the downstream Ypk2 signaling pathway.
EXPERIMENTAL PROCEDURES
Media and Culture Conditions
Yeast cells were cultured at 27 °C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete medium (0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 2% glucose, and appropriate amino acids drop-out mix) for plasmid maintenance. For galactose induction, overnight yeast cultures in a medium containing 2% raffinose were diluted 1:100 into fresh medium and further grown for 4 h before the addition of galactose to a final concentration of 3%; the cells were harvested after 3 h of induction.
Strains and Plasmids
Yeast strains and plasmids used in this study are listed in supplemental Tables S1 and S2, respectively. A chromosome allele expressing Ypk2-Myc was engineered by PCR-based gene tagging (31). Primers Ypk2-TAG-F2 (5′-ACAGTTGGGTGATTCTCCTTCGCAGGGGAGAAGCATTAGTCGGATCCCCGGGTTAATTAA-3′) and Ypk2-TAG-R1 (5′-AAATTCCGTCCGGCTCGGCTCGGCTTGCTTCGGCTTGCTTGAATTCGAGCTCGTTTAAAC-3′) were used to amplify a cassette containing multiple copies of the Myc epitope and the HIS3 selection marker from pFA6a-13Myc-His3MX5 (31). For expressing a constitutively active form of Ypk2, primers Ypk2-S1 (5′-AAAGAGCTCGGAACGATGGAGCAACCAGTG-3′) and Ypk2-A1 (5′-AAAGAGCTCGGTATAAGCGTCCGTCCTTC-3′) were used to amplify the cDNA fragment encoding for aa 224–677 of Ypk2, and the PCR product was subcloned into pRS424 (32). To express GST-tagged full-length and fragments of Ypk2 in yeast cells, plasmids pHC2–9 were constructed by inserting various YPK2 fragments into pGAL1-GST-URA3. For expressing a kinase-dead version of the Ypk2224–677 fragment, primers Ypk2-K373A-S1 (5′-GATTTACGCGTTGGCGGCTCTGAGAAAAG-3′) and Ypk2-K373A-A1 (5′-CTTTTCTCAGAGCCGCCAACGCGTAAATC-3′) were used in the PCR-based site-directed mutagenesis on pHC7. To express GST-Ypk2 and MBP-Ypk2 fusion proteins in Escherichia coli, pHC2 was digested with BamHI, and the YPK2 fragment was subcloned into pGEX2T (Amersham Biosciences) and pMAL-C2 (New England Biolabs), respectively. For in vitro binding assays, variant forms of AVO1 were amplified from yeast genomic DNA and subcloned into yTA (Yeastern). Similarly, full-length LST8 was subcloned into pcDNA3.1 (Invitrogen), and AVO3 was subcloned into pSG5 (Stratagene). For yeast two-hybrid assays, different YPK2 fragments were obtained from plasmids pHC2–9 by BamHI digestion and subcloned into pGBKT7 (Clontech), whereas a PCR fragment encoding aa 600–840 of Avo1 was subcloned into pACT2 (Clontech).
Spot Assay for Yeast Growth
About 107 (0.5 A600) cells from 27 °C overnight cultures were used to make 10-fold serial dilutions over a 10000-fold range. Aliquots (5-μl) of each dilution were spotted onto appropriate plates and incubated at the indicated temperatures until colonies formed.
Trypan Blue Assay
This assay for cell wall integrity was done as described (18). Briefly, overnight cultures were diluted to an A600 of 0.15 and incubated at 27 °C for 3 h. Each culture was then divided into two aliquots; one was shifted to 37 °C, whereas the other was kept at 27 °C and further incubated for 6 h. The cells were harvested, washed in distilled water, and stained with 0.05% trypan blue for 1 h. At least 200 cells from each sample were examined under the microscope.
Actin Staining
Overnight cultures were diluted to an A600 of 0.15, grown at 27 °C for 3 h, and subsequently shifted to 37 °C for 3 h. Harvested cells were fixed in 4% formaldehyde at room temperature for 1 h. Fixed cells were washed twice and resuspended in PBS; Rhodamine-Phalloidin (Sigma) and Triton X-100 were added to the suspensions to 0.66 μm and 0.02%, respectively, and cells were stained at room temperature for 1.5 h. Harvested cells were washed four times in PBS and resuspended in the mounting solution. Actin organization and cell morphology were examined using fluorescence and differential interference contrast microscopy. At least 200 cells from each sample were examined. At least three independent experiments were done.
GST Pulldown Assay
To examine the interaction of TORC2 components with Ypk2 in yeast cells, GST- and GST-Ypk2-expressing plasmids were transformed into yeast strains carrying different HA- or Myc-epitope-tagged TORC2 components, respectively. Cultures of transformants were subjected to galactose induction for 4 h. Harvested cells were lysed by vortexing together with glass beads in a lysis buffer containing 1× PBS, 10% glycerol, 0.5% Tween 20, 10 mm NaF, 10 mm NaN3, 10 mm sodium pyrophosphate, 10 mm ρ-nitrophenylphosphate, 10 mm β-glycerophosphate, 1 mm PMSF, and a protease inhibitor mixture (Millipore). Crude extracts were centrifuged at 4 °C to remove cell debris. Twenty microliters of glutathione-Sepharose 4B beads (GE Healthcare) were added into each 10-mg lysate sample and incubated at 4 °C for 1 h. The beads were collected, washed five times in the wash buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100), and resuspended in 50 μl of sample buffer. The pulled-down proteins were subjected to SDS-PAGE and Western analysis using anti-HA (Covance) or anti-Myc (LTK) antibodies.
In Vitro Binding Assays
GST and GST-Ypk2 proteins were expressed in E. coli (BL21) and purified using glutathione-Sepharose 4B beads. Different TORC2 components were transcribed and translated in vitro using the TNT T7 quick system (Promega). Equal amounts of individual 35S-labeled TORC2 component translated in vitro were incubated with bead-bound GST or GST-Ypk2 for 2 h at 4 °C. After centrifugation, bead-bound proteins were washed five times in the washing buffer, resuspended in 1× sample buffer, and subjected to SDS-PAGE and autoradiography. The amounts of each 35S-labeled protein in GST and GST-YPK2 pulldown samples were compared. To test the Ypk2334–667 fragment for competition with the full-length Ypk2 in Avo1 binding, MBP and MBP-Ypk2 were expressed in E. coli and pulled down using amylose resin. 35S-Labeled Avo1 was prepared by in vitro transcription/translation as above-mentioned. GST and GST-Ypk2334–667 expressed in E. coli were purified following the GST pulldown and glutathione elution procedures. Equal amounts of 35S-labeled Avo1 were mixed with amylose resin-bound MBP and MBP-Ypk2 for in vitro binding at 4 °C for 2 h in the presence of recombinant GST or GST-Ypk2334–667 as the competitor. Resin-bound proteins were collected by centrifugation, washed five times in the binding buffer, and analyzed by SDS-PAGE and autoradiography. The amounts of 35S-labeled Avo1 pulled down with MBP-Ypk2 in the presence of different doses of competitor were compared.
Ypk2 Band Shift Assay and Alkaline Phosphatase Treatment
Cultures of Ypk2-Myc-expressing yeast strains were grown to an A600 of 0.8 and shifted to 37 °C for 3 h. The lysates were prepared as in the GST pulldown assay and subjected to Western analysis with anti-Myc antibodies. For alkaline phosphatase treatment, samples of lysates containing 3 mg of proteins were immunoprecipitated by incubating with anti-Myc antibodies and protein G-agarose beads (Millipore) at 4 °C for 2 h. After centrifugation, the beads were washed three times in the lysis buffer. Each sample was mixed with FastAPTM thermosensitive alkaline phosphatase (Upstates) and incubated for 1 h at 37 °C. The reaction was stopped by resuspending the beads in the sample buffer. The samples were boiled at 100 °C for 5 min and subjected to SDS-PAGE and Western analysis.
RESULTS
Constitutively Active Ypk2 Rescues Multiple Phenotypes of avo3ts Mutants
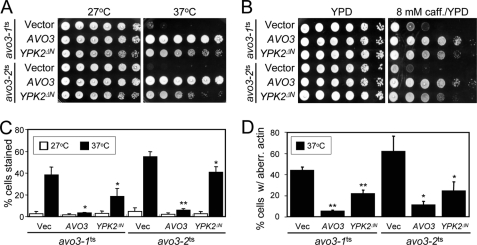
We have previously obtained two temperature-sensitive mutants of the TORC2-specific component Avo3; the growth of these avo3ts mutants is sensitive to high temperatures, and they display defects in cell wall integrity and actin organization if grown at their nonpermissive temperatures such as 37 °C (18). It has been shown that phenotypes of tor2ts mutants can be suppressed by expressing a constitutively active allele of YPK2 (YPK2ΔN), which encodes a truncated form of Ypk2 missing the N-terminal 224 aa (27). We tested the effect of expressing the same Ypk2 variant on the temperature-sensitive growth of our avo3ts mutants. The results demonstrated that overexpressing YPK2ΔN could rescue the growth of both avo3ts mutants at 37 °C (Fig. 1A). The Saccharomyces genome encodes a Ypk2-related Ser/Thr protein kinase, Ypk1 (33). However, overexpressing the YPK1ΔN allele, which can also suppress the tor2ts phenotypes (27), did not have the same effect as YPK2ΔN in our strain background (data not shown).
FIGURE 1.
Constitutively active Ypk2 rescues phenotypes of avo3ts mutants. Two avo3ts mutants carrying the control vector (pRS424), or a plasmid expressing AVO3 (pTSS1) or YPK2ΔN (pHC1) were examined in different assays. A, temperature sensitivity assay. 10-fold serial dilutions of cell suspensions were spotted on SC-Trp plates and incubated at the indicated temperatures until colonies formed. B, caffeine sensitivity assay. Cell suspensions were spotted on YPD plates or YPD plates containing 8 mm caffeine and incubated at 27 °C until colonies formed. C, trypan blue assy. Different cultures were diluted to A600 = 0.15 and shifted to indicated temperatures for 6 h. The cells were stained with trypan blue and observed under the microscope. Shown are results from three independent experiments. **, p < 0.01; *, p < 0.05 (Student's t test). D, actin distribution assay. Log phase cultures were shifted to 37 °C for 3 h, fixed, and stained for F-actin using TRITC-phalloidin. Individual cells were examined by fluorescence and differential interference contrast microscopy. Small-budded cells showing four or more actin patches in the mother cells were scored as cells with aberrant actin distribution. Shown are the data from three independent experiments. **, p < 0.01; *, p < 0.05 (Student's t test). caff., caffeine; Vec, vector; aberr, aberrant.
Because studies have implicated Ypk2 in the maintenance of cell wall integrity and actin organization (27, 29), we investigated the effects of active Ypk2 on the cell wall and actin phenotypes of avo3ts mutants. Many mutants, including the avo3ts mutants, which are defective in the cell wall integrity, display caffeine sensitivity (18, 34). When we tested the cell growth in caffeine-containing media, the results showed that Ypk2ΔN overexpression markedly improved the growth of avo3ts mutants in the presence of 8 mm caffeine (Fig. 1B). In agreement with the caffeine sensitivity experiment, another cell wall assay, i.e. the trypan blue assay, also demonstrated that the cell wall defect of both avo3ts mutants at 37 °C could be suppressed when Ypk2ΔN was overexpressed (Fig. 1C). We also evaluated the effect of Ypk2ΔN on the actin phenotype of avo3ts mutants by staining cellular F-actin with TRITC-phalloidin and observing the cells using fluorescence microscopy. Overexpression of Ypk2ΔN caused a decrease in the percentages of cells with abnormal actin distribution in avo3ts mutants at 37 °C (Fig. 1D). Together, our data demonstrated that the constitutively active form of Ypk2 could rescue multiple phenotypes resulting from defective TORC2 functions in the avo3ts mutants, confirming Ypk2 as the major TORC2 effector.
Ypk2 Physically Associates with TORC2 and Directly Binds to Avo1
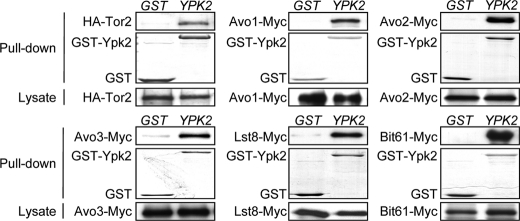
To investigate the mechanism of TORC2 signaling to Ypk2, we examined whether Ypk2 physically associates with TORC2. We expressed GST-Ypk2 in wild-type yeast strains with individual epitope-tagged TORC2 components (supplemental Table S1) and performed GST pulldown assays to assess the TORC2-Ypk2 interaction. The results demonstrated that GST-Ypk2, but not GST, was able to pull down major TORC2 components (Fig. 2), indicating that Ypk2 physically interacts with TORC2 in yeast cells.
FIGURE 2.
Ypk2 physically associates with TORC2. The interaction between Ypk2 and each TORC2 component was determined by the GST pulldown experiment. Lysates were prepared from cells expressing GST or GST-Ypk2 along with one specific epitope-tagged TORC2 component and incubated with glutathione-Sepharose beads. Proteins bound on the beads were separated and detected by Western analysis using anti-HA or anti-Myc antibodies to detect the tagged TORC2 component. GST and GST-Ypk2 in the pulldown samples were visualized by Coomassie Blue staining. The expression levels of the HA- or Myc-tagged TORC2 components in the lysates are shown in the bottom panels.
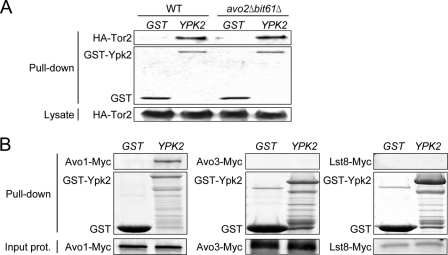
We next investigated which TORC2 component is required for the interaction of Ypk2 with TORC2. Two of the TORC2 components, i.e. Avo2 and Bit61, are not essential for yeast cell growth; therefore we were able to compare the TORC2-Ypk2 interaction in the presence or absence of these two components. The results showed that similar amounts of Tor2 were pulled down with GST-Ypk2 in wild-type and avo2Δbit61Δ yeast cells (Fig. 3A), suggesting that neither Avo2 nor Bit61 was required for the TORC2-Ypk2 interaction. Because other TORC2 components, including Avo1, Avo3, and Lst8, are essential for yeast cell growth and deletion strains could not be generated, we prepared 35S-labeled proteins by in vitro transcription and translation and employed in vitro binding assays to test for direct interaction of these TORC2 components with recombinant GST-Ypk2 from E. coli. We found that, compared with the GST control, GST-Ypk2 specifically pulled down 35S-labeled Avo1, but not Avo3 or Lst8 (Fig. 3B), demonstrating that Ypk2 directly binds to Avo1 in vitro. Taken together, our data suggest that Ypk2 is coupled to TORC2 through a direct physical interaction with Avo1.
FIGURE 3.
Ypk2 directly binds to Avo1. A, Tor2-Ypk2 interaction in the WT or avo2Δbit61Δ background. Lysates from HA-Tor2-expressing yeast cells transformed with a plasmid expressing GST or GST-Ypk2 were subjected to GST pulldown procedures. Pulled down proteins were separated and detected by Western analysis using anti-HA antibodies for HA-Tor2 and by Coomassie Blue staining for GST and GST-Ypk2. Expression levels of HA-Tor2 are shown in the bottom panel. B, in vitro binding assay. 35S-Labeled Avo1, Avo3, and Lst8 were prepared using an in vitro transcription/translation kit and incubated with glutathione-Sepharose bead-bound recombinant GST or GST-Ypk2 produced in E. coli. After washes, the bound 35S-labeled proteins were analyzed by SDS-PAGE and autoradiography; GST fusion proteins were stained with Coomassie Blue. Shown in the bottom panels are input 35S-labeled proteins (one-twentieth of the amount used in pulldown) analyzed by SDS-PAGE and autoradiography. prot., protein.
Avo1 and Avo3 Are Important for TORC2-Ypk2 Interaction in Cells
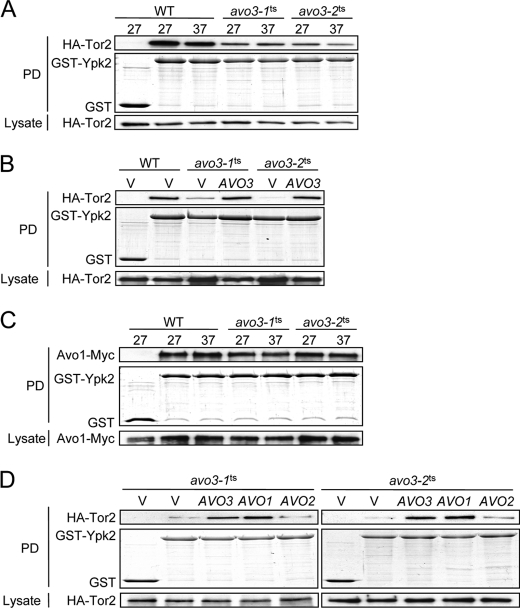
We further investigated whether Avo1 is required for TORC2-Ypk2 coupling in yeast cells. Because AVO1 is an essential gene and conditional avo1 mutants were not available, we took advantage of the avo3ts mutants we previously generated to address this issue. Avo1 and Avo3 bind cooperatively to Tor2 and serve a scaffold-like function (19). In the avo3ts mutants, the composition of TORC2 is greatly altered, and the Tor2-Avo1 interaction is markedly decreased at both permissive and nonpermissive temperatures (20). We figured that if Ypk2 associates with TORC2 through Avo1, then the interaction between Ypk2 and Tor2 would be affected in avo3ts cells. Hence, we examined the interaction by GST pulldown assays in yeast strains expressing GST or GST-Ypk2 and HA-Tor2. It should be noted that the addition of tri-HA tag at the N terminus of Tor2 neither affects the TORC2 complex integrity nor interferes with its kinase activity (19, 27, 35). The GST pulldown results indeed showed a decrease in the Tor2-Ypk2 interaction in both avo3ts strains in comparison with the wild type (Fig. 4A). Expressing AVO3 could restore the Tor2-Ypk2 interaction in avo3ts mutants to wild-type-like levels (Fig. 4B), suggesting that the intact function of Avo3 is required for the optimal TORC2-Ypk2 interaction. Notably, there was no significant change in the Avo1-Ypk2 interaction in avo3ts cells when compared with wild-type cells (Fig. 4C). These data are consistent with a scenario in which Ypk2 directly binds to Avo1 and thereby associates with TORC2, whereas Avo3 plays a scaffolding function and stabilizes the TORC2-Ypk2 interaction by maintaining Avo1 in TORC2.
FIGURE 4.
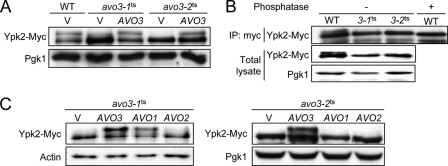
Interaction between Ypk2 and TORC2 is affected in avo3ts mutants. Tor2-Ypk2 or Avo1-Ypk2 interaction was determined by GST pulldown assays. A and C, WT and avo3ts cells expressing HA-Tor2 or Avo1-Myc were transformed with a plasmid expressing GST or GST-Ypk2. Exponentially growing cultures were incubated at indicated temperatures for 3 h and subjected to pulldown procedures. Pulled down proteins were analyzed by Western analysis using anti-HA or anti-Myc antibodies to detect HA-Tor2 or Avo1-Myc, whereas GST and GST-Ypk2 were visualized by Coomassie Blue staining. Expression levels of HA-Tor2 or Avo1-Myc in lysates are shown in the bottom panels. B and D, cells containing the empty vector (V), or an AVO1-, AVO2-, or AVO3-expressing plasmid as indicated were grown at 37 °C, and their lysates were subjected to the GST pulldown assay as in A and C. PD, pulldown.
AVO1 and AVO2 are multicopy suppressors of avo3ts mutants (18); we examined the effect of expressing these suppressors on the Tor2-Ypk2 interaction. HA-Tor2-expressing avo3ts strains were transformed with a plasmid overexpressing Avo1, Avo2, or Avo3, and the interaction of Tor2 with Ypk2 was analyzed at a nonpermissive temperature by GST pulldown assays. The results showed that overexpression of AVO3 or AVO1, but not AVO2, could restore the Tor2-Ypk2 interaction in both avo3ts mutants at 37 °C (Fig. 4D). These observations further support that both Avo1 and Avo3 are important for the TORC2-Ypk2 interaction in yeast cells.
Avo1 and Avo3 Play Important Roles in the TORC2-dependent Post-translational Modification of Ypk2
The contribution of Avo1 or Avo3 or TORC2-Ypk2 signaling was next examined. It has been shown that Ypk2 can be phosphorylated by TORC2 in vitro (27). The phosphorylation status of Ypk2 can be monitored by its electrophoretic mobility (17). In the Western analysis on lysates prepared from Myc-tagged Ypk2-expressing wild-type yeast cells cultured at 37 °C and treated with Zymolyase, we detected a band shift by anti-Myc antibodies; the signal of this up-shifted band of Ypk2-Myc was greatly reduced in lysates of avo3ts cells (Fig. 5A). The band shift disappeared if we treated the wild-type lysates with alkaline phosphatase prior to the Western analysis (Fig. 5B), suggesting that the electrophoretic mobility shift was associated with phosphorylation. These observations together suggest that the up-shifted band is a result of Avo3-dependent phosphorylation of Ypk2.
FIGURE 5.
TORC2-dependent post-translational modification of Ypk2 is affected in avo3ts mutants. A, the control vector (V) or AVO3-expressing plasmid was transformed into WT or avo3ts cells expressing Ypk2-Myc. Log phase cultures were shifted to 37 °C for 3 h and treated with Zymolyase for 1 h before lysis. The lysates were subjected to SDS-PAGE and Western analysis for Ypk2-Myc using anti-Myc antibodies. Pgk1 serves as a loading control. B, lysates from wild-type or avo3ts cells were subjected to immunoprecipitation (IP) with anti-Myc antibodies. Immunoprecipitates were incubated with or without alkaline phosphatase for 1 h and analyzed by Western blotting using anti-Myc antibodies. Ypk2-Myc levels in the lysates detected by Western blotting are also shown. C, two avo3ts mutants carrying the control vector (V) or a plasmid expressing AVO1, AVO2, or AVO3 were subjected to the same assay as in A.
We examined the effect of overexpressing AVO1 or AVO2 on the phosphorylation of Ypk2 in avo3ts mutants. Consistent with its lack of effects on the Tor2-Ypk2 interaction (Fig. 4D), the expression of AVO2 in neither avo3ts strain restored the band shift of Ypk2-Myc (Fig. 5C). Intriguingly, although not as robustly as AVO3 overexpression did, overexpression of AVO1 recovered the Ypk2-Myc band shift in avo3-1ts, but not in avo3-2ts cells (Fig. 5C). The results are in agreement with the finding that AVO1 is an avo3-1ts-specific multicopy suppressor (18). Considering that AVO1 overexpression could rescue the TORC2-Ypk2 interaction in both avo3ts mutants (Fig. 4D), the above observations seem to suggest that, in addition to Avo1, the Avo3 function is also required for the optimal TORC2-dependent post-translational modification of Ypk2.
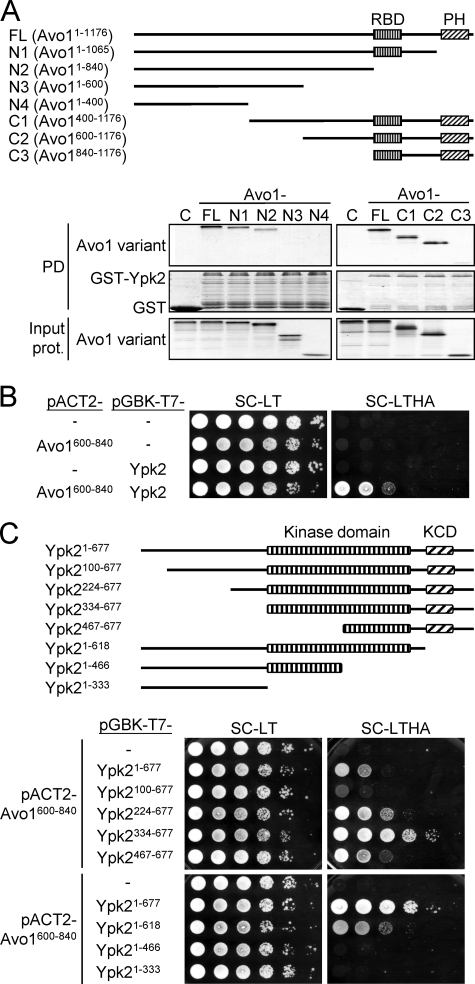
Mapping of the Interaction Regions between Ypk2 and Avo1
To identify the regions in the Avo1 protein that mediate the interaction with Ypk2, lysates prepared from yeast strains expressing different deletion variants of Avo1 (Fig. 6A) were mixed with recombinant GST-Ypk2 from E. coli and subjected to in vitro pulldown assays. Among the C-terminally truncated Avo1 variants tested, Avo11–840 still displayed interaction with Ypk2 in the pulldown assay (Fig. 6A), indicating that the Raf-like Ras-binding and pleckstrin homology domains of Avo1 are dispensable for the interaction with Ypk2. On the other hand, analysis of Avo1 variants with N-terminal truncations showed that Avo1600–1176 could still be pulled down by GST-Ypk2. Together, the results suggest that the region spanning aa 600–840 of Avo1 may serve to interact with Ypk2. We next tested this Avo1 region for Ypk2 interaction using the yeast two-hybrid analysis. A partial AVO1 cDNA fragment encoding this aa 600–840 region and a full-length YPK2 cDNA fragment were subcloned into the prey plasmid pACT2 and the bait plasmid pGBKT7, respectively, and transformed into the AH109 yeast strain to test for reporter gene expression. We found that Avo1600–840 interacted with Ypk2 in the two-hybrid assay (Fig. 6B). We then tested a series of N-terminal and C-terminal truncation mutants of Ypk2 for interaction with Avo1600–840 (Fig. 6C). Among the N-terminally truncated Ypk2 baits, we found that Ypk2100–677, compared with the full-length Ypk21–677, resulted in less growth on the reporter plate, suggesting that a region within the first 99 aa of Ypk2 may contribute to the Ypk2-Avo1 interaction. The C-terminal truncation mutants Ypk21–466 and Ypk21–333 showed no detectable growth on the reporter plate, indicating that the region C-terminal to aa 466 of Ypk2 is essential for interacting with Avo1. The AGC-kinase C-terminal domain, which contains the TORC2 substrate sites Ser641 and Thr659 (27), may also contribute to the interaction with Avo1, as a bait (Ypk21–618) carrying deletion of this domain showed reduced reporter expression compared with the full-length Ypk2.
FIGURE 6.
Mapping of the Avo1-Ypk2 interaction regions. A, GST pulldown assay for the Avo1-Ypk2 interaction. A schematic representation of the tested Avo1 deletion mutants is shown. RBD, Raf-like Ras-binding domain; PH, pleckstrin homology domain. 35S-Labeled Avo1 variants were prepared using an in vitro transcription/translation kit. Equal amounts of each 35S-labeled Avo1 fragment were incubated with glutathione-Sepharose bead-bound GST or GST-Ypk2. Pulled down proteins were analyzed by SDS-PAGE and autoradiography; GST and GST-Ypk2 were visualized by Coomassie Blue staining. One-twentieth amount of the input 35S-labeled proteins used in the pulldown (PD) procedures were analyzed by SDS-PAGE and autoradiography and shown in the bottom panels. B, yeast two-hybrid assay for the interaction between Avo1600–840 and Ypk2. 10-fold serial dilutions of yeast suspensions were spotted on plates and incubated at 27 °C until colonies formed. Yeast cells containing both pACT2- and pGBK-T7-derived plasmids can grow on the Leu- and Trp-dropout SC medium (SC-LT). Growth on a reporter medium lacking His and Ade (SC-LTHA) indicates interaction between the two test proteins. C, yeast two-hybrid assay for the interaction between Avo1600–840 and different regions of Ypk2. A schematic representation of the tested Ypk2 deletion mutants is shown. KCD, AGC-kinase C-terminal domain. The spot assay was performed as in B. The SC-LTHA reporter plate at the bottom was incubated longer than the upper one to better demonstrate cell growth resulting from the interaction between Avo1600–840 and Ypk21–618.
Overexpression of a C-terminal Fragment (aa 334–677) of Ypk2 Perturbs TORC2-Ypk2 Signaling
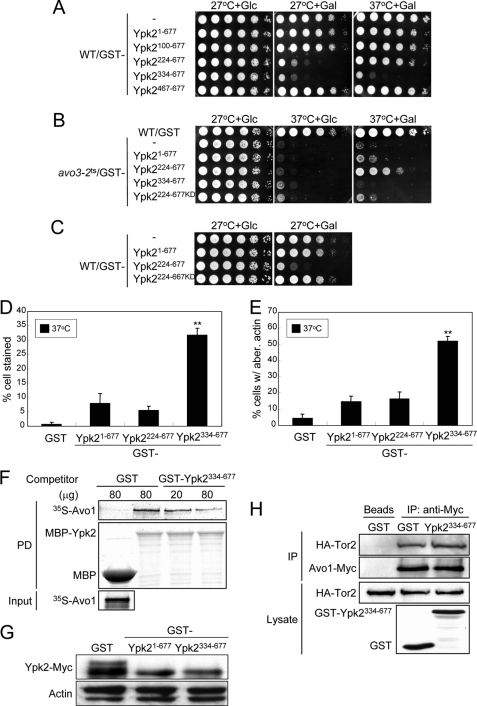
We next sought to investigate whether the direct interaction between Avo1 and Ypk2 is essential for TORC2 signaling to Ypk2. Toward this end, we tested whether overexpression of any of the Avo1-interacting Ypk2 fragments could interfere with the endogenous Avo1-Ypk2 interaction and result in a perturbation of TORC2 to Ypk2 signaling. We transformed plasmids for GAL promoter-driven overexpression of different Avo1-interacting Ypk2 fragments into the wild-type strain and checked for dominant-negative effects under galactose induction. Two fragments, Ypk2224–677 and Ypk2334–677, were found to affect the growth of wild-type cells in the spot assays (Fig. 7A). Ypk2224–677 overexpression in the wild-type cells inhibited growth at 27 °C but not at 37 °C; other TORC2-regulated functions such as cell wall integrity and actin organization were not affected (Fig. 7, D and E). Given these results, we figured it unlikely that Ypk2224–677 caused a perturbation of TORC2 downstream coupling. Because Ypk2224–677 has been shown to be a constitutively active form (27), we suspected that its hyperactivity at 27 °C causes a toxic effect and thus inhibits growth. We prepared a kinase-dead version of the Ypk2224–677 fragment (Ypk2224–677KD) by engineering a K373A point mutation (27). When checked in the avo3-2ts background, Ypk2224–677KD overexpression did not rescue the cell growth at 37 °C as Ypk2224–677 did (Fig. 7B), confirming its loss of function. When overexpressed in the wild-type cells, Ypk2224–677KD exerted no detrimental effects on cell growth at 27 °C (Fig. 7C), suggesting that the toxicity of Ypk2224–677 at 27 °C resulted from the hyperactivity of Ypk2 downstream signaling. Taking these data together, we did not consider Ypk2224–677 a dominant-negative fragment we were looking for. On the other hand, overexpression of Ypk2334–677 in the wild-type background affected cell growth at both 27 and 37 °C (Fig. 7A), caused a cell wall integrity defect and dramatically increased the number of stained cells in the trypan blue assay (Fig. 7D), and perturbed the actin organization (Fig. 7E). In the avo3-2ts background, overexpression of Ypk2334–677 failed to rescue the cell growth at 37 °C (Fig. 7B), suggesting that it may be an inactive form. Among the fragments examined, Ypk2334–677 appeared to interact best with Avo1 (Fig. 6C). It is possible that Ypk2334–677 overexpression in the wild-type cells may have caused a competition with endogenous Ypk2 for Avo1 binding, resulting in decreased TORC2-Ypk2 coupling and the above-mentioned dominant-negative effects. To test this, we first examined whether Ypk2334–677 could influence the Avo1-Ypk2 interaction in an in vitro binding assay. In the assay, 35S-labeled Avo1 prepared by in vitro transcription and translation could be pulled down by recombinant MBP-Ypk2, but not MBP, purified from E. coli (Fig. 7F). When increasing amounts of recombinant GST-Ypk2334–667 were added as the competitor into the binding reactions, decreasing amounts of 35S-labeled Avo1 were pulled down by MBP-Ypk2, demonstrating that Ypk2334–667 could interfere with the interaction between wild-type Ypk2 and Avo1 in vitro. We further checked whether Ypk2334–677 could compete with the endogenous Ypk2 protein for Avo1 interaction and perturb the coupling of TORC2 to Ypk2 in yeast cells. The GST-fusion form of full-length Ypk21–677 or the Ypk2334–677 fragment was overexpressed in a wild-type strain with a chromosomal tag to express Ypk2-Myc endogenously, and the TORC2-dependent electrophoretic mobility shift of Ypk2-Myc was used as a read-out for the TORC2-Ypk2 interaction. Consistent with a competition effect on the endogenous TORC2-Ypk2 interaction, the results showed that overexpression of GST-Ypk2334–677 reduced the band shift of Ypk2-Myc as efficiently as the full-length GST-Ypk21–677 did (Fig. 7G). We excluded the possibility that Ypk2334–677 overexpression may somehow affect the complex integrity of TORC2, because the amount of Tor2 co-immunoprecipitated with Avo1 in cells overexpressing Ypk2334–677 was similar to that observed in cells not overexpressing Ypk2334–677 (Fig. 7H). Taken together, our data are consistent with the conclusion that the Avo1-Ypk2 interaction plays an important role in mediating TORC2 signaling to the downstream Ypk2 pathway.
FIGURE 7.
Expression of Ypk2334–677 affects TORC2-Ypk2 signaling and downstream functions. A, effects of overexpressing Avo1-interacting Ypk2 fragments on the growth of wild-type cells. Plasmids expressing different Ypk2 variants under the control of the GAL1 promoter were transformed into a wild-type strain (MCY142). 10-fold serial dilutions of cell suspensions were spotted onto plates containing glucose (Glc) or galactose (Gal) and incubated at the indicated temperatures until colonies formed. B, effects of overexpressing Ypk2 fragments on the lethality of avo3-2ts cells at the nonpermissive temperature. Plasmids expressing different Ypk2 fragments under the control of the GAL1 promoter were transformed into avo3-2ts cells. Transformants were subjected to spot assays as in A under the indicated conditions. C, the kinase-dependent toxic effect of GST-Ypk2224–667 overexpression on cell growth at 27 °C. Wild-type cells transformed with plasmids expressing the indicated Ypk2 variants were tested in spot assays for growth as in A. GST-Ypk2224–667KD is a kinase-dead version of GST-Ypk2224–667 carrying the K373A point mutation. D and E, trypan blue and actin distribution assays. Wild-type cells overexpressing different Ypk2 fragments were subjected to the trypan blue assay for cell wall integrity (D) and TRITC-phalloidin cell staining for F-actin distribution (E) as in Fig. 1 (C and D, respectively). Shown are data from three independent experiments for each assay. **, p < 0.01 (Student's t test), comparing full-length Ypk21–667 and the Ypk2 fragment. F, in vitro binding competition assay. MBP and MBP-Ypk2 expressed in E. coli were pulled down using amylose resin. 35S-Labeled Avo1 was prepared by in vitro transcription/translation. Recombinant GST and GST-Ypk2334–667 were obtained from E. coli following the GST pulldown (PD) and glutathione elution procedures. In the assay, equal amounts of 35S-labeled Avo1 were mixed with resin-bound MBP and MBP-Ypk2, and different doses of recombinant GST proteins were added into the reactions for competition of binding. After centrifugation and washing, resin-bound proteins were analyzed by SDS-PAGE and autoradiography. The amounts of 35S-labeled Avo1 bound to MBP-Ypk2 in different samples were compared to assess the competition effect of GST-Ypk2334–667. G, Ypk2 band shift assay. Log phase yeast cultures were shifted to a galactose-containing medium and incubated at 37 °C for 3 h for the induction of the GAL promoter-driven overexpression of GST fusion proteins and Ypk2 activation. Lysates prepared after the induction were subjected to SDS-PAGE and Western analysis using anti-Myc antibodies to detect Ypk2-Myc. Actin serves as a loading control. H, co-immunoprecipitation assay for the Tor2-Avo1 interaction. A wild-type strain engineered to express both HA-Tor2 and Avo1-Myc was transformed with a GST or GST-Ypk2334–667-expressing plasmid. Lysates prepared after 6 h of galactose induction were incubated with protein G beads in the absence or presence of anti-Myc antibodies for immunoprecipitation. Proteins in the starting lysates and the immunoprecipitates (IP) were analyzed by SDS-PAGE and Western analysis.
DISCUSSION
TOR responds to diverse environmental cues and regulates a myriad of cellular functions. Although significant progress has been made over the years in understanding the very complex cellular TOR signaling network, we still know little about how different signal inputs are integrated by TOR and how a cell engages specific downstream pathways to differentially respond to various signals. Because TOR is the catalytic core of at least two functionally distinct multi-protein complexes, the subunit composition of each complex must somehow assist in specifying target effectors and regulating downstream signaling. Consistent with this notion, it has been reported that the mammalian TORC1 component Raptor directly binds to the TOR signaling motifs on the mTORC1 substrates S6K and 4EBP1, thus linking them to mTOR and facilitating their phosphorylation (24, 36, 37). Likewise, Mip1, a TORC1-specific protein in Schizosaccharomyces pombe, binds to the RNA-binding protein Mei2p and exerts a TORC1-mediated function (38). The molecular functions of TORC2 components in mediating downstream signaling are only beginning to be discovered. In this study, we provide evidence supporting that TORC2 couples to the downstream signaling pathway through a direct physical interaction between its component Avo1 and its downstream substrate Ypk2. Remarkably, this Avo1-mediated recruitment of TORC2 downstream effectors appears to be a conserved mechanism. Two very recent reports published during the preparation of this manuscript also indicate that mSIN1, the mammalian ortholog of Avo1, directly interacts with mTORC2 substrates SGK1 and PKCϵ (39, 40). The mSIN1-SGK1 interaction is essential for the phosphorylation of SGK1 hydrophobic motif and the SGK1-regulated activation of epithelial sodium channel (39). Direct interaction with mSIN1 is also important for PKC to be phosphorylated by mTORC2 (40). Interestingly, Avo1 appears to use different regions for binding SGK1 and PKC; the SGK1-interacting region is mapped to an N-terminal fragment (aa 1–166), whereas the region important for PKC interaction is a highly conserved central region termed the CRIM domain. Whether SGK1 and PKC can simultaneously bind to mSIN1 remains to be determined. Although the yeast Ypk2 protein is most homologous to SGK (28), the aa 600–840 region that we have mapped in Avo1 for Ypk2 interaction more resembles the PKC-interacting CRIM domain in mSIN1. The mSIN1 mutant with deletion of a 14-aa highly conserved peptide (EDDGEVDTDFPPLD) within the CRIM domain is able to exert a dominant-negative effect on PKC phosphorylation (40). It is of interest to see whether deletion of the corresponding peptide (DEDGEPFEDNFGKLD) in yeast Avo1 can also disrupt the TORC2-mediated Ypk2 phosphorylation.
There are multiple members of the AGC kinase family known to be substrates of mTORC2 (24), and how individual mTORC2 components facilitates selective coupling to these substrates is not completely elucidated. The study by Lu et al. (39) suggests that, at least in kidney epithelial cells, mSIN1 selectively recruits SGK1 but not Akt to mTORC2 for phosphorylation. The mSIN1/Q68H mutant, which can assemble into mTORC2 yet cannot interact with SGK1, exerts a dominant-negative effect on SGK1 phosphorylation and sodium transport via the epithelial sodium channel. However, the Akt binding to mTORC2, Akt phosphorylation, and Akt-dependent glucose uptake are not affected by mSIN1/Q68H. The mTORC2 component responsible for the direct physical interaction with Akt is still not identified. In S. cerevisiae, TORC2 phosphorylates two phospholipid-binding proteins Slm1 and Slm2 in addition to Ypk2 (30). This study has identified Avo1 as the direct Ypk2-binding partner in TORC2; Avo2 and Bit61 are dispensable for the recruitment of Ypk2 to TORC2 (Fig. 3A), and no direct physical interaction of Ypk2 with Avo3 or Lst8 can be detected (Fig. 3B). On the other hand, Slm proteins have not been reported to bind to Avo1, whereas in vitro binding assays have demonstrated the direct interaction between Avo2 and Slm proteins (30). Our previous results have suggested the existence of two separate TORC2 downstream signaling mechanisms, one being Avo1-dependent and the other mediated by Avo2 and Slm proteins (18, 20). Taken together, the existing data are consistent with the hypothesis that the Avo1-mediated recruitment to TORC2 is specific for Ypk2, whereas the coupling of TORC2 to the downstream Slm1/Slm2-mediated pathway might rely on the physical interaction between Avo2 and Slm proteins. Further experimental evidence is warranted to support this notion.
Our co-immunoprecipitation (Fig. 2) and in vitro binding (Fig. 3B) results indicate that although Avo3 in TORC2 does bind Ypk2, the two proteins do not interact directly. However, the reduced Tor2-Ypk2 interaction (Fig. 4A) and the loss of phosphorylation-associated electrophoretic mobility shift of Ypk2 (Fig. 5A) in both avo3ts mutants suggest an essential role of Avo3 for coupling TORC2 to Ypk2 in cells. These observations could be explained by the defective TORC2 integrity in avo3ts cells (20). In this scenario, even though the binding of Ypk2 to Avo1 was not affected in the avo3ts mutants (Fig. 4C), yet the lower amount of Avo1 present in TORC2 (20) would result in less Tor2 being pulled down with Ypk2 and diminished TORC2-mediated Ypk2 phosphorylation. Because expression of AVO3 can restore the TORC2 integrity in avo3ts cells (20), the Tor2-Ypk2 interaction and Ypk2 phosphorylation could be recovered (Figs. 4B and 5A). It is not surprising that overexpressing AVO2 did not recover the physical interaction or signaling between Tor2 and Ypk2 in avo3ts cells (Figs. 4D and 5C), because previous studies have shown that AVO2 overexpression cannot rescue the TORC2 integrity in avo3ts mutants, and Avo2 likely acts as an adaptor for Slm proteins to mediate the Avo1-independent branch of TORC2 downstream signaling (20, 41). Interestingly, although AVO1 overexpression cannot restore the integrity of TORC2 (20), it could enhance the Tor2-Ypk2 interaction in both avo3ts cells (Fig. 4D). It has been suggested that Avo1, like Avo3, can serve a scaffold-like function to maintain the TORC2 structure (19); this function may have helped in bringing Ypk2 and Tor2 together in both avo3ts mutants when AVO1 was overexpressed. However, the TORC2-mediated Ypk2 phosphorylation was only rescued by AVO1 overexpression in avo3-1ts but not avo3-2ts cells (Fig. 5C), which is consistent with AVO1 being an avo3-1ts-specific suppressor (20). The failure of AVO1 to rescue Ypk2 phosphorylation in avo3-2ts cells suggests that the Avo1-Ypk2 interaction is necessary but not sufficient for the TORC2-mediated Ypk2 phosphorylation. The result is in line with the conclusion from previous studies that avo3-2ts cells are defective in a TORC2 function that cannot be restored by AVO1 overexpression (18).
Because AVO1 is an essential gene in yeast and its conditional mutants are not available, we could not test the functional importance of Avo1-Ypk2 interaction in Avo1-deficient cells. We therefore investigated the significance of Avo1-Ypk2 interaction in TORC2 signaling by overexpressing truncation mutants of both proteins in the wild-type background and testing for a dominant-negative effect. Of the Avo1 truncation mutants examined, none was able to cause any perturbation of TORC2 functions when overexpressed in the wild-type cells.3 It is possible that these Avo1 mutants could not compete well with the endogenous Avo1 for Ypk2 interaction because of their relatively weak Ypk2-binding activity (Fig. 6A). On the other hand, among the Ypk2 truncation mutants tested using yeast two-hybrid assays, Ypk2334–677 was found to interact significantly better than the full-length Ypk2 did with the fragment of Avo1 mapped important for Ypk2 interaction (Fig. 6C). When overexpressed in the wild-type cells, Ypk2334–677 did cause phenotypes similar to mutants with TORC2 dysfunction, including defects in growth, cell wall integrity, and actin organization (Fig. 7, A, D, and E). This perturbation of TORC2 function by Ypk2334–677 overexpression is probably not due to an effect on the integrity of TORC2, because the association of Avo1 and Tor2 was similar when either GST or GST-Ypk2334–677 was expressed (Fig. 7H). Given that GST-Ypk2334–677 was able to compete with MBP-Ypk2full-length and inhibit its interaction with Avo1 in the in vitro binding assay (Fig. 7F), it is most likely that the dominant-negative effect of Ypk2334–677 was a result of the interference with the endogenous Avo1-Ypk2 interaction and hence the hindrance of TORC2 coupling to the Ypk2 pathway. The loss of TORC2-mediated phosphorylation of the Myc-tagged endogenous Ypk2 (Fig. 7G) also suggests a defective TORC2-Ypk2 coupling when GST-Ypk2334–677 was overexpressed.
The N-terminal region of Ypk2 has been suggested to be a cis-inhibitory region able to interact with the C-terminal domain containing the turn and hydrophobic motifs and affect Ypk2 phosphorylation (27). The observation that Ypk2224–677 and Ypk2334–677 could interact better than wild-type Ypk2 did with Avo1 (Fig. 6C) may suggest that the N-terminal region also negatively regulates the Avo1-binding of Ypk2. Ypk2334–677 still carries the whole kinase domain as well as the TORC2 and Pkh phosphorylation sites required for the full activation of the wild-type Ypk2; therefore it might not be a kinase-dead form. However, its inability to rescue avo3-2ts cell growth at 37 °C and its dominant-negative effect on TORC2-Ypk2 signaling and downstream functions suggest that Ypk2334–677 is a loss-of-function form of Ypk2. Ypk2334–677 can still interact with Avo1 and thus is presumably able to couple to TORC2 (Fig. 6C). It remains a formal possibility that Ypk2334–677 may not assume an appropriate conformation to serve as a substrate for TORC2 and/or Pkh kinases and consequently cannot be activated in the yeast cells. Another speculated scenario for the loss of function is that maybe Ypk2334–677 is defective in interacting with Ypk2 downstream targets. To date, Ypk2 substrates or downstream effectors involved in regulating actin organization and/or cell wall integrity have remained elusive. One interesting candidate is a yeast type I myosin protein Myo5, which can be phosphorylated by Ypk2 in vitro (42) and has been implicated in regulating the actin assembly (43). Further investigation is required to see whether Ypk2334–677 can phosphorylate Myo5 or to uncover other Ypk2 downstream effectors and examine their interactions with Ypk2334–677.
To sum up, our results suggest specific roles for individual TORC2 components in downstream signaling. Avo1 facilitates specific effector recruitment. Direct binding between Avo1 and Ypk2 is pivotal for TORC2-mediated phosphorylation of Ypk2 and activation of the downstream function(s) in regulating cell integrity and actin organization. Avo3 likely functions as a scaffold to maintain the TORC2 integrity, thereby stabilizing the interaction between Ypk2 and TORC2. Identification of the direct binding partners in TORC2 for other TORC2 substrates/effectors will provide further insights into the mechanisms underlying TORC2 signaling and regulation.
Supplementary Material
This work was supported by Grants NSC94-2311-B-010-010 and NSC95-2311-B-010-013 from the National Science Council, Taiwan, and “Aim for the Top University Plan” grant 100AC-T506 from the Ministry of Education, Taiwan.

This article contains supplemental Tables S1 and S2.
H.-C. Liao and M.-Y. Chen, unpublished data.
- TOR
- target of rapamycin
- TORC
- target of rapamycin complex
- MBP
- maltose-binding protein
- aa
- amino acids.
REFERENCES
- 1. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 2. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR. From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laplante M., Sabatini D. M. (2009) An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 19, R1046–R1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 6. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 7. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 8. Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 9. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 10. Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. (2006) mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16, 1865–1870 [DOI] [PubMed] [Google Scholar]
- 11. Helliwell S. B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., Hall M. N. (1994) TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J. (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inoki K., Ouyang H., Li Y., Guan K. L. (2005) Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 69, 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762 [DOI] [PubMed] [Google Scholar]
- 17. Aronova S., Wedaman K., Aronov P. A., Fontes K., Ramos K., Hammock B. D., Powers T. (2008) Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 7, 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho H. L., Shiau Y. S., Chen M. Y. (2005) Saccharomyces cerevisiae TSC11/AVO3 participates in regulating cell integrity and functionally interacts with components of the Tor2 complex. Curr. Genet. 47, 273–288 [DOI] [PubMed] [Google Scholar]
- 19. Wullschleger S., Loewith R., Oppliger W., Hall M. N. (2005) Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280, 30697–30704 [DOI] [PubMed] [Google Scholar]
- 20. Ho H. L., Lee H. Y., Liao H. C., Chen M. Y. (2008) Involvement of Saccharomyces cerevisiae Avo3p/Tsc11p in maintaining TOR complex 2 integrity and coupling to downstream signaling. Eukaryot. Cell 7, 1328–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma X. M., Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 22. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., Loewith R. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 23. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 24. Jacinto E., Lorberg A. (2008) TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410, 19–37 [DOI] [PubMed] [Google Scholar]
- 25. García-Martínez J. M., Alessi D. R. (2008) mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 26. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25, 7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. (1999) Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9, 186–197 [DOI] [PubMed] [Google Scholar]
- 29. Roelants F. M., Torrance P. D., Bezman N., Thorner J. (2002) Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13, 3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fadri M., Daquinag A., Wang S., Xue T., Kunz J. (2005) The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 16, 1883–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 32. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen P., Lee K. S., Levin D. E. (1993) A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol. Gen. Genet. 236, 443–447 [DOI] [PubMed] [Google Scholar]
- 34. Levin D. E. (2005) Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wedaman K. P., Reinke A., Anderson S., Yates J., 3rd, McCaffery J. M., Powers T. (2003) Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 37. Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003) The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 38. Shinozaki-Yabana S., Watanabe Y., Yamamoto M. (2000) Novel WD-repeat protein Mip1p facilitates function of the meiotic regulator Mei2p in fission yeast. Mol. Cell. Biol. 20, 1234–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu M., Wang J., Ives H. E., Pearce D. (2011) mSIN1 protein mediates SGK1 protein interaction with mTORC2 protein complex and is required for selective activation of the epithelial sodium channel. J. Biol. Chem. 286, 30647–30654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cameron A. J., Linch M. D., Saurin A. T., Escribano C., Parker P. J. (2011) mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 439, 287–297 [DOI] [PubMed] [Google Scholar]
- 41. Audhya A., Loewith R., Parsons A. B., Gao L., Tabuchi M., Zhou H., Boone C., Hall M. N., Emr S. D. (2004) Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23, 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grosshans B. L., Grötsch H., Mukhopadhyay D., Fernández I. M., Pfannstiel J., Idrissi F. Z., Lechner J., Riezman H., Geli M. I. (2006) TEDS site phosphorylation of the yeast myosins I is required for ligand-induced but not for constitutive endocytosis of the G protein-coupled receptor Ste2p. J. Biol. Chem. 281, 11104–11114 [DOI] [PubMed] [Google Scholar]
- 43. Goodson H. V., Anderson B. L., Warrick H. M., Pon L. A., Spudich J. A. (1996) Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133, 1277–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.