Background: The polysialyltransferase, polysialylate, selects a group of proteins.
Results: Substrate recognition and polysialylation are reduced when basic residues in a noncatalytic region of ST8SiaIV/PST are replaced.
Conclusion: Specific residues in a polysialyltransferase polybasic region are critical for substrate recognition.
Significance: Understanding the mechanism of protein-specific polysialylation will allow for the modulation of this process during development and disease.
Keywords: Adhesion, Glycoprotein Biosynthesis, Glycosylation, Sialic Acid, Sialyltransferase, SynCAM 1, Neural Cell Adhesion Molecule, Neuropilin-2, Polysialylation, Polysialyltransferase
Abstract
Polysialic acid on the neural cell adhesion molecule (NCAM) modulates cell-cell adhesion and signaling, is required for proper brain development, and plays roles in neuronal regeneration and the growth and invasiveness of tumor cells. Evidence indicates that NCAM polysialylation is highly protein-specific, requiring an initial polysialyltransferase-NCAM protein-protein interaction. Previous work suggested that a polybasic region located prior to the conserved polysialyltransferase catalytic motifs may be involved in NCAM recognition, but not overall enzyme activity (Foley, D. A., Swartzentruber, K. G., and Colley, K. J. (2009) J. Biol. Chem. 284, 15505–15516). Here, we employ a competition assay to evaluate the role of this region in substrate recognition. We find that truncated, catalytically inactive ST8SiaIV/PST proteins that include the polybasic region, but not those that lack this region, compete with endogenous ST8SiaIV/PST and reduce NCAM polysialylation in SW2 small cell lung carcinoma cells. Replacing two polybasic region residues, Arg82 and Arg93, eliminates the ability of a full-length, catalytically inactive enzyme (PST H331K) to compete with SW2 cell ST8SiaIV/PST and block NCAM polysialylation. Replacing these residues singly or together in ST8SiaIV/PST substantially reduces or eliminates NCAM polysialylation, respectively. In contrast, replacing Arg82, but not Arg93, substantially reduces the ability of ST8SiaIV/PST to polysialylate neuropilin-2 and SynCAM 1, suggesting that Arg82 plays a general role in substrate recognition, whereas Arg93 specifically functions in NCAM recognition. Taken together, our results indicate that the ST8SiaIV/PST polybasic region plays a critical role in substrate recognition and suggest that different combinations of basic residues may mediate the recognition of distinct substrates.
Introduction
The mammalian polysialyltransferases (polySTs),2 ST8Sia II (STX) and ST8Sia IV (PST), catalyze the synthesis of linear homopolymers of α2,8-linked sialic acid, called polysialic acid (polySia), on the termini of N- or O-linked glycans found on a small group of glycoproteins (1–4). These glycoprotein substrates include the neural cell adhesion molecule (NCAM) (5), neuropilin-2 (NRP-2) (6), synaptic cell adhesion molecule 1 (SynCAM 1) (7), the α chain of the voltage-sensitive sodium channel (8), a small proportion of the CD36 scavenger receptor found in human milk (9), and the polySTs themselves, which can polysialylate their own N-glycans (autopolysialylation) (10, 11). The limited number of polyST substrates, the observation that the polySTs polysialylate N-glycans attached to NCAM more efficiently than free glycans (12, 13), the absence of any unique features on the modified core glycan structures (14–16), as well as data from our laboratory (17–19), suggest that polysialylation is highly protein-specific and requires an initial protein-protein interaction between the polySTs and their substrates.
The most abundant polysialylated protein is NCAM. NCAM engages in both heterophilic and homophilic interactions that allow for cell-cell adhesion and signal transduction (reviewed in Refs. 20 and 21). NCAM consists of five immunoglobulin-like domains (Ig1–5), two fibronectin type III repeats (FN1 and FN2), a transmembrane region, and a cytosolic tail (NCAM140 and NCAM180) (supplemental Fig. S1) or a glycosyl-phosphatidylinositol anchor (NCAM120) (22). The polySTs add polySia to two N-glycans in the Ig5 domain of NCAM (23). The addition of long negatively charged polySia chains to NCAM glycans allows for the coordination of water molecules and an increase in the hydrodynamic radius of the protein (24). This leads to an increase in intermembrane repulsion (25) and prevents both NCAM-dependent and NCAM-independent cell-cell adhesion (26, 27). In addition, polySia was recently demonstrated to bind and sequester neurotrophic factors and neurotransmitters and this ability could impact signaling and contribute to its various roles during development and in the adult animal (28, 29).
Polysialylated NCAM is critical for neurite outgrowth, axon guidance and pathfinding, synaptic plasticity, and consequently plays roles in nervous system development, learning, memory formation, and neuronal regeneration (reviewed in Refs. 24 and 30–33). PolySia is first expressed on embryonic day 8–8.5 in the mouse, remains expressed throughout prenatal development (33), then disappears from a majority of neural regions 3 weeks after birth (34), coinciding with expression patterns of PST and STX (35). Simultaneous deletion of STX and PST to generate mice lacking all polySia revealed that polySia is critical for brain development (36). These mice exhibit abnormalities in major brain fiber tracts, hydrocephalus, impaired postnatal growth, and died within 3–4 weeks after birth. Subsequent deletion of NCAM reversed these effects, suggesting that without polySia, there is an aberrant gain of NCAM function causing abnormal neuronal migration and differentiation and altered nervous system development (36). In the adult, polySia expression is restricted to distinct regions of the brain, such as the olfactory bulb, hippocampus, and hypothalamus (37–42), where it allows the proper formation of new neuronal tracts. Transient up-regulation of polySia in damaged peripheral neurons is important for their regeneration and myelination (43). Interestingly, aberrantly high polyST and polySia expression has been noted in the late stages of several human cancers, including neuroblastoma, malignant astrocytoma, and small cell and non-small cell lung carcinoma, where it is suggested to enhance metastatic potential and invasiveness (44–49).
The precise mechanism by which the polySTs add polySia to NCAM is not fully understood. Current evidence suggests that the polysialylation of NCAM is highly protein-specific, requiring an initial recognition step between the enzymes and NCAM. We found that the NCAM FN1 domain is necessary for the polysialylation of N-glycans on the adjacent Ig5 domain (17). Additional studies highlighted the importance of an NCAM FN1 acidic surface patch in polyST recognition and binding (18, 50, 51). These observations led us to ask what complementary sequences of the polySTs mediate NCAM recognition.
The polySTs, PST and STX, are Golgi-localized glycosyltransferases that synthesize polySia chains extending to ∼60 sialic acid units in mouse brain and over 100 units in human tumor cells (52, 53). The polySTs are type II membrane proteins with short N-terminal cytoplasmic tails, single pass transmembrane regions, and large C-terminal regions that include a membrane proximal stem region and a catalytic domain (5). Four conserved sequences have been identified in the catalytic domains of all sialyltransferases, including the polySTs (see Fig. 1). The large sialyl motif (SML) is thought to bind to the donor substrate, CMP-sialic acid (54), whereas the small sialyl motif (SMS) is believed to bind to both CMP-sialic acid and the acceptor glycan (55). The role of the very small sialyl motif (SMVS) is still unclear, however, mutation of a conserved histidine residue in this region (His331 in PST, His346 in STX) abolishes PST catalytic activity (56, 57). Motif III is a sequence of four amino acid residues located between the SMS and SMVS that may also play a role in recognizing the acceptor glycans (58).
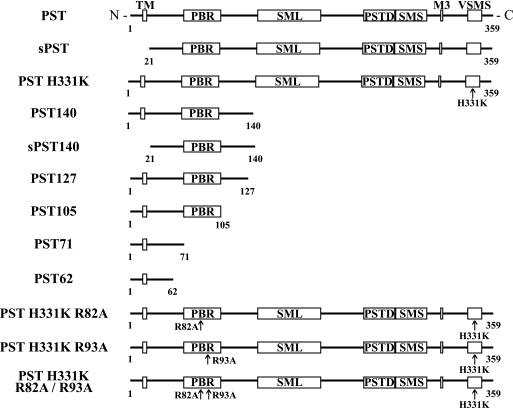
FIGURE 1.
Schematic of ST8Sia IV/PST and PST mutants. Shown are full-length ST8SiaIV/PST and the truncated and mutant proteins used in this study. All truncated PST mutants lack the sialyl motifs (SML, SMS, motif III, SMVS) found in full-length PST, and hence are catalytically inactive. Soluble PST mutants begin at residue 21 and do not contain the cytosolic tail or transmembrane region (TM) (sPST and sPST140). Mutation of His331 in PST to lysine (PST H331K) was previously shown to render a full-length PST protein catalytically inactive (57). The polybasic region (PBR) is found between residues 71 and 105 in PST. Mutation of Arg82 and Arg93 in the PBR was previously shown to greatly reduce NCAM polysialylation (61). The PSTD is a stretch of 32 amino acids (residues 246–277 in PST) that is contiguous with the SMS and is conserved in the two polySTs (60). Previous work demonstrated that selected residues in the PSTD are required for NCAM polysialylation (60, 61).
Angata et al. (59) used chimeric polyST proteins to pinpoint residues critical for NCAM polysialylation. Their work suggested that PST residues 62–127 (prior to SML) and possibly 194–267 (between SML and SMS) are required for NCAM polysialylation and potentially substrate recognition. These results allowed the identification of two conserved polyST regions that are involved in NCAM polysialylation. Troy and colleagues (60) characterized a stretch enriched in basic residues termed the polysialyltransferase domain (PSTD) (residues 246–277 in PST and 261–292 in STX), and found that the overall positive charge of this region was required for NCAM polysialylation. They postulated that the PSTD facilitates processivity of the polysialylation process by interacting with the growing, negatively charged polySia chain (60). However, our analysis of these critical PSTD residues suggested that this region is important for polyST catalytic activity (61). We identified a conserved polybasic region (PBR) we thought might function as the complementary binding region for the FN1 acidic patch (residues 71–105 in PST and 86–120 in STX). Mutation of specific basic residues (Arg82/Arg93 in PST and Arg97/Lys108 in STX) within this region substantially decreased NCAM polysialylation without similarly decreasing enzyme autopolysialylation, suggesting that the PBR is important for recognition of the NCAM substrate and not general catalytic activity (61). Unfortunately, we were unable to consistently detect decreases in the interaction of PST PBR mutants and NCAM in co-immunoprecipitation experiments (data not shown). Consequently, we sought another approach to further evaluate the potential role of the PBR, and specifically Arg82/Arg93, in PST recognition of NCAM.
In this article, we use a competition strategy to evaluate the role of the PBR in PST recognition of NCAM. Our studies indicate that PST sequences between residues 71 and 127 are required for NCAM recognition, and truncated PST proteins containing these sequences bind to NCAM and compete with the endogenous SW2 cell PST to block NCAM polysialylation. We demonstrate that replacing Arg82 and Arg93 with alanine residues in a full-length catalytically inactive PST protein (PST H331K) blocks its ability to compete with endogenous SW2 cell PST to reduce NCAM polysialylation, indicating that these PST residues are essential for NCAM recognition. We also find that the PST R82A/R93A mutant exhibits reduced ability to polysialylate both SynCAM 1 and NRP-2. Interestingly, Arg82 appears to play a greater role in the recognition and polysialylation of these two proteins than does Arg93. This is in contrast to PST recognition of NCAM, where Arg82 and Arg93 seem to play relatively equivalent roles (61). Collectively, our work has identified two residues in a polybasic region prior to the conserved sialyl motifs that are essential for substrate recognition and polysialylation.
EXPERIMENTAL PROCEDURES
Tissue culture media and reagents, including Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), Opti-MEM I, Lipofectin, Lipofectamine, and Lipofectamine 2000 were purchased from Invitrogen. The cDNA for full-length human NCAM140 and the SW2 small cell carcinoma cell line were gifts from Dr. Nancy Kedersha (Brigham and Women's Hospital, Boston, MA). The cDNA for full-length human ST8Sia IV/PST was obtained from Dr. Minoru Fukuda (Sanford Burnham Medical Research Institute, La Jolla, CA). The cDNA for full-length human neuropilin-2 was obtained from Dr. Nicholas Stamatos (University of Maryland School of Medicine, Baltimore, MD). The cDNA for full-length human SynCAM 1 was obtained from Dr. Thomas Biederer (Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT). The QuikChangeTM site-directed mutagenesis kit and Pfu DNA polymerase were purchased from Stratagene (La Jolla, CA). All oligonucleotides were purchased from Invitrogen. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). DNA purification kits were obtained from Qiagen (Valencia, CA). Protease inhibitors were purchased from Roche Applied Science. Poly-l-lysine microscope coverslips were purchased from BD Biosciences. Protein A-Sepharose was purchased from GE Healthcare. Mouse monoclonal anti-Myc epitope tag antibody was purchased from Invitrogen for immunocytochemistry and Cell Signaling Technologies (Danvers, MA) for immunoblotting. Rabbit polyclonal anti-Myc epitope tag antibody for immunoprecipitation was purchased from Abcam (Cambridge, MA). Anti-V5 epitope tag antibody and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) were purchased from Invitrogen. Horseradish peroxidase (HRP)-, fluorescein isothiocyanate (FITC)-, and rhodamine (TRITC)-conjugated goat anti-mouse secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA). Precision PlusTM Protein standards were obtained from Bio-Rad. Nitrocellulose membranes were purchased from Schleicher & Schuell. SuperSignal West Pico chemiluminescence reagent was obtained from Pierce. Blue Ultra Autorad film was obtained from BioExpress (Kaysville, UT). All other chemicals and reagents were purchased from Sigma and Fisher.
Creation of Membrane-associated PST Mutants
The ST8Sia IV/PST cDNA was cloned into the EcoRV and XbaI sites of a previously digested pcDNA3.1/Myc-HisB mammalian expression vector containing a carboxyl-terminal Myc epitope tag and a stop codon prior to the His6 tag. The resulting plasmid containing the DNA sequence for PST-Myc served as the template for the creation of all full-length and truncated PST mutants. All mutations were performed using the QuikChangeTM site-directed mutagenesis kit and Pfu DNA polymerase according to the manufacturer's instructions, utilizing the oligonucleotide primers listed in supplemental Table S1. PST mutants that are truncated from the carboxyl terminus were created in two steps. First, an XbaI restriction site (5′-TCTAGA-3′) was inserted into the PST cDNA immediately downstream of the codon corresponding to the amino acid that will serve as the truncated carboxyl-terminal residue of PST. Second, this plasmid was digested with XbaI at 37 °C for 16 h, to cleave both the newly created XbaI restriction site as well as an XbaI site located between the carboxyl terminus of the full-length PST DNA sequence and the start of the Myc epitope DNA sequence. Following digestion, the expression plasmid was ligated using T4 DNA ligase at 16 °C for 3 h to create an expression vector containing a truncated Myc-tagged PST mutant. All DNA sequences resulting from mutagenesis and digestion/ligation were checked for accuracy by DNA sequencing performed by the DNA Sequencing Facility at the Research Resources Center at the University of Illinois at Chicago.
Construction of Soluble PST-Myc and Soluble PST140-Myc Mutants
Soluble PST (sPST-myc) lacking the cytosolic tail and transmembrane region was generated as described previously by Thompson et al. (50). Soluble PST140 (sPST140-Myc) was then generated from this vector using mutagenesis and XbaI digestion, as described above.
Competition Studies in SW2 Cells and Confocal Immunofluorescence Microscopy to Evaluate Protein Expression and Changes in Polysialylation
SW2 small cell lung carcinoma cells maintained in DMEM, 10% fetal bovine serum (FBS) were plated on poly-l-lysine-coated coverslips and grown in a 37 °C, 5% CO2 incubator until 50–70% confluent. Cells on each plate were then transfected with 1 μg of PST-, sPST-, or a PST-Myc mutant DNA in 300 μl of Opti-MEM I containing 3 μl of Lipofectamine 2000. Cells were incubated with the transfection mixture for 6 h at 37 °C followed by the addition of 1 ml of DMEM, 10% FBS. After 18 h, cells were washed with phosphate-buffered saline (PBS) and permeabilized using −20 °C methanol to view internal structures as well as the cell membrane. After permeabilization, cells were blocked with immunofluorescence blocking buffer (5% normal goat serum in PBS) for 1 h, then incubated with the following primary antibodies diluted in immunofluorescence blocking buffer for 2 h at room temperature: anti-Myc tag antibody (1:250) to detect expression and localization of PST or a mutant PST, and the OL.28 anti-polySia antibody (1:100) to detect polysialylation. After washing with PBS (4 times), cells were then incubated for 1 h with the following secondary antibodies diluted in blocking buffer: FITC-conjugated goat anti-mouse IgG (1:100) to visualize protein localization, and TRITC-conjugated goat anti-mouse IgM (1:100) to visualize polySia. After washing with PBS (4 times), the coverslips were mounted on glass slides using 20 μl of mounting medium (15% Vinol 205 polyvinyl alcohol (w/v), 33% glycerol (v/v), 0.1% azide, pH 8.5). Cells were visualized with a Zeiss Axiovert 200M inverted confocal microscope using a ×63 oil immersion objective.
Co-immunoprecipitation of NCAM with PST and PST Mutants
COS-1 and Lec2 CHO cells were maintained in DMEM and F-12 media, respectively, supplemented with 10% FBS, were plated onto 100-mm tissue culture plates and grown in a 37 °C, 5% CO2 incubator until 50–70% confluent. Cells were then co-transfected with 10 μg of V5-tagged NCAM cDNA and 10 μg of empty pcDNA3.1/Myc-HisB vector (control), Myc-tagged PST, or Myc-tagged PST mutant cDNA in 3 ml of Opti-MEM I containing 30 μl of Lipofectin (COS-1) or Lipofectamine (Lec2 CHO) transfection reagent. Cells were incubated with the transfection mixture for 6 h at 37 °C. Following incubation, 7 ml of DMEM (COS-1) or F-12 (Lec2 CHO) media supplemented with 10% FBS was added and the cells were allowed to grow overnight in a 37 °C, 5% CO2 cell incubator. The following day, cell medium was removed and the cells were washed with 10 ml of PBS. One milliliter of co-immunoprecipitation buffer (50 mm Hepes, 100 mm NaCl, 1% Triton X-100, pH 7.2) was then added and the cells were scraped off the plates using a sterile cell scraper. The cells were incubated on ice for 20 min to allow for lysis, and then pelleted by centrifugation. To immunoprecipitate PST-Myc or a Myc-tagged PST mutant, the resulting supernatant was incubated with 2.5 μl of rabbit polyclonal anti-Myc tag antibody. After 2 h of rotation at 4 °C, 50 μl of protein A-Sepharose beads (50% suspension in PBS) was added to each sample and the samples were rotated for 1 h at 4 °C. The immune complexes bound to Protein A-Sepharose beads were pelleted and washed 4 times with 1 ml of cold co-immunoprecipitation buffer. Samples were resuspended in 100 μl of Laemmli sample buffer containing 5% β-mercaptoethanol and heated to 100 °C for 10 min. Precipitated proteins were separated on a 5% stacking, 10% resolving SDS-polyacrylamide gel and then subjected to immunoblotting as described below. To determine the relative expression of NCAM and PST or PST mutant proteins in the initial samples, a 150-μl sample was removed from each lysate prior to co-immunoprecipitation. Fifty microliters of Laemmli sample buffer containing 5% β-mercaptoethanol was added to each sample. Prior to immunoblotting, the samples for determining NCAM expression levels were separated on a 10% resolving gel, whereas those for determining the PST or PST mutant expression levels were separated on a 15% resolving gel.
Immunoblot Analysis of NCAM Proteins
Following SDS-PAGE, proteins were transferred to nitrocellulose membranes at 450 mA overnight. Membranes were blocked for 1 h at 4 °C in blocking buffer (5% nonfat dry milk in Tris-buffered saline, pH 8.0, 0.1% Tween 20). To detect NCAM co-immunoprecipitated with a Myc-tagged PST or PST mutant and determine relative NCAM expression levels, nitrocellulose membranes were incubated with anti-V5 epitope tag antibody diluted 1:5000 in blocking buffer for 2 h at 4 °C, and for 1 h at 4 °C with HRP-conjugated goat anti-mouse IgG secondary antibody diluted 1:4000 in blocking buffer. To detect relative PST and PST mutant expression levels, membranes were incubated with anti-Myc epitope tag antibody diluted 1:5000 in blocking buffer for 2 h at 4 °C and for 1 h at 4 °C with HRP-conjugated goat anti-mouse IgG secondary antibody diluted 1:4000 in blocking buffer. Membranes were washed twice before and four times after secondary antibody incubation with Tris-buffered saline, pH 8.0, 0.1% Tween 20 for 15 min per wash. Immunoblots were developed using the SuperSignal West Pico chemiluminescence kit and BioExpress Blue Ultra Autorad film.
Triple Transfection of COS-1 Cells with NCAM, PST, and a PST Mutant
COS-1 cells were transfected and prepared as described above. Briefly, COS-1 cells were transfected with 2 μg of V5-tagged NCAM cDNA, 2 μg of PST-Myc cDNA, and 16 μg of a Myc-tagged PST mutant cDNA. For controls, one sample was transfected with 2 μg of V5-tagged NCAM and 18 μg of empty pcDNA3.1/Myc-HisB vector, whereas another sample was transfected with 2 μg each of NCAM-V5 and PST-Myc and 16 μg of empty pcDNA3.1/Myc-HisB vector.
Evaluation of NCAM, Neuropilin-2, and SynCAM 1 Polysialylation by PST and Mutant PST Proteins
Eighteen hours post-transfection, cells were washed with 10 ml of PBS and lysed in 1 ml of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1% SDS). The lysates were pre-cleared with 50 μl of protein A-Sepharose beads, and NCAM-V5, neuropilin-2-V5, and SynCAM 1-V5 were immunoprecipitated with 3 μl of anti-V5 tag antibody for 2 h at 4 °C with rotation, followed by incubation with 50 μl of protein-A-Sepharose beads for 1 h at 4 °C. Beads were then washed 4 times with immunoprecipitation buffer and once with immunoprecipitation buffer containing 1% SDS. Samples were then resuspended in 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol, heated for 10 min at 65 °C, and separated on a 3% stacking, 5% resolving SDS-polyacrylamide gel. The relative expression levels of NCAM, neuropilin-2, SynCAM 1, and PST proteins were evaluated as described above.
The polysialylation of NCAM, SynCAM 1, and NRP-2 by PST and mutant PST proteins was evaluated with OL.28 immunoblotting. Briefly, proteins were transferred to a nitrocellulose membrane at 450 mA overnight. Membranes were blocked for 1 h at 4 °C in blocking buffer (5% nonfat dry milk in Tris-buffered saline, pH 8.0, 0.1% Tween 20). Membranes were then incubated overnight at 4 °C with OL.28 anti-polySia antibody diluted 1:150 in 2% nonfat dry milk in Tris-buffered saline, pH 8.0, followed by a 1-h incubation at 4 °C with HRP-conjugated goat anti-mouse IgM antibody diluted 1:4000 in blocking buffer. Membranes were then washed and developed as described above.
RESULTS
Expression of Truncated PST Proteins Blocks NCAM Polysialylation by Endogenous PST in SW2 Cells
We hypothesize that polysialylation is a highly protein-specific process in which the polySTs must first recognize the substrate protein prior to modifying its glycans. Our data indicate that the polySTs recognize sequences in the NCAM FN1 domain that then allow them to polysialylate two N-glycans in the adjacent Ig5 domain (supplemental Fig. S1). Based on previous studies from our laboratory (61) and Angata et al. (59), we hypothesize that sequences in the membrane proximal region of the polySTs prior to the SML function in the recognition of NCAM. To test this hypothesis we have taken a competition approach using SW2 small cell lung carcinoma cells that contain endogenous PST and polysialylated NCAM. For PST competitors we created a catalytically inactive full-length PST (PST H331K) (57), and a series of catalytically inactive PST mutants truncated prior to the SML (Fig. 1). We expected that the expression of a catalytically inactive PST mutant containing sequences important for NCAM recognition would compete with endogenous, active PST for NCAM recognition and block NCAM polysialylation.
We first examined whether the expression of a truncated PST protein consisting of the first 140 residues of PST (PST140), or a soluble version of PST140 lacking the cytosolic and transmembrane sequences (sPST140) (Fig. 1), altered NCAM polysialylation in SW2 small cell carcinoma cells. We also expressed a full-length catalytically inactive PST mutant (PST H331K) that would be expected to compete with the endogenous catalytically active SW2 cell PST, and catalytically active, full-length PST and soluble PST (sPST) proteins that would be expected to increase NCAM polysialylation to levels greater than those generated by the endogenous enzyme (Fig. 1). Like wild type PST, the membrane-associated PST mutants were found to localize primarily to the Golgi apparatus in COS-1 cells (supplemental Fig. S2, localization). Likewise, sPST and sPST140 are transported through the Golgi and are efficiently secreted (supplemental Fig. S2, localization, and data not shown). Furthermore, when expressed in COS-1 cells, PST and sPST were autopolysialylated as expected for active enzymes, whereas PST H331K, PST140, and sPST140 that lack critical catalytic sequences were not (supplemental Fig. S2, autopolysialylation).
To determine whether PST H331K, PST140, and sPST140 could compete with endogenous PST and decrease NCAM polysialylation, each protein was individually expressed in SW2 cells, which were then stained with anti-Myc antibody to identify cells expressing the Myc-tagged PST proteins (Fig. 2, localization), and the OL.28 anti-polySia antibody to examine the polysialylation of endogenous NCAM (Fig. 2, polysialylation). We found that SW2 cells expressing exogenous PST or sPST displayed an increase in cell surface NCAM polysialylation and Golgi enzyme autopolysialylation relative to surrounding cells that did not express these proteins (Fig. 2, PST and sPST, arrowheads). Our ability to detect enzyme autopolysialylation only when PST or sPST are highly expressed likely reflects the low expression level of endogenous PST. In contrast, cells expressing PST H331K exhibit substantially decreased NCAM polysialylation compared with nonexpressing cells (Fig. 2, PST H331K, arrowheads). Likewise, cells expressing PST140, although fewer in number, exhibited a reduction in surface NCAM polysialylation (Fig. 2, PST140, arrowheads). Expression of sPST140 also decreased NCAM polysialylation, although residual OL.28 staining was observed in some expressing cells (Fig. 2, sPST140, arrowheads). This may be due to the transient residence of sPST140 in the Golgi apparatus. These results support the notion that PST residues 21–140 between the transmembrane region and the beginning of the SML are likely to function in NCAM recognition.
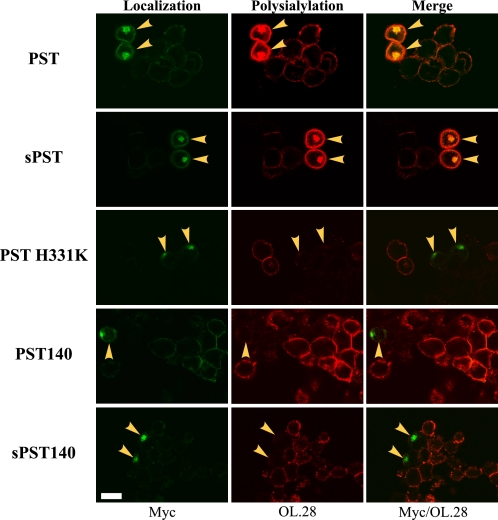
FIGURE 2.
Expression of PST H331K, PST140, and sPST140 inhibits NCAM polysialylation by endogenous PST in SW2 cells. Myc-tagged PST, sPST, PST H331K, PST140, and sPST140 were transiently expressed in SW2 small cell lung carcinoma cells containing endogenous PST and polysialylated NCAM. Eighteen hours post-transfection, cells were fixed and indirect immunofluorescence was performed using the anti-Myc antibody to analyze enzyme localization (Myc, Localization) or the OL.28 anti-polySia antibody to analyze total polysialylation (OL.28, Polysialylation). Overlaying the fluorescent signals for PST protein localization and polysialylation demonstrated the impact that expressing exogenous PST proteins had on the polysialylation of endogenous SW2 cell NCAM (Myc/OL.28, Merge). Cells expressing exogenous PST proteins are marked with yellow arrowheads. Cells were examined using a Zeiss 200M inverted confocal microscope, ×63 oil immersion objective. Scale bar = 10 μm.
To further narrow down the sequences involved in NCAM recognition, we created four additional PST140 truncation mutants (Fig. 1). PST127 and PST105 contain the PBR that extends from residue 71 to 105, whereas PST71 and PST61 do not. We found that PST127, PST71, and PST62 were mainly Golgi-localized and were not autopolysialylated when expressed in COS-1 cells, as expected (supplemental Fig. S2), but PST105 expression was not detected and this construct was not used for further experimentation (data not shown). We expressed these truncated PST mutants in SW2 cells and analyzed the impact on NCAM polysialylation as described above. Notably, as with PST140, PST127, PST71, and PST62 were detected in fewer SW2 cells than full-length PST or PST H331K suggesting some instability or increased turnover of the shorter proteins in these cells. Nevertheless, we found that cells expressing PST127 exhibited a decrease in NCAM polysialylation similar to that seen for PST140 (Fig. 3, PST140 and PST127, arrowheads). In contrast, cells expressing PST71 or PST62 did not show a decrease in NCAM polysialylation (Fig. 3, PST71 and PST62, arrowheads). Rather, the polysialylation of NCAM seemed unchanged from baseline levels, suggesting that PST71 and PST62 do not have sequences required for NCAM recognition and are unable to compete with endogenous PST to block NCAM polysialylation. Collectively, these results suggest that residues 71–127 of PST are important for NCAM recognition.
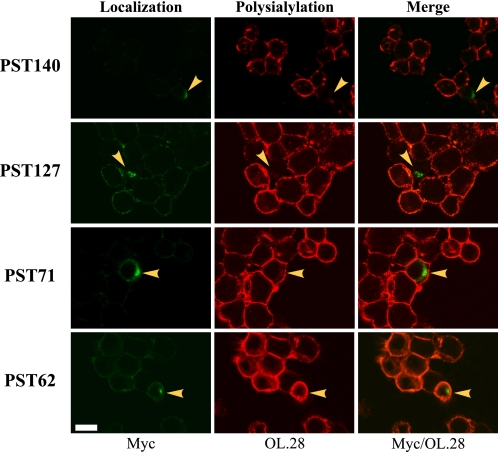
FIGURE 3.
NCAM polysialylation by endogenous PST is inhibited by expression of PST127 but not PST71 or PST62. Myc-tagged PST140, PST127, PST71, and PST62 were transiently expressed in SW2 small cell lung carcinoma cells. Eighteen hours post-transfection, cells were fixed and indirect immunofluorescence was performed to analyze enzyme localization (Myc, Localization) or total polysialylation (OL.28, Polysialylation). Overlaying the fluorescent signals for PST protein localization and polysialylation demonstrated the impact that expressing exogenous PST proteins had on the polysialylation of endogenous SW2 cell NCAM (Myc/OL.28, Merge). Cells expressing exogenous PST proteins are marked with yellow arrowheads. Cells were examined using a Zeiss 200M inverted confocal microscope, ×63 oil immersion objective. Scale bar = 10 μm.
The Ability of PST Truncation Mutants to Bind NCAM Correlates with Their Ability to Competitively Inhibit NCAM Polysialylation
Previous studies demonstrated that PST binds to NCAM (50). We therefore wanted to determine whether the ability of PST H331K, PST140, sPST140, and PST127 to decrease or eliminate NCAM polysialylation in SW2 cells is the result of their ability to bind to NCAM and block access of endogenous, catalytically active PST. To evaluate this possibility we took a co-immunoprecipitation approach. Following co-expression of V5-tagged NCAM with Myc-tagged wild-type PST, sPST, or one of the four truncated PST mutants in COS-1 cells, the enzymes were immunoprecipitated from the cell lysates with an anti-Myc antibody. Co-immunoprecipitated NCAM-V5 was detected by immunoblotting with anti-V5 antibody as detailed under “Experimental Procedures” (Fig. 4A, upper panel). Relative expression levels of NCAM and PST proteins were determined by immunoblotting an aliquot of cell lysate with anti-V5 and anti-Myc antibodies, respectively (Fig. 4A, middle and lower panels). We did detect some nonspecific binding of NCAM to the protein A-Sepharose beads in the absence of co-expressed enzyme (Fig. 4, A and B, lane 1). However, as expected, NCAM-V5 was co-immunoprecipitated with PST (Fig. 4A, lane 2). In addition, NCAM-V5 was co-immunoprecipitated with sPST, suggesting that the cytosolic tail and transmembrane region of PST are not critical for NCAM binding (Fig. 4A, lane 5). We observed that NCAM-V5 also bound to Myc-tagged, membrane-associated PST H331K, PST140, as well as sPST140 (Fig. 4A, lanes 3, 4, and 6).
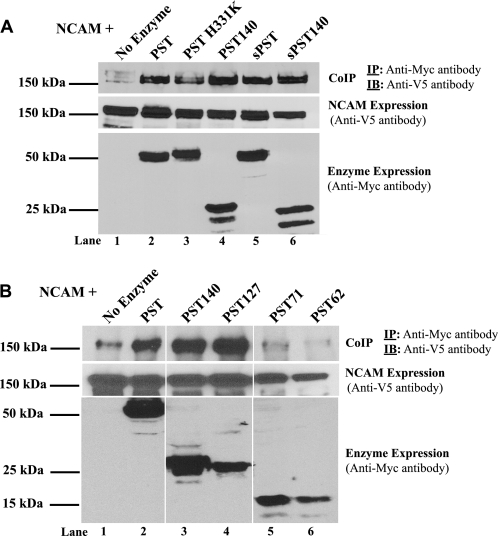
FIGURE 4.
Wild type PST and all PST mutants containing the PBR effectively bind to NCAM, whereas PST71 and PST62 that lack the PBR do not. V5-tagged NCAM was co-expressed with Myc-tagged PST or a PST mutant in COS-1 cells (A) or Lec2 CHO cells (B). After 18 h, cells were lysed and lysates were incubated with anti-Myc antibody to immunoprecipitate (IP) the PST proteins, followed by incubation with protein A-Sepharose beads for 1 h. Immunoprecipitates were subjected to SDS-PAGE and immunoblotting (IB) with anti-V5 antibody to detect co-immunoprecipitated NCAM (CoIP, IP: Anti-Myc antibody, IB: Anti-V5 antibody). To determine the relative expression levels of NCAM and PST proteins, an aliquot of lysate was removed prior to co-immunoprecipitation and subjected to SDS-PAGE and immunoblotting with anti-V5 antibody to detect NCAM (NCAM expression, Anti-V5 antibody) and anti-Myc antibody to detect the PST proteins (Enzyme expression, Anti-Myc antibody).
A similar co-immunoprecipitation analysis was performed in Lec2 CHO cells that compared the binding of NCAM to PST and the truncated PST proteins (Fig. 4B). Lec2 CHO cells cannot (poly)sialylate glycoconjugates due to the absence of a functional CMP-sialic acid transporter (62), and were used here to eliminate the potential effects of polySia on binding of wild type PST and NCAM. We found that PST140 and PST127 bound to NCAM like full-length PST (Fig. 4B, lanes 2–4), consistent with their ability to block NCAM polysialylation in SW2 cells. In contrast, PST71 and PST62 exhibited a significantly reduced ability to bind to NCAM, even when their somewhat lower level of expression is taken into consideration, and this correlates with the inability of these two truncated PST proteins to block NCAM polysialylation (Fig. 4B, lanes 5 and 6).
Arg82 and Arg93 in the PST PBR Are Required for Ability of PST H331K to Block NCAM Polysialylation
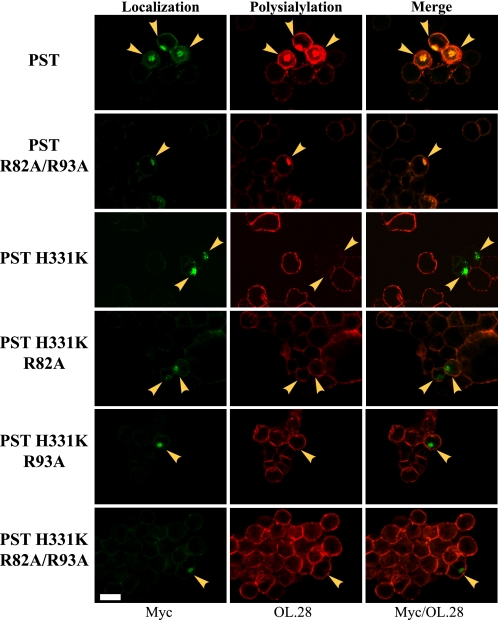
Previous work from our laboratory suggested that Arg82 and Arg93 in the PBR of PST are critical for NCAM polysialylation (61). It was postulated that these residues play a role in NCAM recognition because in the presence of a catalytically active PST R82A/R93A mutant, NCAM polysialylation was substantially decreased without an equal decrease in PST autopolysialylation. Because Arg82 and Arg93 are found within the stretch of PST amino acids (71–127) that our results suggest are important for NCAM recognition, we wondered whether replacing these residues would decrease the ability of the various PST mutants to competitively inhibit NCAM polysialylation in SW2 cells. We found that replacing Arg82 and Arg93 in PST140 and PST127 compromised their folding leading to some retention in the endoplasmic reticulum (data not shown), and so decided to focus on the impact of replacing these residues on the ability of PST H331K to block SW2 cell NCAM polysialylation. As a control we also evaluated the impact of expressing PST R82A/R93A on SW2 cell NCAM polysialylation. PST H331K proteins with the R82A and R93A mutations introduced individually or together were predominantly localized to the Golgi apparatus like the parent protein, wild type PST, and PST R82A/R93A (supplemental Figs. S2 and S3). Following expression of PST, PST H331K, and their mutants in SW2 cells, the cells were stained with anti-Myc antibody and OL.28 anti-polySia antibody as described above (Fig. 5). As shown in Fig. 2, expressing wild type PST enhanced the polysialylation of NCAM (cell surface) and allowed detection of the autopolysialylated enzyme in the Golgi (Fig. 5). However, cells expressing the PST R82A/R93A mutant retained detectable Golgi enzyme autopolysialylation, but exhibited no enhanced cell surface polysialylation consistent with the inability of the PST R82A/R93A mutant to recognize endogenous SW2 cell NCAM (Fig. 5). Most strikingly, whereas PST H331K significantly decreased NCAM polysialylation in SW2 cells as shown above (Figs. 2 and 5, PST H331K), the introduction of the R82A/R93A mutation appeared to abolish the ability of PST H331K to compete with endogenous enzyme (Fig. 5). The impact of the single arginine replacements on the ability of PST H331K to block NCAM polysialylation was more difficult to discern in these experiments, and led us to take another approach.
FIGURE 5.
Replacing Arg82 and Arg93 block the ability of PST to enhance the polysialylation of SW2 cell NCAM and the ability of PST H331K to inhibit the polysialylation of SW2 cell NCAM by endogenous enzyme. Myc-tagged PST, PST R82A/R93A, PST H331K, PST H331K R82A, PST H331K R93A, and PST H331K R82A/R93A were transiently expressed in SW2 small cell lung carcinoma cells. Eighteen hours post-transfection, cells were fixed and indirect immunofluorescence was performed to analyze enzyme localization (Myc, green, Enzyme localization) or total polysialylation (OL.28, red, Polysialylation). Cells expressing exogenous PST and PST mutant proteins are marked with yellow arrowheads. Cells were examined using a Zeiss 200M inverted confocal microscope, ×63 oil immersion objective. Scale bar = 10 μm.
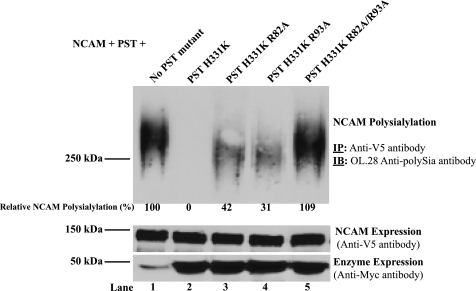
To more precisely quantify the contribution of each of these arginine residues to NCAM recognition, we wanted to establish a more sensitive assay to measure changes in NCAM polysialylation. Initially, we attempted to generate SW2 cells stably expressing the various PST constructs, but found that these cell lines were difficult to establish and that the selection procedure actually led to increased polysialylation (data not shown). In lieu of this, we established a triple expression system in COS-1 cells, in which NCAM and PST would be co-expressed with an excess of competitor. We expressed V5-tagged NCAM, Myc-tagged PST, and either a Myc-tagged PST H331K or a PST H331K PBR mutant in COS-1 cells at a 1:1:6 ratio. NCAM was precipitated from cell lysates with anti-V5 antibody, and the level of NCAM polysialylation was assessed by immunoblotting with the OL.28 anti-polySia antibody (Fig. 6, upper panel). Relative substrate expression (Fig. 6, middle panel), as well as enzyme expression (Fig. 6, bottom panel), were evaluated as described above. Furthermore, using NIH ImageJ we quantified the relative NCAM polysialylation observed in the presence of PST H331K and its mutants in this representative immunoblot. We found that in the presence of PST and no competitor, NCAM was polysialylated (Fig. 6, lane 1). However, when excess PST H331K was co-expressed with wild type PST, NCAM polysialylation was undetectable (Fig. 6, lane 2). Expression of PST H331K with either the R82A or R93A mutation decreased but did not eliminate the ability of PST H331K to block NCAM polysialylation and 42 or 31% of wild type polysialylation was retained, respectively (Fig. 6, lanes 3 and 4). However, when the two arginines were replaced simultaneously (R82A/R93A) the ability of PST H331K to block NCAM polysialylation was eliminated (Fig. 6, lane 5). These results indicate that the PST H331K double mutant is unable to compete with wild type PST, and strongly suggest that Arg82 and Arg93 are important for PST-NCAM recognition.
FIGURE 6.
The ability of PST H331K to inhibit the polysialylation of NCAM by endogenous SW2 PST is eliminated when Arg82 and Arg93 are replaced with alanine residues. V5-tagged NCAM and Myc-tagged PST were expressed in COS-1 cells without (lane 1) or with Myc-tagged PST H331K, PST H331K R82A, PST H331K R93A, or PST H331K R82A/R93A in a 1:1:6 ratio (lanes 2–5). After 18 h, cells were lysed and NCAM was immunoprecipitated (IP) using the anti-V5 antibody. Immunoprecipitated NCAM was subjected to SDS-PAGE and immunoblotting (IB) to analyze NCAM polysialylation (NCAM polysialylation, IP: Anti-V5 antibody, IB: OL.28 Anti-polySia antibody). Relative NCAM, PST, and PST mutant protein expression levels were assessed by removing an aliquot of cell lysate prior to immunoprecipitation and subjecting it to SDS-PAGE and immunoblotting with anti-V5 antibody (NCAM Expression, Anti-V5 antibody) or anti-Myc antibody (Enzyme Expression, Anti-Myc antibody). Note that enzyme expression reflects the expression levels of both wild type and mutant PST proteins.
The Polysialylation of NCAM, NRP-2, and SynCAM 1 by PST Exhibits Different Requirements for PBR Residues Arg82 and Arg93
Our finding above that Arg82 and Arg93 in the PST PBR are required for recognition and polysialylation of NCAM, led us to ask whether this region and these residues are also required for the polysialylation of other polyST substrates, namely, NRP-2 and SynCAM 1. NRP-2 is a widely expressed transmembrane protein that plays roles in vascular development, tumorigenesis, and neuronal patterning (reviewed in Ref. 63). NRP-2 has also been previously shown to be polysialylated by PST on O-linked glycans in mature dendritic cells (6). Currently, the precise function of polySia on NRP-2 is still being debated, as it has been suggested to inhibit dendritic cell-induced T cell activation and proliferation (6), and conversely promote this process by enhancing dendritic cell migration to the lymph nodes through the stimulation of CCR7 receptor signaling (64). SynCAM 1 is a synaptic adhesion molecule that forms homo- and heterophilic adhesive complexes and aids in the formation and maintenance of synapses (65). SynCAM 1 has been found to be polysialylated on a single N-glycan in a small subset of NG2 glial cells in perinatal brain (7). PolySia has been shown to decrease SynCAM 1 homophilic adhesion in vitro, and is postulated to regulate the formation of a neuron-glial synapse (7).
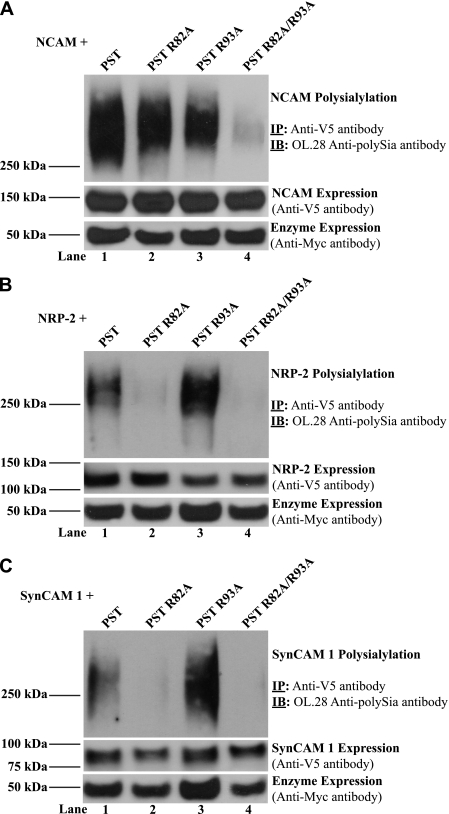
We evaluated the impact of replacing PST Arg82 and Arg93 individually or together on the polysialylation of both NRP-2 and SynCAM 1. Following co-expression of V5-tagged NCAM, NRP-2, or SynCAM 1 with Myc-tagged PST or PST R82A, PST R93A, or PST R82A/R93A in COS-1 cells, the substrate proteins were precipitated with anti-V5 antibody and their polysialylation was analyzed as described above (Fig. 7, A–C, upper panels). Relative substrate expression (Fig. 7, A–C, middle panels), as well as enzyme expression (Fig. 7, A–C, bottom panels), were evaluated as described above. As seen previously, replacing Arg82 and Arg93 with alanine residues decreased the polysialylation of NCAM (Ref. 61 and Fig. 7A, lanes 1–3). Furthermore, a significant decrease in NCAM polysialylation required that both Arg82 and Arg93 be replaced (Ref. 61 and Fig. 7A, lane 4). This result suggests that both arginine residues are required for NCAM recognition. In contrast, mutation of Arg82 to alanine alone, or in combination with Arg93, eliminates NRP-2 and SynCAM 1 polysialylation by PST (Fig. 7, B and C, lanes 1, 2, and 4). However, mutation of Arg93 alone to alanine did not reduce NRP-2 or SynCAM 1 polysialylation (Fig. 7, B and C, lane 3). Taken together, these results suggest that the PST PBR functions specifically in the recognition of substrates for polysialylation, and that Arg82 in the PST PBR may be required for general substrate recognition, whereas Arg93 specifically aids the recognition of NCAM.
FIGURE 7.
The ability of PST to polysialylate NCAM is eliminated when Arg82 and Arg93 are replaced, whereas the ability of PST to polysialylate NRP-2 and SynCAM 1 requires only the presence of Arg82. NCAM-V5 (A), NRP-2-V5 (B), or SynCAM 1-V5 (C) were co-expressed with Myc-tagged PST, PST R82A, PST R93A, or PST R82A/R93A in COS-1 cells. Eighteen hours post-transfection, cells were lysed and substrates were immunoprecipitated using the anti-V5 antibody. Immunoprecipitates (IP) were subjected to SDS-PAGE and immunoblotting (IB) with the OL.28 anti-polySia antibody to analyze the level of substrate polysialylation (Polysialylation, IP: Anti-V5 antibody, IB: OL.28 Anti-polySia antibody). Relative protein expression levels were determined by removing an aliquot of cell lysate prior to immunoprecipitation and subjecting it to SDS-PAGE followed by immunoblotting with anti-V5 antibody (NCAM, NRP-2, or SynCAM 1 Expression, Anti-V5 antibody) or anti-Myc antibody (Enzyme Expression, Anti-Myc antibody).
DISCUSSION
In this study, we sought to identify specific sequences within PST that are required for substrate recognition leading to protein-specific polysialylation. Competition and binding studies demonstrated that PST sequences between residues 71 and 127 are required for NCAM recognition (Figs. 2–4). Previous work suggested that the PBR of both ST8SiaIV/PST and ST8SiaII/STX may play a role in substrate recognition, and two PBR residues (Arg82 and Arg93 in PST) are particularly important for NCAM polysialylation (61). In this study, we found that replacing these residues in the inactive PST H331K protein eliminated its ability to compete with endogenous PST in SW2 cells (Figs. 5 and 6). From this study and analysis of NCAM polysialylation by PST R82A and R93A mutants, it appeared that both residues make substantial contributions to NCAM recognition and polysialylation, with Arg93 possibly playing a slightly larger role (Ref. 61 and Figs. 5 and 6). However, analysis of the polysialylation of NRP-2 and SynCAM 1 by PST and its arginine mutants suggests that Arg82 plays a predominant role in PST recognition and polysialylation of these substrates (Fig. 7, B and C). These results emphasized the role of Arg82 and Arg93 in substrate recognition rather than general enzyme activity, and raised the intriguing possibility that different substrates may be recognized by slightly different binding surfaces of the polySTs.
A particularly interesting and repeatable observation was the consistent increase in NRP-2 and SynCAM 1 polysialylation observed when PST Arg93 is replaced by alanine (Fig. 7, B and C). One possibility is that the presence of Arg93 forms a less optimal binding site for these substrates. If this is the case, then eliminating Arg93 could enhance polysialylation of NRP-2 and SynCAM 1 by optimizing their interaction with the polyST. Additional experiments will be performed to determine whether other PBR residues play roles in the recognition and polysialylation of NRP-2 and SynCAM 1.
How can just one or two arginine residues have such an impact on substrate recognition? Researchers studying the propensity of certain amino acids at protein interfaces suggest that these interfaces favor residues that can engage in hydrophobic and electrostatic interactions, including tryptophan, arginine, and tyrosine (reviewed in Ref. 66). Arginine is favored in “hot spots” of binding energy because it is capable of multiple types of favorable interactions, including forming up to five hydrogen bonds and a salt bridge utilizing the positive charge on its guanidinium moiety (67). In fact, an arginine-aspartate salt bridge is found at the interface of the α- and β-subunits of the GABAA receptor, and removal of this bridge through alanine mutagenesis compromises GABA binding and subunit association (68). Applying these observations to the polyST-NCAM interaction, Arg82 and Arg93 may promote recognition through the formation of hydrogen bonds and salt bridges with residues in NCAM. Previous results from our lab suggest that it is plausible that salt bridges could be formed between the polySTs and NCAM. Earlier work identified an acidic patch on the surface of NCAM FN1 comprised of several aspartate and glutamate residues (18). Mutating these residues to alanine resulted in reduced NCAM polysialylation (18) and reduced PST binding (50), indicating that these residues may form part of a docking site for the polySTs.
It is tempting to try and model the interactions of these basic residues in PST and the NCAM FN1 domain. However, whereas the NCAM Ig5-FN1 structure has been solved by our group (69), the polyST structures are not known and unlikely to precisely match the structure of the bovine ST3Gal I (70) or the Campylobacter jejuni CST enzyme (71) sialyltransferases that show limited overall homology and little to no homology in the stem regions prior to the SML where the critical arginine residues are located in PST. Secondary structure predictions using PSIPRE (72) or JUFO (73) place Arg82 in an α-helix and Arg93 in an adjacent coil but provide no information as to the orientation of their side chains or the relationship of these two structural motifs, making it difficult to predict how these residues might form salt bridges with acidic residues of the NCAM FN1 acidic patch that extends along one face of this domain (18). On the other hand, acidic surfaces are predicted or observed for SynCAM 1 and NRP-2 domains that may be recognized by the polySTs. For example, modeling the SynCAM 1 extracellular portion (Ig1-Ig2-Ig3), based on the NCAM Ig1-Ig2-Ig3 structure (74) (PDB code 1QZ1), predicts an acidic surface patch on Ig2 that may allow polyST docking for the polysialylation of a single N-glycan on Ig1.
Attempts to demonstrate a role for Arg82 and Arg93 in direct protein-protein interaction between polyST and NCAM in co-immunoprecipitation experiments were unsuccessful. We were unable to detect a reduction in polyST-NCAM binding when Arg82 and Arg93 are replaced, despite the profound effect this alteration has on NCAM polysialylation. One possibility is that the polySTs can interact with substrates in more than one way, but only when they are aligned properly with the glycans to be modified can substrate polysialylation take place. In support of this idea, we have demonstrated that deleting NCAM FN1 eliminates polyST binding (50), however, replacing NCAM FN1 with other FN1 domains allows binding, but not polysialylation, suggesting that residues in NCAM FN1 allow the precise positioning of enzyme and substrate and that this is critical for substrate polysialylation.3
In this regard it is notable that replacing Arg82 and Arg93 has different effects on the three substrates. These results suggest that the polySTs use different but overlapping sequences to recognize their substrates and position them for polysialylation. Removing the key PST basic residues may redirect substrates to other polyST surfaces that do not allow optimal alignment of the enzyme and substrate, and consequently prevent polysialylation without greatly altering enzyme-substrate binding. Related to this possibility, previous work in our laboratory demonstrated that changes in the linker region between NCAM Ig5 and FN1 are capable of eliminating NCAM polysialylation, and highlight the importance of a defined Ig5-FN1 relationship for the ability of a polyST interacting with the NCAM FN1 domain to access and polysialylate the Ig5 N-glycans (51).
Another possibility is that the polyST PBR residues are involved in an interaction with the growing polySia chain and that this is critical for chain elongation. Troy and colleagues (60) looked for conserved basic residues in the polySTs, and identified the PSTD, based on the idea that these enzymes would likely engage the growing, negatively charged polySia chain using a stretch of positively charged, basic residues to increase the processivity of the process. Indeed, glycosyltransferases that synthesize bacterial capsular polysaccharides frequently have carbohydrate recognition sequences outside the catalytic site that mediate association with the growing polysaccharide chain. For example, the capsular polysaccharide of Streptococcus pneumoniae is synthesized by a processive glycosyltransferase that requires a “primer” of approximately eight monosaccharides that is engaged by a carbohydrate recognition sequence to initiate the processive phase of synthesis (75).
Our results strongly suggest that the PBR residues, and in particular Arg82 and Arg93, are involved in PST-substrate protein-protein recognition, but we cannot rule out a secondary interaction of the PBR sequences with the growing polySia chain. In fact, it seems likely that mammalian polysialylation may be a two-stage, processive process, where the first stage involves an initial protein-protein interaction between polyST and substrate that facilitates the synthesis of short polySia chains, whereas the second stage is mediated by polyST noncatalytic sequences that interact with the growing polySia chain, allowing the enzyme to maintain association with the substrate and polymerize long polySia chains. Whether polysialylation is a two-stage process, and whether both stages involve the polyST PBR is yet to be determined.
In conclusion, our results have shown that sequences within a polybasic region of the polyST, PST, are involved in substrate recognition. In particular, two PST residues within this region, namely Arg82 and Arg93, are specifically required for efficient recognition and polysialylation of NCAM. In contrast, PST Arg82 plays a predominant role in the recognition and polysialylation of NRP-2 and SynCAM 1. Taken together, these results support the premise that polysialylation is highly protein specific and reveal that the polySTs may use overlapping but distinct surfaces to recognize their different substrates.
Supplementary Material
Acknowledgment
We thank Deirdre A. Foley for help in the preparation of the PST PBR mutant protein constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM063843 (to K. J. C.) and Fellowship F31 GM096739 from the NIGMS (to J. L. Z.).

This article contains supplemental Table S1 and Figs. S1–S3.
M. Thompson, unpublished results.
- polyST(s)
- polysialyltransferase(s)
- STX
- ST8Sia II or sialyltransferase X
- PST
- ST8Sia IV or polysialyltransferase-1
- NCAM
- neural cell adhesion molecule
- NRP-2
- neuropilin-2
- SynCAM 1
- synaptic cell adhesion molecule 1
- Ig
- immunoglobulin-like domain
- FN
- fibronectin type III repeat of NCAM
- polySia
- polysialic acid
- SML
- large sialyl motif
- SMS
- small sialyl motif
- SMVS
- very small sialyl motif
- PSTD
- polysialyltransferase domain
- PBR
- polybasic region
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Nakayama J., Fukuda M. N., Fredette B., Ranscht B., Fukuda M. (1995) Expression cloning of a human polysialyltransferase that forms the polysialylated neural cell adhesion molecule present in embryonic brain. Proc. Natl. Acad. Sci. U.S.A. 92, 7031–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kojima N., Yoshida Y., Kurosawa N., Lee Y. C., Tsuji S. (1995) Enzymatic activity of a developmentally regulated member of the sialyltransferase family (STX). Evidence for α2,8-sialyltransferase activity toward N-linked oligosaccharides. FEBS Lett. 360, 1–4 [DOI] [PubMed] [Google Scholar]
- 3. Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Molecular characterization of eukaryotic polysialyltransferase-1. Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 4. Scheidegger E. P., Sternberg L. R., Roth J., Lowe J. B. (1995) A human STX cDNA confers polysialic acid expression in mammalian cells. J. Biol. Chem. 270, 22685–22688 [DOI] [PubMed] [Google Scholar]
- 5. Angata K., Fukuda M. (2003) Polysialyltransferases. Major players in polysialic acid synthesis on the neural cell adhesion molecule. Biochimie 85, 195–206 [DOI] [PubMed] [Google Scholar]
- 6. Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N. M. (2007) Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 7. Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., Geyer H. (2010) Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. James W. M., Agnew W. S. (1987) Multiple oligosaccharide chains in the voltage-sensitive Na channel from electrophorus electricus. Evidence for α2,8-linked polysialic acid. Biochem. Biophys. Res. Commun. 148, 817–826 [DOI] [PubMed] [Google Scholar]
- 9. Yabe U., Sato C., Matsuda T., Kitajima K. (2003) Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 10. Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 15, 6943–6950 [PMC free article] [PubMed] [Google Scholar]
- 11. Close B. E., Colley K. J. (1998) In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 12. Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul O., Fukuda M. (2000) Differential biosynthesis of polysialic acid on neural cell adhesion molecule (NCAM) and oligosaccharide acceptors by three distinct α2,8-sialyltransferases, ST8Sia IV (PST), ST8Sia II (STX), and ST8Sia III. J. Biol. Chem. 275, 18594–18601 [DOI] [PubMed] [Google Scholar]
- 13. Kojima N., Tachida Y., Yoshida Y., Tsuji S. (1996) Characterization of mouse ST8Sia II (STX) as a neural cell adhesion molecule-specific polysialic acid synthase. Requirement of core α1,6-linked fucose and a polypeptide chain for polysialylation. J. Biol. Chem. 271, 19457–19463 [DOI] [PubMed] [Google Scholar]
- 14. von Der Ohe M., Wheeler S. F., Wuhrer M., Harvey D. J., Liedtke S., Mühlenhoff M., Gerardy-Schahn R., Geyer H., Dwek R. A., Geyer R., Wing D. R., Schachner M. (2002) Localization and characterization of polysialic acid-containing N-linked glycans from bovine NCAM. Glycobiology 12, 47–63 [DOI] [PubMed] [Google Scholar]
- 15. Kudo M., Kitajima K., Inoue S., Shiokawa K., Morris H. R., Dell A., Inoue Y. (1996) Characterization of the major core structures of the α2,8-linked polysialic acid-containing glycan chains present in neural cell adhesion molecule in embryonic chick brains. J. Biol. Chem. 271, 32667–32677 [DOI] [PubMed] [Google Scholar]
- 16. Geyer H., Bahr U., Liedtke S., Schachner M., Geyer R. (2001) Core structures of polysialylated glycans present in neural cell adhesion molecule from newborn mouse brain. Eur. J. Biochem. 268, 6587–6599 [DOI] [PubMed] [Google Scholar]
- 17. Close B. E., Mendiratta S. S., Geiger K. M., Broom L. J., Ho L. L., Colley K. J. (2003) The minimal structural domains required for neural cell adhesion molecule polysialylation by PST/ST8Sia IV and STX/ST8Sia II. J. Biol. Chem. 278, 30796–30805 [DOI] [PubMed] [Google Scholar]
- 18. Mendiratta S. S., Sekulic N., Lavie A., Colley K. J. (2005) Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST. J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 19. Mendiratta S. S., Sekulic N., Hernandez-Guzman F. G., Close B. E., Lavie A., Colley K. J. (2006) A novel α-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation. J. Biol. Chem. 281, 36052–36059 [DOI] [PubMed] [Google Scholar]
- 20. Ditlevsen D. K., Povlsen G. K., Berezin V., Bock E. (2008) NCAM-induced intracellular signaling revisited. J. Neurosci. Res. 86, 727–743 [DOI] [PubMed] [Google Scholar]
- 21. Walmod P. S., Kolkova K., Berezin V., Bock E. (2004) Zippers make signals. NCAM-mediated molecular interactions and signal transduction. Neurochem. Res. 29, 2015–2035 [DOI] [PubMed] [Google Scholar]
- 22. Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. (1987) Neural cell adhesion molecule. Structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science 236, 799–806 [DOI] [PubMed] [Google Scholar]
- 23. Nelson R. W., Bates P. A., Rutishauser U. (1995) Protein determinants for specific polysialylation of the neural cell adhesion molecule. J. Biol. Chem. 270, 17171–17179 [DOI] [PubMed] [Google Scholar]
- 24. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 25. Johnson C. P., Fujimoto I., Rutishauser U., Leckband D. E. (2005) Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J. Biol. Chem. 280, 137–145 [DOI] [PubMed] [Google Scholar]
- 26. Fujimoto I., Bruses J. L., Rutishauser U. (2001) Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function, and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 276, 31745–31751 [DOI] [PubMed] [Google Scholar]
- 27. Horstkorte R., Lessner N., Gerardy-Schahn R., Lucka L., Danker K., Reutter W. (1999) Expression of the polysialyltransferase ST8SiaIV. Polysialylation interferes with adhesion of PC12 cells in vitro. Exp. Cell Res. 246, 122–128 [DOI] [PubMed] [Google Scholar]
- 28. Kanato Y., Kitajima K., Sato C. (2008) Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology 18, 1044–1053 [DOI] [PubMed] [Google Scholar]
- 29. Isomura R., Kitajima K., Sato C. (2011) Structural and functional impairments of polysialic acid by a mutated polysialyltransferase found in schizophrenia. J. Biol. Chem. 286, 21535–21545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brusés J. L., Rutishauser U. (2001) Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie 83, 635–643 [DOI] [PubMed] [Google Scholar]
- 31. Bonfanti L. (2006) PSA-NCAM in mammalian structural plasticity and neurogenesis. Prog. Neurobiol. 80, 129–164 [DOI] [PubMed] [Google Scholar]
- 32. Durbec P., Cremer H. (2001) Revisiting the function of PSA-NCAM in the nervous system. Mol. Neurobiol. 24, 53–64 [DOI] [PubMed] [Google Scholar]
- 33. Hildebrandt H., Mühlenhoff M., Weinhold B., Gerardy-Schahn R. (2007) Dissecting polysialic acid and NCAM functions in brain development. J. Neurochem. 103, 56–64 [DOI] [PubMed] [Google Scholar]
- 34. Seki T., Arai Y. (1993) Distribution and possible roles of the highly polysialylated neural cell adhesion molecule (NCAM-H) in the developing and adult central nervous system. Neurosci. Res. 17, 265–290 [DOI] [PubMed] [Google Scholar]
- 35. Nakayama J., Angata K., Ong E., Katsuyama T., Fukuda M. (1998) Polysialic acid, a unique glycan that is developmentally regulated by two polysialyltransferases, PST and STX, in the central nervous system. From biosynthesis to function. Pathol. Int. 48, 665–677 [DOI] [PubMed] [Google Scholar]
- 36. Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 37. Perera A. D., Lagenaur C. F., Plant T. M. (1993) Postnatal expression of polysialic acid-neural cell adhesion molecule in the hypothalamus of the male rhesus monkey (Macaca mulatta). Endocrinology 133, 2729–2735 [DOI] [PubMed] [Google Scholar]
- 38. Kiss J. Z., Wang C., Rougon G. (1993) Nerve dependent expression of high polysialic acid neural cell adhesion molecule in neurohypophysial astrocytes of adult rats. Neuroscience 53, 213–221 [DOI] [PubMed] [Google Scholar]
- 39. Hu H. (2000) Polysialic acid regulates chain formation by migrating olfactory interneuron precursors. J. Neurosci. Res. 61, 480–492 [DOI] [PubMed] [Google Scholar]
- 40. Hu H., Tomasiewicz H., Magnuson T., Rutishauser U. (1996) The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. Neuron 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 41. Stoenica L., Senkov O., Gerardy-Schahn R., Weinhold B., Schachner M., Dityatev A. (2006) In vivo synaptic plasticity in the dentate gyrus of mice deficient in the neural cell adhesion molecule NCAM or its polysialic acid. Eur. J. Neurosci. 23, 2255–2264 [DOI] [PubMed] [Google Scholar]
- 42. Burgess A., Wainwright S. R., Shihabuddin L. S., Rutishauser U., Seki T., Aubert I. (2008) Polysialic acid regulates the clustering, migration, and neuronal differentiation of progenitor cells in the adult hippocampus. Dev. Neurobiol. 68, 1580–1590 [DOI] [PubMed] [Google Scholar]
- 43. Jungnickel J., Brämer C., Bronzlik P., Lipokatic-Takacs E., Weinhold B., Gerardy-Schahn R., Grothe C. (2009) Level and localization of polysialic acid is critical for early peripheral nerve regeneration. Mol. Cell Neurosci. 40, 374–381 [DOI] [PubMed] [Google Scholar]
- 44. Scheidegger E. P., Lackie P. M., Papay J., Roth J. (1994) In vitro and in vivo growth of clonal sublines of human small cell lung carcinoma is modulated by polysialic acid of the neural cell adhesion molecule. Lab. Invest. 70, 95–106 [PubMed] [Google Scholar]
- 45. Petridis A. K., Wedderhopp H., Hugo H. H., Mehdorn H. M. (2009) Acta Neurochir. 151, 601–604 [DOI] [PubMed] [Google Scholar]
- 46. Suzuki M., Suzuki M., Nakayama J., Suzuki A., Angata K., Chen S., Sakai K., Hagihara K., Yamaguchi Y., Fukuda M. (2005) Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology 15, 887–894 [DOI] [PubMed] [Google Scholar]
- 47. Hildebrandt H., Becker C., Glüer S., Rösner H., Gerardy-Schahn R., Rahmann H. (1998) Polysialic acid on the neural cell adhesion molecule correlates with expression of polysialyltransferases and promotes neuroblastoma cell growth. Cancer Res. 58, 779–784 [PubMed] [Google Scholar]
- 48. Miyahara R., Tanaka F., Nakagawa T., Matsuoka K., Isii K., Wada H. (2001) Expression of neural cell adhesion molecules (polysialylated form of neural cell adhesion molecule and L1-cell adhesion molecule) on resected small cell lung cancer specimens. In relation to proliferation state. J. Surg. Oncol. 77, 49–54 [DOI] [PubMed] [Google Scholar]
- 49. Seidenfaden R., Gerardy-Schahn R., Hildebrandt H. (2000) Control of NCAM polysialylation by the differential expression of polysialyltransferases ST8SiaII and ST8SiaIV. Eur. J. Cell Biol. 79, 680–688 [DOI] [PubMed] [Google Scholar]
- 50. Thompson M. G., Foley D. A., Swartzentruber K. G., Colley K. J. (2011) Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation. J. Biol. Chem. 286, 4525–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Foley D. A., Swartzentruber K. G., Thompson M. G., Mendiratta S. S., Colley K. J. (2010) Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera. J. Biol. Chem. 285, 35056–35067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galuska S. P., Oltmann-Norden I., Geyer H., Weinhold B., Kuchelmeister K., Hildebrandt H., Gerardy-Schahn R., Geyer R., Mühlenhoff M. (2006) Polysialic acid profiles of mice expressing variant allelic combinations of the polysialyltransferases ST8Sia II and ST8Sia IV. J. Biol. Chem. 281, 31605–31615 [DOI] [PubMed] [Google Scholar]
- 53. Livingston B. D., Jacobs J. L., Glick M. C., Troy F. A. (1988) Extended polysialic acid chains (n > 55) in glycoproteins from human neuroblastoma cells. J. Biol. Chem. 263, 9443–9448 [PubMed] [Google Scholar]
- 54. Datta A. K., Paulson J. C. (1995) The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J. Biol. Chem. 270, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 55. Datta A. K., Sinha A., Paulson J. C. (1998) Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J. Biol. Chem. 273, 9608–9614 [DOI] [PubMed] [Google Scholar]
- 56. Geremia R. A., Harduin-Lepers A., Delannoy P. (1997) Identification of two novel conserved amino acid residues on eukaryotic sialytransferase. Implication for the mechanism of action. Glycobiology 7, v-vii [DOI] [PubMed] [Google Scholar]
- 57. Kitazume-Kawaguchi S., Kabata S., Arita M. (2001) Differential biosynthesis of polysialic or disialic acid Structure by ST8Sia II and ST8Sia IV. J. Biol. Chem. 276, 15696–15703 [DOI] [PubMed] [Google Scholar]
- 58. Jeanneau C., Chazalet V., Augé C., Soumpasis D. M., Harduin-Lepers A., Delannoy P., Imberty A., Breton C. (2004) Structure-function analysis of the human sialyltransferase ST3Gal I. Role of N-glycosylation and a novel conserved sialyl motif. J. Biol. Chem. 279, 13461–13468 [DOI] [PubMed] [Google Scholar]
- 59. Angata K., Chan D., Thibault J., Fukuda M. (2004) Molecular dissection of the ST8Sia IV polysialyltransferase. Distinct domains are required for neural cell adhesion molecule recognition and polysialylation. J. Biol. Chem. 279, 25883–25890 [DOI] [PubMed] [Google Scholar]
- 60. Nakata D., Zhang L., Troy F. A. (2006) Molecular basis for polysialylation. A novel polybasic polysialyltransferase domain (PSTD) of 32 amino acids unique to the α2,8-polysialyltransferases is essential for polysialylation. Glycoconj. J. 23, 423–436 [DOI] [PubMed] [Google Scholar]
- 61. Foley D. A., Swartzentruber K. G., Colley K. J. (2009) Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule, NCAM. J. Biol. Chem. 284, 15505–15516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stanley P. (1984) Glycosylation mutants of animal cells. Annu. Rev. Genet. 18, 525–552 [DOI] [PubMed] [Google Scholar]
- 63. Pellet-Many C., Frankel P., Jia H., Zachary I. (2008) Neuropilins, structure, function, and role in disease. Biochem. J. 411, 211–226 [DOI] [PubMed] [Google Scholar]
- 64. Rey-Gallardo A., Escribano C., Delgado-Martín C., Rodriguez-Fernández J. L., Gerardy-Schahn R., Rutishauser U., Corbi A. L., Vega M. A. (2010) Polysialylated neuropilin-2 enhances human dendritic cell migration through the basic C-terminal region of CCL21. Glycobiology 20, 1139–1146 [DOI] [PubMed] [Google Scholar]
- 65. Thomas L. A., Akins M. R., Biederer T. (2008) Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J. Comp. Neurol. 510, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moreira I. S., Fernandes P. A., Ramos M. J. (2007) Hot spots. A review of the protein-protein interface determinant amino acid residues. Proteins 68, 803–812 [DOI] [PubMed] [Google Scholar]
- 67. Bogan A. A., Thorn K. S. (1998) Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 68. Laha K. T., Wagner D. A. (2011) A state-dependent salt-bridge interaction exists across the β/α intersubunit interface of the GABAA receptor. Mol. Pharmacol. 79, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Foley D. A., Swartzentruber K. G., Lavie A., Colley K. J. (2010) Structure and mutagenesis of neural cell adhesion molecule domains. Evidence for flexibility in the placement of polysialic acid attachment sites. J. Biol. Chem. 285, 27360–27371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rao F. V., Rich J. R., Raki B., Buddai S., Schwartz M. F., Johnson K., Bowe C., Wakarchuk W. W., Defrees S., Withers S. G., Strynadka N. C. (2009) Structural insight into mammalian sialyltransferases. Nat. Struct. Mol. Biol. 16, 1186–1188 [DOI] [PubMed] [Google Scholar]
- 71. Chiu C. P., Watts A. G., Lairson L. L., Gilbert M., Lim D., Wakarchuk W. W., Withers S. G., Strynadka N. C. (2004) Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11, 163–170 [DOI] [PubMed] [Google Scholar]
- 72. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 73. Meiler J., Baker D. (2003) Coupled prediction of protein secondary and tertiary structure. Proc. Natl. Acad. Sci. U.S.A. 100, 12105–12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Soroka V., Kolkova K., Kastrup J. S., Diederichs K., Breed J., Kiselyov V. V., Poulsen F. M., Larsen I. K., Welte W., Berezin V., Bock E., Kasper C. (2003) Structure and interactions of NCAM Ig1–2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure 11, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 75. Forsee W. T., Cartee R. T., Yother J. (2006) Role of the carbohydrate binding site of the Streptococcus pneumoniae capsular polysaccharide type 3 synthase in the transition from oligosaccharide to polysaccharide synthesis. J. Biol. Chem. 281, 6283–6289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.