Background: Oxidized derivatives of the plant hormone jasmonoyl-isoleucine accumulate in wounded Arabidopsis leaves.
Results: Cytochromes P450 CYP94C1 and CYP94B3 cooperate to catalyze the formation of 12OH-JA-Ile and 12COOH-JA-Ile.
Conclusion: CYP94C1 and CYP94B3 define a major route for JA-Ile catabolism.
Significance: Elucidation of CYP94-mediated JA-Ile oxidation opens new avenues for understanding jasmonate metabolism and signaling.
Keywords: Cytochrome P450, Metabolism, Plant Defense, Plant Hormones, Signaling, Arabidopsis, CYP94, Jasmonate Metabolism, Metabolic Profiling, Wound Response
Abstract
The jasmonate hormonal pathway regulates important defensive and developmental processes in plants. Jasmonoyl-isoleucine (JA-Ile) has been identified as a specific ligand binding the COI1-JAZ co-receptor to relieve repression of jasmonate responses. Two JA-Ile derivatives, 12OH-JA-Ile and 12COOH-JA-Ile, accumulate in wounded Arabidopsis leaves in a COI1- and JAR1-dependent manner and reflect catabolic turnover of the hormone. Here we report the biochemical and genetic characterization of two wound-inducible cytochromes P450, CYP94C1 and CYP94B3, that are involved in JA-Ile oxidation. Both enzymes expressed in yeast catalyze two successive oxidation steps of JA-Ile with distinct characteristics. CYP94B3 performed efficiently the initial hydroxylation of JA-Ile to 12OH-JA-Ile, with little conversion to 12COOH-JA-Ile, whereas CYP94C1 catalyzed preferentially carboxy-derivative formation. Metabolic analysis of loss- and gain-of-function plant lines were consistent with in vitro enzymatic properties. cyp94b3 mutants were largely impaired in 12OH-JA-Ile levels upon wounding and to a lesser extent in 12COOH-JA-Ile levels. In contrast, cyp94c1 plants showed wild-type 12OH-JA-Ile accumulation but lost about 60% 12COOH-JA-Ile. cyp94b3cyp94c1 double mutants hyperaccumulated JA-Ile with near abolition of 12COOH-JA-Ile. Distinct JA-Ile oxidation patterns in different plant genotypes were correlated with specific JA-responsive transcript profiles, indicating that JA-Ile oxidation status affects signaling. Interestingly, exaggerated JA-Ile levels were associated with JAZ repressor hyperinduction but did not enhance durably defense gene induction, revealing a novel negative feedback signaling loop. Finally, interfering with CYP94 gene expression affected root growth sensitivity to exogenous jasmonic acid. These results identify CYP94B3/C1-mediated oxidation as a major catabolic route for turning over the JA-Ile hormone.
Introduction
Hormonal control is of crucial importance for plant development and their adaptation to ever changing environmental conditions (1, 2). Molecular genetics combined with biochemical and metabolic approaches have revealed major actors and mechanisms in the biosynthesis, perception, and cellular signaling of several plant hormones. As hormonal responses are orchestrated in a temporal and spatial manner, homeostasis of active compounds requires tight control of both biosynthetic and catabolic processes. Plant hormonal metabolism largely involves cytochrome P450 (CYP)3 enzymes to either activate or inactivate a wide range of compounds (3). CYPs form an important family of heme-containing oxygenases that catalyze a wide variety of monooxygenation/hydroxylation reactions in primary and secondary metabolism (4). CYP-catalyzed enzymatic conversions are, therefore, important regulatory points in hormonal pathways (3), and accordingly, misexpression of CYPs in both biosynthetic and catabolic steps perturbs hormone homeostasis and provokes serious plant development defects. For example, overexpression of CYP707 ABA-8′-hydroxylases depletes ABA pools upon germination, resulting in seed hyperdormancy (5). Severe growth defects were observed when overexpressing brassinolide- or gibberellin-oxidizing CYP enzymes (6, 7).
Jasmonic acid (JA) is a well characterized fatty acid-derived hormone playing prominent roles in the reproduction of plants and in their responses to attacks by insects or microbial pathogens (8–11). Significant advances in the knowledge of JA biosynthesis, perception, and signaling has been recently gained, whereas in contrast, our understanding of the termination of the hormonal signal lags behind (8, 12–14). JA-related compounds are common in plants and are collectively referred to as jasmonates (14). JA is a prohormone that needs to be activated by conjugation to isoleucine by the enzyme JAR1. The resulting JA-isoleucine conjugate (JA-Ile) promotes the assembly of a co-receptor composed of the F-box protein COI1 and members of the family of JAZ proteins (15, 16). JAZ proteins are transcriptional repressors that under low JA-Ile levels prevent transcription of JA-responsive genes (17). Upon JA-Ile binding to the receptor, the JAZ repressors undergo ubiquitination by the SCFCOI1 E3 ubiquitin ligase and are targeted for proteolytic degradation by the 26 S proteasome, resulting in the de-repression of downstream responses (18, 19).
Jasmonate metabolism is strongly stimulated upon mechanical wounding or herbivory in Arabidopsis leaves (20, 21). Consistent with its role as a major regulator of induced defense responses, the levels of JA-Ile are controlled dynamically, suggesting the existence of a modification mechanism(s) that rapidly clears the active hormone from stimulated cells when JA signaling must be switched off. Among the numerous wound-induced jasmonates are the hydroxylated derivatives 12OH-JA (known as tuberonic acid) and 12OH-JA-Ile (21–23). In addition, a more oxidized, dicarboxy derivative, 12COOH-JA-Ile, was described recently (Ref. 21; Fig. 1A). Such oxidative conversions likely reflect critical changes in the biological activity of jasmonates as; for example, tuberonic acid has biological properties that are distinct from those of JA (22). The widespread occurrence of oxidized jasmonates raises the questions of the genes/enzymes involved in their formation and of which conversion routes are predominant in planta. For example, JAR1 is essentially inactive in conjugating 12OH-JA to Ile (24); therefore, 12OH-JA-Ile formation likely occurs by oxidizing JA-Ile. In addition to being an intermediate for further oxidation to 12COOH-JA-Ile, 12OH-JA-Ile may represent an alternative source of unconjugated hydroxylated jasmonates. Enzymatic oxidation of the terminal ω-carbon of an aliphatic chain is a typical feature of the action of a subclass of CYP enzymes, acting usually as fatty acid ω-hydroxylases (25). We describe in this paper the involvement of two Arabidopsis CYP genes of the CYP94 family in the wound-induced accumulation of hydroxylated and di-carboxylated JA-Ile. Two recent reports have identified CYP94B3 as a JA-Ile-12-hydroxylase (26, 27), accounting for most of 12OH-JA-Ile accumulating in damaged leaves. Here we used a combination of genetic, enzymatic, and metabolic approaches to show that CYP94C1, a second wound-responsive member of this subclade, is the main enzyme for JA-Ile carboxylation and that deficiency in the two oxidases in the double mutant cyp94b3cyp94c1 prevents the accumulation of 12COOH-JA-Ile. These results highlight the critical role of two CYP94 enzymes in oxidative turnover of the JA-Ile hormone.
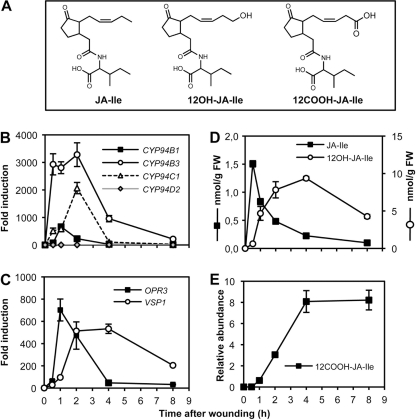
FIGURE 1.
CYP94B1, CYP94B3, and CYP94C1 gene expression precedes the accumulation of 12OH-JA-Ile and 12COOH-JA-Ile upon wounding in Arabidopsis wild-type leaves. A, structures of JA-Ile, 12OH-JA-Ile, and 12COOH-JA-Ile are shown. B and C, real-time PCR analysis of the expression of CYP94, OPR3 and VSP1 genes in mechanically wounded leaves is shown. Expression is represented as -fold induction relative to level at time 0 that was set to 1 for each gene. Expression was normalized with two reference genes. D, shown is the time course of JA-Ile and 12OH-JA-Ile accumulation in wounded leaves. Samples from the same plants as used in B were extracted for jasmonate determination by LC/MS. Note that 12OH-JA-Ile levels are expressed in a specific scale on the right of graph. FW, fresh weight. E, time course of 12COOH-JA-Ile accumulation in wounded leaves is shown. All analyses are the means ± S.E. from three biological samples.
EXPERIMENTAL PROCEDURES
Plant Growth and Treatments
Arabidopsis thaliana genotypes used were all in the Col0 ecotype and were grown under a 12-h/12-h photoperiod in a growth chamber. T-DNA insertion lines used were: coi1-1, jar1-1, cyp94c1-1 (SALK_55455) and cyp94b3-1 (CS302217), all obtained from the Nottingham Arabidopsis Stock Center (NASC). For generating RNAi lines, a CYP94C1 cDNA fragment (nucleotides 451–750) was PCR-amplified and cloned in both orientations in the pFGC5941 vector. Overexpressing lines were generated by cloning the CYP94C1 ORF successively in the Gateway (Invitrogen) vectors pDONR207 and pB7WG2 downstream of the cauliflower mosaic virus 35 S promoter. Binary vector constructs were transferred to Agrobacterium tumefaciens strain GV3101 and introduced by floral dip transformation into WT plants. T3 homozygous plants were used in all experiments. For wounding experiments, between 4 and 6 fully expanded leaves of 6–7-week-old plants were wounded 3 times across the midvein with a hemostat. At increasing time points after mechanical damage, leaf samples were quickly harvested and flash-frozen in liquid nitrogen before storing at −80 °C until use.
For root growth inhibition assays, surface-sterilized seeds were sown on square plates with sucrose-containing (1%) Gamborg B5 medium supplemented or not with JA. Plates were incubated for 48 h at 4 °C and then vertically under standard growth conditions for the remainder of the experiment. Root length was measured 11 days later using the simple neurite tracer tool of Fiji software.
RT-Quantitative PCR Gene Expression Assays
Total RNA was extracted from plant leaves with TRIzol reagent (Molecular Research Center). 1 μg of RNA was reverse-transcribed using the ImProm-II reverse transcription system (Promega, Madison, WI). Real-time PCR was performed on 10 ng of cDNA as described in Berr et al. (28) using a LightCycler 480 II instrument (Roche Applied Science). The housekeeping genes EXP (At4g26410) and GAPDH (At1g13440) were used as internal references. Gene-specific primer sequences used for quantitative PCR are listed in supplemental Table S1.
Recombinant CYP94 Protein Production and Enzymatic Assays
For functional expression of CYP94C1 and CYP94B3 we used a yeast expression system specifically developed for the expression of P450 enzymes. Coding sequences were amplified from an Arabidopsis genomic library and were cloned in the plasmid pYeDP60 and introduced into Saccharomyces cerevisiae WAT11 strain. Yeast has only three cytochromes P450, which are expressed at a negligible level under growth condition used. Yeast cultures were grown and cytochrome P450 expression was induced as described in Pompon (29). After growth, cells were harvested by centrifugation and manually broken with glass beads (0.45 mm diameter) in 50 mm Tris-HCl buffer (pH 7.5) containing 1 mm EDTA and 600 mm sorbitol. The homogenate was centrifuged for 10 min at 10,000 × g. The resulting supernatant was centrifuged for 1 h at 100,000 × g. The pellet consisting of microsomal membranes was resuspended in 50 mm Tris-HCl (pH 7.4), 1 mm EDTA, and 30% (v/v) glycerol with a Potter-Elvehjem homogenizer and stored at −30 °C. The volume of resuspension buffer is proportional to the weight of yeast pellet; microsomes extracted from 6 g of yeast were resuspended in 3 ml of buffer. All procedures for microsomal preparation were carried out at 0–4 °C. The cytochrome P450 content was measured by the method of Omura and Sato (30). For determination of enzymatic activities, the standard assay (0.1 ml) contained yeast microsomes with 0,22 nmol of cytochrome P450 in 20 mm sodium phosphate (pH 7.4), 1 mm NADPH, and substrate (100 μm). The reaction was initiated by the addition of NADPH and was stopped after 20 min by the addition of 150 μl of methanol. The mixture was centrifuged for 1 min at 2000 × g, and the supernatant was cleared by filtration through a 0.22-μm filter. Metabolites in incubation media were directly analyzed by UPLC-MS/MS as described below.
Jasmonate Profiling
Jasmonates were identified and quantified in plant extracts and enzymatic incubations by ultra performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) using MS transitions determined from pure standards (JA-Ile and 12OH-JA-Ile) or published data (21). The synthesis of jasmonic derivatives will be reported as a full paper in a more specialized journal. The relative quantification in samples was achieved by reporting MS peak areas relative to internal standard peak area and mass of biological material. Absolute quantifications of JA-Ile and 12OH-JA-Ile were determined by comparison of sample signal with dose-response curves established with pure compounds. For plant extraction, five volumes of ice-cold 90% methanol containing 9,10-dihydro-JA-Ile as an internal standard were added to one fresh weight (100–150 mg preweighed) of frozen leaf powder in a screw-capped tube containing glass beads. Material was ground twice for 30 s with a Precellys 24 tissue homogenizer (Bertin Technologies, Montigny-Le-Bretonneux, France). Homogenates were cleared by two successive centrifugations at 20,000 × g, and supernatants were saved for UPLC-MS analysis. All analyses were performed using a Waters Quattro Premier XE (Waters, Mildorf, MA) equipped with an electrospray ionization source and coupled to an Acquity UPLC system (Waters). Chromatographic separation was achieved using an Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 μm; Waters) and precolumn. The mobile phase consisted of water (A) and methanol (B), both containing 0.1% formic acid. The run started by 2 min of 95% A, then a linear gradient was applied to reach 100% B at 12 min followed by isocratic run using B during 2 min. Return to initial conditions was achieved in 3 min, with a total run time of 17 min. The column was operated at 35 °C with a flow rate of 0.35 ml/min, injecting 3-μl samples. Nitrogen was used as the drying and nebulizing gas. The nebulizer gas flow was set to 50 liters/h, and the desolvation gas flow was set to 900 liters/h. The interface temperature was set to 400 °C, and the source temperature was set to 135 °C. The capillary voltage was set to 3.2 kV, cone voltage was set to 25 V, the ionization was in positive or negative mode. Low mass and high mass resolution were 14 for the first mass analyzer and 13 for the second, ion energies 1 and 2 were 0.6 V, entrance and exit potential were 2 V, and detector (multiplier) gain was 650 V. Data acquisition and analysis were performed with the MassLynx software. The transitions were, in negative mode, 12-OH-JA-Ile 338 > 130 and 12-COOH-JA-Ile 352 > 130, and in positive mode, JA-Ile 324 > 151. Collision energy was 20 V.
RESULTS
JA-Ile Oxidative Turnover Correlates with CYP94 Gene Expression upon Wounding and Is COI1- and JAR1-dependent
Plant hormone oxidation, essentially through hydroxylation, has been reported to be catalyzed by cytochrome P450 enzymes in several cases (3). We hypothesized that formation of 12OH-JA-Ile and 12COOH-JA-Ile in wounded tissue could possibly be accounted for by enzymes of the CYP94 family, based on two conclusions from our previous work on CYP94C1 (31). First, the CYP94C1 gene was found to be transiently induced by mechanical wounding and by methyl-JA treatment in Arabidopsis leaves. Second, the CYP94C1 enzyme, when prepared as microsomes from recombinant yeast cells, displayed the unique capacity of catalyzing the formation of ω-hydroxy and dicarboxylic derivatives of linear fatty acids, particularly lauric acid (C12:0). The two novel JA-Ile derivatives are also oxidized on the terminal, 12th carbon of the jasmonate moiety (Fig. 1A). By mining genome-wide public expression databases (32, 33), we found that three CYP94 genes, CYP94B1 (At5g63540), CYP94B3 (At3g48520), and CYP94C1 (At2g27690), are wound- and JA-inducible. In addition, three other genes, CYP94B2 (At3g01900), CYP94D1 (At1g34540), and CYP94D2 (At3g56630), belong to the Arabidopsis CYP94 subfamily that branches closest to the CYP86 subfamily of fatty acid hydroxylases (25, 27).
To investigate the possible relevance of CYP94 gene expression to JA-Ile oxidation, we monitored the temporal expression of all six CYP94 genes and the levels of JA-Ile and its two oxidized derivatives in a kinetic analysis of wounded leaves (Fig. 1). CYP94B2 and CYP94D1 expression was undetectable, and CYP94D2 displayed a weak and constant expression in wounded tissues (Fig. 1B). In contrast, CYP94B1, CYP94B3, and CYP94C1 were expressed at very low levels in non-stimulated leaves and exhibited a massive and transient induction concomitant with the JA biosynthetic gene OPR3 whose expression precedes induction of the late wound response gene VSP1 (compare Fig. 1, B and C). CYP94B3 transcripts showed the most ample burst, with high transcript levels maintained between 0.5 to 2 h post-wounding, whereas CYP94B1 and CYP94C1 displayed sharp expression peaks at 1 and 2 h, respectively. Jasmonate conjugates were then quantified by LC-MS/MS in the corresponding plant samples. As expected, JA-Ile and its derivatives were essentially undetectable in unstressed leaves (Fig. 1, D and E). JA-Ile accumulation rose very rapidly to maximal levels at 0.5 h. Its decline was accompanied by a concomitant accumulation of 12OH-JA-Ile that peaked at 4 h before decreasing slowly (Fig. 1D). The levels of 12COOH-JA-Ile were found to increase even later, peaking between 4 and 8 h post-wounding (Fig. 1E), in accordance with a previous report (21). Taken together, these results indicate a clear correlation between the expression of 3 CYP94 genes and the oxidative turnover of JA-Ile upon wounding.
We then examined CYP94 gene expression and accumulation of oxidized JA-Ile derivatives in jar1 and coi1 mutants affected in the JA pathway. The expression of all three CYPs, CYP94B1, CYP94B3, and CYP94C1, was nearly abolished in coi1 (supplemental Fig. S1A, left panels). This was accompanied by a strong reduction in 12OH-JA-Ile and by a near suppression of 12COOH-JA-Ile (supplemental Fig. S1B, left panels) compared with WT plants. Consistent with earlier studies reporting reduced JA-Ile turnover in tobacco or tomato coi1 mutants (23), JA-Ile accumulated to high levels despite of low JA precursor (supplemental Fig. S1B, left panels). In jar1, CYP94 expression was only moderately depressed compared with WT (supplemental Fig. S1A, right panels), but the two oxidized JA-Ile derivatives were at very low levels, probably because of the limited availability of JA-Ile substrate (supplemental Fig. S1B, right panels). Altogether, these results support the simple hypothesis that most 12OH-JA-Ile and 12COOH-JA-Ile directly derive from JAR1-generated JA-Ile through the COI1-dependent expression of CYP94 enzymes.
CYP94C1 and CYP94B3 Specifically Catalyze Two Successive Oxidation Steps of JA-Ile
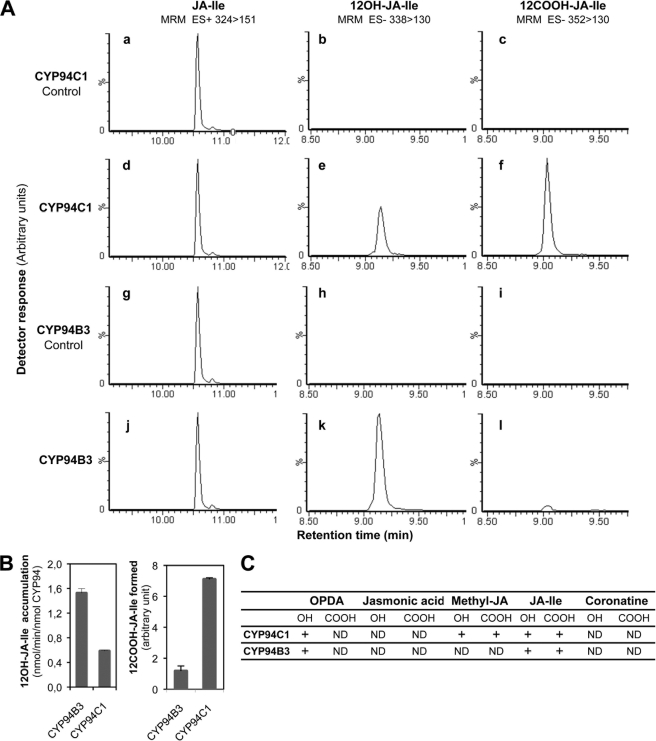
To directly assess the involvement of the candidate CYP94s in the formation of oxidized JA-Ile, biochemical studies of CYP94C1 and CYP94B3 were conducted after heterologous expression in yeast. Fig. 2 shows LC-MS/MS chromatograms obtained after incubation of JA-Ile with microsomes from yeasts expressing CYP94C1 (Fig. 2A, panels a–f) or CYP94B3 (Fig. 2A, panels g–l). After 20 min in presence of NADPH, 12OH-JA-Ile (Fig. 2A, panel e) and 12COOH-JA-Ile (Fig. 2A, panel f) were detected in incubations performed with microsomes containing CYP94C1. NADPH-dependent formation of these two compounds was also catalyzed by CYP94B3 (Fig. 2A, panels k and l). These metabolites were not detected when incubations were performed in the absence of NADPH (Fig. 2A, panels b and c and panels h and i) or with microsomes from yeast transformed with a void plasmid (not shown), confirming involvement of CYP94C1 and CYP94B3 in their formation. Strikingly, CYP94C1 and CYP94B3 generated distinct patterns of JA-Ile oxidation, visualized by different ratios of the three compounds. CYP94C1 was most efficient in catalyzing double oxidation to 12COOH-JA-Ile, but significant levels of 12OH-JA-Ile also accumulated. CYP94B3 produced mainly 12OH-JA-Ile, with minor conversion to 12COOH-JA-Ile. Activities of both enzymes on JA-Ile or 12OHJA-Ile were estimated by separate incubations with either substrate and carbon monoxide spectra quantification of CYP proteins. As shown in Fig. 2B (left panel), when JA-Ile was provided as substrate, 12OH-JA-Ile accumulation was almost 3 times higher with CYP94B3 than with CYP94C1. In contrast, when 12OH-JA-Ile was used as substrate, CYP94C1 was about 6 times more efficient than CYP94B3 in converting 12OH-JA-Ile to 12COOH-JA-Ile (Fig. 2B, right panel). These in vitro results demonstrate a functional specialization of CYP94B3 and CYP94C1 enzymes for each oxidation step.
FIGURE 2.
Catalytic activities of recombinant CYP94C1 and CYP94B3. A, shown are LC chromatograms of residual JA-Ile, 12OH-JA-IIe, and 12COOH-JA-Ile produced in incubations of microsomes of yeast expressing CYP94C1 (panels a–f) or CYP94B3 (panels g–l). Incubations were performed in the absence (panels a–c and g–i) or in presence (panels d–f and j–l) of NADPH. Reaction products were identified by LC-MS/MS based on their retention times and detection in multiple reaction monitoring (MRM, m/z 324 > 124 for JA-Ile, m/z 338 > 130 for 12OH-JA-Ile, and m/z 352 > 130 for 12COOH-JA-Ile). B, shown is a comparison of hydroxylase (left panel) and carboxylase (right panel) activities of CYP94B3 and CYP94C1. Microsomes containing 84 pmol of P450 protein were incubated for 20 min with 100 μm JA-Ile (left panel) or 12OH-JA-Ile (right panel). Metabolite formation was monitored by LC-MS/MS analysis. Values are the means ± S.E. of triplicate determinations. C, microsomes of yeast expressing CYP94C1 or CYP94B3 were incubated with 100 μm concentrations of different jasmonates tested as substrates in the presence of NADPH. Hydroxylated (OH) and carboxylated (COOH) metabolites were searched for by LC-MS/MS analysis. ND, not detected. +, detection of oxidized product. ES+, electrospray positive ionisation; ES−, electrospray negative ionisation.
To further explore substrate specificity of CYP94C1 and CYP94B3, we tested their capacity to oxidize different jasmonates and the bacterial phytotoxin coronatine. As shown in Fig. 2C, unconjugated jasmonic acid was not metabolized, extending a previous study on CYP94B3 (26). However, weak, NADPH-dependent activities were detected with related jasmonates. Methyl-jasmonate was hydroxylated and carboxylated by CYP94C1, whereas it was not metabolized by CYP94B3. The JA precursor OPDA was oxidized by both enzymes to a compound with the expected mass for a hydroxyl derivative. However, no compound with the mass of the corresponding carboxylated derivative was detected. Coronatine is a structural mimic of JA-Ile and a potent agonist of the COI1-JAZ receptor (34, 35). In our conditions, no metabolization of coronatine by CYP94C1 or CYP94B3 was detected.
CYP94C1 Is Involved in Wound-induced Synthesis of 12COOH-JA-Ile in Vivo
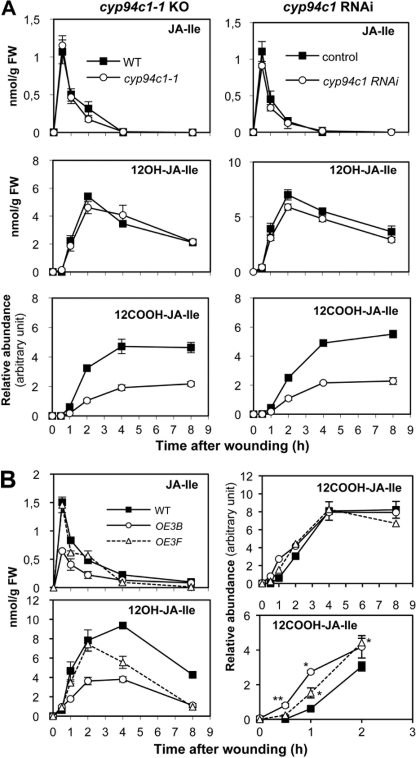
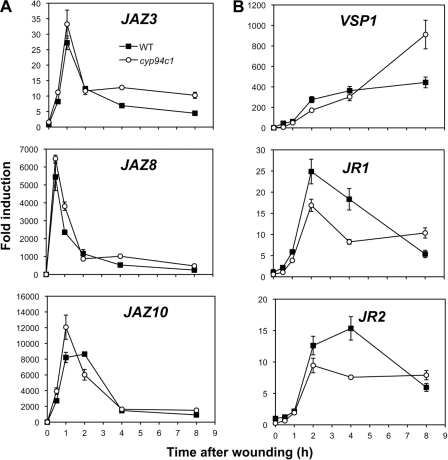
To obtain direct genetic evidence for the involvement of CYP94C1 in JA-Ile oxidation and turnover in planta, we used a T-DNA insertion mutant line (N55455, cyp94c1−1) that is a null allele of the gene (supplemental Fig. S2A). As no second suitable insertion allele was available, we also generated RNAi lines that specifically target CYP94C1 for silencing (supplemental Fig. S2B). JA-Ile and oxidized derivatives were determined for both mutant lines in kinetic wounding experiments. Both cyp94c1−1 (Fig. 3A, left panels) and RNAi plants (Fig. 3A, right panels) showed similar profiles. Essentially identical JA-Ile and 12OH-JA-Ile profiles were recorded in mutant and WT plants. In contrast, accumulation of 12COOH-JA-Ile in CYP94-deficient lines was reduced and never exceeded 40–45% that of control levels between 4 and 8 h post-wounding. We then generated and analyzed plants overexpressing CYP94C1 under the strong cauliflower mosaic virus 35 S promoter. CYP94C1-OE3B (OE3B) and CYP94C1-OE3F (OE3F) lines were selected that represented two levels of transgene expression, which was higher in OE3B than in OE3F line (supplemental Fig. S2C). A reduction in peak accumulation of JA-Ile and 12OH-JA-Ile was marked in OE3B and only detectable at 4–8 h after wounding in OE3F (Fig. 3B, left panels). Consistently, subtle but statistically significant increases in 12COOH-JA-Ile levels relative to WT were measured in CYP94C1-overexpressing lines at early time points (up to at least 2 h post-wounding), but levels were similar to WT in the later response (Fig. 3B, lower panels). These antagonistic effects of CYP94C1 depletion or overexpression on jasmonate levels are consistent with the in vitro catalytic activity and demonstrate that CYP94C1 is responsible for the accumulation of more than half of the 12COOH-JA-Ile detected in wounded leaves. They also suggest the existence of an additional enzyme(s) contributing to the formation of this JA-Ile metabolite in planta.
FIGURE 3.
CYP94C1 is involved in 12COOH-JA-Ile accumulation in wounded leaves. Leaves were harvested at increasing times after wounding and extracted for jasmonate determination. A, shown is the time course of JA-Ile (top panels), 12OH-JA-Ile (middle panels), and 12COOH-JA-Ile (bottom panels) accumulation in wounded leaves of wild-type and T-DNA insertion (cyp94c1−1, left panels) or CYP94C1 RNAi lines (right panels). Similar results were obtained with additional, independent RNAi lines. B, shown is the time course of JA-Ile and oxidized derivative accumulation in wounded leaves of wild-type and CYP94C1-overexpressing lines. The bottom right panel is a close-up of 0–2 h data shown in the top right panel. Data are the means ± S.E. from three biological samples. FW, fresh weight. Asterisks denote a significant difference compared with WT value at same time point using Student's t test (*, p < 0.01; **, p < 0.001).
CYP94B3 and CYP94C1 Cooperate to Control Oxidative JA-Ile Turnover in Response to Wounding
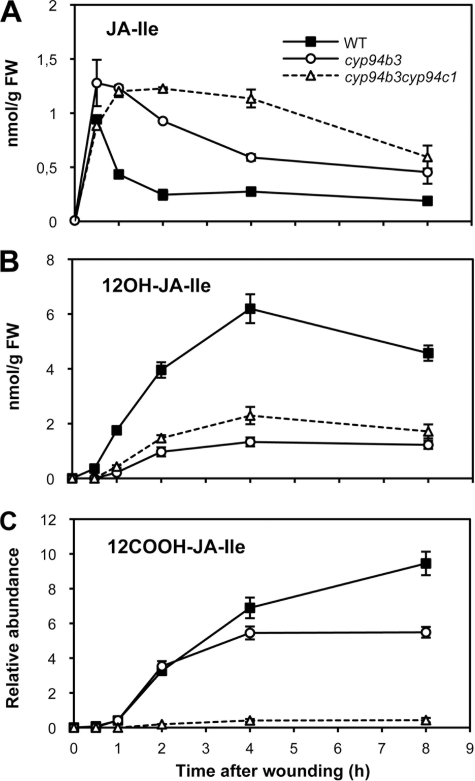
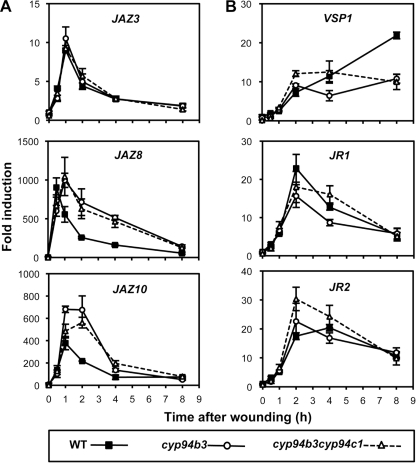
The fact that CYP94B3 and CYP94C1 are both able to catalyze JA-Ile oxidation in vitro led us to address the extent of in vivo redundancy between these two enzymes. We generated a double cyp94b3cyp94c1 mutant and analyzed its wound-induced oxidated JA-Ile profile relative to cyp93b3 mutant and WT plants. The cyp94b3 mutation reduced drastically the levels of 12OH-JA-Ile (Fig. 4B) and consequently increased those of precursor JA-Ile (Fig. 4A), confirming the critical role of CYP94B3 in hydroxylating JA-Ile in planta that was described recently (26, 27). The levels of 12COOH-JA-Ile in cyp94b3 were similar to WT at early time points (up to 2 h post-wounding) but never exceeded 60% of WT between 4 and 8 h. The double mutant presented a remarkable jasmonate profile. cyp94b3cyp94c1 exhibited the most persistent JA-Ile accumulation compared with cyp94b3 and WT plants (Fig. 4A). Double mutant 12OH-JA-Ile levels were intermediate between the low levels in cyp94b3 and WT levels (Fig. 4B). In contrast, the wound-induced increase of 12COOH-JA-Ile was nearly abolished in the double mutant. Taken together, these profiling results indicate that CYP94B3 and CYP94C1 are both involved in oxidative JA-Ile turnover in vivo and that the concerted activities of CYP94B3 and CYP94C1 account for more than 90% 12COOH-JA-Ile accumulation.
FIGURE 4.
CYP94B3 and CYP94C1 cooperate in vivo to control oxidative JA-Ile turnover. Leaves of WT, cyp94b3-1, or cyp94b3cyp94c1 plants that were mechanically wounded and harvested at increasing time points were extracted for jasmonate determination. A, JA-Ile levels. B, 12OH-JA-Ile levels. FW, fresh weight. C, 12COOH-JA-Ile levels. Data are the means ± S.E. from three biological samples.
Impact of CYP94C1 and CYP94B3 Deregulation on Jasmonate-dependent Responses
cyp94b3 plants display elevated wound-induced levels of the hormonally active JA-Ile because of impaired oxidation to less active 12OH-JA-Ile (this study and Koo et al. (27)). This metabolic alteration was reportedly associated with enhanced expression of early JA-responsive genes. As cyp94b3cyp94c1 mutants maintain near maximal JA-Ile levels for a longer time period than cyp94b3 plants (Fig. 4A), we anticipated that hyperinduction of JA-responsive genes may be even more pronounced in the double mutant. Fig. 5A shows that expression of the early response genes JAZ8 and JAZ10 is persistently enhanced in cyp94b3 and cyp94b3cyp94c1 mutants compared with WT plants, whereas JAZ3 expression profile was the same in all three genotypes. When defense genes with slower dynamics (36) were analyzed, a different picture emerged (Fig. 5B). No gain in VSP1, JR1, or JR2 expression was found associated with high JA-Ile persistence in cyp94b3 plants. In the double mutant, a transiently enhanced expression was observed for VSP1 and JR2 2 h after wounding. We similarly analyzed cyp94c1−1 single mutant plants that display reduced 12COOH-JA-Ile levels but near WT JA-Ile and 12OH-JA-Ile contents (Fig. 3A). Expression of the three JAZ genes analyzed was slightly enhanced at early time points (Fig. 6A), but amplitude of defense gene induction was reduced in cyp94c1−1, with the exception of VSP1 expression, which was boosted 8 h post-wounding (Fig. 6B). Very similar trends were observed in an independent experiment with cyp94c1 RNAi plants (supplemental Fig. S3). In all these experiments, the effects of cyp94 inactivation appear at a later time point(s) for VSP1 transcript levels (8 h) than for those of JR1/2 (2–4 h). Therefore, preventing JA-Ile oxidation to various extents in these mutants has differential impacts on the induction of jasmonate-responsive genes upon wounding.
FIGURE 5.
Impaired JA-Ile oxidation in single or double CYP94-deficient plants differentially affects jasmonate-responsive genes upon wounding. Leaves of WT, cyp94b3-1, or cyp94b3cyp94c1 plants (same plant set as used in Fig. 4) were mechanically wounded and harvested for RNA extraction. Gene expression time courses were established by real time PCR. Expression is represented as -fold induction relative to the level at time 0 in WT plants, which was set to 1 for each gene. A, shown are early response JAZ genes. B, shown are wound- and jasmonate-responsive defense genes. Data are the means ± S.E. from three replicate samples.
FIGURE 6.
JA-responsive gene expression in CYP94C1-deficient plants upon wounding. Leaves of WT and cyp94c1−1 plants (same plant set as used in Fig. 3A) were mechanically wounded and harvested for RNA extraction. Gene expression time courses were established by real time PCR. Expression is represented as -fold induction relative to level at time 0 in WT plants that was set to 1 for each gene. taw, time after wounding. A, early responsive JAZ genes are shown. B, wound- and jasmonate-responsive defense genes are shown. Data are the means ± S.E. from three replicate samples.
To begin evaluating physiological consequences of JA-Ile oxidation on jasmonate sensitivity, we conducted root growth inhibition assays on the different CYP94-modified plant lines generated in the study. Single mutations in the two CYP94 genes had only marginal effects on root growth. In contrast, double cyp94b3cyp94c1 mutant was more sensitive to exogenous JA than WT, particularly at 10 μm JA (supplemental Fig. S4). Conversely, roots of the most CYP94C1-overexpressing line, CYP94C1-OE3B, displayed an enhanced resistance to JA by growing longer roots. These data indicate that CYP94C1 acts in concert with CYP94B3 to modulate sensitivity to exogenous JA.
DISCUSSION
In a previous report we described CYP94C1 as a fatty acid hydroxylase that generates dicarboxylic acids from a range of fatty acids, lauric acid (C12:0) being one of the preferred in vitro substrates (31). Dicarboxylic fatty acids are abundant cutin monomers in Arabidopsis; however, cyp94c1 mutants displayed no altered cutin composition.4 The rapid and transient wound induction of CYP94C1 and its co-regulation with the jasmonate pathway argued in favor of its involvement in a biological process not related to cutin synthesis. The description of two hydroxy and carboxy derivatives of JA-Ile in wounded tissues (21) that are also oxidized on the carbon 12 of the JA moiety made these compounds strong candidates for being in vivo products of CYP94C1 activity. During the course of this work, two reports appeared that provided genetic evidence for CYP94B3 being a JA-Ile-12-hydroxylase that contributes to partial inactivation of the JA-Ile hormone (26, 27). We describe here novel biochemical characterization of CYP94C1 and CYP94B3 and provide genetic evidence that these two enzymes are major players in the two-step oxidative turnover of JA-Ile.
Among the six CYP94 genes in the Arabidopsis genome, four were found expressed in mature leaves, CYP94D2 transcript levels being unaffected by wounding. In contrast, as predicted by microarray data and previous analysis (27), CYP94B1, CYP94B3, and CYP94C1 were strongly and transiently induced by mechanical wounding with typical kinetics of early responsive genes in the JA pathway. This gene expression window enclosed the sharp peak accumulation (0.5–1 h post-wounding) of JA-Ile, the putative substrate, and preceded the sequential accumulation of potential reaction products 12OH-JA-Ile and 12COOH-JA-Ile. The use of jar1 and coi1 mutants provided further evidence for a link with the JA pathway. jar1 maintained most CYP94 expression, but its JA-Ile deficiency is the likely cause of the near absence of 12OH-JA-Ile and 12COOH-JA-Ile accumulation. In contrast, the loss of CYP94 expression in coi1 was correlated with a severe reduction in 12OH-JA-Ile and 12COOH-JA-Ile levels, consistent with the reduced capacity of coi1 plants to turnover JA-Ile that was reported in Solanaceous species (23, 37). The remaining 12OH-JA-Ile in coi1 plants may originate from residual CYP94B3 expression (supplemental Fig. S1A, left panels) or alternatively from an unknown, COI1-independent enzyme(s). This correlative evidence strengthens the hypothesis of a causal link between CYP94 genes and oxidative metabolization of JA-Ile.
We used our long-standing established procedures to produce yeast microsomes containing active CYP94 enzymes and investigated their activity on jasmonates. Consistent with its dual activity on linear fatty acids (31), CYP94C1 catalyzed two successive oxidations on JA-Ile, with partial conversion of 12OH-JA-Ile to the carboxy-derivative. We established that CYP94B3 was also able to produce 12COOH-JA-Ile, although to a lesser extent than CYP94C1. This property was missed in previous reports (26, 27), probably because of poor activity of enzymatic preparations. In their work, Koo et al. (27) reported less than 1% of JA-Ile metabolization in their enzymatic assay, meaning that 12OH-JA-Ile was in competition with ∼99% residual JA-Ile for further metabolism. Our results indicate that the two enzymes are not redundant in vitro, as they produce a distinct blend of oxidized compounds. The relative activities of CYP94C1 and CYP94B3 for each oxidation reaction were determined and illustrated the cooperative nature of the two enzymes, although the knowledge of the precise ratio between hydroxy and carboxy derivatives of JA-Ile awaits the availability of the latter pure compound in sufficient amounts. Alternatively, the ancient evolutionary origin of the CYP94 family and high conservation of CYP94C1 and CYP94B3 homologs in plant species (38) could be indicative of specific roles for each enzyme. The substrate specificity of CYP94C1 and CYP94B3 was further addressed in incubations with different jasmonates. Their total lack of activity on free JA indicates that 12OH-JA, which can be abundant in plant tissues (22), must be formed by another enzyme(s). In contrast, the detection of oxidized derivatives of methyl jasmonate or OPDA is intriguing, although the physiological relevance of this finding is unclear. Despite high levels of OPDA, we could not detect hydroxy-OPDA in plant extracts, which is consistent with the distinct cellular localizations of OPDA in chloroplasts and of CYP94 enzymes presumably in the endoplasmic reticulum. Coronatine is a structural mimic of JA-Ile that is used by Pseudomonas bacteria as a virulence factor to promote disease by inducing jasmonate responses that suppress salicylic acid defenses (35, 39). We show that CYP94C1 and CYP94B3, both highly active on JA-Ile, did not evolve the capacity to metabolize coronatine. In addition to its very high affinity for the COI1-JAZ receptor, the potency of this phytotoxin may be strengthened by escaping inactivation by plant enzymes.
It was of particular interest to investigate how the similar yet distinct enzymatic activities recorded for recombinant CYP94C1 and CYP94B3 contribute to the wound-induced accumulation of oxidized JA-Ile derivatives in planta. In the different plant genotypes analyzed, jasmonate profiles were consistent with in vitro data, although we cannot exclude that CYP94C1 and CYP94B3 metabolize additional substrates in planta. Depleting CYP94C1 expression by T-DNA insertion in the cyp94c1−1 line or by silencing the gene in RNAi lines generated similar changes in jasmonate pools. The persistence of about 40% 12COOH-JA-Ile levels in these lines points to the existence of at least a second enzyme involved in its formation. The WT levels of 12OH-JA-Ile in CYP94C1-deficient plants suggest that this compound is equally a substrate and a product of CYP94C1. CYP94C1-overexpressing lines presented opposite changes that were correlated with transgene expression. The low gain in 12COOH-JA-Ile levels observed at early time points may be limited by substrate availability or by further metabolization of 12COOH-JA-Ile. Together, this set of results shows that CYP94C1 is the major enzyme forming 12COOH-JA-Ile from JA-Ile upon wounding and that this enzyme does not affect the apparent size of the 12OH-JA-Ile pool. It was reported previously that CYP94B3 deficiency severely compromises the formation of 12OH-JA-Ile, with a concomitant hyperaccumulation of JA-Ile (27). We extended these studies, and genetic analysis identified CYP94B3 as a second enzyme contributing to in planta 12COOH-JA-Ile accumulation. Combining both mutations confirmed that the concerted action of CYP94C1 and CYP94B3 is required to achieve WT levels of 12COOH-JA-Ile. The slight elevation in 12OH-JA-Ile in the double mutant compared with cyp94b3 single mutant likely reflects the proportion of this compound normally oxidized by CYP94C1. Because they are strongly impaired in JA-Ile turnover, cyp94b3cyp94c1 plants hyperaccumulate the hormone even longer than cyp94b3 single mutants (27). These findings demonstrate that CYP94B3/C1-mediated oxidation is a major catabolic route for clearing JA-Ile and that these conversions shape the transient nature of the JA-Ile peak after leaf wounding. CYP94B1, which is coregulated with the two CYP94 genes described in this study, awaits characterization and may account for the residual JA-Ile oxidizing activity detected in wounded cyp94b3cyp94c1 plants. To our knowledge this is the first case of catabolism of a plant hormone by the successive conversion of a methyl to hydroxy and carboxy derivatives by single enzymes, as CYP-mediated inactivation of other hormones occurs by hydroxylation or epoxidation. In plants, many CYPs belonging mainly to families CYP86, CYP94, CYP704, CYP92, and CYP78 are able to hydroxylate a methyl group. So far, only CYP94C1 and CYP94B3 together with CYP94A5 from tobacco (40) share the unique capacity of catalyzing the complete oxidation of a methyl to a carboxyl group in aliphatic chains, which may proceed through the formation of a gem-diol (41).
It has been demonstrated that the jasmonate pathway is under strong stereochemical control along biosynthetic steps, with a cis configuration introduced into OPDA by allene oxide cyclase (42), which is maintained through the natural (+)-7-iso-JA stereoisomer (14). The highly stereospecific JAR1 then exclusively produces the (+)-7-iso-JA-Ile epimer (43), that is a ligand of the COI1-JAZ coreceptor (15, 34). The rapid jasmonate extraction and UPLC-MS analysis procedures used here for fresh, underivatized plant samples analysis are likely to preserve the endogenous stereochemistry of jasmonates better than more complex GC-MS methods with derivatized samples that were reported previously (43, 44). Therefore, it is likely that the oxidized JA-Ile derivatives measured here are in their native configuration. To formally answer this question, specific studies are required to determine if CYP94B3- and CYP94C1-catalyzed oxidation of JA-Ile maintains the cis stereochemistry or if a change in configuration at oxidation steps could represent a novel regulatory level in jasmonate catabolism. What could be the physiological function of a two-step oxidation of JA-Ile ? The formation of a carboxyl group may prepare the compound for further modifications, like shortening of the side chain by β-oxidation or conjugation. It was established that the introduction of an hydroxyl group on the terminal carbon of the JA moiety by CYP94B3 corresponds to a partial switch-off of biological activity in COI1-JAZ binding assays (27). Accordingly, ectopic CYP94B3 overexpression increases the oxidation status of JA-Ile and recapitulates JA-insensitive phenotypes. We extended this finding and showed that interfering with CYP94C1 expression alone or in combination with CYP94B3 inactivation alters the sensitivity of root growth to exogenous JA. This indicates that CYP94C1-mediated JA-Ile catabolism also impacts JA response physiology. The availability of cyp94c1 and cyp94b3 single and double mutants that display specific oxidized jasmonate profiles also allowed us to monitor the behavior of JA-regulated genes when catabolism of JA-Ile is gradually impaired. In this respect, the high JA-Ile content in cyp94b3 has been previously associated with enhanced expression of wound-responsive genes (27). Our data show that this effect is limited to early genes and that high JA-Ile content has no major enhancing effect on defense responses. The slight and transient enhancement in defense transcripts (see Fig. 5, VSP1 and JR2) may be reflective of an initial positive impact of persistent high JA-Ile levels, but this increased output of the pathway is rapidly dampened by hyperinduction of some, but not all, JAZ genes. The second oxidation step leading to 12COOH-JA-Ile may be needed for full hormonal inactivation by loss of affinity for the receptor. In CYP94C1-deficient plants accumulating less 12COOH-JA-Ile despite near WT JA-Ile and 12OHJA-Ile levels, JAZ genes were slightly overinduced, whereas peak accumulation of defense transcripts was depressed. These alterations suggest that 12COOH-JA-Ile levels are not neutral in terms of jasmonate signaling, and therefore, its formation may not serve only catabolic purposes. The correlation between hyperinduction of JAZ genes and defense attenuation in CYP94-deficient plants points to the existence of a negative feed-back loop exerted by JAZ repressor proteins on transcription of target genes under high JA-Ile levels. In support to this view, JAZ10 that is hyperinduced in CYP94-deficient lines, produces truncated, proteolysis-resistant splice variants (for example JAZ10.4) that mediate target gene repression (45). Further investigations are needed to determine if JAZ hyperinduction under high JA-Ile may also limit the intensity of other responses. This could for example explain why root growth inhibition phenotypes observed in cyp94 mutants are relatively modest (supplemental Fig. S4). One possibility to terminate signaling would be that JA-Ile catabolites and newly synthesized JAZ proteins desensitize the receptor to render high JA-Ile levels temporarily inactive. Complex interactions between multiple JAZ proteins and additional partners appear as crucial for proper regulation of later responses (46). Such a tight control of the jasmonate signaling pathway may complicate the biotechnological strategy aiming at enhancing JA-mediated defense and herbivore resistance by reducing JA-Ile catabolism. It would also illustrate how plant evolution has optimized hormone dynamics and signaling circuits to allow maximal defensive output while minimizing resource allocation costs. Clearly, additional research is needed to understand the physiological meaning of 12COOH-JA-Ile formation and its cellular fate at later phases of jasmonate signaling.
Supplementary Material
Acknowledgments
We are grateful to Michel Legrand for critical reading of the manuscript. We thank Dimitri Heintz for initial jasmonate analysis, Malek Alioua for help with real-time PCR and DNA sequencing, the gardener team for producing the numerous plants used in this study, and the Nottingham Arabidopsis Stock Center for providing seeds of T-DNA insertion lines. The UPLC-MS/MS system was cofinanced by the CNRS, the Université de Strasbourg, the Région Alsace, the Institut National de la Recherche Agronomique, and the Tepral Company.

This article contains supplemental Figs. S1–S4 and Table S1.
F. Pinot, unpublished information.
- CYP
- cytochrome P450
- JA
- Jasmonic acid
- UPLC
- ultra performance liquid chromatography
- OPDA
- 12-oxo-phytodienoic acid.
REFERENCES
- 1. Depuydt S., Hardtke C. S. (2011) Hormone signaling cross-talk in plant growth regulation. Curr. Biol. 21, R365–R3673 [DOI] [PubMed] [Google Scholar]
- 2. Santner A., Estelle M. (2009) Recent advances and emerging trends in plant hormone signaling. Nature 459, 1071–1078 [DOI] [PubMed] [Google Scholar]
- 3. Mizutani M., Ohta D. (2010) Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315 [DOI] [PubMed] [Google Scholar]
- 4. Schuler M. A., Werck-Reichhart D. (2003) Functional genomics of P450s. Annu. Rev. Plant Biol. 54, 629–667 [DOI] [PubMed] [Google Scholar]
- 5. Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., Mizutani M. (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turk E. M., Fujioka S., Seto H., Shimada Y., Takatsuto S., Yoshida S., Wang H., Torres Q. I., Ward J. M., Murthy G., Zhang J., Walker J. C., Neff M. M. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 42, 23–34 [DOI] [PubMed] [Google Scholar]
- 7. Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., Hanada A., Zhou H., Wang R., Li P., Zhu X., Mander L. N., Kamiya Y., Yamaguchi S., He Z. (2006) Elongated Uppermost Internode encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Browse J. (2009) Jasmonate passes muster. A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 [DOI] [PubMed] [Google Scholar]
- 9. Howe G. A., Jander G. (2008) Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 [DOI] [PubMed] [Google Scholar]
- 10. Spoel S. H., Dong X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351 [DOI] [PubMed] [Google Scholar]
- 11. Wu J., Baldwin I. T. (2010) New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24 [DOI] [PubMed] [Google Scholar]
- 12. Chung H. S., Niu Y., Browse J., Howe G. A. (2009) Top hits in contemporary JAZ. An update on jasmonate signaling. Phytochemistry 70, 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fonseca S., Chico J. M., Solano R. (2009) The jasmonate pathway. The ligand, the receptor, and the core signaling module. Curr. Opin. Plant Biol. 12, 539–547 [DOI] [PubMed] [Google Scholar]
- 14. Wasternack C., Kombrink E. (2010) Jasmonates. Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5, 63–77 [DOI] [PubMed] [Google Scholar]
- 15. Sheard L. B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T. R., Kobayashi Y., Hsu F. F., Sharon M., Browse J., He S. Y., Rizo J., Howe G. A., Zheng N. (2010) Jasmonate perception by inositol phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009) The Arabidopsis coronatine-insensitive 1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E. E. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chini A., Fonseca S., Fernández G., Adie B., Chico J. M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F. M., Ponce M. R., Micol J. L., Solano R. (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, 666–671 [DOI] [PubMed] [Google Scholar]
- 19. Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S. Y., Howe G. A., Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 448, 661–665 [DOI] [PubMed] [Google Scholar]
- 20. Chung H. S., Koo A. J., Gao X., Jayanty S., Thines B., Jones A. D., Howe G. A. (2008) Regulation and function of Arabidopsis jasmonate ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glauser G., Grata E., Dubugnon L., Rudaz S., Farmer E. E., Wolfender J. L. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 283, 16400–16407 [DOI] [PubMed] [Google Scholar]
- 22. Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127 [DOI] [PubMed] [Google Scholar]
- 23. Paschold A., Bonaventure G., Kant M. R., Baldwin I. T. (2008) Jasmonate perception regulates jasmonate biosynthesis and JA-Ile metabolism. The case of COI1 in Nicotiana attenuata. Plant Cell Physiol. 49, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 24. Guranowski A., Miersch O., Staswick P. E., Suza W., Wasternack C. (2007) Substrate specificity and products of side reactions catalyzed by jasmonate-amino acid synthetase (JAR1). FEBS Lett. 581, 815–820 [DOI] [PubMed] [Google Scholar]
- 25. Pinot F., Beisson F. (2011) Cytochrome P450 metabolizing fatty acids in plants. Characterization and physiological roles. FEBS J. 278, 195–205 [DOI] [PubMed] [Google Scholar]
- 26. Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 27. Koo A. J., Cooke T. F., Howe G. A. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berr A., McCallum E. J., Alioua A., Heintz D., Heitz T., Shen W. H. (2010) Arabidopsis histone methyltransferase set domain group 8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pompon D., Louerat B., Bronine A., Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 30. Omura T., Sato R. (1964) The Carbon Monoxide Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 31. Kandel S., Sauveplane V., Compagnon V., Franke R., Millet Y., Schreiber L., Werck-Reichhart D., Pinot F. (2007) Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS J. 274, 5116–5127 [DOI] [PubMed] [Google Scholar]
- 32. Ehlting J., Sauveplane V., Olry A., Ginglinger J. F., Provart N. J., Werck-Reichhart D. (2008) An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009) (+)-7-iso-jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 [DOI] [PubMed] [Google Scholar]
- 35. Katsir L., Schilmiller A. L., Staswick P. E., He S. Y., Howe G. A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. León J., Rojo E., Titarenko E., Sánchez-Serrano J. J. (1998) Jasmonic acid-dependent and -independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol. Gen. Genet. 258, 412–419 [DOI] [PubMed] [Google Scholar]
- 37. VanDoorn A., Bonaventure G., Schmidt D. D., Baldwin I. T. (2011) Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum. COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 190, 640–652 [DOI] [PubMed] [Google Scholar]
- 38. Nelson D., Werck-Reichhart D. (2011) A P450-centric view of plant evolution. Plant J. 66, 194–211 [DOI] [PubMed] [Google Scholar]
- 39. Brooks D. M., Bender C. L., Kunkel B. N. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6, 629–639 [DOI] [PubMed] [Google Scholar]
- 40. Le Bouquin R., Skrabs M., Kahn R., Benveniste I., Salaün J. P., Schreiber L., Durst F., Pinot F. (2001) CYP94A5, a new cytochrome P450 from Nicotiana tabacum, is able to catalyze the oxidation of fatty acids to the ω-alcohol and to the corresponding diacid. Eur. J. Biochem. 268, 3083–3090 [DOI] [PubMed] [Google Scholar]
- 41. Scheller U., Zimmer T., Becher D., Schauer F., Schunck W. H. (1998) Oxygenation cascade in conversion of n-alkanes to α,ω-dioic acids catalyzed by cytochrome P450 52A3. J. Biol. Chem. 273, 32528–32534 [DOI] [PubMed] [Google Scholar]
- 42. Ziegler J., Stenzel I., Hause B., Maucher H., Hamberg M., Grimm R., Ganal M., Wasternack C. (2000) Molecular cloning of allene oxide cyclase. The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J. Biol. Chem. 275, 19132–19138 [DOI] [PubMed] [Google Scholar]
- 43. Suza W. P., Rowe M. L., Hamberg M., Staswick P. E. (2010) A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-l-isoleucine in wounded leaves. Planta 231, 717–728 [DOI] [PubMed] [Google Scholar]
- 44. Wasternack C., Xie D. (2010) The genuine ligand of a jasmonic acid receptor. Improved analysis of jasmonates is now required. Plant Signal. Behav. 5, 337–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chung H. S., Cooke T. F., Depew C. L., Patel L. C., Ogawa N., Kobayashi Y., Howe G. A. (2010) Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pauwels L., Goossens A. (2011) The JAZ proteins. A crucial interface in the Jasmonate signaling cascade. Plant Cell 23, 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.