Background: Ultraviolet (UV) light causes genetic instability, suggesting chromosome fragmentation.
Results: UV light induces chromosomal fragmentation, which is independent of excision of UV lesions but requires replication forks, homologous strand exchange, and Holliday junction resolution.
Conclusion: Fragmentation is a result of replication fork reversal and breakage.
Significance: Breakage is the first step in rescuing stalled replication forks.
Keywords: Chromosomes, DNA Damage, DNA Enzymes, DNA Helicase, DNA Nucleotide Excision Repair, DNA Repair, DNA Replication, Double Strand Breaks, Ultraviolet Light
Abstract
Ultraviolet (UV) irradiation is not known to induce chromosomal fragmentation in sublethal doses, and yet UV irradiation causes genetic instability and cancer, suggesting that chromosomes are fragmented. Here we show that UV irradiation induces fragmentation in sublethal doses, but the broken chromosomes are repaired or degraded by RecBCD; therefore, to observe full fragmentation, RecBCD enzyme needs to be inactivated. Using quantitative pulsed field gel electrophoresis and sensitive DNA synthesis measurements, we investigated the mechanisms of UV radiation-induced chromosomal fragmentation in recBC mutants, comparing five existing models of DNA damage-induced fragmentation. We found that fragmentation depends on active DNA synthesis before, but not after, UV irradiation. At low UV irradiation doses, fragmentation does not need excision repair or daughter strand gap repair. Fragmentation absolutely depends on both RecA-catalyzed homologous strand exchange and RuvABC-catalyzed Holliday junction resolution. Thus, chromosomes fragment when replication forks stall at UV lesions and regress, generating Holliday junctions. Remarkably, cells specifically utilize fork breakage to rescue stalled replication and avoid lethality.
Introduction
UV radiation-induced DNA damage causes genetic instability in bacteria (1, 2), lower eukaryotes (3, 4), and higher eukaryotes (5, 6) and is the leading cause of skin cancer in humans (7, 8). However, the mechanisms linking UV radiation-induced DNA lesions to genetic instability are unclear. UV irradiation causes formation of pyrimidine dimers in DNA, but these lesions are efficiently mended by nucleotide excision repair, represented in Escherichia coli by the UvrABC excinuclease (9). At the same time, UV irradiation also inhibits DNA replication, which resumes after a lag period (10–12). Encounters of replication forks with unrepaired pyrimidine dimers lead to several complex phenomena, explained by a variety of models, that include replication fork inhibition (13–15), formation of daughter strand gaps (16, 17), and double strand breaks (18–20). However, the current consensus on the processing and restart of stalled replication forks (12, 21, 22) does not explain UV radiation-induced genetic instability, leaving our understanding of UV damage processing incomplete.
Chromosomal fragmentation kills cells of any type if the double strand breaks are not repaired (23–25). Repair of fragmented chromosomes induces genetic instability (26–28). In fact, by the magnitude of these effects, chromosomal fragmentation is the most consequential of all DNA lesions and an important contributor to cancerous transformation (29, 30). Endogenous chromosomal fragmentation is caused by a variety of mechanisms (31, 32), including contamination of the DNA precursor pools (33, 34) and malfunctioning of the replisome (35, 36), but whether exogenous one-strand DNA lesions, like those induced by UV irradiation, cause chromosomal fragmentation in biologically relevant (sublethal) doses is still not settled (12, 18, 21, 37, 38).
We hypothesized that sublethal UV irradiation doses trigger genetic instability by inducing chromosomal fragmentation, which avoided previous detection in wild type cells (18, 19) because of efficient double strand break repair or linear DNA degradation. Through our interest in low level spontaneous chromosomal fragmentation induced by endogenous DNA damage and following the lead of Michel et al. (39), we developed a sensitive technique based on pulsed field gel electrophoresis to detect and quantify chromosomal fragmentation in E. coli (31, 40). Use of recBC mutants allows us to block the recombinational repair of double strand breaks on the one hand and linear DNA degradation on the other, thus dramatically increasing the sensitivity of our measurements. When we used our sensitive assay to measure chromosome instability in E. coli after UV irradiation, we found highly fragmented chromosomes. Genetic analysis of this fragmentation in combination with the sensitive measurements of the DNA synthesis rate ruled out all the current models of DNA damage-induced fragmentation except the one in which stalled replication forks actively regress to form Holliday junctions, which are then resolved to break the forks.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The E. coli strains (all derivatives of K12) used in this study are described in supplemental Table S1. All strains were grown in LB (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl/liter of broth, pH to 7.4 with 250 μl of 4 m NaOH; LB agar contained 15 g of agar/liter of LB broth) at 28 °C unless stated otherwise. When required, antibiotics were added to the following final concentrations: ampicillin, 100 μg/ml; spectinomycin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 10 or 30 μg/ml; and tetracycline, 10 μg/ml. Alleles were moved among the strains by P1 transduction as described (41). Various mutants were confirmed by Southern hybridization, polymerase chain reaction (PCR), or functional analysis. The pGB-ruvABC1 plasmid is pGB2 expressing ruvABC+ genes (B. Michel). The pGB-ruvAB plasmid is a derivative of pGB-ruvABC1 from which we deleted the ruvC gene, so the plasmid harbors only the ruvAB+ genes.
Chromosomal Fragmentation
In our protocol, quantification of chromosomal fragments is facilitated by 32P labeling of DNA; other 32P-labeled species, such as RNA, polyphosphates, LPS, and phospholipids are removed either during plug preparation or during the electrophoretic run (42). The amount of 32P label that we use is not enough to affect the viability of even the most DNA damage-sensitive mutants, like uvrA recA double mutants. To radiolabel the chromosomal DNA, the overnight-grown cultures were diluted to an initial A600 of 0.03–0.05 in fresh LB medium (with antibiotics when required) containing 2–4 μCi/ml [32P]orthophosphoric acid (MP Biochemicals) and grown aerobically at 28 °C. Once the cultures reached an A600 of 0.3–0.4, the suspensions were transferred to sterile polypropylene centrifuge tubes, and cells were harvested at 6,000 rpm in a Sorvall RC 5B centrifuge (4,000 × g) operating at room temperature. The cell pellets were suspended in volumes of sterile 1% NaCl containing 0.01% Triton X-100 (Sigma) to yield an absorbance of approximately 0.600. The irradiation was performed at 28 °C in a dark room under yellow lamps (F15T8-GO, General Electric) to avoid photoreactivation. The labeled cells (in a volume of 1.5 ml) were spread in a thin, uniform layer (∼0.25 mm) on the back surface of a sterile 100 mm × 15 mm polystyrene Petri plate with a ridge (Fisher Scientific), transferred to a Hoefer UVC 500 UV cross-linker, and irradiated with the indicated UV irradiation dosages. Following irradiation, the cultures from the plate were collected in sterile glass tubes and processed as required. In experiments requiring large culture volumes, multiple plates were irradiated, and the UV radiation-treated cells were pooled together before processing. The general processing involved diluting the irradiated cells 1:1 with a sterile no-salt 2× LB solution and growing them in the dark for various periods of time at 28, 37, or 42 °C as described in various experiments. After UV irradiation treatments and incubations, the cells were collected and made into agarose plugs as described below.
Pulsed Field Gel Electrophoresis
The optical densities of the cultures were normalized to 0.350 with sterile LB, and cells from 1.5 ml of cultures were harvested to make plugs for pulsed field gel electrophoresis. The cell pellets were resuspended in 1 ml of sterile TE buffer (25 mm Tris-HCl, 1 mm EDTA, pH 7.4), and cells were repelleted and resuspended in 60 μl of TE buffer. To make the agarose plugs, 5 μl of 5 mg/ml proteinase K (Invitrogen or Roche Applied Science; final concentration in plugs, 200 μg/ml) and 65 μl of molten agarose in lysis buffer (1.2% agarose in 1% lauroylsarcosine, 50 mm Tris-HCl, 25 mm EDTA, pH 8.0) were mixed with the cells, and the suspensions were transferred to plug molds (Bio-Rad). The solidified agarose plugs were submerged in 1 ml of the lysis buffer (1% lauroylsarcosine, 50 mm Tris-HCl, 25 mm EDTA, pH 8.0) and incubated overnight at 60 °C. The plugs were loaded onto a 1% agarose gel in 0.5% Tris borate-EDTA buffer and electrophoresed at 12 °C in a Bio-Rad CHEF-DRII pulsed field gel electrophoresis system operating at 6 V/cm for 24–26 h with initial and final switch times of 60 and 120 s, respectively. Following the electrophoresis, the gel was dried under a vacuum, exposed to a phosphorimaging screen and scanned by an FLA-3000 series fluorescent image analyzer (FujiFilm). The data were processed using Image Gauge version 3.41 software (FujiFilm). The percentage of chromosomal fragmentation was calculated as the signal in the lane below the well divided by the combined signal of the lane plus the well and multiplied by 100.
Quantitative UV Irradiation Sensitivity
Strains were grown, processed, and exposed to various doses of UV irradiation essentially as described for the chromosomal fragmentation assay with the exception that the cultures were not radioactively labeled. Following UV irradiation treatments, samples of exposed cultures were serially diluted into 1% sterile NaCl and spotted by 5 μl on LB agar. The plates were incubated at 37 °C for 12 h, and colonies were counted under a stereomicroscope while still small.
Rate of DNA Synthesis
To measure the rate of DNA synthesis, the strains were grown in LB medium at the indicated temperatures with or without treatments as detailed under “Results.” At appropriate times, 200 μl of the culture to be tested was mixed with an equal volume of prewarmed LB containing 1 μCi of [methyl-3H]thymidine (MP Biomedicals) and 0.4 μg of thymine. The reaction was incubated at 37 °C for either 1 or 5 min (depending on the experiment) after which the incorporation of thymidine was stopped by the addition of 5 ml of freshly made ice-cold 5% trichloroacetic acid (TCA). The TCA-killed cells were kept on ice and filtered through a 1.6-μm glass fiber filter (Fisher Scientific) using a vacuum manifold. The filters were sequentially washed with 5 ml each of 5% TCA and ethanol and allowed to dry on a stack of paper towels. On each dried filter, 100 μl of 100 mm KOH was spotted to quench fluorescence, and the filters were redried and transferred to scintillations vials. The filters were incubated with scintillation mixture in the dark for 18 h, and incorporation of thymidine was counted in an LS 6500 multipurpose scintillation counter (Beckman Coulter).
RESULTS
UV Irradiation Induces Chromosomal Fragmentation
Some models of UV irradiation damage processing in E. coli envision no chromosomal fragmentation (12, 16, 21, 22), and indeed, when we treated wild type E. coli with 36 J/m2 UV irradiation, the dose that under our conditions kills about 85% of wild type cells, we detected no fragmentation even after 2 h of incubation in growth medium (Fig. 1, A, B, and E). However, when we blocked the repair and degradation of double strand ends with the recBC defect, we found that the same dose of UV irradiation (36 J/m2) followed by identical growth conditions fragments up to 30% of the chromosomal DNA (Fig. 1, A and B), suggesting that the low levels of fragmentation in the wild type cells are due to the efficient repair of double strand breaks and to linear DNA degradation, the two processes catalyzed by the RecBCD enzyme (43), and that RecBCD needs to be saturated to reveal fragmentation. Indeed, fragmentation in the wild type cells starts right after 40 J/m2 and continues rising through 100 J/m2 (Fig. 1, C and D); at these doses of DNA damage, the cells become RecBCD− phenocopies due to titration of this limited enzyme by DNA damage (44–46).
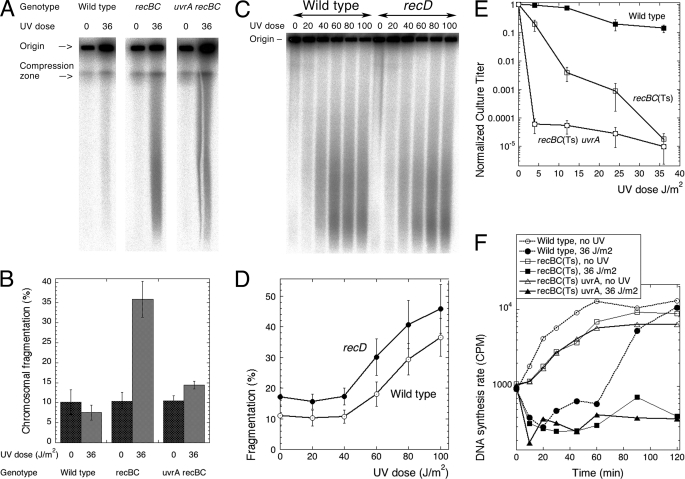
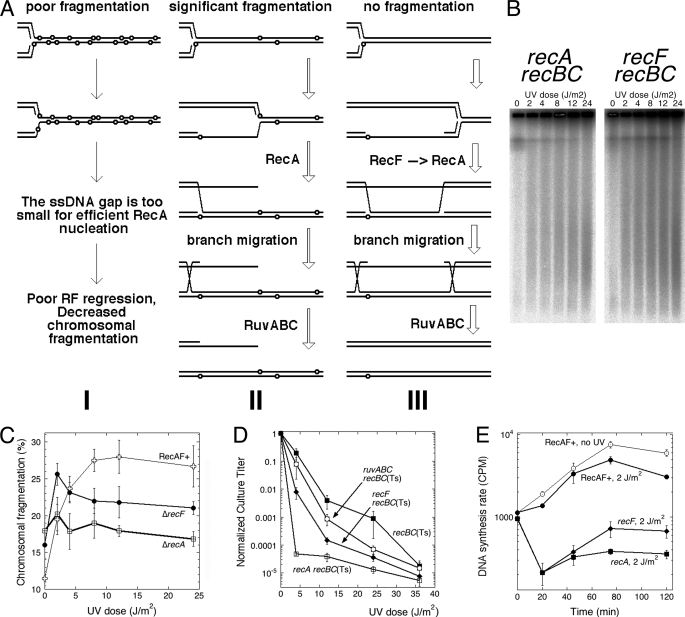
FIGURE 1.
UV radiation-induced chromosomal fragmentation. A, a representative pulsed field gel showing chromosomal fragmentation after no UV irradiation versus 36 J/m2 UV irradiation with subsequent 2 h of shaking in the growth medium at 37 °C. Strains are as follows: wild type, AB1157; recBC(Ts), SK129; uvrA recBC(Ts), SRK301. B, quantification of UV radiation-induced chromosomal fragmentation in wild type (AB1157), recBC(Ts) (SK129), and uvrA recBC(Ts) (SRK301) strains after 2 h of post-UV irradiation incubation in the growth medium. In this and in all subsequent quantifications, the values are means of at least three independent measurements (four on average) done on different days ±S.E. C, a representative pulsed field gel of the dose response of chromosomal fragmentation in wild type (AB1157) and recD mutant (AK3) cells. D, quantification of the dose response of chromosomal fragmentation in wild type and the recD mutant cells from five to 10 independent measurements like that in C ±S.E. E, the UV irradiation survival curves of the three strains. F, the DNA synthesis rates in the three strains. Incorporation was for 1 min. Filled symbols, irradiated cells; open symbols, unirradiated controls.
Linear DNA degradation by RecBCD enzyme could also prevent formation of double strand breaks by degrading the double strand ends generated by reversed replication forks (36, 47). This reasoning predicts that if degradation alone is compromised by the recD mutation (48) then chromosomal fragmentation after UV irradiation will be elevated even at low UV irradiation doses as in recBC mutants. However, the recD mutant shows an essentially wild type shape of dose dependence of UV radiation-induced chromosomal fragmentation, its slightly elevated background levels perhaps reflecting the linear DNA degradation defect of the recD mutant (Fig. 1, C and D). Thus, the increased fragmentation in the recBC mutant reflects the inability to repair and degrade linear DNA rather that the inability to prevent formation of double strand breaks. The UV irradiation survival curves for the wild type and the recBC mutant strains were as expected (Fig. 1E), the decreased titer of the recBC mutant illustrating the importance of double strand break repair for survival of UV-irradiated cells. The recBC mutants also showed a major defect in resumption of DNA synthesis after UV irradiation (Fig. 1F), illustrating another important aspect of double strand break repair. We conclude that UV light induces chromosomal fragmentation in E. coli, but this fragmentation is eliminated by efficient double strand break processing/repair. The models of UV irradiation damage processing that avoid chromosomal fragmentation are inconsistent with this result.
Five Models
Five distinct models of DNA damage-induced chromosomal fragmentation are potentially applicable to UV radiation-induced fragmentation (Fig. 2A). Historically, the first idea was the “clustered excision model” (17, 18) according to which simultaneous excision of two UV lesions in opposite strands leads to a double strand break (Fig. 2A, path 1). A conceptually related idea is the “daughter strand gap instability” model (20, 49) according to which double strand breaks form when the lesion that caused the formation of the daughter strand gap is eventually removed by excision from ssDNA2 (Fig. 2A, path 2). The third is the “replication fork breakage” model (36, 50, 51) (Fig. 2A, path 3) that envisions a replication fork stalling at or past the lesion with subsequent replication fork regression, breakage via Holliday junction resolution, and ligation of the resolution nicks. The fourth is the “replication fork collapse” model (52, 53) according to which a replication fork encounters an excision intermediate and comes apart (collapses) with subsequent repair of the single strand interruption in the template DNA (Fig. 2A, path 4). Finally, the fifth idea is the “replication fork collapse at daughter strand gaps” model (53) (Fig. 2A, path 5). Its distinction from other models is that it requires significant DNA synthesis because it postulates that replication forks of the next round collapse at the ssDNA gaps opposite UV lesions left by the previous replication round. The five different models of chromosomal fragmentation make distinct predictions about the role of DNA synthesis, excision repair, and recombinational repair that we tested.
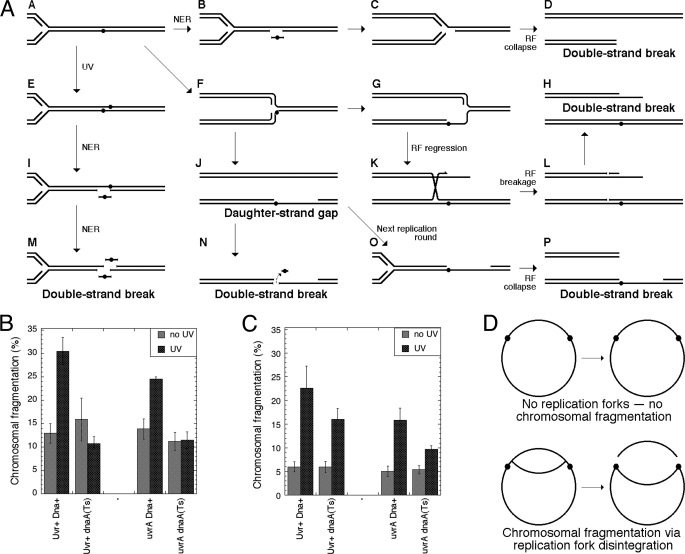
FIGURE 2.
UV radiation-induced chromosomal fragmentation models and its dependence on DNA replication. A, possible models of UV radiation-induced chromosomal fragmentation. UV radiation-induced lesions are shown as filled circles in one strand of DNA duplexes. UV, ultraviolet light; NER, nucleotide excision repair; RF, replication fork. Path 1, A → E → I → M, the clustered excision model: simultaneous excision of two UV lesions in opposite strands leads to a double strand break. Path 2, A → F → J → N, the daughter strand gap instability model. Path 3, A → F → G → K → L → H, the replication fork breakage model. Stalling occurs at or past the lesion (F or G) with subsequent replication fork regression (K), breakage via Holliday junction resolution (K → L), and ligation of the resolution nicks (H). Path 4, A → B → C → D, the replication fork collapse model. The replication fork encounters an excision intermediate (B → C) and collapses (C → D) with subsequent repair of the single strand interruption in the template DNA (D). Path 5, A → F → J → O → P, “the replication fork collapse at daughter strand gaps” variation. B, UV radiation-induced chromosomal fragmentation is eliminated by prior block to replication initiation. Cells were grown at 28 °C until A600 reached 0.1 and shifted to 42 °C for 2 h before UV irradiation. UV irradiation doses were 24 J/m2 for Uvr+ recBC(Ts) cells and 2 J/m2 for uvrA recBC(Ts) double mutant cells. Here and in C, the lower UV irradiation dose for excision-deficient cells ensured higher fragmentation (see Fig. 3B). After UV irradiation, incubation continued at 42 °C for an additional 2 h. In B and C, values are means of five independent measurements ± S.E. Strains are as follows: DnaA+, SK129; DnaA+ uvrA, SRK315; dnaA(Ts), SRK309; dnaA(Ts) uvrA, SRK310. C, UV radiation-induced chromosomal fragmentation is decreased, but not eliminated, by concurrent block to replication initiation. Cells were grown at 28 °C until A600 reached 0.3, UV-irradiated, and then incubated at 42 °C for 2 h. UV irradiation doses and strains were as in B. D, a scheme explaining the findings. The chromosome is depicted as a circle (a single line stands for duplex DNA) with UV lesions represented by small filled circles. Chromosomal fragmentation is (at least partially) independent of the lesion processing but completely depends on the presence of replication forks, suggesting replication fork disintegration at DNA lesions as the fragmentation mechanism.
Only Replicating Chromosomes Fragment
Although UV radiation-induced fragmentation was readily detectable in wild type cells (Fig. 1, C and D), it was at background levels at moderate UV irradiation doses, so we decided to begin by studying its mechanisms in the recBC(Ts) background in which fragmentation levels are much higher. From this point onward, all strains in this study are recBC(Ts) in case there is no mention of this fact. First, it was important to know whether UV radiation-induced chromosomal fragmentation is dependent on concurrent DNA replication. Pretreatment of cells with chloramphenicol and preincubation of dnaA(Ts) mutant cells at the non-permissive temperature for 2 h before UV irradiation to prevent new initiations of chromosomal replication both blocked UV radiation-induced fragmentation completely (Fig. 2B and supplemental Fig. S1), indicating that the double strand DNA breaks happen either at replication forks or in the newly synthesized DNA. Thus, the “clustered excision” model (Fig. 2A, path 1) according to which chromosomal fragmentation is induced by simultaneous excision of lesions from both strands of the duplex in the same location does not explain the UV radiation-induced fragmentation we observed in this work. When we blocked initiation of new replication rounds only after UV irradiation, we observed a reduced fragmentation (Fig. 2C), suggesting that both the existing replication forks and the new ones initiated after DNA damage contribute to fragmentation. We conclude that UV irradiation damage fragments chromosomes via replication forks (Fig. 2D) for example after replication forks run into UV lesions or intermediates of their excision or at daughter strand gaps that were left behind (Fig. 2A).
There are total of four models that explain how DNA lesions can break chromosomes via disintegration of replication forks or at daughter strand gaps. Of the four models, the daughter strand gap instability idea (Fig. 2A, path 2) and the replication fork collapse idea (Fig. 2A, path 4) both predict that fragmentation is dependent on the concurrent excision repair and will be inhibited if excision of UV lesions is inactivated. At the same time, the replication fork regression/breakage idea (Fig. 2A, path 3) and replication fork collapse at daughter strand gap idea (Fig. 2A, path 5) both predict that fragmentation is independent of excision repair and should be in fact stimulated by its inactivation due to more frequent replication fork encounters with unrepaired UV lesions or their consequences (daughter strand gaps). When we irradiated a uvrA mutant with 36 J/m2 UV irradiation, the level of chromosomal fragmentation was reduced at least 3-fold compared with the UvrA+ strain (Fig. 1, A and B) (as expected, there were no survivors at this dose in uvrA mutant (Fig. 1E)). The reduced fragmentation was still dependent on the concurrent DNA replication as the experiments with chloramphenicol treatment (supplemental Fig. S1) and in the ΔuvrA dnaA(Ts) at the non-permissive temperature (Fig. 2, B and C) demonstrated. At face value, such a reduction suggested that the fragmentation in excision-proficient cells has a complex nature with most of it dependent on excision of UV lesions but some of it independent of excision.
Fragmentation Is Independent of Excision and Coincides with Resumption of Replication
We tested the complex nature of UV radiation-induced fragmentation by determining its onset in excision repair-proficient versus excision-deficient cells. The daughter strand gap instability model (Fig. 2A, path 2) and the replication fork collapse model (Fig. 2A, path 4) both predict that fragmentation should happen early after UV irradiation and coincide with inhibition of DNA synthesis, the former because daughter strand gaps should form right away and should be accessible to excision repair, and the latter because the fragmentation is a direct result of replication forks running into excision intermediates. These single stage models both predict no fragmentation in excision-deficient cells. In contrast, the replication fork regression/breakage idea (Fig. 2A, path 3) and replication fork collapse at daughter strand gap idea (Fig. 2A, path 5) both predict that chromosomal fragmentation as a multistage process should take some time to develop and should coincide with restoration of replication at the forks that avoided breakage. In contrast to the single stage models, these multistage models also predict fragmentation to be independent of excision. Based on these predictions and on results in Fig. 1B, we expected that the bulk of the chromosomal fragmentation in excision-proficient cells would happen early and would coincide with inhibition of DNA synthesis, whereas a small part would be delayed, coinciding with both the restoration of DNA synthesis and the residual chromosomal fragmentation in the excision-deficient cells.
To accurately determine the timing of replication restart in both excision repair-proficient and -deficient strains, we had to lower the dose of UV irradiation because 36 J/m2 UV irradiation severely inhibited DNA synthesis in both the recBC(Ts) and in the recBC(Ts) uvrA mutants (Fig. 1F). To find an optimal dose for both strains, we determined the dose response of chromosomal fragmentation and found it to be (as expected) a smooth saturating curve in the excision-proficient cells (Fig. 3, A and B). Unexpectedly, in the excision-deficient mutant, chromosomal fragmentation peaked around 20% at 2 J/m2 and then decreased with higher doses (Fig. 3, A and B). Thus, fragmentation was in fact independent of excision, whereas the decrease of fragmentation in the excision-deficient cells at 36 J/m2 was the consequence of a high density of UV lesions. This finding was inconsistent with both the daughter strand gap instability model and the replication fork collapse model, which both depend on excision.
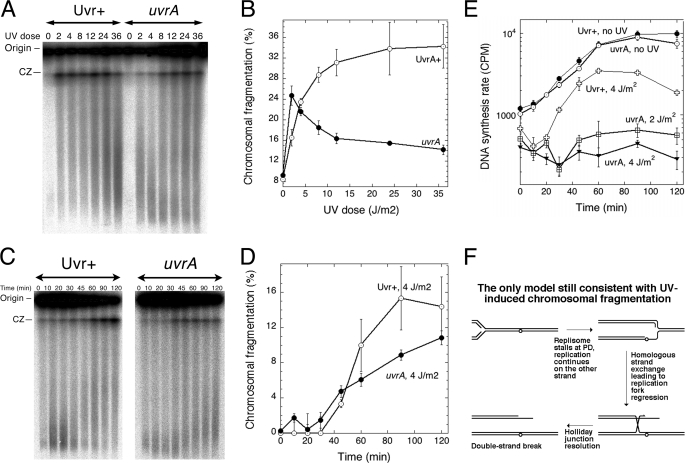
FIGURE 3.
Replication forks first stall and then either restart or disintegrate. A, a representative pulsed field gel of the dose dependence of chromosomal fragmentation in Uvr+ (SK129) and uvrA mutant (SRK301) cells. CZ, compression zone. The UV irradiation dose is expressed in J/m2. Note that the higher the UV irradiation dose, the more radioactivity is in the lane (both the well and the gel). This does not reflect UV radiation-induced replication; rather, it is a trivial consequence of our protocol of preparation of genomic DNA in the plug that calls for a specific density of cell suspension. As a result, the better the culture grows after labeling, the more it is diluted for genomic DNA preparation. B, quantification of the dose dependence of chromosomal fragmentation in Uvr+ and uvrA mutant cells from four independent experiments like that in A (means ± S.E.). C, a representative pulsed field gel of the kinetics of chromosomal fragmentation in Uvr+ (SK129) and uvrA mutant (SRK301) cells irradiated with 4 J/m2 UV irradiation. CZ, compression zone. D, quantification of the kinetics of chromosomal fragmentation in Uvr+ and uvrA mutant cells from three independent experiments like that in C (means ± S.E.). The unirradiated background is subtracted in this case to help reveal the rise of fragmentation. E, the kinetics of DNA synthesis restart after 4 J/m2 delivered at time 0. Incorporation was for 1 min. Unirradiated cultures are shown in parallel. For the uvrA mutant, the resumption after 2 J/m2 is also shown. The values are means of three independent measurements ± S.E. The strains are as follows: Uvr+, SK129; uvrA mutant, SRK301. F, the only model still consistent with the results is the replication fork stalling, reversal, and breakage model. Open circle, UV lesion. PD, pyrimidine dimer.
Because of the different shapes of the two dose dependence curves, there was a particular dose (4 J/m2) that caused similar levels of fragmentation in both excision-proficient and -deficient cells (Fig. 3B) and even similar kinetics of fragmentation (somewhat faster in excision-deficient cells) (Fig. 3, C and D), so we used this low dose to determine the timing of replication resumption. We found that replication resumes in the excision repair-proficient cells around 30 min (Fig. 3E) and thus shortly precedes or coincides with the first detectable chromosomal fragmentation (Fig. 3D). In the excision-deficient cells, replication recovered only weakly after 4 J/m2 UV irradiation (recovery is better after 2 J/m2) (Fig. 3E). The severe inhibition of replication recovery but high fragmentation in excision-deficient mutants indicates that restoration of replication and chromosomal fragmentation are independent, if not competing, events. The independence of fragmentation of the replication restart is inconsistent with the idea of collapse at daughter strand gaps, so the only mechanism still consistent with the data is replication fork regression and breakage (Fig. 3F) as elaborated further below.
Fate of Blocked Replication Forks
Resumption of replication is severely affected in excision repair-deficient mutants, making it possible that the residual replication after UV irradiation (Fig. 3E) all comes from new initiations at the origin, whereas the blocked replication forks are permanently disabled (13, 14). On the other hand, it was proposed that when replication forks encounter a UV lesion they restart downstream, leaving behind daughter strand gaps (16). To test the idea of replication fork restart at UV lesions, we sought to block initiations of new replication rounds from the origin in UV-irradiated excision repair-deficient cells, restricting post-UV irradiation replication to the forks present at the time of irradiation. Under these conditions, any replication above background would indicate restart at the forks originally blocked at the (non-removable) UV lesions.
One way to block initiation of new replication rounds is to shift the dnaA(Ts) mutants, which are defective in the origin recognition protein, to the non-permissive temperature. Without UV irradiation, such a shift resulted in the inhibition of the replication rate to background levels within 1 h (Fig. 4A). Interestingly, if uvrA dnaA(Ts) cells were UV-irradiated before the shift to the non-permissive temperature, then the DNA synthesis rate stabilized (Fig. 4A), suggesting that blocked replication forks are capable of restart. This conclusion is not weakened by the known stimulation of stable DNA replication in dnaA− conditions by UV irradiation (12, 54) because such inducible stable DNA replication is not observed in the recBC(Ts) mutant at 42 °C (55).
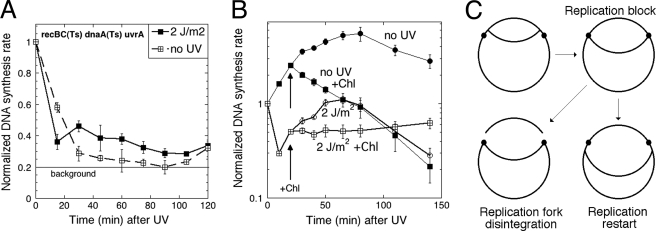
FIGURE 4.
Replication restart from blocked forks. The rate of DNA synthesis was measured by incorporation for 5 min. Normalization was to the average count of the corresponding unirradiated culture at time 0 min. A, replication restart at blocked forks in the dnaA(Ts) uvrA strain (recBC(Ts) dnaA uvrA, SRK310). The cells were grown at 28 °C before UV irradiation but shifted to 42 °C (non-permissive for dnaA(Ts)) right after irradiation to block initiations of new replication rounds from the origin. The values are means of four independent measurements ±S.E. Background was the average of several measurements of the same cells but treated with 500 μg/ml kanamycin. B, replication restart at blocked replication forks in the uvrA mutant cells (recBC(Ts) uvrA, SRK301). Irradiation, where applied, was at time 0. At 20 min after irradiation, chloramphenicol was added to a 40 μg/ml final concentration to one-half of the culture to prevent initiations of new replication rounds from the origin. The values are means of three independent measurements ±S.E. C, explanation of the findings until this point. The chromosome is depicted as a circle (a single line stands for duplex DNA) with UV lesions represented by small filled circles.
Because replication resumption in excision-deficient cells irradiated with 2 J/m2 was clearly detectable after 30 min post-UV irradiation (Fig. 3E), we also blocked initiation of new replication rounds with chloramphenicol added at 20 min post-UV irradiation. This 20-min period after UV irradiation should allow for SOS induction of any function required for replication resumption (56). Again, in unirradiated cells, chloramphenicol addition caused a gradual decrease in the replication rate consistent with prevention of new initiations (Fig. 4B, compare “no UV” with “no UV +Chl” curves). Interestingly, pre-UV irradiation of chloramphenicol-treated culture again stabilized the replication rate (Fig. 4B, compare “2 J/m2” and “2 J/m2 +Chl” curves), suggesting a robust restart of replication forks blocked at UV lesions. On the basis of our observations so far, we conclude that replication forks first stall at UV lesions and then either disintegrate or restart with neither process being dependent on the ability to remove UV lesions by excision repair (Fig. 4C).
Fragmentation Depends on Homologous Strand Exchange
As already mentioned above, the prominent late UV radiation-induced fragmentation in the excision-deficient mutant combined with minimal after-replication (at higher UV irradiation doses) strongly distinguished between the four remaining models of DNA damage-induced fragmentation. The absence of excision requirement for fragmentation rules out the daughter strand gap instability model (Fig. 2A, path 2) and the replication fork collapse model (Fig. 2A, path 4), whereas the replication fork collapse at daughter strand gap idea (Fig. 2A, path 5) is ruled out because there was no need for significant DNA synthesis after UV irradiation. The only idea still consistent with our results at this point was the replication fork regression/breakage idea (Fig. 2A, path 3, and Fig. 3F), which we decided to test further by challenging it to explain the peculiar inhibition of fragmentation at higher densities of UV lesions in excision-deficient mutants (Fig. 3B).
We interpreted this observation to mean that fragmentation depends not only on the original encounter of the replisome with a UV lesion but also on the probability of the second replisome stalling at the lesion in the complementary strand, which in the case of the uvrA mutants will be a function of the UV lesion density in the downstream template DNA. We reasoned that the extent of replication past the lesion was critical for subsequent fragmentation (Fig. 5A). If there is limited extension past the first lesion (comparable with the size of the replisome footprint; ∼50 bp), the amount of stable ssDNA is insufficient to allow RecA polymerization, there is no replication fork regression, and chromosomal fragmentation is depressed (Fig. 5A, scenario I). Significant continuation of replication on the unblocked strand generates a substrate for recombinational repair, a stable single-stranded DNA opposite an intact sister duplex. RecA-catalyzed strand exchange with subsequent branch migration in such a substrate reverses the replication fork, turning it into a Holliday junction that can be resolved to break the fork (Figs. 3F and 5A, scenario II). The prediction then is that UV radiation-induced chromosomal fragmentation should be dependent on RecA. If the lesion in the opposite strand is not encountered before the next Okazaki fragment is initiated, the replication fork moves away, leaving behind a daughter strand gap (Fig. 5A, scenario III). RecA polymerization on the daughter strand gaps is efficiently promoted by RecF (57), and no fragmentation follows. Thus, another prediction of this reasoning linking fragmentation to a particular lesion density is that fragmentation should be independent of RecF. Because UV lesions are induced in DNA almost at random (whenever there are side-by-side pyrimidines), the length of DNA stretches between neighboring lesions follows an exponential distribution similar to the distribution of restriction fragment length in long genomes (58, 59). In general, although scenario III will always predominate, it does not stall replication forks and is therefore “invisible” in our analysis. Replication fork stalling scenarios II and I are less frequent at low density of UV lesions with scenario II predominating but will become more frequent at higher density of UV lesions with scenario I gradually reducing chromosomal fragmentation.
FIGURE 5.
UV radiation-induced fragmentation in recA and recF mutants. A, a scheme describing a hypothetical dependence of the fragmentation outcome on the density of UV lesions. RF, replication fork. UV lesions are shown as small open circles. For the sake of clarity, their density is shown constant in time as in the uvrA mutants (no excision), although the same logic equally applies to excision-proficient conditions. The three scenarios start exactly the same; the only difference between them is the density of UV lesions, which is highest in scenario I and lowest in scenario III. B, a representative pulsed field gel of the dose dependence of chromosomal fragmentation in the recA mutant (SRK311) and the recF mutant (SRK312) cells. C, quantification of the dose dependence of chromosomal fragmentation in RecAF+ and the recA and recF mutant cells from four or five independent experiments like that in A (means ± S.E.). D, the UV irradiation survival curves of the recBC(Ts) mutant and its recA (SRK311), recF (SRK312), and ruvABC (SRK302) derivatives. In D and E, the values are means of three independent measurements ± S.E. E, the effect of the recA and recF mutations on the resumption of DNA synthesis after 2 J/m2 UV irradiation. Incorporation was for 1 min.
When we measured the dose dependence of UV radiation-induced fragmentation in the recBC(Ts) recA double mutant, we found an unusually high background (∼18%) in the absence of UV irradiation but no additional fragmentation over this background with UV irradiation doses from 2 to 24 J/m2 (Fig. 5, B and C). Thus, UV radiation-induced fragmentation at least in this range of doses is indeed completely dependent on the RecA-catalyzed strand exchange. This RecA dependence of UV radiation-induced fragmentation creates a paradox because the recA mutants are exquisitely sensitive to UV irradiation (Fig. 5D) and are inhibited for post-UV irradiation DNA synthesis (Fig. 5E), indicating that UV radiation-induced chromosomal fragmentation is (or at least reflects) the cellular repair pathway to restart the stalled replication forks and save the cells. In addition, the inability of the recA mutant to restart blocked forks does not automatically translate into a higher chromosomal fragmentation, indicating that it is the RecA processing of stalled forks that makes possible the subsequent decision to either restart or break the forks.
The dose dependence of UV radiation-induced fragmentation in the recBC(Ts) recF double mutant showed a more complex pattern (Fig. 5, B and C). Although the background without UV irradiation was equally high, there was up to 8% UV radiation-induced fragmentation, but the peak of it was at the lowest dose (2 J/m2), decreasing to about half this amount at higher doses. In fact, the fragmentation increase from 0 to 2 J/m2 was the same for RecF+ and recF mutant strains (Fig. 5C). Thus, UV radiation-induced fragmentation appears only partially dependent on RecF and only at higher UV irradiation doses, suggesting that the fragmentation may be in fact completely independent of RecF, but the lack of another factor that depends on RecF and at the same time promotes fragmentation at higher doses limits the overall fragmentation in the recF mutants. Again, this factor may have been the extent of DNA replication past the blocking lesion. Although in the uvrA mutants this extent is limited physically by the next lesion, in the recF mutant, this extent may be limited by a defect in the replisome release (reactivation) from the blocking lesion (57). The resumption of DNA replication after 2 J/m2 UV irradiation in the recF mutants was severely inhibited (Fig. 5E) as reported before (10, 11), which could explain the peculiar dose dependence of UV radiation-induced fragmentation of the recF mutant. We conclude that UV radiation-induced fragmentation completely depends on the RecA-catalyzed homologous strand exchange but is mechanistically distinct from the RecF-catalyzed RecA polymerization at daughter strand gaps behind the replication fork (which are also important for UV irradiation survival (Fig. 5D)).
Role of Holliday Junction Resolution
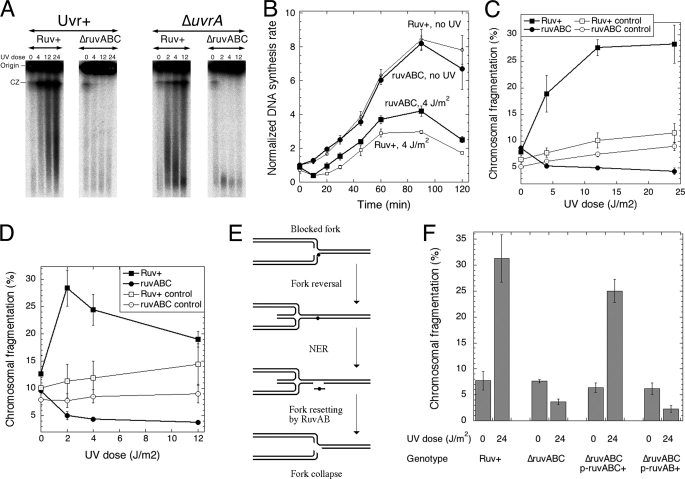
Our findings about UV radiation-induced chromosomal fragmentation in recBC mutants, namely 1) the stimulation of fragmentation by inactivation of excision repair (Fig. 3B), 2) the significant lag preceding fragmentation (Fig. 3D), 3) the concurrence with resumption of DNA synthesis (Fig. 3, D versus E) but independence of post-UV DNA synthesis, and 4) the dependence on RecA-catalyzed homologous strand exchange, were all consistent with the replication fork reversal/breakage model (Fig. 3F). As the definitive genetic test, we inquired whether a defect in the Holliday junction resolvasome, RuvABC, would block chromosomal fragmentation as predicted by this model (Fig. 5A). We determined the dose dependence of UV radiation-induced fragmentation in both excision-proficient and -deficient cells carrying an additional ruvABC defect. We found that in both cases UV radiation-induced chromosomal fragmentation after 2 h in growth conditions is completely dependent on RuvABC (Fig. 6A, C, and D). At the same time, replication resumed in the ruvABC mutant at an even faster rate and more strongly than in the Ruv+ cells (Fig. 6B), again suggesting that resumption competes with fragmentation. This result strongly supports the replication fork reversal/breakage model to describe UV radiation-induced chromosomal fragmentation. Paradoxically and as in the case of the recA mutants above, the UV irradiation sensitivity of ruv mutants (Ref. 60 and Fig. 5D) indicates that breaking replication forks is the repair pathway to rescue stalled replication in UV-irradiated cells.
FIGURE 6.
UV radiation-induced chromosomal fragmentation depends on RuvABC resolvasome and specifically on RuvC resolvase. A, a representative pulsed field gel showing the dose response of the chromosomal fragmentation in Ruv+ cells versus ΔruvABC cells (both Uvr+ and uvrA mutant). CZ, compression zone. The UV irradiation dose is expressed in J/m2. B, the effect of the ruvABC mutation on the restart of DNA synthesis after UV irradiation revealed as the normalized rate of DNA synthesis. Incorporation was for 1 min. Normalization was to the average count of the corresponding unirradiated culture at time 0 min. The curves of the Ruv+ strain are (the normalized curves) from Fig. 3E. The values are means of three independent measurements ± S.E. C, quantification of the dose dependence of chromosomal fragmentation in Ruv+ (SK129) and ΔruvABC mutant (SRK302) cells from three independent experiments like that in A (means ± S.E.). “Control” means that cells were processed immediately after UV irradiation in contrast to the experiment in which cells were processed after 2 h at 37 °C in growth medium. D, quantification of the dose dependence of chromosomal fragmentation in uvrA mutant (SRK301) and in its ΔruvABC derivative (SRK303) from three independent experiments like that in A (means ± S.E.). Conditions are the same as in C. E, a scheme of nucleotide excision repair (NER)-dependent, RuvAB-dependent, but RuvC-independent, replication fork collapse. F, complementation with the RuvABC+ plasmid restores UV radiation-dependent chromosomal fragmentation in the ΔruvABC mutant, but complementation with the RuvAB+ (RuvC−) plasmid does not.
RuvAB can function separately from RuvC as a Holliday junction translocase (61). We noticed that there is a specific version of the replication fork collapse model in excision-proficient cells that is dependent on the RuvAB-catalyzed translocation of Holliday junctions but independent of the RuvC-catalyzed resolution. According to this model, Holliday junctions at reversed replication forks could be “resolved” by RuvAB pushing them back into the excision intermediates of UV lesion removal (Fig. 6E). Because in the previous experiment we used a triple ΔruvABC deletion mutant, this unique model was not specifically tested. To test this model, we used either ΔruvC single gene deletion (using ΔruvA single deletion as a control) or complemented the triple ΔruvABC deletion with a plasmid carrying only ruvAB+ genes so the strain would remain a ruvC mutant. The results were still unequivocal: there was no chromosomal fragmentation after UV irradiation in the ruvC mutants (Fig. 6F and supplemental Fig. S2), meaning that the Holliday junction cleavage by RuvC is specifically required for UV radiation-induced chromosomal fragmentation.
Role of RecG Helicase
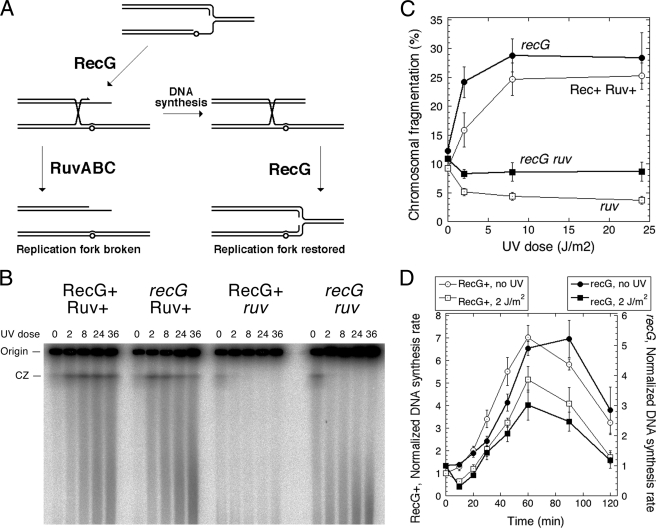
It was proposed on the basis of genetic and biochemical data that the RecG helicase plays a major role in handling replication forks stalled at UV lesions (62). These proposals imagined RecG driving both the fork transformation into Holliday junctions (reversal) and subsequent branch migration of the Holliday junctions to restore the replication forks (resetting) (62), making it hard to predict what effect a recG defect would have on RuvABC-promoted chromosomal fragmentation (Fig. 7A). If RecG mostly catalyzes reversal, the recG defect should decrease fragmentation, whereas if RecG mostly catalyzes resetting, then the recG defect should lead to accumulation of Holliday junctions and to a higher fragmentation. We found that the recG mutation does not change the overall shape and power of the dose-response curve with the exception of the statistically significant elevation of fragmentation at the dose of 2 J/m2 (Fig. 7, B and C). Interestingly, although this fragmentation in recG mutants is still completely dependent on Ruv, the level of fragmentation in the recG ruv double mutant was consistently 2 times higher than the low fragmentation in ruv single mutants (Fig. 7C), confirming the overall impression of the mild fragmentation elevation in the recG mutants. The replication restart after 2 J/m2 UV irradiation was the same in RecG+ cells versus recG mutants (Fig. 7D). We conclude that the recG defect does not influence the restart after UV irradiation and has only a mild increasing effect on UV radiation-induced fragmentation and only at low doses, making it unlikely that RecG plays a major role in processing of replication forks stalled at UV lesions.
FIGURE 7.
Effect of recG inactivation. A, a scheme of how RecG can possibly act at a replication fork stalled at a UV lesion. One possibility is regression of the stalled replication fork, turning it into a Holliday junction. The other possibility is branch migration of the Holliday junction to regenerate the replication fork (note that the originally blocking lesion is now “bridged over,” and the replication fork is no longer blocked). B, a representative pulsed field gel showing the dose response of the chromosomal fragmentation in recG mutants either in combination with Ruv+ or ΔruvABC alleles. CZ, compression zone. The UV irradiation dose is expressed in J/m2. C, quantification of the dose dependence of chromosomal fragmentation in RecG+ Ruv+ (SK129), recG mutant (SRK316), ΔruvABC mutant (SRK302), and double recG ruv mutant (SRK317) cells from three independent experiments like that in B (means ± S.E.). D, the kinetics of DNA synthesis restart in recG mutant and RecG+ cells after 2 J/m2 delivered at time 0. Incorporation was for 1 min. Normalization was to the average count of the corresponding unirradiated culture at time 0 min. Unirradiated cultures are shown as controls. Note the difference between the left and right y axes: even without UV irradiation, DNA synthesis in the recG mutant is 25% lower than in the RecG+ cells. The values are means of three independent measurements ± S.E. The strains are as follows: RecG+, SK129; recG mutant, SRK316.
Location of Holliday Junctions
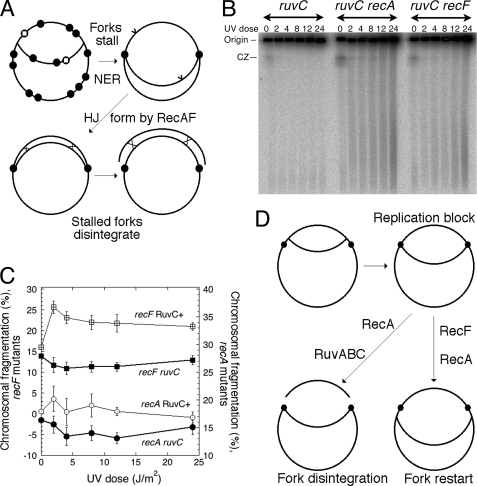
Our demonstration of a continuous post-UV irradiation replication in uvrA mutants with blocked initiations from the origin (Fig. 4) confirmed restart of blocked replication forks and suggested formation of daughter strand gaps whose subsequent RecF-dependent repair should generate Holliday junctions behind replication forks. Such Holliday junctions, which will remain unresolved in ruv mutants, would suppress chromosomal fragmentation by trapping the subchromosomal fragments together with the rest of the chromosome in unresolved intermediates of daughter strand gap repair (Fig. 8A). We blocked formation of Holliday junctions behind replication forks by using the recF single mutants and asked whether the recF defect would suppress the dependence of chromosomal fragmentation on RuvC. We also asked whether the dependence of chromosomal fragmentation on RuvC could be suppressed by the recA defect. Both the recF and the recA mutations appeared to suppress the lack of UV radiation-induced chromosomal fragmentation in the ruvC mutant (Fig. 8B); however, careful quantification of this fragmentation revealed no UV radiation-dependent increase in either mutant (Fig. 8C). The dependence of the chromosomal fragmentation in the recF mutants on Holliday junction resolution strongly argues that Holliday junctions do form at stalled replication forks, and their resolution breaks the chromosome. However, resumption of blocked replication forks without breakage is an alternative pathway also catalyzed by RecA (Fig. 8D). Overall, we conclude that the various characteristics of UV radiation-induced chromosomal fragmentation at least in the lower range of UV irradiation doses and after long incubation in growth conditions in the absence of linear DNA degradation fully support the model of reversal and subsequent breakage of blocked replication forks due to Holliday junction resolution (Fig. 3F), whereas they are inconsistent with the four other models of DNA damage-induced chromosomal fragmentation.
FIGURE 8.
RuvC is still required for chromosomal fragmentation even when recombination behind replication forks is blocked. A, a scheme of how unresolved Holliday junctions behind replication forks prevent release of the subchromosomal fragments even if replication forks disintegrate independently of Holliday junction (HJ) resolution. NER, nucleotide excision repair. Filled circles, UV lesions; empty circles, UV lesions whose excision repair will trigger RecAF-promoted strand exchange. B, a representative pulsed field gel showing the dose response of chromosomal fragmentation in ruvC (SRK308), ruvC recA (SRK313), and ruvC recF (SRK314) mutants. CZ, compression zone. C, quantification of the dose dependence of chromosomal fragmentation in recA (SRK311), recA ruvC (SRK313), recF (SRK312), and recF ruvC (SRK314) mutants. The values are means of four independent measurements ± S.E. D, the overall conclusion: both replication fork restart and disintegration require RecA, only disintegration requires RuvABC, and only restart requires RecF.
DISCUSSION
Excision repair of DNA lesions is known in detail (9), but the consequences of replication forks encountering one-strand DNA lesions, including formation of double strand breaks, continue to present experimental challenges because of their intrinsic complexity and fleeting nature due to efficient repair. We inactivate double strand end processing in E. coli by using the recBC mutants, stabilizing fragmented chromosomes in time. Our study of the mechanism of UV radiation-induced chromosomal fragmentation in the recBC mutants shows that this fragmentation 1) depends on the presence of replication forks in the chromosome and in fact is concurrent with resumption of UV radiation-inhibited DNA synthesis; 2) at very low UV irradiation doses is stimulated by inactivation of excision repair and is not influenced by inactivation of daughter strand gap repair; 3) absolutely depends on RecA, which catalyzes homologous strand exchange; 4) absolutely depends on the RuvABC resolvasome and specifically on the Holliday junction resolvase RuvC; 5) remains RuvC-dependent in the absence of RecF-catalyzed daughter strand gap repair (the latter generates Holliday junctions behind forks), suggesting that Holliday junctions, whose resolution fragments the chromosome, are generated at the forks. We conclude that UV irradiation induces significant chromosomal fragmentation in E. coli that in wild type cells is masked by efficient double strand break processing, and the mechanism of this fragmentation is consistent with the “breakage of regressed replication fork” model (Figs. 2A, path 3, and 3F). It should be noted, however, that our conclusion that replication fork breakage is the major, if not the only, mechanism of chromosomal fragmentation under our experimental conditions does not exclude the possibility that other mechanisms operate under different conditions.
By its RecA/RuvABC dependence, but RecF independence, UV radiation-induced chromosomal fragmentation mirrors chromosomal fragmentation induced by malfunctioning of the replicative helicase DnaB (63). Interestingly, a recent demonstration that branch migration at Holliday junctions in vitro is promoted by repeated RecA polymerization at a partially single-stranded arm of such a junction (64) suggests a possibility that in vivo a blocked replication fork regresses spontaneously to produce an ss-tail, whereas subsequent RecA polymerization on this tail converts the partially regressed replication fork into a Holliday junction. Our data cannot tell whether RecA catalyzes strand exchange and replication fork reversal to produce Holliday junction or simply polymerizes on the ss-tail and promotes branch migration to achieve the same result.
Significance of Findings
The mechanisms of chromosomal damage formation and repair are highly relevant to both human health and basic chromosomal metabolism. Specifically, both UV irradiation and endogenous chromosomal fragmentation induce genetic instability, suggesting that UV irradiation causes chromosomal fragmentation, but the conditions for generation of these double strand breaks as well as their biological relevance are not clear. On the other hand, there are reports (confirmed in this study) that high doses of UV irradiation (survival, 1% or less) in wild type cells do induce chromosomal fragmentation, which is completely dependent on excision repair, both in E. coli (18, 19) and in mammalian cells (65). The classic explanation of this fragmentation is the clustered excision model that envisions simultaneous excision of nearby lesions from opposite DNA strands (17, 18) (Fig. 2A, path 1). A conceptually related explanation is the “instability of daughter strand gaps” idea (Fig. 2A, path 2). Although confirmed for base excision repair, especially for DNA uracil excision (66, 67), these two models lack support for UV lesions because there is no published evidence that the nucleotide excision repair can recognize UV lesions in ssDNA as demanded by these models. Alternatively, there are three more models of UV radiation-induced chromosomal fragmentation linking it to replication fork encounters with DNA lesions (Fig. 2A, paths 3–5). Importantly, none of the five ideas have ever been tested. At the same time, it is widely assumed that sublethal doses of UV irradiation do not break DNA. We show in this work that this apparent absence of UV radiation-induced fragmentation at low doses is a result of efficient double strand break processing by RecBCD enzyme. We also rule out all but one existing model as an explanation for UV radiation-induced chromosomal fragmentation in recBC mutants.
Two features of our experimental system are critical for the high sensitivity of detection of the fragmented chromosomes. Chromosomal fragmentation generates subchromosomal fragments, which are sometimes hard to distinguish from multiple, intact linear chromosomes in eukaryotic cells but are cleanly separated from the circular bacterial chromosomes by pulsed field gel electrophoresis, the procedure that keeps circular chromosomes in the wells (39). The other feature is our ability to specifically inactivate double strand break processing with a single mutation, recBC, which unmasks chromosomal fragmentation induced by very low doses of DNA-damaging agents. In addition to repair, another factor lowers fragmentation in the wild type cells: once the subchromosomal fragment is attached by one of its ends to the circular chromosome (or even to another linear fragment), it becomes invisible in our analysis. In other words, our chromosomal fragmentation values, especially for double strand break repair-proficient cells, are underestimations by a factor of at least 2. To paraphrase, when looking at the uncut chromosomes, we can only detect pairs of double strand ends, and we miss single ends (for example, the tails of σ replication structures). One way to detect all double strand ends (in a specific chromosomal segment) is to cut the chromosome with rare cutter restrictases, such as NotI, as we did before (68), and reveal the resulting subfragmental smear by a probe to this entire fragment.
One should keep in mind, however, that RecBCD also suppresses chromosomal fragmentation in some experimental systems (36), and therefore its inactivation may elevate the level of fragmentation artificially. The relative contribution of this suppression pathway to keeping chromosomal DNA “unfragmented” is still unknown, however, because the repair pathway is so efficient. At the least, comparison of the shapes of dose dependence curves of the recBC mutant versus the recD mutant (Fig. 1D versus 3B) suggests that the reduction in chromosomal fragmentation due to the double strand break prevention pathway can only be minor. However, it is not even important for our argument whether the double strand DNA ends are processed via prior degradation or via subsequent repair as long as such ends are formed. In fact, the way the double strand ends are processed in E. coli depends on the extent of DNA damage as high doses of DNA damage turn WT E. coli into ExoV− phenocopies (44–46). Thus, at low doses of UV irradiation damage, the ends are first significantly degraded and only then repaired, whereas at high doses, the ends may just be repaired, whereas some of the ends are even left unrepaired as we apparently observed in WT and recD mutant cells irradiated with high doses of UV irradiation.
Finally, in eukaryotes and archaea, there is no such powerful linear DNA degradation activity, so using recBC mutants in E. coli makes our studies of chromosomal fragmentation broadly relevant to all kingdoms. What is important is that UV radiation-inhibited replication forks are processed to generate substrates for RecBCD, which are therefore double strand ends of linear subchromosomal fragments. Whether or not these subchromosomal fragments are still attached to the circular chromosome via Holliday junctions (similar to what is drawn in Fig. 7A) is of lesser importance at this point and will be addressed later.
RecA (but Not RecF) Dependence of Fragmentation Suggests Direct RecA Loading by Replisome
RecA protein, the central activity of recombinational repair, forms a spiral filament on ssDNA to find, homologously align, and catalyze strand exchange with an intact sister duplex. This homologous strand exchange generates an intermediate characterized by DNA junctions to facilitate recombinational repair of chromosomal lesions, such as daughter strand gaps and double strand breaks (57). By itself, RecA does not recognize chromosomal lesions and requires licensing factors that target it to specific DNA structures in need of recombinational repair (for a review, see Ref. 69). The two major classes of chromosomal lesions that are mended by recombinational repair in E. coli are double strand ends and daughter strand gaps (for a review, see Ref. 57). The recognition/licensing factor for double strand ends is the RecBCD helicase/nuclease, whereas for daughter strand gaps, it is the RecFOR complex (57, 69). The fact that UV radiation-induced fragmentation is stimulated by the recBC defect and is independent of the recF defect while still being completely dependent on RecA suggests the existence of yet another pathway to license RecA polymerization. This hypothetical licensing factor should operate at stalled replication forks and may well be a component of the replisome itself. Interestingly, the ϵ subunit of DNA polymerase III, the DnaQ proofreading nuclease, was recently found to be required for SOS induction by nalidixic acid (70). Nalidixic acid stalls replication forks (71) while the SOS response can be taken as an indirect, but sensitive, measure of in vivo RecA polymerization triggered by various DNA lesions that interfere with replication (57). We speculate that DnaQ might be the RecA licensing factor of the stalled replisome.
Fragmentation, Replication Fork Repair, and Genetic Death
The complete dependence of UV radiation-induced fragmentation on RecA in our experimental system, although predicted by the replication fork breakage model, is surprising in two different ways. First, it appears to contradict the known observation of “reckless DNA degradation” after UV irradiation in recA mutants catalyzed by the RecBCD enzyme (72) and therefore indicative of the formation of double strand ends, the only starting point of RecBCD-catalyzed DNA degradation (73, 74). The absence of this degradation in the recA recBC triple mutant was taken to mean that chromosomes still fragment, but the linear DNA degradation activity is absent. Our reports of no fragmentation in the recA recBC triple mutant provide an alternative explanation for the absence of DNA degradation after UV irradiation: in the recA mutant conditions, chromosomal fragmentation somehow depends on RecBCD enzyme. This discrepancy needs to be resolved.
Second, the RecA dependence of UV radiation-induced fragmentation also means that production of a double strand DNA ends via replication fork reversal is the only way for the cell to restart blocked replication forks and thus survive. A similar paradoxical situation was described by Michel and co-workers (75) in the dnaE(Ts) and dnaN(Ts) mutants at semipermissive temperature where increased fragmentation due to additional mutations was associated with a better viability. Recently, Long and Kreuzer (76, 77) reported that regression and breakage are used to reset stalled forks during bacteriophage T4 replication, so this turn of events may not be so paradoxical after all. DNA damage in general and UV irradiation in particular cause cancerous transformation through induction of genetic instability (7, 8, 29, 30); our results make it clear that at least in the case of UV irradiation a major contributor to this instability is chromosomal fragmentation (or at the least production of double strand DNA ends) catalyzed by the cell itself in response to blocked replication forks.
From the logic of the scheme in Fig. 5A, no dramatic measures like chromosome fragmentation are needed when only one of the two nascent DNA strands is blocked. Indeed, in scenario III, a daughter strand gap is formed by reinitiation downstream of the blocking lesion, the blocked replisome is reactivated (by RecA polymerization), and the replication fork moves away while the daughter strand gap is repaired via the RecFOR pathway (57). Scenarios I and II are different in that both nascent strands are blocked. In scenario II, the distance between the two blocking lesions allows formation of a sufficiently long (several turns?) RecA filament that then catalyzes strand exchange to effect the fork regression with subsequent breakage. The fork is then reassembled by the RecBCD pathway of recombinational repair and restarted (57). The assumption is that the original blocking UV lesions are by that time removed by excision. In scenario I, the two strands are blocked almost opposite each other (for example, the DNA stretch between them is mostly covered by the replisome), so there may be no efficient mechanism to drive fork regression in this case. Such a non-regressible fork may be irreversibly inactivated (as was suggested before (13, 14)).
Remarkably, an irreversible inactivation of a replication fork will translate into inactivation of the entire chromosome and, if this is the only chromosome in the cell, into genetic death (the inability to divide) of the cell. We wonder whether this hypothetical mechanism of genetic death is behind the well known but still unexplained observation that any type of repairable DNA damage if delivered in sufficient density will kill fully repair-proficient replicating cells long before their capacity to completely remove the induced lesions (or even their consequences) is saturated. Specifically, the LD37 of UV irradiation in our conditions is reached around 30 J/m2, which translates into ∼900 pyrimidine dimers per genome equivalent (16), the amount that from the published kinetics of nucleotide excision repair (12, 78) will be mostly removed within 1 h. However, instead of restarting replication forks after 1 h when almost no pyrimidine dimers remain in the DNA, two-thirds of the cells die. The nature of irreversible replication fork inactivation as well as characterization of chromosome fragmentation in wild type cells awaits future experimentation.
Supplementary Material
Acknowledgments
We thank Stephen Kowalczykowski (University of California-Davis), Bénédicte Michel (CNRS), and Elena Kouzminova and Kawai Kuong (both from this laboratory) for helpful suggestions and discussion of results.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 073115. This work was also supported by American Cancer Society Grant RSG-05-135-01-GMC.

This article contains supplemental Figs. S1 and S2 and Table S1.
- ss
- single-stranded.
REFERENCES
- 1. Seigneur M., Ehrlich S. D., Michel B. (1997) Blocking rolling circle replication with a UV lesion creates a deletion hotspot. Mol. Microbiol. 26, 569–580 [DOI] [PubMed] [Google Scholar]
- 2. Yi T. M., Stearns D., Demple B. (1988) Illegitimate recombination in an Escherichia coli plasmid: modulation by DNA damage and a new bacterial gene. J. Bacteriol. 170, 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fasullo M., Sun M. (2008) UV but not X rays stimulate homologous recombination between sister chromatids and homologs in a Saccharomyces cerevisiae mec1 (ATR) hypomorphic mutant. Mutat. Res. 648, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schiestl R., Wintersberger U. (1983) Induction of mating type interconversion in a heterothallic strain of Saccharomyces cerevisiae by DNA damaging agents. Mol. Gen. Genet. 191, 59–65 [DOI] [PubMed] [Google Scholar]
- 5. Dworaczek H., Xiao W. (2007) Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Crit. Rev. Oncog. 13, 159–177 [DOI] [PubMed] [Google Scholar]
- 6. Schnipper L. E., Chan V., Sedivy J., Jat P., Sharp P. A. (1989) Gene activation by induced DNA rearrangements. Cancer Res. 49, 6640–6644 [PubMed] [Google Scholar]
- 7. Epstein J. H. (1983) Photocarcinogenesis, skin cancer, and aging. J. Am. Acad. Dermatol. 9, 487–502 [DOI] [PubMed] [Google Scholar]
- 8. Rass K., Reichrath J. (2008) UV damage and DNA repair in malignant melanoma and nonmelanoma skin cancer. Adv. Exp. Med. Biol. 624, 162–178 [DOI] [PubMed] [Google Scholar]
- 9. Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2006) DNA Repair and Mutagenesis, pp. 227–266, ASM Press, Washington, D. C [Google Scholar]
- 10. Courcelle J., Carswell-Crumpton C., Hanawalt P. C. (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 94, 3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khidhir M. A., Casaregola S., Holland I. B. (1985) Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol. Gen. Genet. 199, 133–140 [DOI] [PubMed] [Google Scholar]
- 12. Rudolph C. J., Upton A. L., Lloyd R. G. (2007) Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 21, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Billen D. (1969) Replication of the bacterial chromosome: location of new initiation sites after irradiation. J. Bacteriol. 97, 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doudney C. O. (1973) Chloramphenicol effects on DNA replication in UV-damaged bacteria. Mutat. Res. 17, 1–12 [DOI] [PubMed] [Google Scholar]
- 15. Kelner A. (1953) Growth, respiration, and nucleic acid synthesis in ultraviolet-irradiated and in photoreactivated Escherichia coli. J. Bacteriol. 65, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rupp W. D., Howard-Flanders P. (1968) Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31, 291–304 [DOI] [PubMed] [Google Scholar]
- 17. Sedgwick S. G. (1975) Genetic and kinetic evidence for different types of postreplication repair in Escherichia coli B. J. Bacteriol. 123, 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonura T., Smith K. C. (1975) Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J. Bacteriol. 121, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thoms B., Wackernagel W. (1998) Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J. Bacteriol. 180, 5639–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang T. C., Smith K. C. (1986) Postreplicational formation and repair of DNA double-strand breaks in UV-irradiated Escherichia coli uvrB cells. Mutat. Res. 165, 39–44 [DOI] [PubMed] [Google Scholar]
- 21. Courcelle J., Hanawalt P. C. (2003) RecA-dependent recovery of arrested DNA replication forks. Annu. Rev. Genet. 37, 611–646 [DOI] [PubMed] [Google Scholar]
- 22. Higgins N. P., Kato K., Strauss B. (1976) A model for replication repair in mammalian cells. J. Mol. Biol. 101, 417–425 [DOI] [PubMed] [Google Scholar]
- 23. Bonura T., Town C. D., Smith K. C., Kaplan H. S. (1975) The influence of oxygen on the yield of DNA double-strand breaks in x-irradiated Escherichia coli K-12. Radiat. Res. 63, 567–577 [PubMed] [Google Scholar]
- 24. Dugle D. L., Gillespie C. J., Chapman J. D. (1976) DNA strand breaks, repair, and survival in x-irradiated mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 73, 809–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Resnick M. A. (1976) The repair of double-strand breaks in DNA; a model involving recombination. J. Theor. Biol. 59, 97–106 [DOI] [PubMed] [Google Scholar]
- 26. Bender M. A., Griggs H. G., Walker P. L. (1973) Mechanisms of chromosomal aberration production. I. Aberration induction by ultraviolet light. Mutat. Res. 20, 387–402 [DOI] [PubMed] [Google Scholar]
- 27. Difilippantonio M. J., Petersen S., Chen H. T., Johnson R., Jasin M., Kanaar R., Ried T., Nussenzweig A. (2002) Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Limoli C. L., Kaplan M. I., Phillips J. W., Adair G. M., Morgan W. F. (1997) Differential induction of chromosomal instability by DNA strand-breaking agents. Cancer Res. 57, 4048–4056 [PubMed] [Google Scholar]
- 29. Pierce A. J., Stark J. M., Araujo F. D., Moynahan M. E., Berwick M., Jasin M. (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol. 11, S52–S59 [DOI] [PubMed] [Google Scholar]
- 30. Rothkamm K., Löbrich M. (2002) Misrepair of radiation-induced DNA double-strand breaks and its relevance for tumorigenesis and cancer treatment (review). Int. J. Oncol. 21, 433–440 [PubMed] [Google Scholar]
- 31. Kouzminova E. A., Rotman E., Macomber L., Zhang J., Kuzminov A. (2004) RecA-dependent mutants in Escherichia coli reveal strategies to avoid chromosomal fragmentation. Proc. Natl. Acad. Sci. U.S.A. 101, 16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vilenchik M. M., Knudson A. G. (2003) Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. U.S.A. 100, 12871–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Budke B., Kuzminov A. (2010) Production of clastogenic DNA precursors by the nucleotide metabolism in Escherichia coli. Mol. Microbiol. 75, 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kouzminova E. A., Kuzminov A. (2004) Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol. Microbiol. 51, 1279–1295 [DOI] [PubMed] [Google Scholar]
- 35. Flores M. J., Bierne H., Ehrlich S. D., Michel B. (2001) Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J. 20, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seigneur M., Bidnenko V., Ehrlich S. D., Michel B. (1998) RuvAB acts at arrested replication forks. Cell 95, 419–430 [DOI] [PubMed] [Google Scholar]
- 37. Bradley M. O., Taylor V. I. (1983) Repair-induced DNA double strand breaks after ultraviolet-light and either aphidocolin or 1-β-d-arabinofuranosylcytosine/hydroxyurea. Carcinogenesis 4, 1513–1517 [DOI] [PubMed] [Google Scholar]
- 38. Michel B., Boubakri H., Baharoglu Z., LeMasson M., Lestini R. (2007) Recombination proteins and rescue of arrested replication forks. DNA Repair 6, 967–980 [DOI] [PubMed] [Google Scholar]
- 39. Michel B., Ehrlich S. D., Uzest M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J. 16, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kouzminova E. A., Kuzminov A. (2006) Fragmentation of replicating chromosomes triggered by uracil in DNA. J. Mol. Biol. 355, 20–33 [DOI] [PubMed] [Google Scholar]
- 41. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 42. Kuong K. J., Kuzminov A. (2009) Cyanide, peroxide and nitric oxide formation in solutions of hydroxyurea causes cellular toxicity and may contribute to its therapeutic potency. J. Mol. Biol. 390, 845–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dillingham M. S., Kowalczykowski S. C. (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brci-Kosti K., Salaj-Smic E., Marsi N., Kaji S., Stojiljkovi I., Trgovcevi Z. (1991) Interaction of RecBCD enzyme with DNA damaged by γ radiation. Mol. Gen. Genet. 228, 136–142 [DOI] [PubMed] [Google Scholar]
- 45. Marsden H. S., Pollard E. C., Ginoza W., Randall E. P. (1974) Involvement of recA and exr genes in the in vivo inhibition of the recBC nuclease. J. Bacteriol. 118, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rinken R., Wackernagel W. (1992) Inhibition of the recBCD-dependent activation of Chi recombinational hot spots in SOS-induced cells of Escherichia coli. J. Bacteriol. 174, 1172–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miranda A., Kuzminov A. (2003) Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163, 1255–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rinken R., Thomas B., Wackernagel W. (1992) Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J. Bacteriol. 174, 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang T. C., Smith K. C. (1983) Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J. Bacteriol. 156, 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuzminov A. (1995) Instability of inhibited replication forks in E. coli. BioEssays 17, 733–741 [DOI] [PubMed] [Google Scholar]
- 51. Morgan A. R., Severini A. (1990) Interconversion of replication and recombination structures: implications for terminal repeats and concatemers. J. Theor. Biol. 144, 195–202 [DOI] [PubMed] [Google Scholar]
- 52. Hanawalt P. C. (1966) The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem. Photobiol. 5, 1–12 [PubMed] [Google Scholar]
- 53. Kuzminov A. (1995) Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16, 373–384 [DOI] [PubMed] [Google Scholar]
- 54. Jonczyk P., Ciela Z. (1979) DNA synthesis in UV-irradiated Escherichia coli K-12 strains carrying dnaA mutations. Mol. Gen. Genet. 171, 53–58 [DOI] [PubMed] [Google Scholar]
- 55. Magee T. R., Kogoma T. (1990) Requirement of RecBC enzyme and an elevated level of activated RecA for induced stable DNA replication in Escherichia coli. J. Bacteriol. 172, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Courcelle J., Khodursky A., Peter B., Brown P. O., Hanawalt P. C. (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158, 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63, 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Churchill G. A., Daniels D. L., Waterman M. S. (1990) The distribution of restriction enzyme sites in Escherichia coli. Nucleic Acids Res. 18, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fedorova L. V., Dizadex I., Fedorov A. N., Ryskov A. P. (2001) In silico analysis of the restriction fragment length distribution in the human genome. Rus. J. Genet. 37, 358–367 [PubMed] [Google Scholar]
- 60. Otsuji N., Iyehara H., Hideshima Y. (1974) Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J. Bacteriol. 117, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. West S. C. (1997) Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31, 213–244 [DOI] [PubMed] [Google Scholar]
- 62. Briggs G. S., Mahdi A. A., Weller G. R., Wen Q., Lloyd R. G. (2004) Interplay between DNA replication, recombination and repair based on the structure of RecG helicase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Seigneur M., Ehrlich S. D., Michel B. (2000) RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol. Microbiol. 38, 565–574 [DOI] [PubMed] [Google Scholar]
- 64. Rossi M. J., Mazina O. M., Bugreev D. V., Mazin A. V. (2011) The RecA/RAD51 protein drives migration of Holliday junctions via polymerization on DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 6432–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bradley M. O., Taylor V. I. (1981) DNA double-strand breaks induced in normal human cells during the repair of ultraviolet light damage. Proc. Natl. Acad. Sci. U.S.A. 78, 3619–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. D'souza D. I., Harrison L. (2003) Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 31, 4573–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dianov G. L., Timchenko T. V., Sinitsina O. I., Kuzminov A. V., Medvedev O. A., Salganik R. I. (1991) Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 225, 448–452 [DOI] [PubMed] [Google Scholar]
- 68. Kouzminova E. A., Kuzminov A. (2008) Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Mol. Microbiol. 68, 202–215 [DOI] [PubMed] [Google Scholar]
- 69. Kuzminov A. (2011) in EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology (Böck A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Slauch J. M., Squires C. L., eds) ASM Press, Washington, D. C., doi 10.1128/ecosal.7.2.6 [DOI] [Google Scholar]
- 70. Pohlhaus J. R., Long D. T., O'Reilly E., Kreuzer K. N. (2008) The ϵ subunit of DNA polymerase III Is involved in the nalidixic acid-induced SOS response in Escherichia coli. J. Bacteriol. 190, 5239–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Goss W. A., Deitz W. H., Cook T. M. (1965) Mechanism of action of nalidixic acid on Escherichia coli. II. Inhibition of deoxyribonucleic acid synthesis. J. Bacteriol. 89, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Willetts N. S., Clark A. J. (1969) Characteristics of some multiply recombination-deficient strains of Escherichia coli. J. Bacteriol. 100, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Prell A., Wackernagel W. (1980) Degradation of linear and circular DNA with gaps by the recBC enzyme of Escherichia coli. Effects of gap length and the presence of cell-free extracts. Eur. J. Biochem. 105, 109–116 [DOI] [PubMed] [Google Scholar]
- 74. Taylor A. F., Smith G. R. (1985) Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 185, 431–443 [DOI] [PubMed] [Google Scholar]
- 75. Florés M. J., Sanchez N., Michel B. (2005) A fork-clearing role for UvrD. Mol. Microbiol. 57, 1664–1675 [DOI] [PubMed] [Google Scholar]
- 76. Long D. T., Kreuzer K. N. (2008) Regression supports two mechanisms of fork processing in phage T4. Proc. Natl. Acad. Sci. U.S.A. 105, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Long D. T., Kreuzer K. N. (2009) Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep. 10, 394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Courcelle J., Crowley D. J., Hanawalt P. C. (1999) Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol. 181, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.