Abstract
Molecular mechanisms by which fertilization competent acrosome-reacted sperm bind to the oolemma remain uncharacterized. To identify oolemmal binding partner(s) for sperm acrosomal ligands, affinity panning was performed with mouse oocyte lysates using sperm acrosomal protein, SLLP1 as a target. An oocyte specific membrane metalloproteinase, SAS1B (Sperm Acrosomal SLLP1 Binding), was identified as a SLLP1 binding partner. cDNA cloning revealed six SAS1B splice variants, each containing a zinc binding active site and a putative transmembrane domain, with signal peptides in three variants. SAS1B transcripts were ovary specific. SAS1B protein was first detected in early secondary follicles in day 3 ovaries. Immunofluorescence localized SAS1B to the microvillar oolemma of M2 oocytes. After fertilization, SAS1B decreased on the oolemma and became virtually undetectable in blastocysts. In transfected CHO-K1 cells SAS1B localized to the surface of unpermeabilized cells. Recombinant and native SLLP1 co-localized with SAS1B to the microvillar domain of ovulated M2 oocytes. Molecular interactions between mouse SLLP1 and SAS1B were demonstrated by surface plasmon resonance, far-western, yeast two-hybrid, recombinant- and native- co-IP analyses. SAS1B bound to SLLP1 with high affinity. SAS1B had protease activity, and SAS1B protein or antibody significantly inhibited fertilization. SAS1B knockout female mice showed a 34% reduction in fertility. The study identified SAS1B-SLLP1 as a pair of novel sperm-egg binding partners involving the oolemma and intra-acrosomal compartment during fertilization.
Keywords: Oocyte, Sperm, Fertilization, SLLP1, SAS1B, sperm-egg binding
Introduction
Among the events of fertilization few are more important, but as enigmatic, as interactions between the sperm and egg membranes. Although a few sperm proteins that bind to the mammalian oolemma have been identified, there has been no success in identifying the complimentary oocyte binding partners for these sperm ligands. Molecules that are posited to be involved in sperm-oolemmal binding and fusion include the ADAM family ligands and their oocyte integrin receptors (Almeida et al., 1995; Evans et al., 1995; Evans et al., 1997; Yuan et al., 1997). However, gene targeting studies have demonstrated that the sperm ADAMs including fertilin α (ADAM1), fertilin β (ADAM2) and cyritestin (ADAM3) are important primarily for the process of zona pellucida binding and oviduct migration rather than for gamete fusion (Cho et al., 1998; Shamsadin et al., 1999; Nishimura et al., 2001). Attention has also focused on tetraspanins (e.g., CD9, CD81), GPI-anchored proteins, and PIG-A all of which are expressed on oocytes. However, data suggest that these proteins are important for the sperm–oocyte fusion step, not for the binding process (Coonrod et al., 1999; Miyado et al., 2000; Alfieri et al., 2003). Although CD9−/− female mice produced eggs that matured normally, sperm-egg fusion failed in these animals (Kaji et al., 2000; Le Naour et al., 2000). Targeted disruption of CD81 resulted 40% reduction in fertility of female mice and female mice lacking both CD9 and CD81 were completely infertile, indicating their complementary roles in sperm–egg fusion (Rubinstein et al., 2006). It is noteworthy that sperm ligands that interact with oolemmal tetraspanins have not been identified.
Epididymal protein DE (CRISP1) has also been implicated in sperm-oocyte fusion (Cohen et al., 2000) and a specific binding region within CRISP1 was mapped. However, CRISP1 knockout male and female mice showed no differences in fertility compared to controls (Da Ros et al., 2008). Recently, Izumo, an Ig-domain molecule localized within the acrosome was shown to be essential for sperm-egg fusion (Inoue et al., 2005) although its oolemmal binding partner remains unknown.
In view of the very limited knowledge on oolemmal binding partners for sperm ligands and with the aim of characterizing molecular targets for contraception, it is necessary to identify both sperm and oolemma specific interacting proteins involved in the process of fertilization. The unique testis-specific c lysozyme-like, intra-acrosomal transmembrane protein SLLP1, was reported to lack bacteriolytic activity (Mandal et al., 2003), localize to mouse sperm acrosomal membranes, and have oolemma binding properties (Herrero et al., 2005). SLLP1 antibody and recombinant protein blocked in vitro fertilization and sperm-egg binding in mice, suggesting that SLLP1 may play a role in sperm-egg adhesion. To identify oocyte specific binding partners for sperm ligands, sperm acrosomal SLLP1 was used as a target in affinity panning. This study characterizes a SLLP1 binding partner, SAS1B, on the oolemma, and defines the timing and pattern of its expression in developing and adult ovaries, oocytes and early embryos. SAS1B appears to be the only oocyte specific oolemmal metalloprotease yet implicated in sperm-oolemma binding during mammal fertilization.
Materials and methods
Identification of SAS1B by surface plasmon resonance (SPR)
Cumulus and zona free mouse oocytes (n= ~1000) were suspended in 500 μl of Dulbecco’s PBS, freeze–thawed 3 times (−80°C and 37°C), and mixed. The mixtures were spun for 5 min at 13000 xg, and the supernatants were passed over a Biacore Sensor Chip CM5 (GE healthcare, Piscataway, NJ) containing bound mouse soluble recombinant (r) SLLP1 (binding concentration used at 1 μg/μl, 200 μl) or no SLLP1 (negative control) at a flow rate of 10 μl/min at 25°C. The chips were then washed in PBS and eluted in 5 μl of 0.1% trifluoroacetic acid at a flow rate of 12 μl/min at 25°C. For each SPR analysis, 7 to 10 samples bound to the chip were collected and pooled. The pooled samples (size ~35 to 50 μl), were digested with trypsin and analyzed by mass spectrometry in the Biomolecular Research Facility (BRF) at the University of Virginia. Two oocyte specific SAS1B peptides were identified.
Expression and purification of mouse SAS1B, SLLP1, and Ecat1
BLAST analysis of the 2 peptides identified by SPR matched a hypothetical 414 amino acid mouse protein, GenBank accession NP_766127. A mouse ovarian cDNA library (Ambion, Austin, TX) was amplified with gene-specific forward (5′ GGA ATT CCA TAT GGG AGC ACC CTC AGC ATC CAG A 3′) and reverse primers (5′ ATA GTT TAG CGG CCG CGT CTC TGG GCA CCT CTC TAA TGT G 3′) containing NdeI and NotI restriction sites, respectively (Invitrogen, Carlsbad, CA). The 40 cycle PCR product (~1200 bp) was subcloned in pCR2.1-TOPO vector (Invitrogen). The insert was used to express recombinant (r) protein from pET28b+ vector following transformation of BL21DE3 cells and the expressed protein was purified by affinity column chromatography as described earlier (Mandal et al., 2003; Herrero et al., 2005). Mouse rSLLP1 was purified as described previously (Herrero et al., 2005).
Mouse Ecat1 (Filia / 2410004A20Rik) specific forward (5′ CAT GCC ATG GCC TCT CTG AAG AGG TTT CAG 3′) and reverse primers (5′ CCG CTC GAG TAA TAT GAT AAA TGA TTC CCA GGC 3′) containing NcoI and XhoI restriction sites, respectively were used to amplify cDNA encoding the full length protein (440 amino acids) from mouse ovarian cDNA (Ambion). The PCR amplified product was sequenced, and recombinant mouse Ecat1 protein was expressed and purified as described above and used as a negative control in IVF assays.
Polyclonal antibody production and Western analyses
Following collection of pre-immune sera by heart puncture, adult male guinea pigs were immunized with 150 μg of purified rSAS1B in complete Freund’s adjuvant, boosted twice at intervals of 21 days with 150 μg of rSAS1B in incomplete Freund’s adjuvant, and serum was collected as described previously (Herrero et al., 2005). Specificity of the antibody was tested against rSAS1B and mouse oocyte extracts. rSAS1B protein (125 ng/lane) or oocytes (~150/lane) were solubilized in 2x Laemmli buffer, resolved on 12.5% SDS-PAGE Criterion gels (Bio-Rad, Hercules, CA), blotted to nitrocellulose membrane (0.2 μm, Bio-Rad) and blocked with 5% nonfat dry milk in phosphate buffered saline (PBS) with 0.05% Tween 20 (PBST) for 45 min at room temperature. The blots were then probed with primary antibody (1:5000 for rSAS1B, 2h; 1:1000 for oocytes, overnight), washed 10 min in PBST (3x), and incubated with peroxidase-conjugated donkey or goat anti-guinea pig IgG secondary antibody (1:5000 for rSAS1B, 1:2500 for oocytes) for 1 h. The blots were washed in PBST (10 min, 2x), in PBS (10 min, 3x) and then developed in TMB peroxidase substrate (KPL, Gaithersburg, MD).
Protease activity assay of rSAS1B
Protease activity was assayed in serial dilutions (0–2000 ng) of purified bacterial recombinant SAS1B using the EnzChek® Peptidase/Protease Assay Kit (Molecular Probes, Eugene, OR), which provides a FRET (fluorescence resonance energy transfer) -based method for quantitation of a wide range of protease activities. This assay was performed in 100 μl size reactions using a constant substrate concentration and repeated three times with varying concentrations of recombinant SAS1B. Fluorescence was measured after 24 h of incubation at room temperature (23–25°C) following the manufacturer’s protocol. Protease activity of recombinant SAS1B appeared within 1 h and increased within 24 h. rSLLP1 was used as a negative control. The assay method was validated using purified trypsin as a positive control (Sigma).
Immunohistochemistry (IHC) of SAS1B
Newborn (days 0–7), pubertal (day 14) and adult mouse (day ≥56) ovaries were fixed in Bouin’s fixative and embedded in paraffin. Sections were heated at 56°C, deparaffinized in xylene and endogenous peroxidase activity was quenched followed by rehydration. Sections were then blocked with 5% non-fat dry milk in PBS (NFDM-PBS) containing 5% normal goat serum for 1 h at room temperature and probed with immune or pre-immune antibody (dilution 1:200) in blocking solution at 4°C overnight in a humid chamber. The slides were washed in PBS (x3) and probed with HRP-labeled goat anti-guinea pig 2 secondary antibody (Jackson ImmunoResearch, West Grove, PA) diluted (1:1000) in 1% NFDM-PBS for 1 h at room temperature in a humid chamber. After washing, the immunoperoxidase color reaction was developed by using 3,3′-diaminobenzidine substrate (Sigma), counterstained with haematoxylin for 3 min, dehydrated through an increasing series of alcohol concentrations, cleared in xylene and mounted in DPX medium (Sigma). Slides were examined on a Zeiss Axioscope microscope (Carl Zeiss, GmbH, Germany).
Indirect Immuno-Fluorescence (IF) microscopy of oocytes and early embryos
Metaphase II eggs and subsequent embryonic stages including 2, 4, 8 cells and blastocyst stages were collected and blocked in 5% NGS/TYH media for 30 min. Samples were then incubated with SAS1B antibody (1:200) in 0.1%NGS/media for 1 h at 37°C, 5% CO2. Preparations were washed five times and incubated with goat anti-guinea pig/Cy3 red antibody (1:200, Jackson ImmunoResearch) in 0.1%NGS/media for 1 h at 37°C, 5% CO2, washed again, mounted in media on glass slides, and visualized under a Zeiss Standard 18 ultraviolet microscope.
For rSLLP1 co-localization, oocytes were incubated with 10 μg/ml of rSLLP1 for 1 h, washed 5x and incubated with guinea pig SAS1B antibody (1:200) and rat SLLP1 antibody (1:200), simultaneously, in 0.1%NGS/media for 1 h. Oocytes were then washed 5x and incubated with goat anti-guinea pig/Cy3 red antibody (1:200) and goat anti-rat/FITC green antibody (1:200; Jackson ImmunoResearch), in 0.1%NGS/media for 1 h. Oocytes were washed and mounted in media on glass slides. SLLP1 and SAS1B images were captured separately with MrGrab 1.0 (Carl Zeiss Vision) on the Zeiss Standard 18 ultraviolet microscope and were merged digitally to evaluate their respective localizations.
For native (n) SLLP1 co-localization with SAS1B, oocytes were incubated with acrosomal extract prepared from cauda sperm capacitated for 90 min and acrosome reacted for 15 min in 5 μM calcium ionophore A23187 (Herrero et al., 2005). Exocytosed acrosomal supernatant material was collected at 16000 x g for 10 min and dialysed in PBS for 4 h. Ten μl were used in 100 μl fertilization media containing zona-intact oocytes as described above for 1 h at 37°C in 5% CO2. The eggs were then washed and probed with rat SLLP1 and guinea pig SAS1B antibodies and respective 2 secondary antibodies (anti-rat/Cy3 red; anti-guinea pig/FITC green).
Scanning confocal microscopy of eggs and early embryos
Mouse oocytes and early embryos were also studied by scanning confocal microscopy. The stained preparations were washed three times in PBS containing 1% BSA (PBS/BSA) and then fixed in 4% paraformaldehyde in PBS-polyvinylalcohol (PVA) for 20 min at room temperature. Following fixation, oocytes and embryos were washed 5 times in PBS/BSA and permeabilized with 0.5% Triton X-100 in PBS for 20 min at room temperature. Specimens were washed five times in PBS/BSA, placed in 0.4 mg/ml RNase (Sigma) in PBS/BSA for 30 min, and stained with 20 nM Sytox (Molecular Probes) for 10 min. Oocytes and embryos were then extensively washed, placed in slow-fade equilibration media (Molecular Probes) for ~1 min, and mounted on slides in slow fade mounting media. Images were obtained on a Zeiss 410 Axiovert 100 microsystems LSM confocal microscope. For each panel, 4-sec scans were averaged four times per line using a 63x oil lens equipped with a zoom factor of two. Attenuation, contrast, brightness, and pinhole aperture remained constant.
Northern analysis of SAS1B mRNA
Mouse multiple tissues Northern blots (total RNA, ~20 μg/lane) were obtained commercially (Zyagen, San Diego, CA). The Northern membrane was blocked in ExpressHyb hybridization solutions (Clontech, Mountain View, CA) at 68°C for 1h in hybridization oven (GE Healthcare). The blot was then probed with fresh hybridization solution containing 32P-dCTP labeled SAS1B ORF cDNA (4.3 ng/ml) for 2h at 68°C. The membrane was washed in wash solution I and II as described earlier (Naaby-Hansen et al., 2002). The membrane was then exposed to phosphor screen for 24 to 72 h, imaged using PhosphorImager (Storm 820, GE Healthcare) and analyzed by ImageQuant software. The same membrane was stripped at 0.5% SDS in sterile water for 10 min at 90 – 98°C and probed with 32P-dCTP labeled GAPDH cDNA as a positive control.
Localization of SAS1B in transformed CHO-K1 Cells
Full length SAS1B was cloned into pcDNA3.1/V5/His-TOPO vector (Invitrogen) from a PCR product generated from a mouse ovary cDNA library (Ambion) using gene-specific forward (5′ATG GGT ATC ATG GGA AGC CTG TGG 3′) and reverse primers (5′GTC TCT GGG CAC CTC TCT AAT GTG 3′). Adherent Chinese hamster ovary (CHO-K1) cells maintained in Ham’s F-12 nutrient medium (F12-Ham 1X; Invitrogen, containing 10% fetal calf serum, 1mM sodium pyruvate) at 37°C in humidified 5% CO2 were used for transfection at ~50–70% confluence. Before transfection, cells were seeded on 6-well plates containing poly-D-lysine coated 12 mm round coverslips and transfected with Lipofectamine™ 2000 for 5 h at 37°C (pDNA: lipofectamine; 4μg:10μl, per well) in 500 μl of Opti-MEM (Invitrogen) followed by incubation in fresh F12 media for 48 h.
The cells were fixed briefly with 4% paraformaldehyde in PBS, blocked with 10% FBS (Fetal Bovine Serum) in PBS for 30 min, and a cohort was permeabilized with 0.05% NP-40. Cells were incubated with SAS1B antibody for 1 h or mouse anti-V5 tag monoclonal antibody, and immune-complexes were visualized with FITC-donkey anti-guinea pig or mouse antibody (Jackson ImmunoResearch). Cover slips were mounted with slowfade (Molecular Probes), sealed with nail polish, and visualized by ultraviolet microscopy as noted above.
Far-Western analysis
Purified recombinant SAS1B was resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membrane was probed with c-terminal anti-his monoclonal antibody and developed with TMB. Duplicate SAS1B blotted strips were blocked and overlaid with mouse rSLLP1 or not, 5 μg/ml in blocking buffer i.e., 5% milk in PBS with 0.05% Tween 20 [PBST] for 90 min at room temperature. Following overlay, the strips were washed in PBST and incubated with anti-SLLP1 (ADD2) monoclonal antibody for 1h in blocking buffer. After washing in PBST (x4), the strips were probed with goat anti-mouse HRP secondary antibody in blocking buffer for 1h, washed in PBST (x4) followed by PBS (x1), and developed with TMB.
In vitro translation and co-immunoprecipitation (co-IP)
The pGADT7 and pGBKT7 expression plasmids (Clontech), which encode N-terminal HA and c-Myc epitopes, respectively, were used to produce rSLLP1 and rSAS1B proteins in vitro, utilizing the Quick Coupled Transcription/Translation (TNT) system (Promega, Madison, WI). The TNT reactions were performed for 90 min at 30°C using plasmid (≤ 2 μg), TNT reagents, rabbit reticulocyte lysate, and 35S-methionine (GE Healthcare) in 50 μl aliquots as per manufacturer’s protocol. The radiolabeled proteins (5 μl of TNT product in 2x reducing sample buffer) were resolved by 15% SDS-PAGE Criterion gels, fixed (50% methanol, 10% glacial acetic acid) for 30 min, equilibrated in 10% glycerol (with 20% ethanol) for 30 min, air dried for 1h (GelAir drying system, Bio-Rad) and exposed to phosphor screens.
For Co-IP reactions, the specific TNT products, ranging from 7 to 23 μl were gently mixed for 1h at 4°C. Simultaneously the anti-c-Myc and anti-HA antibody coupled agarose beads (Profound IP/Co-IP system, Pierce) were blocked in 5% milk in PBST for 1h at 4°C in Handee spin columns (Pierce). The antibody beads were incubated with IP or Co-IP samples overnight at 4°C with gentle end-over-end mixing. The beads were then washed 3x in 500 μl of PBST at 4000 x g for 10 sec with 3 gentle mixes during each wash. The IP and Co-IP products were eluted from beads in 23 μl of 2x non-reducing sample buffer, heated (99°C, 5 min), centrifuged, mixed with 2 μl of β-mercaptoethanol, and 20 μl loaded per lane for SDS-PAGE analysis. The gels were fixed, air dried, exposed to phosphor screens, and imaged as described above.
Yeast Two-Hybrid (Y2H) Analysis
The mature mSLLP1 protein (C-terminal 128 residues) was cloned as a fusion to the GAL4 activation domain in the pGADT7 vector while full length SAS1B (414 residues), N-terminal SAS1B (121 residues), and C-terminal SAS1B (210 residues) proteins were each fused to the GAL4 DNA-binding domain in the pGBKT7 vector utilizing EcoRI and BamHI sites (Yeast two-hybrid system 3, Clontech). Competent strain AH109 yeast cells were transformed with single, double or empty constructs, and plated on low (LS) and high stringency (HS) plates. Selection on LS plates confirmed the co-transformation by both plasmids, which supplied the deficient amino acids (tryptophan and leucine from pGBKT7 and pGADT7 vectors, respectively). The HS plates, deficient in adenine and histidine in addition to tryptophan and leucine, were used for evaluation of interaction between the test plasmids. This Y2H system 3 uses the expression of three reporter genes – ADE2, HIS3, and MEL1 (or LacZ) – under the control of GAL4 upstream activating sequences and TATA boxes. The ADE2 and HIS3 reporters provide strong nutritional selections to the yeast strain, AH109. Furthermore, the expression of MEL1 gene, which encodes a secretory α-galactosidase, was used to screen and visualize the blue colonies indicative of positive interaction by incorporating X-α-gal into the culture plates. The cells were grown at 30°C for 5 to 6 days. The experiments were repeated 3 times.
Native SAS1B co-IP
Mouse cauda (n=10) epididymal sperm and cumulus free zona-intact (55%) and zona-free (45%) oocytes (n=2069) were extracted in RIPA buffer in presence of complete protease inhibitors (Roche, Indianapolis, IN). The sperm (450 μl) and egg extracts (95 μl) were mixed (3 h, 4°C, x2), incubated with rat SLLP1 preimmune or immune antibody (3 h) and treated with 70 μl of protein G-agarose beads (Roche) overnight at 4°C. The beads were then washed in the presence of protease inhibitors in wash buffer 1 (20 min, x2), wash buffer 2 (20 min, x2) and wash buffer 3 (20 min, x1). The IP complexes were then eluted in 75 μl 2x Laemmli sample buffer, divided equally, resolved in 15% or 10% SDS-PAGE, blotted and probed with guinea pig SLLP1 or SAS1B antibodies. Additionally, the IP blots were stripped (Pierce) and again probed with a rabbit anti-human SAS1B pro-peptide antibody (Abcam, Cambridge, MA).
Affinity between SLLP1 and SAS1B
The protein-protein interaction affinity between SLLP1 and SAS1B in terms of dissociation constant (KD) was determined by surface plasmon resonance with a Biacore 3000 system (GE healthcare). A CM-5 sensor (carboxylated dextran matrix) chip was activated with a 1:1 mixture of 75 mg/ml 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 11.5 mg/ml N-hydroxysuccinimide for 7 min. Briefly, mouse rSLLP1 (60 μg/ml in 10 mM sodium acetate pH 4.0) was injected over an activated CM-5 chip for 7 min at a flow rate of 10 μl/min at 25 °C. Remaining active groups on the matrix were blocked in 1 M ethanolamine at pH 8.5. Coupling of SLLP1 on CM-5 sensor chip resulted in a surface concentration of ~10 ng/mm2. Protein solutions of SAS1B (0, 25, 50, 100, 200, 300, 400 nM) were prepared in 10 mM HEPES buffer, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.0025% surfactant P20 (running buffer) and injected for 3 min at a flow rate of 30 μl/min. After 15 min for dissociation, the surface was regenerated with 10 mM glycine-HCl buffer, pH 3.0, for 60 sec followed by a 15 min wash in running buffer at 30 μl/min. The fitting of association and dissociation curves according to a 1:1 binding model was used for the calculation of kinetic constants (Biaevaluation software version 4.1).
In vitro fertilization (IVF)
ICR mice were used in all experiments. All animals were handled according to institutional guidelines. Epididymal sperm and oocytes were collected as described previously (Herrero et al., 2005). For evaluation of rSAS1B effect on IVF, cauda sperm were placed for 5 min in 200 μl drops of fertilization medium under paraffin oil. The sperm suspension was diluted to a concentration of 106 sperm/ml in a volume of 200 μl and then incubated for 120 min in a humidified tissue culture incubator (37°C, 5% CO2 in air) to allow capacitation. The spermatozoa were incubated with varying concentrations of purified rSAS1B (0–100 μg/ml PBS) or purified mouse Ecat1 (50 μg/ml PBS) or PBS only for the last 60 min of capacitation. One cumulus mass was placed in 135 μl drop of fertilization medium under paraffin oil, and 15 μl of the sperm suspension (final: 105 sperm/ml) was added to each drop. Six hours following insemination, oocytes were relocated to 100 μl drops of fertilization medium under mineral oil. Following overnight incubation, eggs were stained in 10 μg/ml Hoechst dye for 10 min, washed 3 times in fertilization medium and examined as described earlier (Herrero et al., 2005). Two-cell embryos and eggs with double pronuclei (representing only 1 to 2% of fertilized eggs) were scored as fertilized, while one-nucleus oocytes were scored as unfertilized. For evaluation of antibody effects, cumulus free zona-intact oocytes were treated with SAS1B preimmune or immune antibody at 1:20 or 1:60 dilutions for 1 h followed by insemination with capacitated sperm and scored for fertilization as described above.
Generation of SAS1B deficient mice
The SAS1B protein is encoded by the Astl gene (astacin-like metalloendopeptidase). The Astl knockout mice were obtained from Lexicon Pharmaceuticals (The Woodlands, TX). Briefly, the employed methods were as follows. The Astl targeting vector was derived using long-range PCR to generate the 5′and 3′ arms of homology using 129S5 ES cell DNA as a template. The 4940 bp 5′ arm was generated using primers Astl-3 (5′-AAT GGC GCG CCT CAA GAT AAT TAG CAT ATC CAT CGG -3′) and Astl-2 (5′-TAA ATG GCC GCT ATG GCC GAG AGA GGG CAG CTC AGA GTT AAA T -3′) and cloned using the TOPO (Invitrogen) cloning kit. The 2869 bp 3′ arm was generated using primers Astl-7 (5′-TTA ATG GCC AGC GAG GCC CTC AGG CCA GGG CTG GAG TTG AGG A -3′) and Astl-9 (5′-AAT GGC GCG CCC CCA TAA TGC ATC ACA GAT GAG TAG -3′) and cloned using the TOPO cloning kit. The 5′ arm was excised from the holding plasmid using AscI and SfiI. The 3′ arm was excised from the holding plasmid using SfiI and AscI. The arms were ligated to an Sfi I prepared selection cassette containing a negative selection marker thymidine kinase (TK), a Betagalactosidase (LacZ) marker along with a MC1 promoter driven Neomycin (Neo) resistance marker and inserted into an AscI cut pKO Scrambler vector (Stratagene) to complete the Astl targeting vector which resulted in the deletion of exons 5, 6 and 7 (NCBI genomic accession NT_039207.7) encoding a total of 131 residues (SPF – ILP), including the putative transmembrane and the catalytic domains. The Not I linearized targeting vector was electroporated into 129S5 ES cells. G418/FIAU resistant ES cell clones were isolated, and correctly targeted clones were identified and confirmed by Southern analysis using a 243 bp 5′ external probe (18/19), generated by PCR using primers Astl-18 (5′-AGG CCT TTG ACT TTG TTA AGC A -3′) and Astl-19 (5′-CCA TAT CAG AGC AGC CGT CAT C -3′) and a 484 bp 3′ external probe (20/21), amplified by PCR using primers Astl-20 (5′-TTG CCA AGC ATC TGT GAT CCT A -3′) and Astl-21 (5′-CAG GTC TGC ATT GCC ATA CCA G -3′). Southern analysis using 5′-probe detected a 10.1 kb wild type band and 8.4 kb mutant band in ApaLI digested genomic DNA while 3′-probe detected a 7 kb wild type band and 12 kb mutant band in ApaI digested genomic DNA. Six targeted ES cell clones were identified and microinjected into C57BL/6 (albino) blastocysts to generate chimeric animals which were bred to C57BL/6 (albino) females, and the resulting heterozygous offspring were interbred to produce homozygous Astl deficient mice.
Genotyping and fertility evaluation of SAS1B knockout mice
Genomic DNA was extracted from tail tips of mice using Sigma Redextract-N-amp reagents (Sigma-Aldrich) and their genotypes were determined by PCR with a mixture of three primers (exon 7 forward, WF: 5′-GCT TCT GGC ATG AGC ATT CA -3′; neo forward, NF: 5′-CGT TGG CTA CCC GTG ATA TTG -3′; intron 7 reverse, WR: 5′-GGA CAC TGC CAA CCT CAC ATT -3′). The PCR conditions were 35 cycles of 94 °C for 30 sec, 59 °C for 30 sec and 72 °C for 90 sec. In vivo fertility of was evaluated by caging 2 females (−/−, +/+) with 1 male (+/+) for two weeks. The females were then caged separately for fetal or litter size count. For determination of fetal count, the female mice were sacrificed on day 15 of pregnancy.
Statistical methods
All in vitro fertilization assays were done at least 3 or more times. Experimental and control groups were reported as mean ± standard error of mean. Statistical significance between the groups was analyzed by unpaired Student’s t-test assuming equal variances and differences were reported at P ≤0.05 as the level of significance.
Results
Identification of SAS1B
To identify putative oolemmal partner(s) for SLLP1, ligand affinity panning was performed with mouse egg lysate using immobilized rSLLP1 on a Biacore sensor chip. Peptide analysis of eluted proteins matched to several proteins including an un-annotated gene which showed a pattern of ESTs in the GenBank database indicating restricted expression in ovary, oocytes and zygote. The protein was designated sperm acrosomal SLLP1 binding, SAS1B. Six SAS1B splice variants were cloned and deposited to GenBank (Suppl. Fig. 1). All six variants contained the zinc binding active site motif, a characteristic of zinc dependent metalloproteases, and a putative transmembrane domain. Three SAS1B variants (V1, V4, V6) revealed a putative signal sequence followed by a predicted cleavage site between residues SMG-AP. SAS1B variants revealed deletions of 34 residues from exons 4 and 5 (V3, V4) or deletion and insertion in exon 5 (V5, V6). SAS1B was found to be conserved among mammals (Suppl. Fig. 2, 67% identity to human) and its homologues can be traced to lower invertebrates (identity, 42% zebrafish; 36% nematode). Alignment revealed conservation of signal peptide and the zinc binding signatures.
SAS1B isoforms and protease assay
The rSAS1B (V2), expressed in E. coli with a histidine tag and purified by affinity column chromatography, revealed two major bands (~50 & ~25 kD) and several minor bands (Fig. 1A, L4). Each of the affinity purified bands identified by Coomassie was confirmed by anti-his tag monoclonal antibody Western (Fig. 1A, L5). The ~25 kD band was found by Edman degradation to be a 210 residue C-terminal fragment that begins at aa 205 (V2, Suppl. Fig. 1). The C-terminal fragments appear to result from proteolysis during purification.
Figure 1.
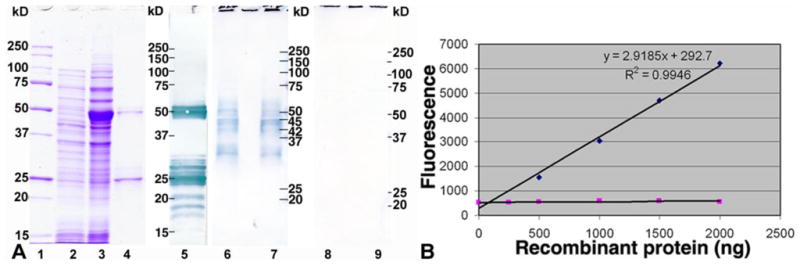
Expression, purification, Western and protease activity of rSAS1B. (A) The mature protein was expressed in E. coli, uninduced (L2), induced (L3), purified (L4, 3 μg) and stained with Coomassie, protein standards (L1). All purified bands were confirmed as rSAS1B by anti-His tag Western analysis (L5). SAS1B antibody recognized a range of native proteins from ~54 to ~31 kDa in zona intact (L6) and zona free (L7) oocyte protein extracts but no proteins were identified with preimmune antibody in the identical extracts (L8 and L9). (B) Assay of protease activity of rSAS1B (◆) using a fluorescent tagged synthetic peptide as substrate. Varying concentrations of purified proteins were used in 100 μL assay system. rSLLP1 (■) was used as a negative control.
SAS1B antibody recognized the recombinant (Suppl. Fig. 3A, L2), and the native SAS1B in egg protein extracts. Zona-intact and zona-free mouse egg extracts showed essentially identical microheterogeneity of immunoreactive bands of native SAS1B between ~54 and ~31 kD (Fig 1A, L6, L7), with major bands of ~45/44 (V2, V6, / V4) and ~31 kD, and minor bands of ~54, ~51, ~48 (V1), and ~41/42 kD (V3, V5). Pre-immune control antibody showed no immunoreactivity with egg protein extracts (Fig 1A, L8, L9) or rSAS1B (Suppl. Fig. 3A, L1). SAS1B protein micro-heterogeneity likely corresponds to the six splice variants, which are predicted to encode proteins of 47.5 (V1), 45.2 (V2), 41.4 (V3), 43.7 (V4), 42.4 (V5) and 44.8 (V6) kD, while the lower ~31 kD protein may be a processed form. Protein heterogeneity of the splice variants was confirmed by comparing the masses of three SAS1Bisoforms (V2, V3, and V5) expressed in E. coli, resulting in the expected molecular weight differences (data not shown). The presence of masses higher than those deduced from primary sequences may suggest possible post-translational modifications of SAS1B isoforms. All SAS1B variants revealed two putative N-glycosylation sites (aa 266, 280, V1).
Purified rSAS1B was tested for protease activity using a fluorescent conjugated synthetic peptide substrate (Fig. 1B). rSAS1B exhibited concentration dependent proteolytic activity assayed by fluorescence resonance energy transfer (FRET) over the range 200–2000 ng. rSAS1B demonstrated protease activity within one hour (Suppl. Fig. 3B) and fluorophore conjugated substrate hydrolysis was linear with respect to concentration at 24 h (Fig. 1B). Mouse rSLLP1 purified by identical procedures to rSAS1B from the same E. coli strain showed no enzymatic activity. This result indicated that E. coli expressed rSAS1B refolded sufficiently to retain proteolytic activity.
SAS1B expression in ovary and oocyte
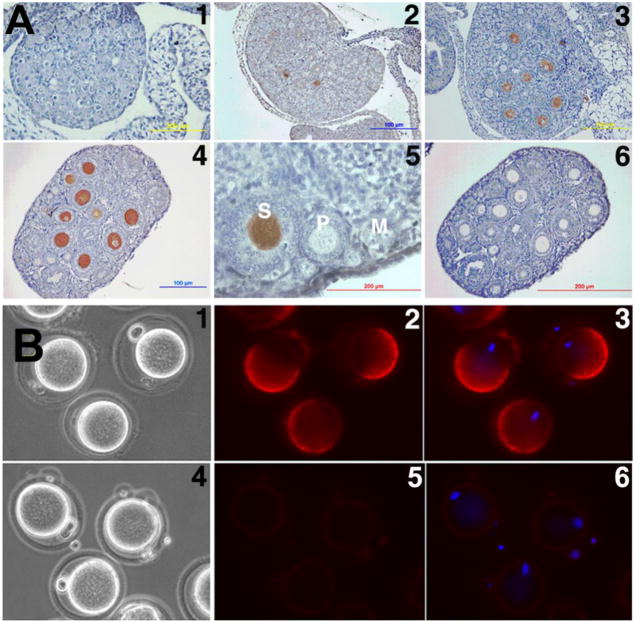
To follow SAS1B ontogeny, ovaries from postnatal (PN) days 0, 1.5, 2, 3, 4, 7, 14, 28 and 56 were studied by immunohistochemistry (IHC). SAS1B protein was first detected in the ooplasm of early bilaminar secondary follicles beginning with PN day 3 (Fig 2A2). SAS1B was not detected in naked oocytes, oocytes in primordial or unilaminar primary follicles, or in any other ovarian cells at pre-pubertal, pubertal or adult stages (Fig 2A). SAS1B expression was localized only in secondary, antral and Graafian follicle oocytes.
Figure 2.
Localization of SAS1B in developing ovary by IHC and in M2 oocytes by IF. (A) SAS1B expressed in ooplasm of secondary (S) follicles beginning in day 3 ovary (A2). No SAS1B was detected in naked oocytes (A1), primordial (M) and primary (P) follicles (A5). Panels 1, 2, 3, 4, 5 and 6 represent postpartum ovaries at day 0, 3, 7, 14, 56 and 14 respectively. Preimmune control, A6. (B) In unpermeablized zona-intact ovulated M2 oocytes, SAS1B was concentrated in a dome shaped microvillar domain on the surface of the oocyte plasma membrane. Eccentric nuclei (blue) were antipodal to the SAS1B positive domain. Preimmune control panels: 4, 5 and 6. Panels: 1 and 4, phase; 2 and 5, fluorescence; 3 and 6, merged image of DAPI/fluorescence.
Immunofluorescence (IF) of live, zona-intact and zona-free ovulated M2 secondary oocytes prior to fertilization showed that SAS1B was localized asymmetrically on the oocyte surface (Fig. 2B and Suppl. Fig. 4A), being concentrated in a dome corresponding to the microvillar region of the oolemma (Evans et al., 1997) which is antipodal to the eccentric nucleus.
Tissue specificity of SAS1B mRNA
A 32P labeled SAS1B ORF cDNA of V2 was used to probe Northern blot containing total RNA from 15 mouse tissues. A single band of ~3.0 kb size was observed only in ovary indicating ovary specific expression of SAS1B transcript (Fig. 3A). Reprobing of the blot with mouse GAPDH cDNA confirmed integrity of RNA samples.
Figure 3.
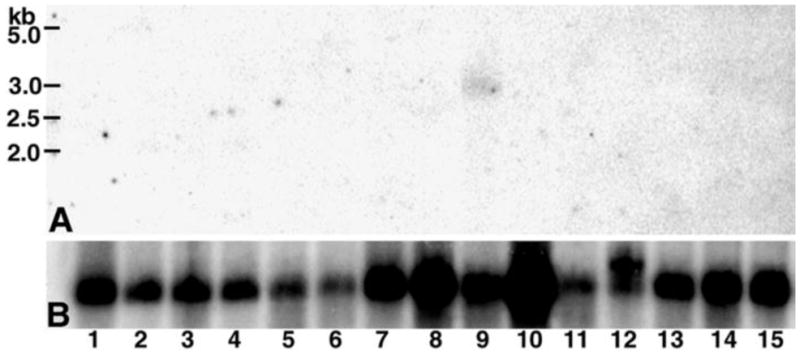
Tissue expression of SAS1B transcript. A 15 mouse tissue Northern blot containing total RNA probed with 32P labelled SAS1B cDNA revealed a SAS1B mRNA of ~3.0 kb (A) only in ovary (lane 9). Same blot probed with GAPDH cDNA as control (B). Tissues: 1, brain; 2, stomach; 3, intestine; 4, colon; 5, liver; 6, lung; 7, kidney; 8, heart; 9, ovary; 10, muscle; 11, spleen; 12, testis; 13, thymus; 14, uterus; 15, placenta.
SAS1B confocal localization in oocyte and early embryo
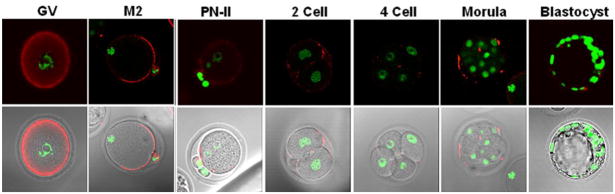
In ovulated GV stage oocytes, SAS1B was observed throughout the ooplasm with enrichment at the oocyte periphery (Fig. 4, GV). In M2 oocytes, SAS1B was concentrated in the microvillar domain of the oolemma antipodal to the nucleus and in the membrane of the first polar body. These observations imply that during the GV to M2 transition a re-orientation of membrane components including SAS1B takes place. Observations of zygotes with two pronuclei revealed that SAS1B was no longer polarized in the microvillar domain but now showed only weak staining of the plasma membrane (PN-II). In 2-cell through morulae stages diffuse, punctuate SAS1B staining was observed mainly in the perivitelline space (PVS), but occasionally on the oolemma. This low level of punctate staining remained mainly in the PVS through the early blastocyst stage and then disappeared in late blastocyst stages. Thus, SAS1B demonstrated the greatest intensity of staining on the plasma membranes of GV and M2 oocytes.
Figure 4.
Confocal localization of SAS1B before and after fertilization. Oocytes and cultured early embryos stained with SAS1B antibody (red) and with Sytox (for nucleus, green, upper panel; merged on phase images, lower panel). SAS1B was localized throughout the cytoplasm of the GV oocyte where it concentrated at the cell periphery. After polar body formation, in M2 oocyte SAS1B localized mainly in the microvillar domain of oolemma. In fertilized oocyte (PN-II), SAS1B was located only in punctate regions at the cell periphery. These small patches persisted from 2-cell to morula stages when SAS1B appeared within the PVS. SAS1B was virtually undetectable in blastocyst stages.
SAS1B is a membrane protein
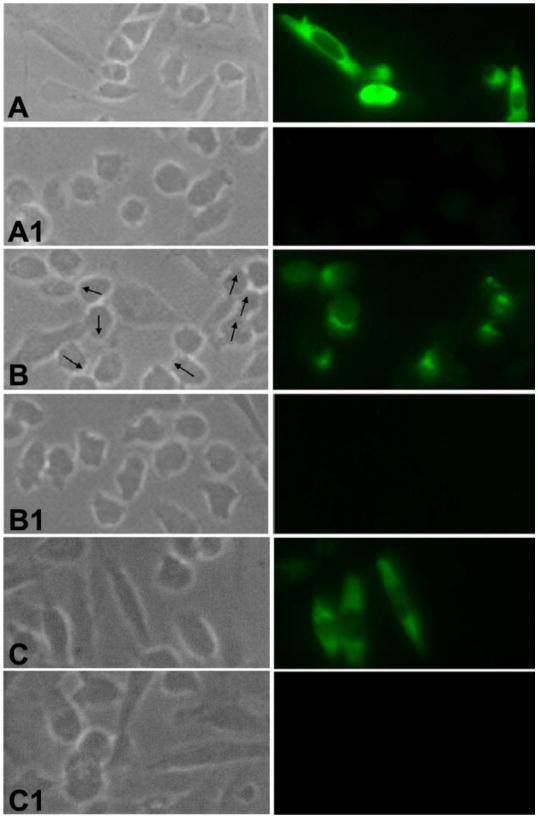
Full length SAS1B with a C-terminal V5 tag was expressed in mammalian CHO-K1 cells and the protein was localized in permeabilized and unpermeabilized transfected cells by IF. SAS1B antibody localized SAS1B in the cytoplasm and on the cell membranes of permeabilized cells and at the cell membrane of unpermeabilized cells (Fig. 5A, 5B). Interestingly, SAS1B was concentrated asymmetrically in unpermeabilized transfected cells, where SAS1B localized in regions where blebs and lamellipodia were noted by phase contrast microscopy (arrows). Unpermeable CHO-K1 cells did not stain with C-terminal V5 antibody (data not shown), whereas permeable cells did stain (Fig. 5C). The results suggested that the C- terminus of SAS1B is cytoplasmic and the N-terminus is extracellular in orientation. Together, these observations support the conclusion that SAS1B is a protein that translocates to the plasmalemma. The respective control cells transfected with blank vector showed no staining (Fig. 5, A1, B1, C1).
Figure 5.
SAS1B expression on the cell surface of transfected CHO-K1 cells. (A) In fixed and permeabilized cells SAS1B localized to cytoplasmic and surface domains. (B) Polarized localization (arrows) of SAS1B at the cell surface in fixed, unpermeabilized cells confirmed that SAS1B is a membrane protein. (C) Fixed and permeabilized cells probed with SAS1B C-terminal V5 tag antibody showing its presence in the cytoplasm. Panels: left, phase; right, fluorescence. The control cells (A1, B1, C1) that were transfected with the respective pcDNA3.1-TOPO vector lacking a SAS1B construct were probed with SAS1B specific antibodies revealing no immunofluorescence.
Protein-protein interactions between SAS1B and SLLP1
SPR and Far-western (FW) analyses
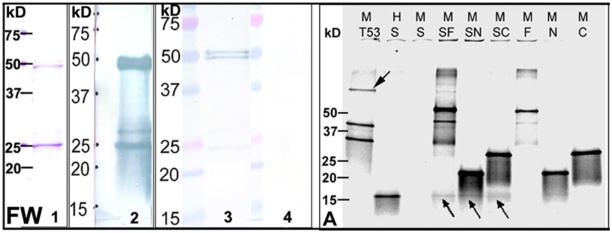
As noted earlier, SAS1B was first identified by SPR from egg protein extract as a putative partner protein for the sperm acrosomal ligand SLLP1 (Suppl Fig. 1). Next, affinity between SAS1B and SLLP1 was studied by FW analysis with purified rSAS1B and rSLLP1. SAS1B was transferred to nitrocellulose membranes. A rSAS1B blot probed with anti-His tag monoclonal antibody identified the specific rSAS1B bands (Fig. 6, FW, L2), including the full length and truncated proteins. Additional blots were either overlaid or not with rSLLP1, washed and probed with SLLP1 monoclonal antibody (Fig. 6, FW). The FW revealed that SLLP1 interacted strongly with the full length SAS1B doublet (~51, ~50 kD) but very weakly with the C-terminal fragment (~25 kD) indicating that the SAS1B N-terminus is more important to the conformation responsible for the SLLP1 binding domain than the C-terminus alone.
Figure 6.
SAS1B-SLLP1 interactions by Far-Western (FW) and rCo-IP (A) analyses. (FW) Profile of purified rSAS1B stained with Coomassie used for FW analysis (L1). Western of rSAS1B probed with anti-his tag antibody (L2). rSAS1B blot either overlaid (L3) or not overlaid (L4) with purified soluble rSLLP1 (5 μg/ml) and probed with anti-SLLP1 monoclonal antibody. Full length SAS1B (~51, ~50 kD) bound to rSLLP1; however, the C-terminal ~25 kD protein band of similar intensity bound rSLLP1 only very weakly. (A) rCo-IP of SLLP1 using SAS1B myc-tag (M) antibody. Proteins were synthesized by in vitro translation in presence of S35-methionine and analyzed by SDS-PAGE. Translated SAS1B full length (F), N-terminus (N), C-terminus (C) and p53 (53) had myc-tag (M) while T-antigen (T) and SLLP1 (S) had HA-tag (H). Co-IP of T-antigen or SLLP1 (arrows) was done using anti-myc antibody from partner proteins. Co-IP reactions: T53, T-antigen + p53; SF, SLLP1 + SAS1B-F; SN, SLLP1 + SAS1B-N; SC, SLLP1 + SAS1B-C.
rCo-IP analyses
To further study interactions between SAS1B and SLLP1, co-IP was performed using 35S-methionine labeled recombinant constructs. In vitro translated N-term, C-term, full length SAS1B, and p53 had myc-tags (M) while SLLP1 and T-antigen had HA-tags (H). Each produced a major band at the expected mass by SDS-PAGE analyses (Suppl. Fig. 7A). SLLP1 (~16 kD) was co-immunoprecipitated with an anti-myc tag antibody to SAS1B full length, N- and C-terminal fragments (Fig. 6A). In the reverse co-IP experiment, SAS1B full length (~50 kD), N- (~21 kD) and C-terminal (~26 kD) fragments were pulled down by anti-HA tag antibody to SLLP1 (Suppl. Fig. 7B). As a positive control, large T-antigen was co-immunoprecipitated with p53 taged myc antibody (Fig. 6A). Together the results indicate that SLLP1 has interacting domains with SAS1B.
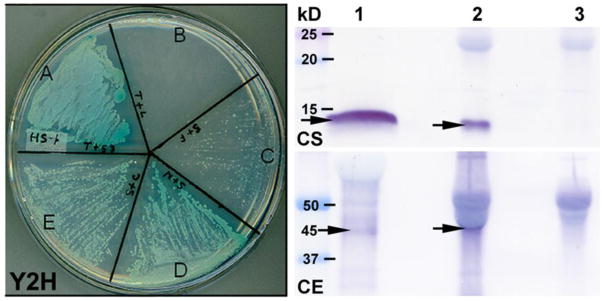
Yeast two hybrid (Y2H) analyses
The Y2H system with stringent selection conditions was used to study affinity between rSLLP1 and three rSAS1B constructs: full length 414 residues (V2), N-term 121 residues, and C-term 210 residues. Successful co-transfection by both plasmid constructs was confirmed by survival on the low stringency plate (Suppl. Fig. 6). Protein-protein interactions were confirmed by survival and formation of blue colonies on high stringency plates. Yeast cells co-transfected with SLLP1 and N-terminal SAS1B showed the fastest rescue with strongest blue color by 3 and 5 days (Fig. 7D and Suppl. Fig. 6D). Co-transfection of C-terminal SAS1B and SLLP1 also rescued the cells within three days (Suppl. Fig. 6E, HS), however co-transfection of full length SAS1B with SLLP1 did not rescue the yeast cells in 3 days but only weakly in 5 days (Fig. 7C and Suppl. Fig. 6C, HS). The relative lack of full length SAS1B interaction with SLLP1 is likely due to the presence of the putative transmembrane domain in SAS1B (Luban and Goff, 1995; Li et al., 1996). The results in the Y2H system confirm the strong molecular interaction between N-terminal SAS1B and SLLP1.
Fig. 7.
Y2H and native Co-IP analyses between SAS1B and SLLP1. Y2H assay showed affinity between SLLP1 and SAS1B fragments. Growth of yeast cells on high stringency plate forming blue colonies indicated interaction between the proteins. (A) Positive interaction between T-antigen and its partner p53. (B) Negative interaction between T-antigen and human lamin C. (C) Very weak interaction between full length SAS1B and SLLP1 with few small colonies but none visible at 3 days (Suppl. Fig. 6, HS). Positive interactions were noted between SLLP1 and N-terminal (D) or C-terminal forms (E). Native Co-IP of SAS1B from mixture of sperm and egg extracts using SLLP1 antibody. Equally divided eluted IP products were probed with SLLP1 (CS) or SAS1B antibody (CE). Sperm-egg extract IP by SLLP1 antibody revealed predominant ~45 kD SAS1B. L1, sperm or egg extract; L2, immune IP; L3, preimmune IP. Arrows indicate SLLP1 (~14 kD) or SAS1B (~45 kD).
Native (n) Co-IP analyses
To evaluate interaction between the native proteins, rat SLLP1 antibody was used to precipitate nSAS1B from a mixture of mouse sperm and egg extracts. Equal aliquots of the IP complexes were resolved, blotted and probed with guinea pig SLLP1 or SAS1B antibodies. SLLP1 antibody precipitated both ~14 kD SLLP1 and the predominant ~45 kD SAS1B (V2, V6; Fig. 7CS, CE, arrows) while the pre-immune antibody did not (Fig. 7, L3). The SAS1B band was reconfirmed with a human SAS1B propeptide antibody (Suppl. Fig. 7C). The results confirmed interacting domains in native SAS1B and SLLP1 molecules.
Co-localization of SLLP1 and SAS1B
To determine if both recombinant and native SLLP1 co-localize with SAS1B, zona intact M2 oocytes were incubated with rSLLP1 or nSLLP1, washed, and probed with SAS1B antibody. Strong SAS1B signals were present in the microvillar region of the oolemma and weakly in the perivitelline space [PVS] of the oocyte, and co-localized with the major recombinant or native SLLP1 signal, which also diffusely stained the PVS (Fig. 8A, B). The diffuse staining of SLLP1 and SAS1B in the PVS is consistent with the presence of oolemmal microvilli and previous localization of CD9 in PVS of M2 oocytes (Miyado et al., 2008). In sum, native SAS1B co-localizes with rSLLP1 and nSLLP1 binding sites on the oocyte membrane indicating interactions within a shared domain.
Figure 8.
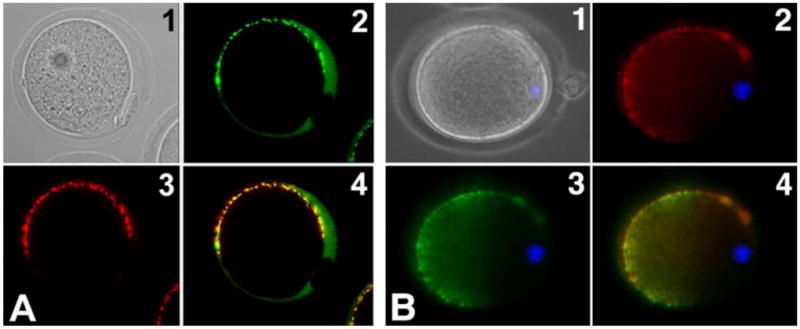
Co-localization of recombinant SLLP1 and native SLLP1 with SAS1B. M2 oocytes incubated with rSLLP1 (A) or nSLLP1 (B), washed and probed with SLLP1 and SAS1B antibodies. SLLP1 (2) co-localized predominantly to the microvillar region of the mouse oolemma marked by SAS1B localization (3). Panels: 1, phase; 2, SLLP1; 3, SAS1B; 4, merge of 2 & 3; blue, nucleus.
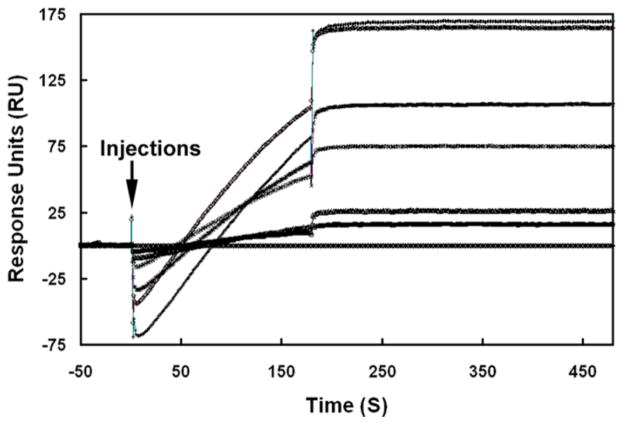
Affinity between SAS1B and SLLP1
To further study affinity between SAS1B and SLLP1, surface plasmon resonance was used to determine a dissociation constant. As observed in Fig. 9, SAS1B was capable of binding with immobilized SLLP1 in a concentration dependent manner, and very little dissociation was apparent. Kinetic evaluation of SAS1B interaction with six different analyte concentrations (400, 300, 200, 100, 50, 25 nM) using a 1:1 binding model, revealed a sub-nanomolar dissociation constant (average KD = 0.32 nM). The sub-nanomolar dissociation constant indicated a high affinity binding between SAS1B and SLLP1 (Felder et al., 1993; Xu et al., 2004).
Figure 9.
Interaction affinity between SAS1B and SLLP1. Binding curves were observed when six different analyte concentrations of SAS1B (400 - top, 300, 200, 100, 50, 25 and 0 nM - bottom) were injected (arrow) for 3 min over immobilized SLLP1 at a flow rate of 30 μl/min in running buffer using Biacore 3000 system. Following the binding phase, the sensograms showed a very low dissociation of SAS1B from the chip.
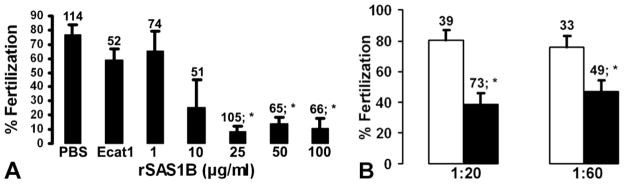
rSAS1B or its antibody inhibit in vitro fertilization (IVF)
To determine the effect of rSAS1B during IVF, capacitated spermatozoa were pre-incubated with rSAS1B prior to insemination. A statistically significant reduction (~77% – 89%, P ≤0.008, df ≥4) in fertilization was observed in samples treated with 25 μg/ml or more of SAS1B (Fig. 10A) when compared with purified recombinant Ecat1, an egg cytoplasmic protein, (Suppl. Fig. 5, L4) or no protein PBS controls. The difference in fertilization between PBS and Ecat1 controls was not statistically significant. Similarly, when zona-intact oocytes (n) were incubated with SAS1B immune antibody followed by insemination, fertilization was significantly reduced by 51% at 1:20 dilution (P ≤0.005; df = 6) or by 38% at 1:60 dilution of immune antibody (P ≤0.02, df = 4) compared to preimmune control (Fig. 10B). These observations are in agreement with the previous inhibition of mouse IVF by rSLLP1 or its antibody (Herrero et al., 2005) and suggest interactions of SAS1B and SLLP1 during fertilization.
Figure 10.
SAS1B mediated inhibition of mouse in vitro fertilization. (A) Capacitated mouse sperm were incubated with varying concentrations of rSAS1B prior to fertilization of cumulus intact oocytes. The percentage of fertilized eggs decreased with increased concentration of SAS1B compared to a controls including the oocyte cytoplasmic protein recombinant Ecat1 or PBS without proteins. (B) Inhibition of fertilization of zona-intact eggs in presence of SAS1B antibody (black bar) compared to pre-immune (white bar) controls. Bars represent mean ± SEM; numbers, eggs per group. Significant P-value differences from no protein (PBS), recombinant Ecat1 (50 μg/ml) or preimmune controls were marked with asterisk (*, P ≤ 0.02 to 0.008).
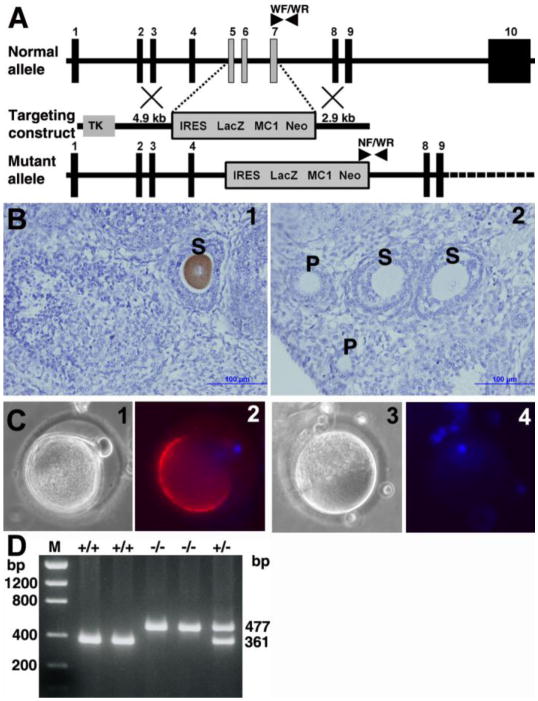
Fertility reduction in SAS1B knockout mice
To determine the role of SAS1B in fertility in vivo, SAS1B protein deficient mice were produced by homologous recombination by gene targeting (Fig. 11A). Knockout mice were genotyped using exon 7 (WF) forward, neo forward (NF), and intron 7 reverse (WR) primers (Fig. 11D). Mating of 18 pairs of heterozygous males and females produced 113 pups (55 males, 58 females) from which 26 (23%) were Sas1b+/+, 59 (52%) were Sas1b+/−, and 28 (25%) were Sas1b−/−, an allele distribution found to be consistent with expected Mendelian ratios of 1:2:1 with an average of 6.3 pups per litter. Both male and female Sas1b−/− mice were viable, grew to adulthood and appeared grossly normal. Sas1b−/− males were fertile, consistent with oocyte-specific expression of SAS1B. In homozygous knockout females, no SAS1B protein was detected by immunohistochemistry in any growing follicle stage using guinea pig anti-SAS1B antibody in ovary sections (Fig. 11B). Immunofluorescence analysis of homozygous knockout ovulated M2 eggs also confirmed lack of SAS1B protein in the microvillar domain (Fig. 11C). Thus homozygous mutant Sas1b mice were functionally null for SAS1B protein in ovaries and oocytes. To study the effect of gene deletion on fertility, Sas1b+/+ and Sas1b−/− females were mated with one Sas1b+/+ male per cage and cohorts evaluated for numbers of fetuses and live born pups. As shown in Table 1, both fetuses and live pups in Sas1b−/− mice were reduced by 36% and 32%, respectively, compared to wild type Sas1b+/+ mice. This subfertile phenotype in mice lacking SAS1B suggested that the Sas1b gene is, in part, involved in female fertility.
Figure 11.
Generation of SAS1B deficient mice. (A) Schematic representation of the targeting strategy of the Astl gene encoding SAS1B by homologous recombination. The targeting construct used thymidine kinase (TK) as a negative selection, MC1 Neo as a positive selection and IRES LacZ as a marker along with 4.9 kb and 2.9 kb homologous arms. The selection cassette targeted 3 coding exons (light vertical bars) 5, 6 and 7 encoding 131 residues (SPF – ILP) of the normal allele. Genotyping primer positions in normal and mutant alleles were indicated by arrow heads. (B) Immunohistochemical localization of SAS1B in wild type (1) and null (2) mice ovaries using anti-SAS1B guinea pig antibody confirmed the loss of SAS1B protein in oocytes within growing follicles of knockout animals. S, secondary follicles; P, primary follicles. (C) Immuno-fluorescence localization of SAS1B in wild type (1 and 2) and knockout (3 and 4) ovulated eggs showed lack of SAS1B in the microvillar domain of mature M2 egg in null mice. Panels: 1 and 3 phase; 2 and 4 merge image of DAPI/fluorescence. (D) Genotype PCR of genomic tail DNA using 3 primers (WF, WR, NF) producing a 361 bp product in wild type (+/+), a 477 bp product in null (−/−) and both products in heterozygous mice. M, DNA mass ladder.
Table 1.
Fertility reduction in SAS1B knockout mice.
| Parent Genotype | Male, wild-type (+/+)
|
|||
|---|---|---|---|---|
| No. / group | Total fetuses1 or pups2 | Average fetuses or pups / female3 | Fertility‡ (%) | |
| Female, wild-type (+/+) | 20 | 124 | 6.2 ± 0.65 | 100 |
| Female, knockout (−/−) | 20 | 82 | 4.1 ± 0.64* | 66.1 |
Each male was paired with a wild-type and a knockout female for two weeks and then separated for fetal or pups count.
Breeding done by Merck Research Laboratories. From 8 breeding pairs, the (+/+) and the (−/−) females produced 56 and 36 fetuses (36% reduction), respectively.
Breeding done by University of Virginia. From 12 breeding pairs, the (+/+) and the (−/−) females produced 68 and 46 pups (32% reduction), respectively.
Values are mean ± SEM.
Fertility was determined as a percentage of average fetal or litter size.
Significantly different from wild-type mice, P ≤ 0.01 (Student’s t-test).
Discussion
SAS1B is a SLLP1 binding partner during fertilization
Six lines of biochemical evidence indicate that SAS1B is an oolemmal binding partner for sperm intra-acrosomal ligand SLLP1. First, native SAS1B in oocyte extracts specifically bound to and was eluted from rSLLP1 bait on the SPR chip (Suppl. Fig. 1). Second, rSLLP1 bound to rSAS1B in far western analyses (Fig. 6). Third, molecular interaction between SAS1B and SLLP1 was demonstrated in the yeast two hybrid assay system (Fig. 7). Fourth, recombinant SAS1B-SLLP1 interactions were shown by co-IP of SLLP1 with antibody to a SAS1B tag, and the reverse, using eukaryotic in vitro translation (Fig. 6). Fifth, native SAS1B co-immunoprecipitated with SLLP1 antibody from a mixture of sperm and egg extracts (Fig. 7). Sixth, a sub-nanomolar dissociation constant (KD = 0.32 nM) revealed a high affinity interaction between these molecules (Fig. 9). The restricted microvillar localization of SAS1B in mature oocytes and co-localization of SLLP1 to this region (Fig. 8), known to be involved in sperm-egg binding and fusion (Evans et al., 2000), provides correlative physiological support for in-vivo SAS1B-SLLP1 interaction. An earlier report showed that SLLP1 is retained in 90% of acrosome reacted sperm, binds to oocyte microvillar domain and that SLLP1 protein or antibody inhibited fertilization and sperm-egg binding at the level of the oolemma in a dose dependent manner in mouse IVF studies (Herrero et al., 2005). The present study also revealed a dose dependent inhibition of fertilization in the presence of SAS1B antibody or protein (Fig. 10). Notably, both SLLP1 (Herrero et al., 2005) and SAS1B are exposed prior to sperm-egg fusion. Together these observations suggest that SAS1B is an oolemmal binding partner that interacts with the sperm intra-acrosomal ligand SLLP1 prior to gamete fusion during fertilization.
SAS1B is an oocyte specific protein
Bio-informatic analyses of mouse EST databases [NCBI] show expression of SAS1B exclusively in ovary, oocyte and zygote. Northern analysis of multiple tissues revealed that SAS1B mRNA is ovary specific. In mouse ovary, SAS1B protein is confined to ooplasm of secondary, pre-antral, and Graafian follicles. Following fertilization, SAS1B assumes a reduced patchy distribution on the surface of the zygote and blastomeres of early embryos, but is virtually undetectable by late blastocyst stage. Thus, both at protein and mRNA levels SAS1B is specifically expressed only in the oocyte and early embryo, suggesting it is a maternal effect gene expressed during oogenesis.
Dynamic re-localization of SAS1B to the microvillar domain
SAS1B was localized throughout the ooplasm of germinal vesicle stage oocytes and was concentrated symmetrically in a corona at the oocyte periphery. SAS1B subsequently localized to the microvillar domain of the oolemma in M2 arrested ovulated secondary oocytes. These observations indicate that a dynamic re-organization of the SAS1B domain occurs as the oocyte matures following meiosis I resulting in two distinct regions of the oocyte plasma membrane; i.e., the region that lacks microvilli and overlies the eccentrically located meiotic spindle, and the microvillar region (Evans et al., 1997). The development of this polarity with respect to SAS1B is particularly noteworthy, as sperm bind to and fuse with the oolemma in the microvillar region.
Mouse SAS1B has a putative transmembrane domain (Suppl. Fig. 1). SAS1B antibody localized the protein on the microvillar domain of ovulated live M2 oocytes and inhibited fertilization, suggesting that the molecule is exposed at the cell surface. Further, CHO-K1 cells transfected with full length SAS1B (Fig. 5) showed localization to plasma membrane protrusions rather than the planar portions of the cell surface. This observation directly correlates with published reports regarding targeting of membrane proteins to sites of membrane insertion in microvilli of epithelial cells and to plasma membrane protrusions of non-epithelial cells (Weigmann et al., 1997).
SAS1B is an active protease
Bioinformatic analysis predicted that SAS1B belongs to a metalloprotease family due to its characteristic zinc binding active site signature. The observation of in vitro proteolytic activity of rSAS1B confirmed this prediction (Fig. 1 and Suppl. Fig. 3). Several other metalloproteases have been reported in the ovary, and their role in ovulation and formation and regression of the corpus luteum has been noted (Goldman and Shalev, 2003). These metalloproteases were localized either in theca interna or externa, interstitial glands, germinal epithelium, granulosa or theca-interstitial cells and oocyte cytoplasm (Robinson et al., 2001). Unlike GDF-9 (Dong et al., 1996) SAS1B is the first metalloprotease reported to be localized on the oolemmal surface. These properties along with the known drugability of metalloproteases suggest this molecule as a candidate contraceptive target.
Metalloproteases are known to be involved in cellular fusion events and fertilization. In yeast, cell-cell fusion requires the zinc metalloprotease gene, AXL1 (Elia and Marsh, 1998). In sea urchin, the zinc chelator (1,10-phenanthroline) showed no effect on the binding of acrosome reacted sperm to the oolemma but blocked (>95%) subsequent membrane fusion (Roe et al., 1988). In mice, inhibitors of the aspartic, cysteine, and serine protease classes had no effect on sperm egg binding or fusion. However, 1,10-phenanthroline, a metalloprotease inhibitor, inhibited mouse sperm-egg fusion without reducing sperm-egg binding, suggesting a critical role of membrane metalloproteases in mammalian gamete fusion (Correa et al., 2000).
SAS1B conservation in mammals and role in fertilization
Orthologous and homologous genes were identified in other mammals and in lower organisms, respectively. In lower organisms, homologous proteins were characterized as hatching enzymes (Hiroi et al., 2004). In a previous report an astacin metalloproteinase detected at the RNA level in human and mouse ovaries was tentatively called ovastacin and was predicted to have a role in hatching (Quesada et al., 2004). However, the expression of SAS1B begins to decrease after fertilization and is virtually undetectable in blastocysts prior to hatching (Fig. 4) and SAS1B RNA was not detected beyond 1.5 days postcoitum in preimplantation embryos (Quesada et al., 2004). Together, the results point to a role of this protein in fertilization, rather than in zonal hatching, suggesting that the name SAS1B conveys its biological interactions better than ovastacin. The intra-acrosomal protein SLLP1 is exposed and retained on the inner acrosomal membrane of 90% acrosome reacted sperm (Herrero et al., 2005). Thus, following zona penetration, acrosome reacted sperm will display SLLP1 on the inner acrosomal membrane where SLLP1 is expected to bind tightly with microvillar SAS1B to facilitate fertilization. This hypothesis is strengthened by gene knockout studies in which deletion of SAS1B caused 34% reduction in fertility in female mice. This subfertile phenotype supports the hypothesis that SAS1B-SLLP1 is a sperm-egg interaction partner pair involved in fertilization. Because mice ovulate multiple oocytes each cycle and ovulate nearly eight times as frequently as humans, the subfertile phenotype seen in mice may be magnified in a normally uni-ovular primate, such as humans (Vjugina and Evans, 2008). The role of SAS1B in primate models and in human infertility and subfertility should therefore be further studied.
Conclusions
The study identified SAS1B as a novel oolemmal binding partner (receptor) for sperm intra-acrosomal protein SLLP1 (ligand) and sheds new insight into the molecular mechanism of sperm-egg interaction at the level of the oolemma prior to fusion and sperm internalization.
Supplementary Material
Research Highlights.
This study identified sperm-egg interaction partners during mammalian fertilization.
SPR analysis with sperm acrosomal SLLP1 identified the oolemmal partner, SAS1B.
SAS1B is an oocyte specific metalloprotease.
The gamete partners revealed high affinity interactions.
SAS1B deletion in female mice showed 34% reduction in fertility.
Acknowledgments
We thank Dr Nena Fox of the Tissue Culture Facility at UVA for providing CHO-K1 cells. This research was supported by the Kenneth A Scott Trust, a Keybank Trust, NIH R03 HD055129, D43 TW/HD 00654 from the Fogarty International Center and a grant from Merck Research Laboratories, Oss, The Netherlands.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfieri JA, Martin AD, Takeda J, Kondoh G, Myles DG, Primakoff P. Infertility in female mice with an oocyte-specific knockout of GPI-anchored proteins. J Cell Sci. 2003;116:2149–2155. doi: 10.1242/jcs.00430. [DOI] [PubMed] [Google Scholar]
- Almeida EA, Huovila AP, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG. Mouse egg integrin α6β1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- Cho C, Bunch DO, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG. Fertilization defects in sperm from mice lacking fertilin β. Science. 1998;281:1857–1859. doi: 10.1126/science.281.5384.1857. [DOI] [PubMed] [Google Scholar]
- Cohen DJ, Ellerman DA, Cuasnicu PS. Mammalian sperm-egg fusion: evidence that epididymal protein DE plays a role in mouse gamete fusion. Biol Reprod. 2000;63:462–468. doi: 10.1095/biolreprod63.2.462. [DOI] [PubMed] [Google Scholar]
- Coonrod SA, Naaby-Hansen S, Shetty J, Shibahara H, Chen M, White JM, Herr JC. Treatment of mouse oocytes with PI-PLC releases 70-kDa (pI 5) and 35- to 45-kDa (pI 5.5) protein clusters from the egg surface and inhibits sperm-oolemma binding and fusion. Dev Biol. 1999;207:334–349. doi: 10.1006/dbio.1998.9161. [DOI] [PubMed] [Google Scholar]
- Correa LM, Cho C, Myles DG, Primakoff P. A role for a TIMP-3-sensitive, Zn(2+)-dependent metalloprotease in mammalian gamete membrane fusion. Dev Biol. 2000;225:124–134. doi: 10.1006/dbio.2000.9825. [DOI] [PubMed] [Google Scholar]
- Da Ros VG, Maldera JA, Willis WD, Cohen DJ, Goulding EH, Gelman DM, Rubinstein M, Eddy EM, Cuasnicu PS. Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1) Dev Biol. 2008;320:12–18. doi: 10.1016/j.ydbio.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk M. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Elia L, Marsh L. A role for a protease in morphogenic responses during yeast cell fusion. J Cell Biol. 1998;142:1473–1485. doi: 10.1083/jcb.142.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Schultz RM, Kopf GS. Mouse sperm-egg plasma membrane interactions: analysis of roles of egg integrins and the mouse sperm homologue of PH-30 (fertilin) beta. J Cell Sci. 1995;108:3267–3278. doi: 10.1242/jcs.108.10.3267. [DOI] [PubMed] [Google Scholar]
- Evans JP, Kopf GS, Schultz RM. Characterization of the binding of recombinant mouse sperm fertilin beta subunit to mouse eggs: evidence for adhesive activity via an egg beta1 integrin-mediated interaction. Dev Biol. 1997;187:79–93. doi: 10.1006/dbio.1997.8611. [DOI] [PubMed] [Google Scholar]
- Evans JP, Foster JA, McAvey BA, Gerton GL, Kopf GS, Schultz RM. Effects of perturbation of cell polarity on molecular markers of sperm-egg binding sites on mouse eggs. Biol Reprod. 2000;62:76–84. doi: 10.1095/biolreprod62.1.76. [DOI] [PubMed] [Google Scholar]
- Felder S, Zhou M, Hu P, Urena J, Ullrich A, Chaudhuri M, White M, Shoelson SE, Schlessinger J. SH2 domains exhibit high-affinity binding to tyrosine phosphorylated peptides yet also exhibit rapid dissociation and exchange. Mol Cell Biol. 1993;13:1449–1455. doi: 10.1128/mcb.13.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Shalev E. The role of the matrix metalloproteinases in human endometrial and ovarian cycles. Eur J Obstet Gynecol Reprod Biol. 2003;111:109–121. doi: 10.1016/s0301-2115(03)00341-5. [DOI] [PubMed] [Google Scholar]
- Herrero MB, Mandal A, Digilio LC, Coonrod SA, Maier B, Herr JC. Mouse SLLP1, a sperm lysozyme-like protein involved in sperm–egg binding and fertilization. Dev Biol. 2005;284:126–142. doi: 10.1016/j.ydbio.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Hiroi J, Maruyama K, Kawazu K, Kaneko T, Ohtani-Kaneko R, Yasumasu S. Structure and developmental expression of hatching enzyme genes of the Japanese eel Anguilla japonica: an aspect of the evolution of fish hatching enzyme gene. Dev Genes Evol. 2004;214:176–184. doi: 10.1007/s00427-004-0397-1. [DOI] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Li X, McDermott B, Yuan B, Goff SP. Homomeric interactions between transmembrane proteins of Moloney murine leukemia virus. J Virol. 1996;70:1266–1270. doi: 10.1128/jvi.70.2.1266-1270.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Goff SP. The yeast two-hybrid system for studying protein-protein interactions. Curr Opin Biotechnol. 1995;6:59–64. doi: 10.1016/0958-1669(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Mandal A, Klotz KL, Shetty J, Jayes FL, Wolkowicz MJ, Bolling LC, Coonrod SA, Black MB, Diekman AB, Haystead TA, Flickinger CJ, Herr JC. SLLP1, a unique, intra-acrosomal, non-bacteriolytic, c lysozyme-like protein of human spermatozoa. Biol Reprod. 2003;68:1525–1537. doi: 10.1095/biolreprod.102.010108. [DOI] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci USA. 2008;105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naaby-Hansen S, Mandal A, Wolkowicz MJ, Sen B, Westbrook A, Shetty J, Coonrod SA, Klotz K, Kim YH, Bush LA, Flickinger CJ, Herr JC. CABYR, a novel calcium-binding tyrosine phosphorylation regulated fibrous sheath protein involved in capacitation. Dev Biol. 2002;242:236–254. doi: 10.1006/dbio.2001.0527. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol. 2001;233:204–213. doi: 10.1006/dbio.2001.0166. [DOI] [PubMed] [Google Scholar]
- Quesada V, Sanchez LM, Alvarez J, Lopez-Otın C. Identification and Characterization of Human and Mouse Ovastacin. J Biol Chem. 2004;279:26627–26634. doi: 10.1074/jbc.M401588200. [DOI] [PubMed] [Google Scholar]
- Robinson LL, Sznajder NA, Riley SC, Anderson RA. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in human fetal testis and ovary. Mol Hum Reprod. 2001;7:641–648. doi: 10.1093/molehr/7.7.641. [DOI] [PubMed] [Google Scholar]
- Roe JL, Farach HA, Jr, Strittmatter WJ, Lennarz WJ. Evidence for involvement of metalloendoproteases in a step in sea urchin gamete fusion. J Cell Biol. 1988;107:539–544. doi: 10.1083/jcb.107.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf J, Levy S, Naour FL, Boucheix C. Reduced fertility of female mice lacking CD81. Dev Biol. 2006;290:351–358. doi: 10.1016/j.ydbio.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Shamsadin R, Adham IM, Nayernia K, Heinlein UAO, Oberwinkler H, Engel W. Male mice deficient for germ-cell cyritestin are infertile. Biol Reprod. 1999;61:1445–1451. doi: 10.1095/biolreprod61.6.1445. [DOI] [PubMed] [Google Scholar]
- Vjugina U, Evans JP. New insights into the molecular basis of mammalian sperm-egg membrane interactions. Front Biosci. 2008;13:462–476. doi: 10.2741/2693. [DOI] [PubMed] [Google Scholar]
- Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvillispecific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci USA. 1997;94:12425–12430. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- Yuan R, Primakoff P, Myles DG. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm-egg plasma membrane adhesion and fusion. J Cell Biol. 1997;137:105–112. doi: 10.1083/jcb.137.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.