Abstract
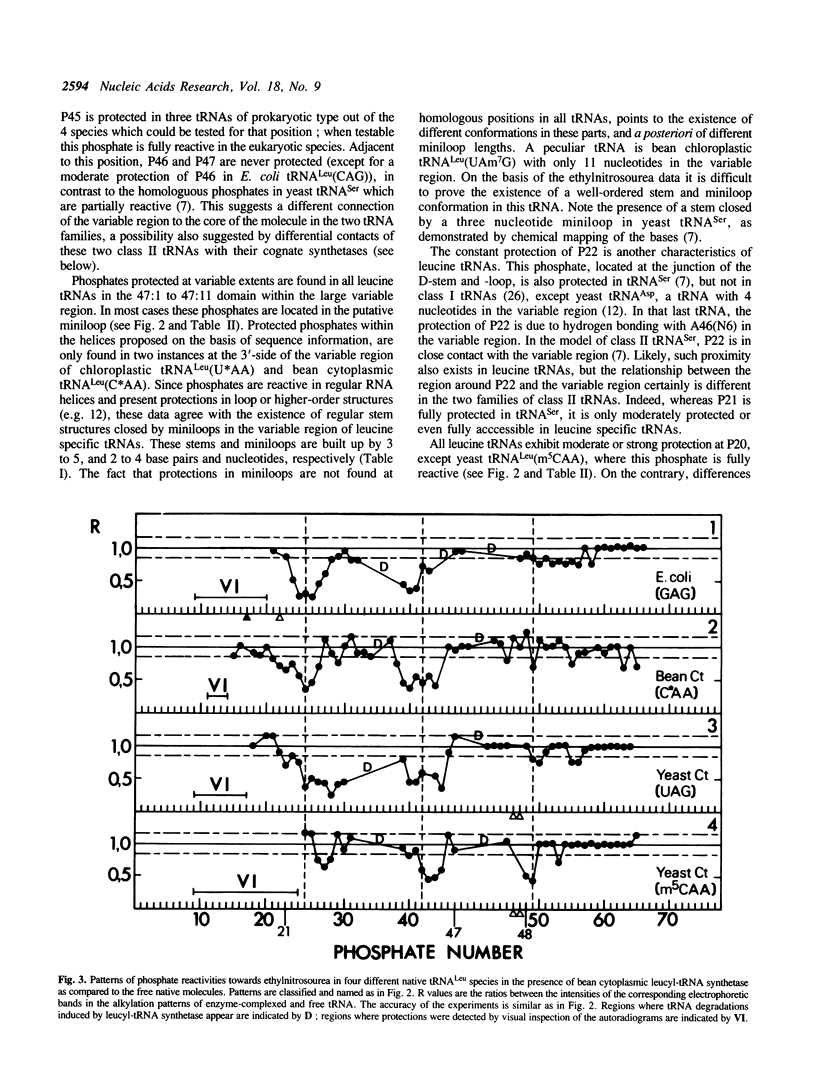
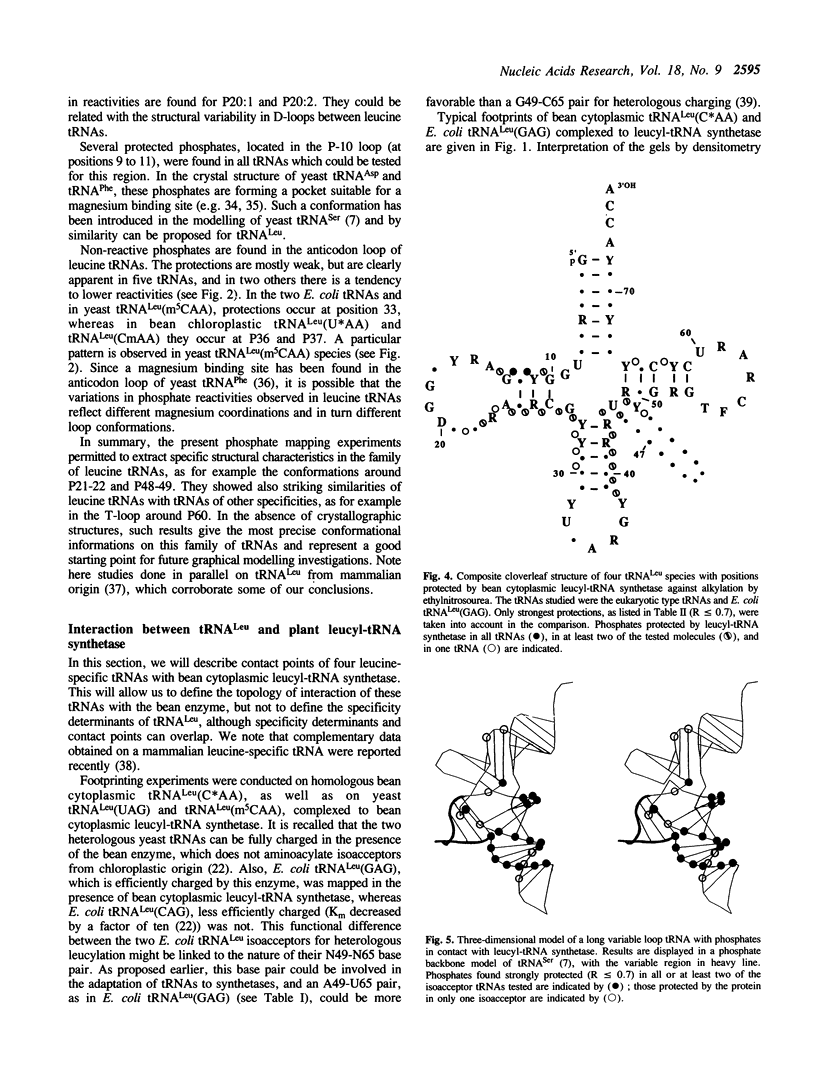
The solution conformation of eight leucine tRNAs from Phaseolus vulgaris, baker's yeast and Escherichia coli, characterized by long variable regions, and the interaction of four of them with bean cytoplasmic leucyl-tRNA synthetase were studied by phosphate mapping with ethylnitrosourea. Phosphate reactivities in the variable regions agree with the existence of RNA helices closed by miniloops. At the junction of these regions with the T-stem, phosphate 48 is strongly protected, in contrast to small variable region tRNAs where P49 is protected. The constant protection of P22 is another characteristics of leucine tRNAs. Conformational differences between leucine isoacceptors concern the anticodon region, the D-arm and the variable region. In several parts of free tRNALeu species, e.g. in the T-loop, phosphate reactivities are similar to those found in tRNAs of other specificities, indicating conformational similarities among tRNAs. Phosphate alkylation of four leucine tRNAs complexed to leucyl-tRNA synthetase indicates that the 3'-side of the anticodon stem, the D-stem and the hinge region between the anticodon and D-stems are in contact with the plant enzyme.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. L., Birkenmeier C. S. Inhibition of intractable nucleases with ribonucleoside--vanadyl complexes: isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979 Nov 13;18(23):5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- Chang S. H., Kuo S., Hawkins E., Miller N. R. The corrected nucleotide sequence of yeast leucine transfer ribonucleic acid. Biochem Biophys Res Commun. 1973 Apr 16;51(4):951–955. doi: 10.1016/0006-291x(73)90019-3. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Souciet G., Colas B., Weil J. H. Phaseolus vulgaris cytoplasmic leucyl-tRNA synthetase. Purification and comparison of its catalytic, structural, and immunological properties with those of the chloroplastic enzyme. J Biol Chem. 1983 Oct 25;258(20):12386–12393. [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Moras D. Conformational changes and dynamics of tRNAs: evidence from hydrolysis patterns. Cold Spring Harb Symp Quant Biol. 1987;52:113–121. doi: 10.1101/sqb.1987.052.01.016. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Dock A. C., Lorber B., Moras D., Pixa G., Thierry J. C., Giégé R. Crystallization of transfer ribonucleic acids. Biochimie. 1984 Mar;66(3):179–201. doi: 10.1016/0300-9084(84)90063-4. [DOI] [PubMed] [Google Scholar]
- Florentz C., Briand J. P., Romby P., Hirth L., Ebel J. P., Glegé R. The tRNA-like structure of turnip yellow mosaic virus RNA: structural organization of the last 159 nucleotides from the 3' OH terminus. EMBO J. 1982;1(2):269–276. doi: 10.1002/j.1460-2075.1982.tb01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz C., Giegé R. Contact areas of the turnip yellow mosaic virus tRNA-like structure interacting with yeast valyl-tRNA synthetase. J Mol Biol. 1986 Sep 5;191(1):117–130. doi: 10.1016/0022-2836(86)90427-4. [DOI] [PubMed] [Google Scholar]
- Garret M., Labouesse B., Litvak S., Romby P., Ebel J. P., Giegé R. Tertiary structure of animal tRNATrp in solution and interaction of tRNATrp with tryptophanyl-tRNA synthetase. Eur J Biochem. 1984 Jan 2;138(1):67–75. doi: 10.1111/j.1432-1033.1984.tb07882.x. [DOI] [PubMed] [Google Scholar]
- Guillemaut P. The N49-N65 base-pair could play an important role in adaptation between tRNAs and aminoacyl-tRNA synthetases. Nucleic Acids Res. 1988 Jul 25;16(14B):7194–7194. doi: 10.1093/nar/16.14.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L. L. The role of the anticodon in recognition of tRNA by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1985;32:237–266. doi: 10.1016/s0079-6603(08)60350-5. [DOI] [PubMed] [Google Scholar]
- Kowalski S., Yamane T., Fresco J. R. Nucleotide sequence of the "denaturable" leucine transfer RNA from yeast. Science. 1971 Apr 23;172(3981):385–387. doi: 10.1126/science.172.3981.385. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Herbert C. J., Dujardin G., Slonimski P. P. Three suppressor mutations which cure a mitochondrial RNA maturase deficiency occur at the same codon in the open reading frame of the nuclear NAM2 gene. EMBO J. 1987 Mar;6(3):713–721. doi: 10.1002/j.1460-2075.1987.tb04812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Nicholas H. B., Jr Differences between transfer RNA molecules. J Mol Biol. 1987 Apr 20;194(4):635–642. doi: 10.1016/0022-2836(87)90240-3. [DOI] [PubMed] [Google Scholar]
- Moras D., Lorber B., Romby P., Ebel J. P., Giegé R., Lewit-Bentley A., Roth M. Yeast tRNAAsp-aspartyl-tRNA synthetase: the crystalline complex. J Biomol Struct Dyn. 1983 Oct;1(1):209–223. doi: 10.1080/07391102.1983.10507435. [DOI] [PubMed] [Google Scholar]
- Mougel M., Eyermann F., Westhof E., Romby P., Expert-Bezançon A., Ebel J. P., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987 Nov 5;198(1):91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- Nicholas H. B., Jr, McClain W. H. An algorithm for discriminating sequences and its application to yeast transfer RNA. Comput Appl Biosci. 1987 Sep;3(3):177–181. doi: 10.1093/bioinformatics/3.3.177. [DOI] [PubMed] [Google Scholar]
- Osorio-Almeida M. L., Guillemaut P., Keith G., Canaday J., Weil J. H. Primary structure of three leucine transfer RNAs from bean chloroplast. Biochem Biophys Res Commun. 1980 Jan 15;92(1):102–108. doi: 10.1016/0006-291x(80)91525-9. [DOI] [PubMed] [Google Scholar]
- Petrushenko Z. M., Tukalo M. A., Matsuka G. Kh. Opredelenie ostatkov fosfornoi kisloty, uchastvuiushchikh v obrazovanii prostranstvennoi struktury tRNK- Leu IAG molochnoi zhele korov. Bioorg Khim. 1986 Nov;12(11):1492–1497. [PubMed] [Google Scholar]
- Pillay D. T., Guillemaut P., Weil J. H. Nucleotide sequences of three soybean chloroplast tRNAsLeu and re-examination of bean chloroplast tRNA2Leu sequence. Nucleic Acids Res. 1984 Mar 26;12(6):2997–3001. doi: 10.1093/nar/12.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rether B., Bonnet J., Ebel J. P. Studies on tRNA nucleotidyltransferase from baker's yeast. 1. Purification of the enzyme. Protection against thermal inactivation and inhibition by several substrates. Eur J Biochem. 1974 Dec 16;50(1):281–288. doi: 10.1111/j.1432-1033.1974.tb03896.x. [DOI] [PubMed] [Google Scholar]
- Riehl N., Giegé R., Ebel J. P., Ehresmann B. Effect of elongation factor Tu on the conformation of phenylalanyl-tRNAPhe. FEBS Lett. 1983 Apr 5;154(1):42–46. doi: 10.1016/0014-5793(83)80871-0. [DOI] [PubMed] [Google Scholar]
- Romby P., Carbon P., Westhof E., Ehresmann C., Ebel J. P., Ehresmann B., Giegé R. Importance of conserved residues for the conformation of the T-loop in tRNAs. J Biomol Struct Dyn. 1987 Dec;5(3):669–687. doi: 10.1080/07391102.1987.10506419. [DOI] [PubMed] [Google Scholar]
- Romby P., Carbon P., Westhof E., Ehresmann C., Ebel J. P., Ehresmann B., Giegé R. Importance of conserved residues for the conformation of the T-loop in tRNAs. J Biomol Struct Dyn. 1987 Dec;5(3):669–687. doi: 10.1080/07391102.1987.10506419. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Souciet G., Dietrich A., Colas B., Razafimahatratra P., Weil J. H. Purification and properties of chloroplast leucyl-tRNA synthetase from a higher plant: Phaseolus vulgaris. J Biol Chem. 1982 Aug 25;257(16):9598–9604. [PubMed] [Google Scholar]
- Spencer M., Neidle S., Jones T. A. Crystallisation of tRNALeuCUG from Escherichia coli after purification with hydroxyapatite. Biochem Biophys Res Commun. 1979 Jan 15;86(1):66–70. doi: 10.1016/0006-291x(79)90382-6. [DOI] [PubMed] [Google Scholar]
- Theobald A., Springer M., Grunberg-Manago M., Ebel J. P., Giege R. Tertiary structure of Escherichia coli tRNA(3Thr) in solution and interaction of this tRNA with the cognate threonyl-tRNA synthetase. Eur J Biochem. 1988 Aug 15;175(3):511–524. doi: 10.1111/j.1432-1033.1988.tb14223.x. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Giegé R., Ebel J. P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur J Biochem. 1981 Sep;119(1):51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Kern D., Romby P., Giegé R., Ebel J. P. Interaction of tRNAPhe and tRNAVal with aminoacyl-tRNA synthetases. A chemical modification study. Eur J Biochem. 1983 May 16;132(3):537–544. doi: 10.1111/j.1432-1033.1983.tb07395.x. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Huizinga J. D., Gaastra W., Welling G. W., Beintema J. J. Affinity chromatography of porcine pancreatic ribonuclease reinvestigation of the N-terminal amino acid sequence. FEBS Lett. 1973 Apr 15;31(2):181–185. doi: 10.1016/0014-5793(73)80098-5. [DOI] [PubMed] [Google Scholar]
- Young J. D., Bock R. M., Nishimura S., Ishikura H., Yamada Y., RajBhandary U. L., Labanauska M., Connors P. G. Structural studies on transfer RNA: crystallization of formylmethionine and leucine transfer RNA's. Science. 1969 Dec 19;166(3912):1527–1528. doi: 10.1126/science.166.3912.1527. [DOI] [PubMed] [Google Scholar]
- Zaccaï G., Morin P., Jacrot B., Moras D., Thierry J. C., Giegé R. Interactions of yeast valyl-tRNA synthetase with RNAs and conformational changes of the enzyme. J Mol Biol. 1979 Apr 15;129(3):483–500. doi: 10.1016/0022-2836(79)90508-4. [DOI] [PubMed] [Google Scholar]




