Abstract
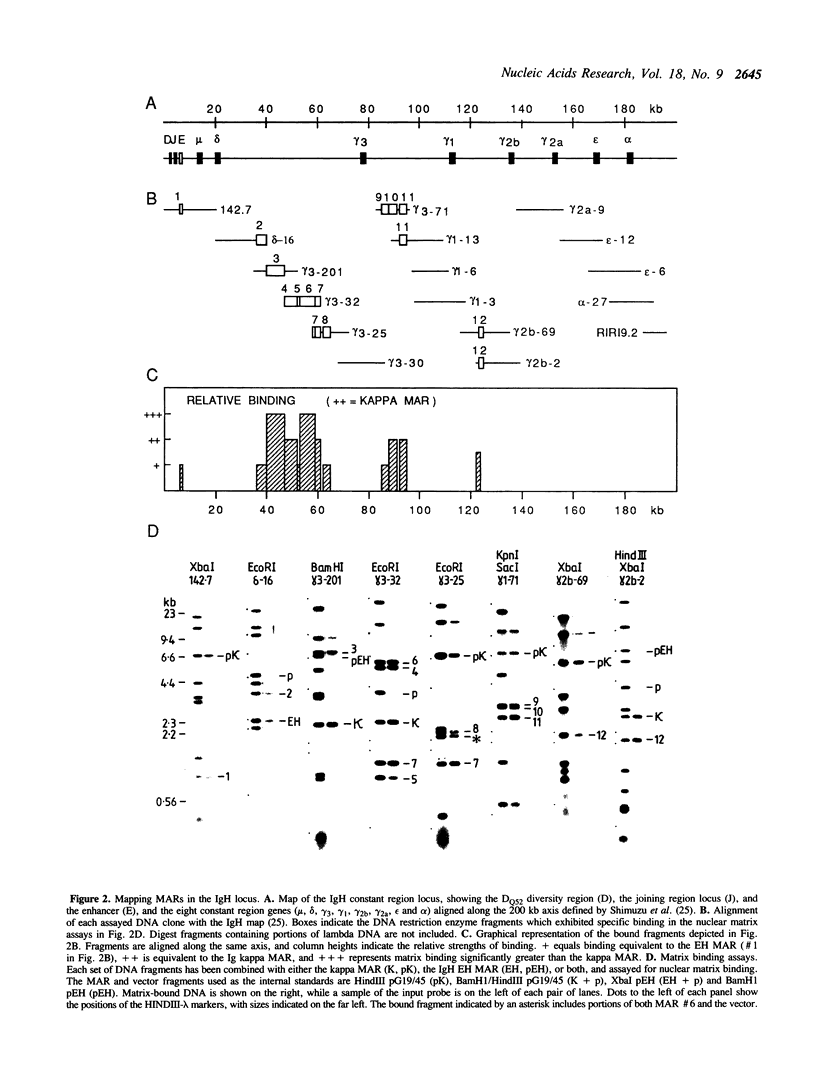
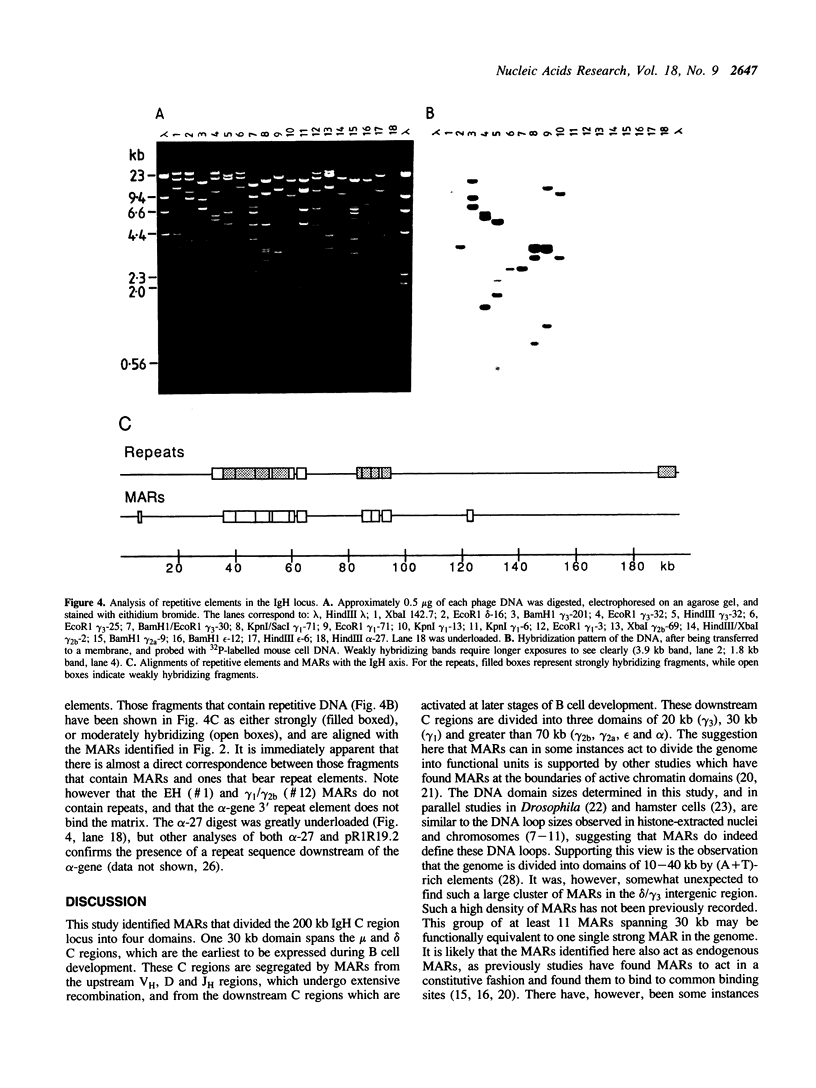
The genome is thought to be divided into domains by DNA elements which mediate anchorage of chromosomal DNA to the nuclear matrix or chromosome scaffold. The positions of nuclear matrix anchorage regions (MARs) have been mapped within the 200 kb mouse immunoglobulin heavy chain constant region locus, thereby allowing an estimate of the size of DNA domains within a segment of the genome. MARs were identified in four regions, which appear to divide the locus into looped DNA domains of 30, 20, 30 and greater than 70 kb in length. These DNA domain sizes fall within the range of DNA loop sizes observed in histone-extracted nuclei and chromosomes. In two regions, large clusters of MARs were identified, and many of these MARs lie on DNA fragments that include repetitive DNA elements, perhaps indicating that repetitive DNA integrates into the genome close to MARs, or that some classes of repeats could themselves act as MARs.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolph K. W., Cheng S. M., Laemmli U. K. Role of nonhistone proteins in metaphase chromosome structure. Cell. 1977 Nov;12(3):805–816. doi: 10.1016/0092-8674(77)90279-3. [DOI] [PubMed] [Google Scholar]
- Aelen J. M., Opstelten R. J., Wanka F. Organization of DNA replication in Physarum polycephalum. Attachment of origins of replicons and replication forks to the nuclear matrix. Nucleic Acids Res. 1983 Feb 25;11(4):1181–1195. doi: 10.1093/nar/11.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A. S., Sedat J. W., Agard D. A. A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J Cell Biol. 1987 Jul;105(1):77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Blasquez V. C., Xu M., Moses S. C., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J Biol Chem. 1989 Dec 15;264(35):21183–21189. [PubMed] [Google Scholar]
- Bode J., Maass K. Chromatin domain surrounding the human interferon-beta gene as defined by scaffold-attached regions. Biochemistry. 1988 Jun 28;27(13):4706–4711. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- Brown E. H., Iqbal M. A., Stuart S., Hatton K. S., Valinsky J., Schildkraut C. L. Rate of replication of the murine immunoglobulin heavy-chain locus: evidence that the region is part of a single replicon. Mol Cell Biol. 1987 Jan;7(1):450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capco D. G., Wan K. M., Penman S. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982 Jul;29(3):847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Yuen M. H., Garrard W. T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987 Apr 15;262(11):5394–5397. [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Heck M. M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G., Singer M. F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987 Dec 8;910(3):203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. doi: 10.1002/j.1460-2075.1987.tb02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO J. 1988 Dec 1;7(12):3667–3677. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkis E., Mercola M., Calame K. Strong transcriptional activation of translocated c-myc genes occurs without a strong nearby enhancer or promoter. Nucleic Acids Res. 1988 Jan 11;16(1):77–96. doi: 10.1093/nar/16.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A., Marcaud L., Moreau J., Scherrer K. Conservation and variation in the large scale organisation of the globin gene domains of duck and chicken. Mol Gen Genet. 1986 May;203(2):193–201. doi: 10.1007/BF00333954. [DOI] [PubMed] [Google Scholar]
- Levy-Wilson B., Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5' and 3' boundaries of the gene. J Biol Chem. 1989 Dec 15;264(35):21196–21204. [PubMed] [Google Scholar]
- Lewis C. D., Laemmli U. K. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982 May;29(1):171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Liu C. P., Tucker P. W., Mushinski J. F., Blattner F. R. Mapping of heavy chain genes for mouse immunoglobulins M and D. Science. 1980 Sep 19;209(4463):1348–1353. doi: 10.1126/science.6774414. [DOI] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Spierer P., Laemmli U. K. Genes and loops in 320,000 base-pairs of the Drosophila melanogaster chromosome. J Mol Biol. 1986 Jul 20;190(2):255–258. doi: 10.1016/0022-2836(86)90296-2. [DOI] [PubMed] [Google Scholar]
- Moreau J., Marcaud L., Maschat F., Kejzlarova-Lepesant J., Lepesant J. A., Scherrer K. A + T-rich linkers define functional domains in eukaryotic DNA. Nature. 1982 Jan 21;295(5846):260–262. doi: 10.1038/295260a0. [DOI] [PubMed] [Google Scholar]
- Neuer-Nitsche B., Lu X. N., Werner D. Functional role of a highly repetitive DNA sequence in anchorage of the mouse genome. Nucleic Acids Res. 1988 Sep 12;16(17):8351–8360. doi: 10.1093/nar/16.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. W., Forbes D. J. The nucleus: structure, function, and dynamics. Annu Rev Biochem. 1987;56:535–565. doi: 10.1146/annurev.bi.56.070187.002535. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Shimizu A., Takahashi N., Yaoita Y., Honjo T. Organization of the constant-region gene family of the mouse immunoglobulin heavy chain. Cell. 1982 Mar;28(3):499–506. doi: 10.1016/0092-8674(82)90204-5. [DOI] [PubMed] [Google Scholar]
- Small D., Nelkin B., Vogelstein B. Nonrandom distribution of repeated DNA sequences with respect to supercoiled loops and the nuclear matrix. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5911–5915. doi: 10.1073/pnas.79.19.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry A. O., Blasquez V. C., Garrard W. T. Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5497–5501. doi: 10.1073/pnas.86.14.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Xu M., Hammer R. E., Blasquez V. C., Jones S. L., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J Biol Chem. 1989 Dec 15;264(35):21190–21195. [PubMed] [Google Scholar]