Abstract
The complete 13 site AvrII restriction map of the genome of E coli strain MG1655 is presented and compared with several other E. coli strains. The map was determined primarily by isolating individual AvrII fragments from pulsed-field gels, and hybridizing these large probes to a battery of mapped E. coli clones in lambda vectors. AvrII restriction patterns for eight other laboratory strains were determined and maps for seven of them deduced from the gel and comparisons between the strain genotypes, the MG1655 map, and AvrII sites in E. coli sequences taken from Genbank.
Full text
PDF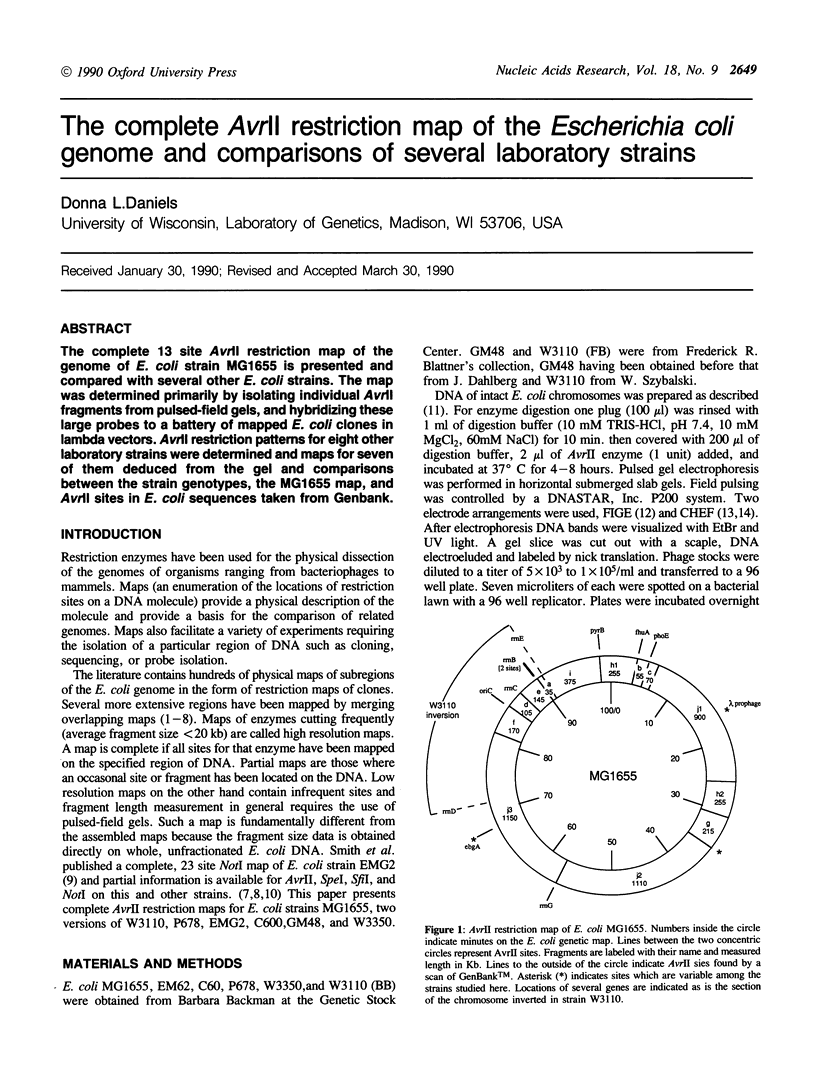


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birren B. W., Lai E., Clark S. M., Hood L., Simon M. I. Optimized conditions for pulsed field gel electrophoretic separations of DNA. Nucleic Acids Res. 1988 Aug 11;16(15):7563–7582. doi: 10.1093/nar/16.15.7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Carrano A. V., Lamerdin J., Ashworth L. K., Watkins B., Branscomb E., Slezak T., Raff M., de Jong P. J., Keith D., McBride L. A high-resolution, fluorescence-based, semiautomated method for DNA fingerprinting. Genomics. 1989 Feb;4(2):129–136. doi: 10.1016/0888-7543(89)90291-7. [DOI] [PubMed] [Google Scholar]
- Church G. M., Kieffer-Higgins S. Multiplex DNA sequencing. Science. 1988 Apr 8;240(4849):185–188. doi: 10.1126/science.3353714. [DOI] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S. H., Little P. F. A cosmid vector for systematic chromosome walking. Gene. 1986;49(1):9–22. doi: 10.1016/0378-1119(86)90381-1. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Blattner F. R. Mapping using gene encyclopaedias. 1987 Feb 26-Mar 4Nature. 325(6107):831–832. doi: 10.1038/325831a0. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Labat I., Brukner I., Crkvenjakov R. Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics. 1989 Feb;4(2):114–128. doi: 10.1016/0888-7543(89)90290-5. [DOI] [PubMed] [Google Scholar]
- Evans G. A., Lewis K. A. Physical mapping of complex genomes by cosmid multiplex analysis. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5030–5034. doi: 10.1073/pnas.86.13.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley R. G., Hu M., Timmons M., Yun K., Deonier R. C. A partial restriction map of the proA-purE region of the Escherichia coli K12 chromosome. Gene. 1983 May-Jun;22(2-3):281–287. doi: 10.1016/0378-1119(83)90113-0. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Barlow D. P., Lehrach H. A large inverted duplication allows homologous recombination between chromosomes heterozygous for the proximal t complex inversion. Cell. 1987 Mar 13;48(5):813–825. doi: 10.1016/0092-8674(87)90078-x. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D., Nizetic D., Lehrach H. Control of partial digestion combining the enzymes dam methylase and MboI. Nucleic Acids Res. 1989 Dec 11;17(23):9571–9582. doi: 10.1093/nar/17.23.9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Blake D. J., Brownlee G. G. Completion of the detailed restriction map of the E. coli genome by the isolation of overlapping cosmid clones. Nucleic Acids Res. 1989 Aug 11;17(15):5901–5912. doi: 10.1093/nar/17.15.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Rees D. J., Cheng Z., Brownlee G. G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988 Mar 25;16(6):2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Maas R., Silva R. M., Gomes T. A., Trabulsi L. R., Maas W. K. Detection of genes for heat-stable enterotoxin I in Escherichia coli strains isolated in Brazil. Infect Immun. 1985 Jul;49(1):46–51. doi: 10.1128/iai.49.1.46-51.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Michiels F., Craig A. G., Zehetner G., Smith G. P., Lehrach H. Molecular approaches to genome analysis: a strategy for the construction of ordered overlapping clone libraries. Comput Appl Biosci. 1987 Sep;3(3):203–210. doi: 10.1093/bioinformatics/3.3.203. [DOI] [PubMed] [Google Scholar]
- Muramatsu S., Kato M., Kohara Y., Mizuno T. Insertion sequence IS5 contains a sharply curved DNA structure at its terminus. Mol Gen Genet. 1988 Nov;214(3):433–438. doi: 10.1007/BF00330477. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Dutchik J. E., Graham M. Y., Brodeur G. M., Helms C., Frank M., MacCollin M., Scheinman R., Frank T. Random-clone strategy for genomic restriction mapping in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C., Botstein D. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Poustka A., Pohl T., Barlow D. P., Zehetner G., Craig A., Michiels F., Ehrich E., Frischauf A. M., Lehrach H. Molecular approaches to mammalian genetics. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):131–139. doi: 10.1101/sqb.1986.051.01.016. [DOI] [PubMed] [Google Scholar]
- Schoner B., Kahn M. The nucleotide sequence of IS5 from Escherichia coli. Gene. 1981 Aug;14(3):165–174. doi: 10.1016/0378-1119(81)90112-8. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]



