Abstract
The past decade has highlighted the central role of epigenetic processes in cancer causation, progression and treatment. Next-generation sequencing is providing a window for visualizing the human epigenome and how it is altered in cancer. This view provides many surprises, including linking epigenetic abnormalities to mutations in genes that control DNA methylation, the packaging and the function of DNA in chromatin, and metabolism. Epigenetic alterations are leading candidates for the development of specific markers for cancer detection, diagnosis and prognosis. The enzymatic processes that control the epigenome present new opportunities for deriving therapeutic strategies designed to reverse transcriptional abnormalities that are inherent to the cancer epigenome.
The past decade has seen a remarkable acceleration in the validation of the concept that cancer is a disease of epigenetic, as well as genetic, abnormalities. Exploration of these connections constitutes one of the most exciting areas in basic cancer biology — with rich potential for clinical translation. Cancer research in epigenetics in the 1990s was dominated by a focus on understanding and extending the discoveries in the 1980s of DNA methylation abnormalities1. During the past 10 years, this focus has merged with an explosion of knowledge about the role of chromatin covalent modifications and organization and their relevance to gene expression2–6, resulting in an emerging view of what may now be called ‘the cancer epigenome’, which harbours myriad abnormalities that are based on somatically heritable alterations that are not due to primary DNA sequence changes7,8 (TIMELINE). Each year, new surprises arise regarding how interactions between epigenetic and genetic changes help to drive the initiation and progression of cancer. This knowledge fosters new potential cancer biomarker strategies and therapeutic opportunities. We highlight examples of these recent advances and what the future holds for them.
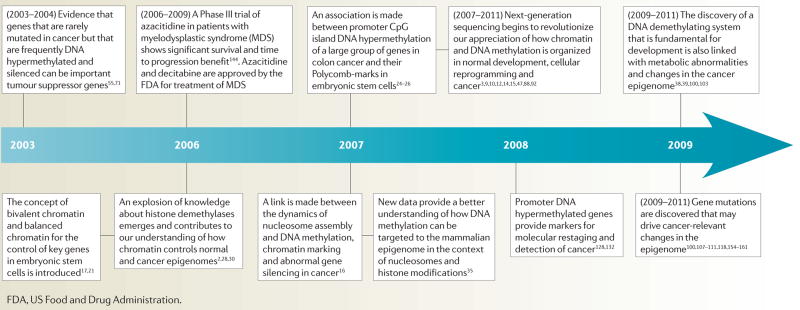
Timeline. Example of key advances in epigenetics and cancer over the past decade.
Functional organization of the genome
Few would have predicted how our view of the human epigenome has expanded over the past 10 years. Next-generation sequencing techniques, as applied to mapping chromatin and DNA methylation in normal, cancer and induced pluripotent stem cells (iPSs), have revolutionized our knowledge of chromatin states, nucleosome positioning and how alterations in these contribute to disease3,9–12. The architecture of gene expression states is being clarified. Nucleosome positions are dynamic and change during cell replication and with gene expression changes11. Active gene promoters, particularly those that are CpG-rich and that normally lack DNA methylation, have nucleosome-depleted regions (NDRs) just upstream of their transcription start sites (TSSs). The nucleosomes that flank these NDRs are marked by the histone modification H3 trimethylated on lysine 4 (H3K4me3), have extensive lysine acetylation and harbour the histone variant H2A.Z, which may destabilize nucleosomes to facilitate transcriptional initiation (FIG. 1). The transcribed regions or gene bodies of active genes also show enrichment of specific covalent marks, including H3K36me3, which may facilitate transcriptional elongation13. These regions normally have dense cytosine methylation, even in downstream CpG islands14, which might also promote transcription elongation rather than repress transcription initiation — as methylation does in promoter regions14. Importantly, structural features of enhancers are being defined (FIG. 1), including deoxyribonuclease 1 (DNase1) sensitivity, nucleosome depletion, and the presence of H3K4me1 and H3 acetylated on lysine 27 (H3K27ac) in the active state15. By contrast, DNA methylation stabilizes epigenetic gene silencing in promoters that lack H2A.Z, that have nucleosomes positioned over the TSS and that harbour repressive histone modifications, such as H3K9me2 or H3K9me3 marks16. Long-term silencing of genes with promoter CpG islands by DNA methylation is normally only associated with inactive X-linked genes, imprinted genes and germ cell-specific genes, but it is also common in many abnormally silenced genes in cancer7,8.
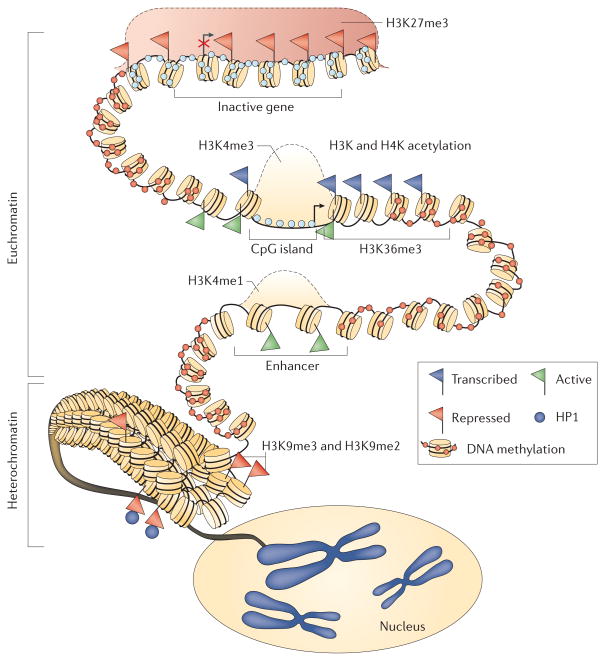
Figure 1. Model of the overall structure of the epigenome in normal human cells.
This diagram shows the balanced state of chromatin, nucleosome positioning and DNA methylation, which maintains the normal packaging state of DNA. A silenced gene (indicated by a red X over the transcription start site designated by the arrow) at the top of the figure has its promoter CpG island occupied by a Polycomb group (PcG) complex (indicated by a red shaded area) that mediates chromatin changes that include the repressive histone modification trimethylation of lysine 27 on histone 3 (H3K27me3). There is no CpG DNA methylation within the gene promoter CpG island (shown by pale blue circles) and nucleosomes are positioned over the transcription start site. Sites upstream from the promoter are heavily DNA methylated (shown by red circles). The gene promoter illustrated below the silenced gene has been signalled to adopt a fully active transcription state and retains the active H3K4me3 marks at the promoter. It also has acetylation of key H3 and H4 lysines, the presence of the variant histone, H2A.Z (not shown) and H3K36me3 in the gene body to facilitate transcriptional elongation. The transcription start region (indicated by an arrow) is not occupied by nucleosomes. Just below, a distal enhancer is shown for this gene with an active nucleosome configuration, and the signature histone modification for enhancers, H3K4me1, is present. Finally, towards the bottom of the figure, the packaging of the majority of the cellular DNA into a transcriptionally repressed configuration is depicted, with compacted nucleosomes, the presence of H3K9me2 and H2K9me3, which are signature repressive marks for constitutive heterochromatin, the presence of heterochromatin protein 1 (HP1; also known as CBX5) and extensive DNA methylation. The folding of the heterochromatin into chromosomal locations in the nucleus is shown. Image is adapted, with permission, from REF. 166© (2008) Macmillan Publishers Ltd. All rights reserved.
One exciting recent advance in our understanding has been that it is the balance between transcriptionally permissive and transcriptionally repressive chromatin modifications that maintains genome-wide gene expression states17,18. In contrast to the strong localization of active marks to TSSs, the H3K27me3 inhibitory mark applied by the Polycomb group proteins (PcGs) can extend over many nucleosomes around genes that typically lack DNA methylation but that have a DNase1-insensitive state owing to the presence of nucleosomes at the TSS19. The unexpected finding is that PcG occupancy can also coexist around the TSS with the active mark, H3K4me3, to form what has been termed a bivalent state3,17,18,20,21 for key developmental and lineage-specific genes in embryonic stem cells (ESCs). This bivalency may allow regulatory flexibility by keeping these genes quiescent to maintain ESC pluripotency but allowing for their rapid activation when needed during differentiation3,17,18. Intriguingly, as discussed below, these gene promoters are prone to undergoing an epigenetic switch22,23 and become de novo DNA methylated in cancer and pre-cancerous cells24–26.
There has also been an explosion of knowledge regarding the molecular determinants of global-scale chromatin architecture. Insulator proteins such as CCCTC-binding factor (CTCF) and others27, together with PcG occupancy19, organize DNA into loops of transcriptionally repressive heterochromatin or into active euchromatin, which facilitates blocks or which connects distal enhancers and proximal promoters. As addressed below, the accessibility of CTCF to DNA is linked to the DNA methylation status of target regions. A dizzying array of enzymes is now known to not only catalyse the addition of transcriptionally activating and repressing histone marks, but also to remove them28–30. The discovery of the demethylases that can remove lysine methylation marks on histones is a particularly exciting advance that is proving to be pivotal for understanding the normal epigenome and several key aspects of cancer biology30–34. Similarly, we are refining our view of how DNA methylation patterns are established. For example, an elegant tetrameric complex has been defined that can neatly surround a nucleosome and presumably initiate methylation in this context35. This complex comprises two molecules of the de novo DNA methyltransferase DNMT3A and two of DNMT3L, a catalytically inactive isomer that is expressed in ESCs. Nucleosomes containing the active histone mark H3K4me3 cannot be encompassed by this complex35, thus targeting DNA methylation to regions such as inactive promoters, intragenic regions, gene bodies and non-control regions.
Cooperation between DNMT1 (a maintenance DNA methyltransferase) and DNMT3A and DNMT3B, is being defined in normal and cancer cells36. It has been suggested that DNMT3A and DNMT3B repair the errors made by DNMT1 after DNA synthesis. Unlike DNMT1, DNMT3A and DNMT3B are firmly anchored to nucleosomes, perhaps allowing these enzymes to remain closely associated with the DNA methylation that they produce to facilitate epigenetic inheritance37. Excitingly, enzyme systems that remove DNA methylation, such as the TET family proteins, which form 5-hydroxylmethyl cytosine from methylated cytosine, have been discovered to be crucially important during development and tumorigenesis38–41.
Visualizing the cancer epigenome
The recent findings outlined above now frame observations from the 1980s that multiple types of cancers abnormally gain and lose DNA methylation1, and these findings are providing a greater understanding of the importance of these abnormalities to tumorigenesis. Juxtaposing these changes with other abnormalities of chromatin organization is rapidly reshaping our view of the cancer epigenome.
Promoter CpG island DNA hypermethylation
Abnormal gains of DNA methylation in normally unmethylated gene promoter CpG islands, and associated stabilization of transcriptional repression and loss of gene function, are the most extensively studied epigenetic alterations in cancer1,7,8. Rapid advances that have enabled the precise mapping of DNA methylation across normal and cancer genomes confirms that almost all cancer types harbour hundreds of genes with these abnormal gains in methylation1,7 (FIG. 2), affecting some 5–10% of the thousands of promoter CpG islands that never normally contain DNA methylation from embryonic development onwards42. Some genes do, however, show changes in CpG island DNA methylation with ageing43,44. Although most genes that become DNA hypermethylated in cancer are usually affected on an individual basis45, subchromosomal domains with multiple hypermethylated loci have recently been discovered46,47.
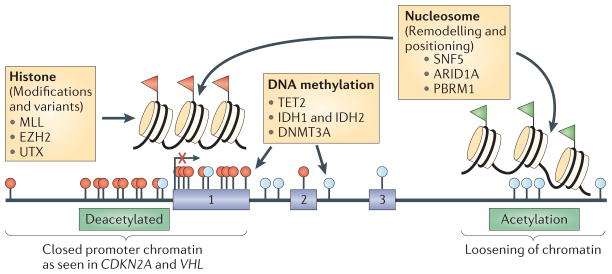
Figure 2. The cancer epigenome and relevant gene mutations.
The cancer epigenome is characterized by simultaneous global losses in DNA methylation (indicated by pale blue circles) with hundreds of genes that have abnormal gains of DNA methylation (indicated by red circles) and repressive histone modifications (indicated by red flags) in promoter region CpG islands. The hypomethylated regions have an abnormally open nucleosome configuration and abnormally acetylated histone lysines (indicated by green flags). Conversely, abnormal DNA hypermethylation in promoter CpG islands is associated with nucleosomes positioned over the transcription start sites of the associated silenced genes (indicated by an arrow with a red X). Recent whole-exon sequencing of human cancers has shown a high proportion of mutations in genes in leukaemias, lymphomas, and ovarian, renal and pancreatic cancers, and rhabdomyosarcoma109–111,154–156 (indicated in yellow boxes), which are depicted as helping to mediate either abnormal DNA methylation, histone modifications and/or nucleosome remodelling100,107,108,118,155,157–165. ARID1A, AT-rich interactive domain-containing protein 1A; DNMT3A, DNA methyltransferase 3A; EZH2, ehancer of zeste 2; IDH1, isocitrate dehydrogenase 1; MLL, mixed lineage leukaemia; PBRM1, protein polybromo 1; SNF5, SWI/SNF-related, matrix associated, actin-dependent regulator of chromatin, subfamily B, member 1; VHL, Von Hippel–Lindau.
Understanding current views of genetic changes in cancers may provide a perspective on how this large number of epigenetic changes contributes to the cancer phenotype. Similar to DNA mutations that frequently occur in specific genes, such as TP53 or KRAS, which act as major drivers for cancer initiation and progression48–50, there are a few rare, high-frequency epigenetic mutations, or ‘mountains’, being discovered in specific genes. For example, the tumour suppressor genes Von Hippel–Lindau (VHL) in renal cancer and CDKN2A (encoding the tumour suppressors INK4A and ARF) in several tumour types are DNA hypermethylated, and these have emphasized the importance of epigenetic changes mediating the loss of gene function in cancer1,7.
What then are the roles, if any, for the hundreds of other DNA hypermethylated cancer genes? Just as the majority of gene mutations are increasingly recognized as low frequency and have been termed ‘hills’, as opposed to mountains48, their importance may be their aggregation in the same signalling pathways, thus helping in the derivation of the cancer phenotype48. Importantly, epigenetic changes can be much more common for many of the same infrequently mutated genes51–53. For example, on average, one gene in the extracellular matrix construction pathway is mutated compared with four being DNA hypermethylated in an individual colon or breast tumour53. Frequent simultaneous hypermethylation occurs in both the GATA4 and GATA5 transcription factors and their downstream targets in colon cancer: GATA4 and GATA5 function in a pathway that is crucial for proper epithelial differentiation54. DNA hypermethylation also affects multiple genes that negatively regulate the WNT pathway from the cell membrane to the nucleus in this same tumour type55. This finding shows how epigenetic changes can complement single driver mutations, such as those occurring in adenomatous polyposis coli (APC) or β-catenin that activate the WNT pathway56,57. Additionally, DNA hypermethylation of a range of genes can collectively contribute to the disruption of the p53 pathway.
One recently recognized and growing important new role for abnormal, promoter DNA hypermethylation is the fostering of pathway disruption that is associated with the transcriptional repression of multiple microRNAs (miRNAs). This can result in the upregulation of oncogenic targets of the microRNAs, such as BCL-6 (REF. 58), and the constitutive activation of signalling that promotes invasiveness and metatstatic activities59,60. Intriguingly, downregulation of the miRNA-29 family has also been linked to overexpression of DNA methyltransferases61,62, possibly creating a scenario that is permissive for gene promoter DNA hypermethylation. miR-101 is often down-regulated in cancer, leading to overexpression of enhancer of zeste 2 (EZH2)63,64 — a key component of the PcG system, which, as previously noted, marks genes that are prone to cancer-specific DNA hypermethylation24–26. Cancer-specific DNA hypermethylation has also been reported to silence other non-coding RNAs65. Intriguingly, non-coding RNAs and anti-sense RNA sequences have also been linked to the development of cancer-specific DNA hypermethylation at gene promoters, and to the recruitment of silencing complexes, such as PcGs, which may subsequently make these regions vulnerable to DNA methylation changes66–69.
Experimentally, the importance of the above gene changes, many involving epigenetically altered cancer genes not affected by base pair mutations, is illustrated by mouse knockout studies of the DNA binding transcriptional repressor hypermethylated in cancer 1 (Hic1). Homozygous knockout mice die from developmental effects70 but heterozygotes develop a range of tumour types that are common in humans as they age71. In these cancers, the wild-type Hic1 allele almost always acquires promoter DNA methylation71. When crossed with mice in which other tumour suppressors have been knocked out, many tumour phenotypes emerge from an initial expansion of progenitor cells and increased transcriptional repression of HIC1 targets72–75. Most recently, targeted DNA hypermethylation of the HIC1 promoter has been found to transform human mesenchymal stem cells76.
Some 20 years ago DNA hypermethylation was implicated in the expansion of progenitor cells that can initiate tumour development in the kidney through a process termed loss of imprinting (LOI)77,78. In this process, a gain of CpG methylation (hypermethylation not hypomethylation) in the insulator region immediately upstream of the imprinted H19 gene on chromosome 11 is associated with loss of CTCF binding to the region79,80. This, in turn, results in the activation of the imprinted insulin-like growth factor 2 (IGF2) promoter on the same chromosome by providing access of this region to a downstream enhancer78,81. Mosaicism for the epigenetic lesion has been shown in pre-neoplastic kidneys ahead of the development of Wilms’ tumour, and in some instances Wilms’ tumour can arise in this setting through DNA microdeletions that are upstream of H19 (REF. 82). This scenario may well illustrate that epigenetic changes can be the earliest initiating factor in a human cancer83. More recently, this change in IGF2 has been linked to the expansion of abnormal stem and progenitor cells in the progression of colon cancer84.
Other DNA methylation changes
Key roles for DNA methylation are emerging for regions other than the most proximal promoter areas. DNA methylation patterns for conserved gene sequences several kilobases upstream or downstream of promoter CpG islands, and termed shores, are tightly linked with the tissue of origin85,86. Alterations of this methylation pattern in cancer cells have been reported to be more frequent than those in nearby CpG islands86, but the importance of these changes is not yet known. Most recent studies indicate that, in cancer, such shores are isolated targets for DNA hypermethylation much less frequently than the CpG islands87,88.
Although regional gains in DNA methylation in cancer are important, simultaneous losses of DNA methylation have long been recognized and are even more widespread1,7,89. Recent genome-wide sequencing of CpG sites in small numbers of colon cancer versus normal colon samples is beginning to reveal intriguing insights into the relationships between these abnormal losses of DNA methylation and the previously discussed gains in many CpG islands. The large regions of DNA methylation losses are not randomly distributed but rather span megabase regions on multiple chromosomes. In turn, these losses, in predominantly CpG-poor regions, are punctuated with narrow gains of DNA methylation, which are those predominantly involving the regional gains in promoter, and other non-annotated, CpG islands87,88,90. Importantly, these domains of losses and gains are associated with late-replicating, lamin-associated nuclear regions and this is profoundly important. It is these regions in ESCs that contain the majority of the genes with bivalent, chromatin promoter domains, which, as discussed earlier, are those that are highly vulnerable to abnormal CpG island DNA hypermethylation in cancer87,88,90. Also, this change and the methylation losses that occur within the bodies of many genes could be functionally related to reduced expression in cancer14. These regional changes in cancer DNA methylation will undoubtedly be the subject of intense investigation over the next few years.
Origins of the cancer epigenome
Clues from developmental biology
Increasingly, the understanding of the cancer epigenome has been linked to the epigenetic dynamics of development. Abnormal DNA hypermethylation in cancer involves many genes that are important to embryonic development. Three studies of colon cancer in 2007 (REFS 24–26,91) revealed that about 50% of genes with cancer-specific promoter CpG island hypermethylation were among the approximately 10% of genes that are controlled by the PcGs in ESCs. As noted earlier, PcG-mediated transcriptional repression, often in the setting of bivalent chromatin, seems to mediate a low, poised transcription state of CpG island promoters for genes in ESCs that are important for regulating lineage determination3,17,18. Importantly, these regulatory events during embryonic development occur in the absence of the abnormal promoter DNA methylation seen in multiple types of cancer92. A working model93 hypothesizes a molecular progression during tumorigenesis that starts with abnormally expanding adult stem or progenitor cell compartments in which vulnerable genes with promoter CpG islands undergo quantitative replacement of flexible, PcG-mediated gene silencing with the more stable silencing that is associated with DNA methylation91,93. The mechanisms, however, remain to be fully characterized. Abnormal retention or imposition of PcGs may occur to the exclusion of DNA methylation for some genes22. In others, once DNA methylation evolves, the PcG complex and the H3K27me3 histone mark may be quantitatively and/or qualitatively replaced91,93 (FIG. 3). These possibilities are consistent with recent data that methylated DNA in a nucleosomal context actually repels components of PcGs94. Indeed, a specific gene marked by bivalent chromatin in the embryonic setting has reduced levels of H3K27me3 when the gene undergoes DNA methylation in cancer95.
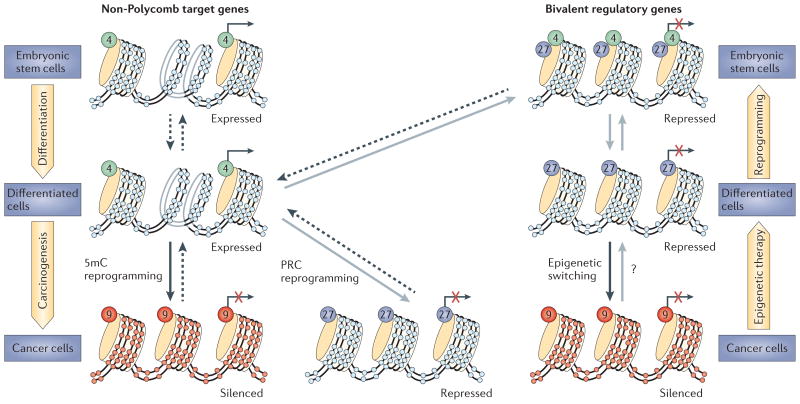
Figure 3. Modes of abnormal gene silencing in cancer.
The currently suggested routes to abnormally silenced genes in cancer are shown. Genes that are active in cells throughout development and adult cell renewal initially have active promoter chromatin that is characterized by the presence of the histone modification, H3K4me (indicated by green circles and dashed arrows), and a lack of DNA methylation (indicated by pale blue circles). Genes that become silenced (indicated by a red X) can do so either by the acquisition of DNA methylation (indicated by red circles) and the presence of the repressive mark, H3K9me (indicated by orange circles and black arrows), or by the presence of Polycomb-mediated repressive chromatin (PRC) marks, H3K27me (purple circles and grey arrows). DNA methylation and H3K9me marks during tumour progression are shown. The wide yellow arrows at the sides of the figure depict movements that link stem and progenitor cells and differentiated cells and which can be impeded by epigenetic abnormalities in cancer or which can be corrected by epigenetic therapy.
What might these changes and switches from PcG marking to DNA methylation mean for tumorigenesis? In stem and/or progenitor cell renewal systems in an adult, such adoption of tight transcriptional repression might prevent their subsequent activation, which is needed for signal transduction events and cell differentiation. In the setting of increased cancer risk states, such as chronic inflammation, where there is an abnormal stress for tissue renewal and a challenge to cell survival, such tight transcriptional repression may favour the abnormal expansion of stem and progenitor cell populations96,97. In turn, this expansion may further select for cell addiction to oncogenic gene mutations, which can help to foster abnormal growth and survival and progression to invasive states and malignancy98. The exploration of these possibilities provides fertile ground for future research.
Genetics meets epigenetics
We have previously reviewed how CpG methylation directly causes genetic changes in cancer by generating mutational hotspots in somatic tissues99 or abnormally silencing DNA repair genes such as MLH1 and O-6-methylguanine-DNA methyltransferase (MGMT) leading to microsatellite instability and a failure to repair DNA lesions, respectively1. However, recent whole-exome sequencing has unexpectedly revealed a high frequency of cancer-specific mutations in genes that are known to directly participate in epigenome organization in multiple tumour types. Thus, many abnormal epigenetic events may lie downstream of genetic abnormalities (FIG. 2). The precise phenotypic consequences of these mutations remain to be determined but important clues are emerging. For example, mutations in the TET2, isocitrate dehydrogenase 1 (IDH1) and IDH2 genes occur in gliomas and leukaemia100,101. In gliomas101, IDH1 mutations correlate with a DNA CpG island hypermethylator phenotype (CIMP)102, an important concept that is continually being advanced and recognized for multiple tumour types. Strikingly, mutations in TET2 and IDH1 (or IDH2) are mutually exclusive in leukaemias100, although each may correlate with increased numbers of DNA hypermethylated genes. This suggests a possible mechanism for the DNA methylation changes100. The TET proteins are hydroxylating enzymes that generate 5-hydroxymethylcytosine from methylated cytosines and may thus help to protect against unwanted DNA methylation in normal cells38,39,103. These enzymes require α-ketoglutarate as a cofactor, and mutant IDH1 and IDH2, through interactions with their wild-type proteins, use α-ketoglutarate to generate a profound build up of the metabolite 2-hydroxyglutarate, which inhibits TET2 (REF. 100). Therefore, focal excesses of 5-methyl-cytosine in the tumours could result from the failure to reduce its accumulation through the TET hydroxylation pathway. The localization of the TET enzymes, and increased levels of 5-hydroxymethylcytosine, at CpG island promoters of the genes that are most vulnerable to cancer-specific DNA hypermethylation (those with bivalent chromatin39,40,104) make this an attractive hypothesis. Furthermore, TET proteins may help to direct transcriptional repression39,40,104,105 that could precede abnormal DNA methylation.
Although the above recent findings for TET enzymes provide insightful leads explaining how regions of the genome, such as promoter CpG islands, could acquire abnormal DNA methylation in cancer, much remains to be resolved about how this may actually evolve. More direct evidence for how the loss of TET, through mutations in these genes, or through the inhibition in the setting of IDH mutations, actually generates abnormal DNA methylation during tumour progression must be provided. Also, it remains to be precisely clarified how the proteins cause DNA demethylation. Current evidence is that this is not solely due to the formation of 5-hydroxymethylcytosine, which instead may be an intermediate that must subsequently be converted to cytosine through the action of the DNA repair processes that are mediated by proteins such as activation-induced cytidine deaminase (AID; also known as AICDA) and apolipoprotein B mRNA-editing enzyme 1 (APOBEC1)105,106. It is clear that future research will concentrate on all of these events over the next few years and the result will be a much clearer understanding of at least some of the events that contribute to the cancer epigenome.
Mutations in DNMT3A have also been recently defined with high frequency in aggressive forms of acute myelogenous leukaemia (AML), although altered DNA methylation patterns have not yet been found to correlate with this107,108. Intriguingly, one mutational hotspot is at a CpG site in DNMT3A, suggesting that the enzyme might be causing its own demise by methylating this site and increasing the likelihood for an inactivating C to T transition creating a base mismatch, which is not repaired.
Mutations in the histone-modifying enzyme EZH2, the catalytically active component of the PRC2 complex, have also been reported109,110. Initially, this was a surprising finding as EZH2 was considered to be an oncogene and the mutations were thought to be inactivating. However, follow-up studies reveal that several of the mutations actually lead to increased levels of H3K27me3, which is consistent with the predicted role of EZH2 in cancer111. Chromosomal translocations that lead to the oncogenic misstargeting of the mixed lineage leukaemia (MLL) histone methyltransferase are increasingly being characterized112–115. Interestingly, leukaemias can have hypermethylation of DNA sequences targeted by MLL owing to the absence of H3K4me3 (REF. 116), which, as discussed above, may repel DNA methyltransferases during de novo methylation. Importantly, some of the MLL translocations seem to occur in utero as they are observed in newborns, suggesting that this is the driver event in this type of cancer.
It has been known for some time that a component of the chromatin remodelling SNF5 complex BAF47, encoded by SMARCB1, is altered in particularly aggressive cases of rhabdomyosarcomas117,118. A recent mouse knockout model for this mutation has shown a tumour phenotype that seems to be entirely dependent on the activity of the PcG pathway for downstream effects119. Perhaps most excitingly, frequent mutations in related chromatin remodelling-encoding genes are increasingly being found in ovarian tumours, kidney tumours and leukaemias (FIG. 2). The frequency of these mutations strongly hints at their having a major causative or driver role. These new discoveries firmly establish that interference in epigenetic processes can lead to cancer and add credence to the idea that epigenetics is a major player in the disease process. A final exciting development linking genetics, epigenetics and cancer origins has been findings that firmly suggest that the inheritance of certain single nucleotide promoter variants, or other regional genetic alterations, can increase the probability of the de novo methylation of key genes such as MLH1, thus contributing to familial cancers and early onset disease120–123.
Translational advances
Biomarker development
In the 1990s, the detection of abnormal promoter CpG island DNA hypermethylation emerged as a potential biomarker strategy for assessing cancer risk, early detection, prognosis and predicting therapeutic responses124. During the past decade, such strategies have moved towards actual clinical practice. The list of potential marker genes, knowledge of their position in cancer progression, and the development of ever more sensitive detection strategies, including nanotechnology approaches are all expanding125,126. Recent key developments include the use of hypermethylated genes in stool and blood DNA as highly sensitive and specific markers for colon cancer risk and detection127,128, as well as the detection of glutathione S-transferase PI (GSTP1) hypermethylation in tumour biopsy samples and urine samples for prostate cancer129,130. Assays for these are commercially offered to clinicians, although the final realization of their value awaits approval by regulatory agencies, such as the US Food and Drug Administration (FDA). Mapping the patterns of DNA methylation has also recently been proposed to help in the identification of cancers of unknown primary site131.
DNA methylation biomarkers can be used for the molecular prognosis of potentially curable, stage I non-small-cell lung cancer132. The concurrent hypermethylation of four genes, CDKN2A and CDH13 in particular, in primary tumour and mediastinal lymph node biopsy samples, strongly correlates with early recurrence and death132. The validation of these findings might lead to a potentially powerful molecular re-staging strategy and might thus identify high risk patients who require special adjuvant treatments.
DNA methylation patterns can robustly predict response to chemotherapy. Importantly, hypermethylation marks that silence the DNA repair gene MGMT, which removes alkyl groups added to guanine in DNA, predict the best response and survival times after standard-of-care treatment with the alkylating agent temozolomide and radio-therapy in patients with glioma133,134. This clinical utility has recently been documented in a large, international Phase III cooperative group trial135, and these data should now lead to FDA approval for using this marker in clinical practice. One caution engendered from recent findings in The Cancer Genome Atlas (TCGA) project, is that gliomas that harbour MGMT methylation in patients who have received alkylating agent treatment have a markedly increased frequency of mutations of the type predicted by DNA alkylation136. It seems that selection in treated tumours for mismatch repair deficiency allows cells to survive with a hypermutator phenotype136.
Other chromatin abnormalities in cancer that we have discussed are also emerging as having important biomarker potential for cancer. For example, global and specific changes in patterns of histone acetylation and methylation have been reported to be hallmarks of cancer and/or to predict risk of tumour recurrence137,138. These changes are presumably due to abnormalities in histone-modifying proteins, which can sometimes be linked to the important problem of treatment resistance, as discussed below. The degree to which these potential biomarkers progress towards the clinic will crucially depend on work over the next several years to expand on these initial findings.
The potential for epigenetic therapy for cancer
There is growing excitement regarding the reversal of epigenetic abnormalities for cancer therapy139,140. 5-azanucleosides, which have long been known to have DNA demethylating activities141, inhibit all three biologically active DNA methyltransferases, and initially proved too toxic for clinical use. However, pioneering clinical work by Silverman and colleagues142, Issa and colleagues143, and others144 generated a remarkable oncology paradigm — therapeutic efficacy could be achieved at low drug doses. Such reduced doses were used in a large trial in patients with myelodisplastic syndrome (MDS), which can lead to leukaemia, and showed an increase in the time of conversion of MDS to frank leukaemia, as well as increased overall survival144. Now, two inhibitors, azacitidine (Vidaza; Celgene) and decitabine (Dacogen; Eisai), have gained recent approval by the FDA for MDS, and this paves the way for refining the use of low-dose regimens not only for leukaemia but also for solid tumours.
Histone deacetylase (HDAC) inhibitors are receiving intense trial activity145. Although HDACs targeted by these compounds affect many protein targets, they clearly also inhibit histone deacetylation that accompanies gene silencing states145. A milestone in therapy using agents that modify the epigenome is the recent FDA approval of vorinostat (Zolinza; Merck) and romidepsin (Istodax; Celgene) for their remarkable efficacy in cutaneous T cell lymphoma146,147. However, the precise molecular mechanisms for response in these patients have yet to be determined. The concept of combination epigenetic therapy has evolved from laboratory findings in 1999 that an HDAC inhibitor, which alone had little heritable effects on reversing the silencing of genes with dense CpG island DNA methylation, could augment gene reactivation following cell treatment with DNA methyl-transferase inhibitors148. Combination of these two classes of drugs is receiving attention in clinical trials for many cancers, but final efficacy is still to be determined149,150.
Another recently raised fascinating possibility regarding the use of HDAC inhibitors is in overcoming the overwhelming problem of resistance to cancer therapy41. Multiple HDAC inhibitors were shown to reverse therapeutic resistance in selected subpopulations of cancer stem-like cells in culture, which are characterized by the overexpression of the histone demethylase JARID1A, which removes the active transcription mark H3K4me3 (REF. 41). A related protein has also just been shown to be overexpressed in stem-like subpopulations of melanoma cells32.
This link between epigenetic changes and drug resistance, and the in vitro findings that an HDAC inhibitor can reverse the process, is important for the future development of drugs that are currently in use for epigenetic therapy. Epigenetic therapy used alone, even in diseases such as MDS and AML, in which efficacy has been greatest to date, is usually not curative. The best use of such drugs may be in combination with other chemotherapeutic strategies. Both HDAC inhibitors, and inhibitors of DNA methylation, could theoretically be used in patients to sensitize responses to standard agents that are being used in cancer management and/or to delay or reverse resistance to such agents and to targeted therapies. Much preclinical work is needed to sort out these possibilities and to derive the best design for future clinical trials to test these possibilities.
Finally, there is growing excitement that some of the mutated enzymes, such as EZH2 in the PcG system and the JARID histone demethylases, or other overexpressed proteins associated with the chromatin abnormalities discussed in this Perspective could be therapeutic targets for specific DNA methylation and histone deacetylation changes. One proof of principle linking genetics and epigenetics, involves translocations in leukaemia that result in altered chromatin landscapes and resultant gene expression changes. For example, translocation-mediated fusion of MLL with the protein AF10, and other partners, in infant leukaemias, leads to the abnormal recruitment of the histone methyltransferase DOT1L to gene targets such as HOXA9 (REF. 114). These are leukaemia-causing events that are dependant on the DOT1L interaction with AF10 and other fusion partners with resultant targeting of hypermethylation of H3K79 and overactivation of HOXA9 and other MLL target genes114,115. Very recently, selective inhibitors of DOT1L have been developed that inhibit the H3K79 methylation, block overexpression of leukaemogenic genes and selectively inhibit, in vitro and in vivo, leukaemic cells harbouring the MLL gene translocations114. Most recently, translocations and/or overexpression of the bromodomain protein BRD4 have been linked to a driver role in a lethal form of paediatric epithelial cancer and to AML151–153. This factor seems to be intimately involved in the overactivation of the MYC oncogene in these settings151–153. Inhibiting BRD4 with small molecules blunts MYC overactivity and has antitumour effects151–153. This provides a potentially unique way of targeting MYC through an epigenetic abnormality and could accomplish the long sought-after goal of suppressing this oncogene. All of the above findings are tremendously compelling for the development of selective cancer therapies that are based on reversing chromatin abnormalities.
Conclusions
The past decade has seen exponential increases in interest and progress in the field of cancer epigenetics. Basic work on defining the structure of the mammalian epigenome in both normal and diseased states is leading to an unprecedented definition of the cancer epigenome. The field is now ripe for the elucidation of the topography of these epigenomes with respect to the precise cellular origins of cancer and how to use epigenetic classifications to better define cancer subcategories. Key remaining questions relate to how enhancer structures are regulated; the role of non-coding RNAs in specifying chromatin structure and the control of normal and abnormal gene expression; how boundaries are maintained and abnormally lost between regions of active versus repressed transcription; and in defining the three-dimensional maintenance and regulation of chromatin regions and how these function in cancer. The full range of epigenetic abnormalities in cancer will be clarified by this research. Rather than separating genetics from epigenetics, or trying to decide which is more important for cancer initiation and progression, the past few years have emphasized that these fields are merging. This is leading to an understanding of how mutations and epigenetic alterations work together to cause this disease. The next challenge lies not only in chronicling the mutations but also in defining the phenotypic ramifications for each of the epigenetic abnormalities in cancer. Exciting doors have been opened for translating all of the emerging knowledge for developing cancer biomarkers and new prevention and therapeutic strategies. Challenges for the next decade include bringing DNA methylation markers to full clinical use and mining the latest knowledge of chromatin abnormalities to obtain new biomarkers. Epigenetic therapies, although clearly now a reality, must be markedly refined and we must better understand how currently available agents are actually producing their clinical efficacies. As we have discussed, we still have much to learn about the best strategies for maximally using such drugs in terms of what combinations to use, how to synchronize them with other treatment modalities and especially whether they can sensitize patients to other agents or be used to overcome the paramount issue of drug resistance. Finally, the many new epigenetic therapeutic targets that are emerging must be intensely and creatively pursued. The future is exciting in terms of the benefits that may emerge for the prevention and management of all types of cancer.
Acknowledgments
The authors thank all of their present and former colleagues in their laboratories for their provision of data and discussions. They also thank all of their colleagues in the field who have contributed the data discussed in this article and apologize to all those whose work has not been included because of space constraints. They are grateful to K. Bender and S. Olivo for help in preparation of the manuscript. Portions of work cited from the authors’ laboratories were supported by grants from the US National Cancer Institute (NCI) and National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH), and by The Cancer Genome Atlas Project (TCGA) funded by the National Genome and National Cancer Institutes. They are also grateful for the support of the Entertainment Industry Foundation (EIF) and the American Association for Cancer Research (AACR), via the Stand up to Cancer (SU2C) project, for work aimed at bringing epigenetic therapy to the forefront of cancer management.
Footnotes
Competing interests statement
The authors declare competing financial interests. See Web version for details.
Contributor Information
Stephen B. Baylin, Email: sbaylin@jhmi.edu, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, Maryland 21231, USA.
Peter A. Jones, Email: pjones@med.usc.edu, The USC Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, California 90089, USA.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Allis C, Jenuwein T, Reinberg D. In: Epigenetics. Caparros M, editor. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 3.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotech. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther MG, Young RA. Transcription. Repressive transcription Science. 2010;329:150–151. doi: 10.1126/science.1193995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly TK, et al. H2A.Z. maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Chi AS, Bernstein BE. Developmental biology. Pluripotent chromatin state. Science. 2009;323:220–221. doi: 10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimura Y, et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development. 2006;133:2371–2381. doi: 10.1242/dev.02405. [DOI] [PubMed] [Google Scholar]
- 20.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nature Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 21.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo Y, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 24.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nature Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 26.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 27.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 28.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 29.Allis CD, Jenuwein T, Reinberg D. Overview and concepts. Cold Spring Harbor Laboratory Press; New York: 2007. [Google Scholar]
- 30.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 41.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 43.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 45.Easwaran HP, et al. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Res. 2010;70:8015–8024. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nature Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 47.Coolen MW, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nature Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 49.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan TA, et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 2008;5:e114. doi: 10.1371/journal.pmed.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuebel KE, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi JM, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17:1535–1545. doi: 10.1158/1078-0432.CCR-10-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akiyama Y, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nature Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki H, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barekati Z, et al. Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum Mol Genet. 2010;19:2936–2946. doi: 10.1093/hmg/ddq199. [DOI] [PubMed] [Google Scholar]
- 58.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 61.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 65.Lujambio A, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carter MG, et al. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum Mol Genet. 2000;9:413–419. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- 71.Chen WY, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nature Genet. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 72.Chen W, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6:387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 73.Briggs KJ, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W, et al. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29:2467–2476. doi: 10.1038/onc.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohammad HP, et al. Loss of a single Hic1 allele accelerates polyp formation in Apc(Delta716) mice. Oncogene. 2011;30:2659–2669. doi: 10.1038/onc.2010.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teng IW, et al. Targeted methylation of two tumor suppressor genes is sufficient to transform mesenchymal stem cells into cancer stem/initiating cells. Cancer Res. 2011;71:4653–4663. doi: 10.1158/0008-5472.CAN-10-3418. [DOI] [PubMed] [Google Scholar]
- 77.Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nature Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 78.Steenman MJ, Rainier S, Dobry CJU, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nature Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. erratum 8, 203 (1994) [DOI] [PubMed] [Google Scholar]
- 79.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 80.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nature Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 81.Moulton T, et al. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nature Genet. 1994;7:440–447. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 82.Sparago A, et al. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nature Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 83.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nature Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 84.Sakatani T, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 85.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Issa JP. Epigenetic variation and cellular Darwinism. Nature Genet. 2011;43:724–726. doi: 10.1038/ng.897. [DOI] [PubMed] [Google Scholar]
- 88.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 90.Berman BP, et al. Regions of focal DNA hypermethylation and long-range 1 hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genet. doi: 10.1038/ng.969. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 94.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tiwari VK, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meng X, Riordan NH. Cancer is a functional repair tissue. Med Hypotheses. 2006;66:486–490. doi: 10.1016/j.mehy.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 98.Baylin SB. Stem cells, cancer, and epigenetics. Stembook. 2009:2–14. [PubMed] [Google Scholar]
- 99.Rideout WM, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 100.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Issa JP. CpG island methylator phenotype in cancer. Nature Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 103.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carey N, Marques CJ, Reik W. DNA demethylases: a new epigenetic frontier in drug discovery. Drug Discov Today. 2011;15–16:683–690. doi: 10.1016/j.drudis.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 106.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamashita Y, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 109.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 110.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 111.Yap D, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stumpel DJ, et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood. 2009;114:5490–5498. doi: 10.1182/blood-2009-06-227660. [DOI] [PubMed] [Google Scholar]
- 113.Schafer E, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood. 2010;115:4798–4809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 116.Erfurth FE, et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci USA. 2008;105:7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 118.Uno K, et al. Aberrations of the hSNF5/INI1 gene are restricted to malignant rhabdoid tumors or atypical teratoid/rhabdoid tumors in pediatric solid tumors. Genes Chromosomes Cancer. 2002;34:33–41. doi: 10.1002/gcc.10052. [DOI] [PubMed] [Google Scholar]
- 119.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2011;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hitchins MP, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 121.Hesson LB, Hitchins MP, Ward RL. Epimutations and cancer predisposition: importance and mechanisms. Curr Opin Genet Dev. 2010;20:290–298. doi: 10.1016/j.gde.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 122.Chan TL, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nature Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 123.Ligtenberg MJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nature Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 124.Laird PW. The power and the promise of DNA methylation markers. Nature Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 125.Bailey VJ, et al. MS-qFRET: a quantum dot-based method for analysis of DNA methylation. Genome Res. 2009;19:1455–1461. doi: 10.1101/gr.088831.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nature Biotech. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Glockner SC, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lofton-Day C, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 129.Cairns P, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–2730. [PubMed] [Google Scholar]
- 130.Rosenbaum E, et al. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 131.Fernandez AF, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2011 May 25; doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brock MV, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 133.Esteller M, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 134.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 135.Gilbert MR, et al. ASCO Annual Meeting 2011 Abstr. University of Texas M. D. Anderson Cancer Center; Chicago: 2006. p. 2011. [Google Scholar]
- 136.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 138.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 139.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nature Biotech. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Issa JP, Kantarjian HM. Introduction: emerging role of epigenetic therapy: focus on decitabine. Semin Hematol. 2005;42:S1–S2. doi: 10.1053/j.seminhematol.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 141.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 142.Silverman LR, Mufti GJ. Methylation inhibitor therapy in the treatment of myelodysplastic syndrome. Nature Clin Pract Oncol. 2005;2:S12–S23. doi: 10.1038/ncponc0347. [DOI] [PubMed] [Google Scholar]
- 143.Issa JP, Kantarjian H. Azacitidine. Nature Rev Drug Discov. 2005;5:S6–S7. doi: 10.1038/nrd1726. [DOI] [PubMed] [Google Scholar]
- 144.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nature Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 146.Duvic M, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 148.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 149.Gore SD, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 150.Griffiths EA, Gore SD. DNA methyltransferase and histone deacetylase inhibitors in the treatment of myelodysplastic syndromes. Semin Hematol. 2008;45:23–30. doi: 10.1053/j.seminhematol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schwartz BE, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011;71:2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 Aug 3; doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Caligiuri MA, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54:370–373. [PubMed] [Google Scholar]
- 155.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Banine F, et al. SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res. 2005;65:3542–3547. doi: 10.1158/0008-5472.CAN-04-3554. [DOI] [PubMed] [Google Scholar]
- 158.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2 and DNMT3A. Blood. 2011 Aug 9; doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wartman LD, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121:1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nature Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]