Abstract
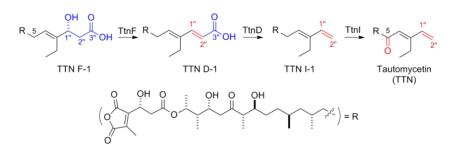
The tautomycetin (TTN) biosynthetic gene cluster has been recently cloned and sequenced from Streptomyces griseochromogenes, unveiling four genes, ttnCDFI, as candidates to encode the tailoring steps for TTN biosynthesis. It is reported that (i) TtnC plays no essential role in TTN biosynthesis, (ii) TtnI catalyzes C-5 oxidation, and (iii) combining the previous findings with TtnFD, the tailoring steps from TTN F-1 to TTN take place in the order of TtnF-catalyzed C-1”/C-2” dehydration, TtnD-catalyzed C-3” decarboxylation, and TtnI-catalyzed C-5 oxidation.
Tautomycetin (TTN), first isolated from Streptomyces griseochromogenes as an antifungal antibiotic,1 is best known for its potent serine/threonine protein phosphatase (PP) inhibitory activity.2 With its 40-fold preference for PP-1 inhibition over PP-2A, TTN is the most selective PP-1 inhibitor known to date.2 Recently, TTN was also found to be a potent immunosuppressant, a property unique to TTN but not found among the other known PP-1/PP-2 inhibitors.3 The latter observation suggested that the immunosuppressive activity of TTN is unlikely related to its PP-1/PP-2A inhibitory activity and prompted us to search for an alternative target for TTN. By screening TTN and its engineered analogues against a panel of protein tyrosine phosphatses (PTPs), we showed that TTN and TTN D-1, one of the engineered analogues, were potent inhibitors of the Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).4 The crystal structure of the SHP2-TTN D-1 complex revealed that TTN D-1 occupied the SHP2 active site in a manner similar to that of the peptide substrate.4 Since SHP2 has been identified as a bona fide oncogene from the PTP superfamily, it represents an exciting target for multiple cancers.5 Therefore, small molecule inhibitors of SHP2 could be promising novel anticancer drug leads.4,5
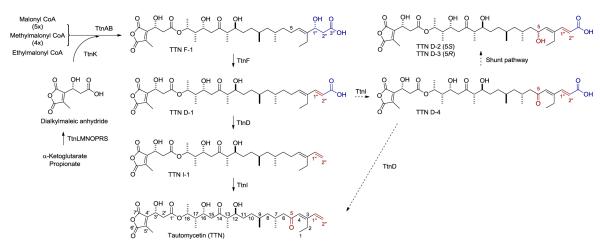
The ttn biosynthetic gene cluster has been cloned and sequenced by us6 and others7 from two producers. On the basis of functional characterization of the genes within the ttn cluster from S. griseochromogenes, we have previously established that (i) biosynthesis of the dialkylmaleic anhydride moiety from one molecule each of α-ketoglutarate and propionate and its subsequent incorporation into the TTN polyketide scaffold are catalyzed by eight proteins of TtnKLMNOPRS, (ii) assembly of the TTN polyketide scaffold from five molecules of malonyl CoA, four molecules of methylmalonyl CoA, and one molecule of ethylmalonyl CoA is catalyzed by the type I polyketide synthase (PKS) proteins of TtnAB, affording TTN F-1 as the nascent intermediate, and (iii) TtnF and TtnD catalyze the C-1”/C-2” dehydration and C-3” decarboxylation, respectively, tailoring TTN F-1 en route to TTN (Figure 1).6
Figure 1.
Proposed biosynthetic pathway for TTN in S. griseochromogenes featuring (i) TtnKLMNOPRS-catalyzed biosynthesis of the dialkylmaleic anhydride moiety from one molecule each of α-ketoglutarate and propionate and its subsequent incorporation into the TTN polyketide scaffold, (ii) TtnAB-catalyzed assembly of the TTN polyketide scaffold from five malonyl CoAs, four methylmalonyl CoAs, and one ethylmalonyl CoA, and (iii) TtnFDI-catalyzed tailoring steps of TTN F-1 to TTN in the order of C-1”/C-2” dehydration by TtnF, C-3” decarboxylation by TtnD, and C-5 oxidation by TtnI. Solid arrows, main pathway; dashed line arrows, side steps due to substrate promiscuity of TtnD and TtnI. TTN D-2 and TTN D-3 are shunt metabolites most likely resulted from reduction of TTN D-4 by adventitious activities in S. griseochromogenes fermentation.
The conversion of TTN F-1 to TTN requires minimally three steps of C-1”/C-2” dehydration, C-3” decarboxylation, and C-5 oxidation. Bioinformatics analysis of the genes within the ttn cluster unveiled four tailoring enzymes of TtnCDFI.6a While the involvement of TtnD and TtnF in tailoring TTN F-1 en route to TTN were confirmed by inactivating ttnD and ttnF in vivo,6b the roles of TtnCI in TTN biosynthesis have not been studied. Furthermore, while the ΔttnF mutant strain S. griseochromogenes SB13014 accumulated TTN F-1 exclusively, the ΔttnD mutant strain S. griseochromogenes SB13013 accumulated four metabolites TTN D-1, -2, -3, and -4, an observation that complicated the determination of the precise timing of the tailoring steps in TTN biosynthesis.6b
Here we now report in vivo functional characterization of ttnCI in S. griseochromogenes for TTN biosynthesis. Our findings revealed that (i) TtnC played no essential role in TTN biosynthesis, (ii) TtnI catalyzed C-5 oxidation, (iii) the tailoring steps from TTN F-1 to TTN proceeded in the order of TtnF-catalyzed C-1”/C-2” dehydration, TtnD-catalyzed C-3” decarboxylation, and TtnI-catalyzed C-5 oxidation, and (iv) TtnI-catalyzed oxidation was rate-limiting for TTN biosynthesis, and overexpression of ttnI in S. griseochromogenes resulted in at least three-fold increase in TTN titer.
Sequence analysis of the ttn cluster indicated that TtnC is a putative flavoprotein decarboxylase.6a Since we have previously established that C-3” decarboxylation was catalyzed by TtnD,6b it was not apparent what role TtnC could play in TTN biosynthesis. To address this dilemma, we inactivated ttnC in S. griseochromogenes using the PCR-targeting and γ-RED-mediated mutagenesis method as described previously6 to afford the ΔttnC mutant strain SB13012, whose genotype was confirmed by PCR analysis [Supporting Information (SI)]. Unexpectedly, when cultured under the standard conditions for TTN production6 using the wild-type strain as a control, SB13012 still produced TTN, and the TTN titter in SB13012 was very similar to that of the wild-type strain (Figure S1). These results suggested that TtnC played no essential role in TTN biosynthesis or its function can be compensated by other genes within the ttn cluster or the S. griseochromogenes genome under the conditions examined. Genes within a natural product biosynthetic gene cluster that have been experimentally confirmed to play no essential role for its biosynthesis are rare but known.8
TtnI was predicted to be a P-450 oxygenase,6a serving as a candidate for C-5 oxidation in TTN biosynthesis. To directly probe its predicted role, ttnI was similarly inactivated in S. griseochromogenes using the PCR-targeting and γ-RED-mediated mutagenesis method6 to afford the ΔttnI mutant strain SB13017 (SI), the genotype of which was subsequently confirmed by Southern analysis (Figure S2). When cultured under the standard conditions6 for TTN production using the wild-type strain as a control, SB13017 failed to produce TTN but instead accumulated a new metabolite (Figure 2, panels I and II vs. III and IV). To eliminate the concern of potential polar effect, genetic complementation to the ΔttnI mutation in SB13017 was carried out by expressing a functional copy of ttnI in trans to afforded the recombinant strain SB13018, in which constitutive expression of ttnI is under the control of the strong promoter ErmE* (SI). Upon fermentation of SB13018, with both the wild-type and SB13017 as controls, under the standard TTN production conditions,6 TTN production was fully restored in SB13018 (Figure 2, panels V and VI vs. III and IV) consistent with the intermediacy of this new compound in TTN biosynthesis (Figure 1).
Figure 2.
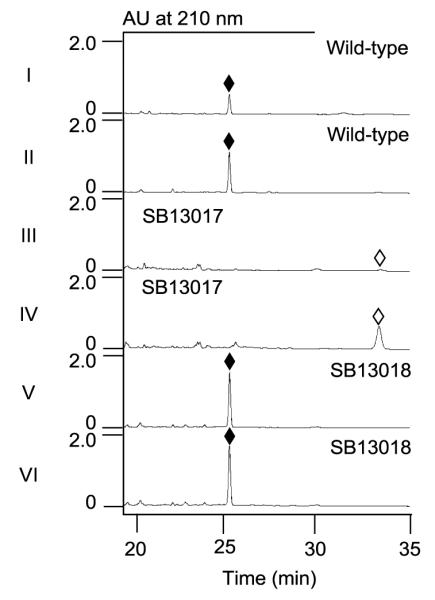
HPLC chromatograms of metabolite profiles from S. griseochromogenes wild-type and recombinant strains: wild-type (I) supernatant and (II) mycelia; SB13017 (ΔttnI mutant) (III) supernatant and (IV) mycelia; and SB13018 (ΔttnI complemented) (V) supernatant and mycelia (VI). TTN, (◆); TTN I-1 (◇).
Large scale fermentation of SB13017 allowed the production and isolation of this new metabolite, named TTN I-1, in quantities sufficient for complete structural characterization (SI). TTN I-1 was obtained as a yellowish gum, and its structure was elucidated on the basis of comprehensive MS and NMR spectroscopic data interpretation (Table 1 and Figure 3). The molecular formula of C33H52O9 for TTN I-1 was established by HR-ESI-MS analysis in positive mode, which afforded an [M + H]+ ion at m/z 593.3688 (calcd m/z 539.3684). Comparison of the 1H and 13C NMR data of TTN I-1 (Table 1) with those of TTN D-16b suggested that the two compounds are closely related. The 1H and 13C NMR data for the C-5 to C-18 fragment, as well as the data for the dialkylmaleic anhydride moiety, were almost superimposable between the two compounds. The only differences were the signals for the fragment comprising of C-1 to C-4 and C-1” to C-3”, resulting from C-3” decarboxylation in TTN I-1 in comparison with TTN D-1.6b TTN I-1 was hence established as 2”-decarboxy TTN D-1 (Figure 1), whose complete 1H and 13C NMR assignments were further confirmed by careful analysis of key COSY and HMBC correlations (Figure 3).
Table 1.
1H and 13C NMR data of TTN I-1 (CDCl3, † in ppm, J in Hz)a
| no. | 1H NMR | 13C |
|---|---|---|
| 1 | 1.01 (3H, t, 7.6) | 13.64 |
| 2 | 2.24 (2H, q, 7.6) | 19.21 |
| 3 | 140.03 | |
| 4 | 5.40 (1H, t, 7.2) | 132.91 |
| 5 | 2.11 (2H, o) | 25.46 |
| 6 | 1.21 (1H, m) | 37.75 |
| 6 | 1.34 (1H, o)b | |
| 7 | 1.31 (1H, o) | 29.84 |
| 7-Me | 0.85 (3H, d, 6.5) | 19.35 |
| 8 | 1.11 (2H, o) | 44.82 |
| 9 | 1.50 (1H, m) | 30.07 |
| 9-Me | 0.83 (3H, d, 6.5) | 19.25 |
| 10 | 1.29 (2H, o) | 33.17 |
| 11 | 1.38 (1H, m) | 31.88 |
| 11 | 1.54 (1H, m) | |
| 12 | 3.73 (1H, m) | 73.42 |
| 13 | 2.65 (1H, m) | 52.68 |
| 13-Me | 1.08 (3H, d, 7.1) | 13.64 |
| 14 | 215.76 | |
| 15 | 2.46 (1H, dd, 2.5, 16.8) | 46.48 |
| 15 | 2.78 (1H, o) | |
| 16 | 4.32 (1H, br d, 9.8) | 66.55 |
| 17 | 1.69 (1H, m) | 42.74 |
| 17-Me | 0.94 (3H, d, 7.1) | 10.31 |
| 18 | 5.01 (1H, m) | 73.91 |
| 18-Me | 1.29 (3H, d, 6.4) | 18.40 |
| 1′ | 170.07 | |
| 2′ | 2.87 (1H, dd, 3.6, 16.1) | 40.66 |
| 2′ | 2.77 (1H, o) | |
| 3′ | 5.19 (1H, m) | 63.82 |
| 4′ | 142.10 | |
| 5′ | 143.02 | |
| 5′-Me | 2.27 (3H, s) | 10.12 |
| 6′ | 165.70 | |
| 7′ | 164.83 | |
| 1″ | 6.24 (1H, dd, 17.5, 10.5) | 140.03 |
| 2″ | 5.10 (1H, d, 17.5) | 110.13 |
| 2″ | 4.91 (1H, d, 10.5) |
1H NMR at 700 MHz, 13C NMR at 175 MHz, and assignments were based on HSQC, COSY, and HMBC experiments
‘o’ denotes overlapping signals
Figure 3.
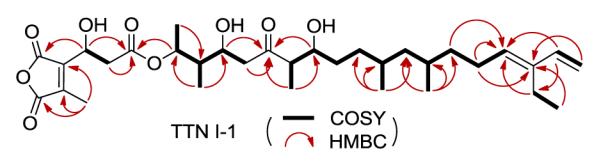
Key COSY and HMBC correlations for TTN I-1.
The current studies completed the characterization of the tailoring steps for TTN biosynthesis. Previously, the observation that the ΔttnD mutant strain SB13013 accumulated four metabolites, TTN D-1, -2, -3, and -4, with mixed oxidation status at C-5, has complicated the determination of the precise timing of the tailoring steps.6 In this study, we have now excluded the participation of TtnC from playing any essential role in TTN biosynthesis. The findings that the ΔttnI mutant strain SB13017 accumulated TTN I-1 as the sole metabolite (Figure 2, panel III and IV) and the ΔttnI complementation recombinant strain SB13017 fully restored TTN biosynthesis (Figure 2, panel V and VI) confirmed the bioinformatics-based prediction of TtnI as a P-450 oxygenase,6 catalyzing C-5 oxidation for TTN biosynthesis (Figure 1). Taken together, we now propose that the tailoring steps for TTN biosynthesis from TTN F-1 to TTN procceds in the order of TtnF-catalyzed C-1”/C-2” dehydration, TtnD-catalyzed C-3” decarboxylation, and TtnI-catalyzed C-5 oxidation (Figure 1). The natural substrates for the TtnF dehydratase, TtnD decarboxylase, and TtnI P-450 oxygenase therefore are TTN F-1, TTN D-1, and TTN I-1, respectively, all of which can now be readily isolated, setting the stage to characterize these enzymes in vitro. Both the TtnD decarboxylase and TtnI P-450 oxygenase also shown some degree of substrate promiscuity, as exemplified by the accumulation of TTN D-4 in the ΔttnD mutant strain SB13013 and subsequent conversion of TTN D-4 to TTN in the ΔttnD complementation strain SB13013.6b Substrate promiscuity is common for enzymes from secondary metabolite biosynthetic machinery,9 a property that has been extensively exploited for natural product structural diversity by combinatorial biosynthesis methods. In the absence of the TtnD carboxylase activity, TTN D-4 apparently could undergo C-5 reduction to afford TTN D-2 and TTN D-3, shunt metabolites whose formation most likely resulted from adventitious activities in the ΔttnD mutant SB13013 fermentation.
Finally, quantitative comparison of TTN production in the wild-type and SB13018 strains showed that TTN titer was increased at least by three-fold in SB13018 (Figure 2, panels I and II vs. V and VI). This finding suggested that TtnI must have been rate-limiting for TTN biosynthesis in the S. griseochromogenes wild-type strain. Given the exciting activity of TTN as the most selective PP-1 inhibitor2 and its newly discovered inhibitory activity against SHP2,4 overexpression of ttnI now joins the previsouly reported strategies of manipulating pathway regulation,7 offering another opportunity to engineer TTN overproducer, thereby facilitating TTN production, isolation, and follow-up mechanistic and preclinical studies. Equally exciting would be the prospect of engineering TTN D-1 overproducer for further study. TTN D-1 was previously produced as a mixture, together with TTN D-2, -3, and -4, by the ΔttnD mutant strain SB13013,6b but TTN D-1 was the only analogue that was shown to retain the SHP2 inhibitory activity.4 Introduction of a ΔttnDI dual mutation into S. griseochromogenes could afford a recombinant strain that produces TTN D-1 exclusively.
Supplementary Material
Acknowledgment
We thank Dr. Hiroyuki Osada, RIKEN, Japan for the S. griseochromogenes wild-type strain. This work was supported in part by NIH grants CA113297.
Footnotes
Supporting Information Available. Detailed experimental procedures, 1D and 2D NMR spectra for TTN I-1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1) (a).Cheng XC, et al. Isono K. J. Antibiot. 1989;42:141–144. [Google Scholar]; (b) Cheng XC, Uramoto M, Isono K. J. Antibiot. 1990;43:890–896. doi: 10.7164/antibiotics.43.890. [DOI] [PubMed] [Google Scholar]
- (2) (a).Mitsuhashi S, Matsuura N, Ubukata M, Oikawa H, Shima H, Kikuchi K. Biochem. Biophys. Res. Commun. 2001;287:328–331. doi: 10.1006/bbrc.2001.5596. [DOI] [PubMed] [Google Scholar]; (b) Oikawa H. Curr. Med. Chem. 2002;9:2033–2054. doi: 10.2174/0929867023368818. [DOI] [PubMed] [Google Scholar]; (c) Chen X, Zheng Y, Shen Y. Chem. Rev. 2007;107:1777–1830. doi: 10.1021/cr050029r. [DOI] [PubMed] [Google Scholar]
- (3).Shim JH, Lee HK, Chang EJ, Chae WJ, Han JH, Han DJ, Morio T, Yang JJ, Bothwell A, Lee SK. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10617–10622. doi: 10.1073/pnas.162522099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Liu S, et al. Zhang Z-Y. Chem. Biol. 2011;18:101–110. doi: 10.1016/j.chembiol.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5) (a).Tartaglia M, Gelb BD. Annu. Rev. Genomics Hum. Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]; (b) Chan RJ, Kalaitzidis D, Neel BG. Cancer Metastasis Rev. 2008;27:179–192. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- (6) (a).Li W, Luo Y, Ju J, Rajski SR, Osada H, Shen BJ. Nat. Prod. 2009;72:450–459. doi: 10.1021/np8007478. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Luo Y, Li W, Ju J, Yuan Q, Peters NR, Hoffmann FM, Huang S-X, Bugni TS, Rajski SR, Osada H, Shen BJ. Am. Chem. Soc. 2010;132:6663–6671. doi: 10.1021/ja9082446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7) (a).Choi S, Hur Y, Sherman DH, Kim E. Microbiology. 2007;153:1095–1102. doi: 10.1099/mic.0.2006/003194-0. [DOI] [PubMed] [Google Scholar]; (b) Scaglione JB, Akey DL, Sullivan R, Kittendorf JD, Rath CM, Kim E, Smith JL, Sherman DH. Angew. Chem. Eng. Ed. 2010;122:5862–5866. doi: 10.1002/anie.201000032. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hur Y, Choi S, Sherman DH, Kim E. Microbiology. 2008;154:2912–2919. doi: 10.1099/mic.0.2008/018903-0. [DOI] [PubMed] [Google Scholar]; (d) Park S, Choi S, Kim Y, Chang Y, Sherman DH, Kim EJ. Ind. Microbiol. Biotechnol. 2009;36:993–998. doi: 10.1007/s10295-009-0580-5. [DOI] [PubMed] [Google Scholar]
- (8) (a).Gallo MA, Ward J, Hutchinson CR. Microbiology. 1996;142:269–275. doi: 10.1099/13500872-142-2-269. [DOI] [PubMed] [Google Scholar]; (b) Chen Y, Yin M, Horsman GP, Shen BJ. Nat. Prod. 2011;74:420–424. doi: 10.1021/np100825y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9) (a).Chen Y, Wendt-Pienkowski E, Rajski SR, Shen BJ. Biol. Chem. 2009;284:24735–24743. doi: 10.1074/jbc.M109.014191. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Carlson JC, Li S, Gunatilleke SS, Anzai Y, Burr DA, Podust LM, Sherman DH. Nat. Chem. 2011;3:628–633. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.