Abstract
Spinocerebellar ataxia type 10 (SCA10) is an autosomal dominant neurodegenerative disorder manifested by ataxia and seizure. SCA10 is caused by a large expansion of an intronic ATTCT pentanucleotide repeat in the ATXN10 gene. We have recently postulated a toxic RNA-mediated gain of function in the pathogenesis of Spinal Cerebellar Ataxia type 10 (SCA10). The spliced intron-9 RNA containing the expanded AUUCU repeat aggregates in SCA10 cells and sequesters hnRNP K. hnRNP K sequestration triggers the translocation of protein kinase C delta (PKCδ) to mitochondria, leading to activation of caspase-3 and apoptosis. To further confirm the toxic RNA-mediated gain of function, we generated a new transgenic mouse model in which the expanded pentanucleotide repeats are constructed in 3′-untranslated region to ensure transcription without translation of the repeat.
We constructed an artificial transgene containing the SCA10 (ATTCT)500 track within the 3′ untranslated region (3′UTR) of the LacZ gene driven by the rat prion promoter (PrP) and used this to generate a new transgenic mouse model for SCA10. We then examined these mice for neurological phenotypes and histopathological, molecular and cellular changes.
The transgenic mice showed irregular gait and increased seizure susceptibility at the age of 6 months, resembling the clinical phenotype of SCA10. The cerebral cortex, hippocampus and pontine nuclei showed neuronal loss. The brains of these animals also showed molecular and cellular changes similar to those previously found in a SCA10 cell model.
Expression of the expanded SCA10 AUUCU repeat within the 3′ UTR of a gene results in neuronal loss with associated gait abnormalities and increased seizure susceptibility phenotypes which resemble those seen in SCA10 patients. Moreover, these results bolster the idea that the SCA10 disease mechanism is mediated by a toxic RNA gain-of-function mutation of the expanded AUUCU repeat.
Keywords: Nucleotide repeat, neurodegenerative disorder, RNA gain-of-function
Introduction
Spinocerebellar ataxia type 10 (SCA10) is an autosomal dominant cerebellar ataxia initially identified in patients from several Mexican families with progressive ataxia and epilepsy (Grewal et al. 1998; Zu et al. 1999). The genetic mutation of SCA10 is the expansion of an ATTCT repeat in the 9th intron of the ATXN10 gene on chromosome 22q13.3. The number of repeats in this locus ranges from 10 to 29 in the normal population but expands up to 4,500 in affected patients (Matsuura et al. 2000). However, the underlying mechanism by which the expansion of this untranslated pentaneucleotide repeat leads to ataxia and seizure is not fully understood.
The protein product of the ATXN10 gene, ataxin-10, plays important roles in the survival of cerebellar neurons in primary culture (Marz 2004) as well as in promoting neuronal differentiation of PC12 cells (Waragai et al. 2006). However, mature mRNA from the mutant ATXN10 gene is unaltered, suggesting that a loss of function of ATXN10 is unlikely to be the pathogenic mechanism of SCA10 (Wakamiya et al. 2006). In addition, expression levels of neighboring genes are unaltered (Wakamiya et al. 2006). Importantly, a recent case report of asymptomatic individuals with a balanced translocation that disrupts the ATXN10 gene strongly argues against the pathogenic role of ATXN10 haploinsufficiency in SCA10 (Keren et al. 2010). Thus, we postulate that a toxic RNA-mediated gain-of-function by the mutant transcript containing the expanded AUUCU repeat contributes to the pathogenic mechanism of SCA10.
A previous study from our lab demonstrated that the AUUCU RNA repeats remain intact and are deposited as discrete insoluble aggregates in primary fibroblasts and lymphoblasts derived from SCA10 patients (White et al. 2010). We also determined that the expanded AUUCU RNA can bind and sequester hnRNPK. In neuronal cultures, hnRNPK sequestration by AUUCU repeats leads to massive translocation of PKCδ to mitochondria, triggering caspase-3 mediated apoptosis (White et al. 2010). Preliminary studies of a transgenic mouse model expressing the expanded SCA10 ATTCT repeat in the rabbit β-globin intron of a LacZ transgene recapitulated these findings; however, behavioral and neuropathological phenotypes of these transgenic mice were not described (White et al. 2010).
To investigate the toxic RNA-mediated gain-of-function, we constructed a new transgenic mouse model in which the expanded SCA10 repeat resides in the 3′UTR of the transgene, which ensures transcription of the repeat in RNA without translation. This new transgene was generated in part due to the poor reproductive success of our previous transgenic model. Here, we describe behavioral and neuropathological phenotypes of this new 3′UTR-ATTCT repeat mouse model as well as demonstrate molecular and cellular changes similar to those found in SCA10 patient fibroblasts as well as the previous intronic transgenic mouse model.
Materials and Methods
Reagents
The β-galactosidase monoclonal antibody and the GFAP polyclonal antibody (rabbit) were obtained from Abcam (Cambridge, MA). The hnRNP K antibody was obtained from Acris Antibodies (Herford, Germany). Monoclonal anti-PKCδ antibody (G-9) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). COX I-complex IVa and fluorescent secondary antibodies (goat anti-mouse Alexa Fluor488, -568, and -633, and goat anti-rabbit Alexa Fluor488, -568, and -633) were obtained from Invitrogen Molecular Probes (Eugene, OR). Pentylenetetrazol (PTZ) and all other reagents were obtained from Sigma Chemicals Co. USA (St. Louis, MO) unless otherwise specified.
Establishment of SCA10 Transgenic Mice
The animal study protocol was approved by UTMB Institutional Animal Care and Use Committee (IACUC). The cytomegalovirus (CMV) promoter sequence in plasmid pcDNA3.1-hygro-lacZ (Invitrogen) was replaced with the ~3.5kb-MfeI/BamHI fragment of the rat prion promoter (PrP) sequence. The expanded ATTCT repeat sequence from the SCA10 somatic cell hybrid line was derived from a Mexican SCA 10 patient who had clinical presentation of ataxia and epilepsy and was PCR amplified with forward primer 5′-CCAAGGATGCAGGTGCCACAGCATCTC-3′ and reverse primer 5′-ATATGCATCCAGCTTCTGATTACATGGACT-3′. A synthetic polylinker sequence was cloned into the XhoI site downstream of LacZ and upstream of the BGH polyA sequences of the plasmid. The synthetic polylinker sequence provided a SwaI site in which the repeat was subsequently cloned, using T4 DNA ligase (Figure 1A). The presence of the expanded ATTCT repeat was confirmed by digestion of the plasmid with SfiI, flanking the SwaI site. The plasmid encoding the expanded ATTCT repeats was transfected into E. coli SURE bacteria (Agilent Technologies, Santa Clara, CA) and grown at 16°C to minimize deletion of repeat sequences.
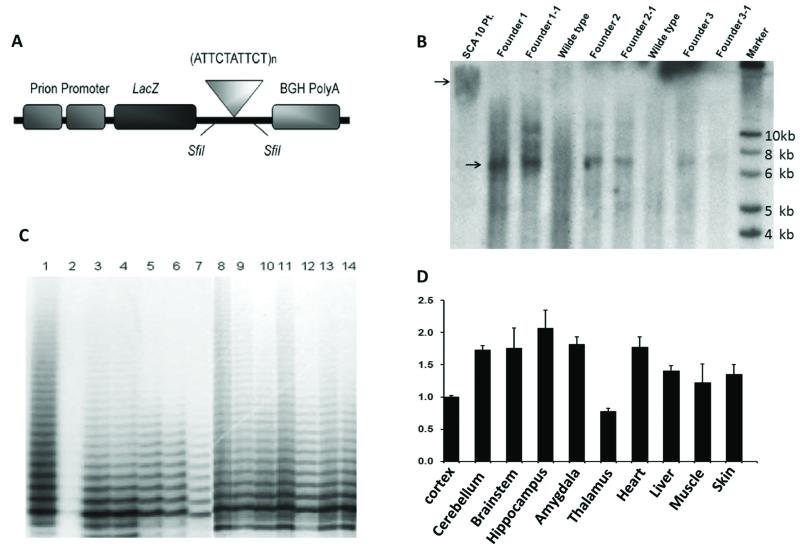
Figure 1.
Establishment of transgenic mouse model and instability of the pentanucleotide repeats.
A. The cytomegalovirus (CMV) promoter sequences in the plasmid pcDNA3.1-hygro-lacZ (Invitrogen) were replaced with the MfeI /BamHI fragment of the rat prion promoter sequences. 500 ATTCT repeats were inserted in the 3′ UTR of LacZ.
B. Presence and integrity of 500 ATTCT repeats as analyzed by Southern blot (arrowhead). The smear phenomenon in the patient lane is due to somatic mosaicism of the expanded allele. Three founders and their F1 animals had the expanded allele.
C. Repeat size analysis of repeat primed PCR (RP-PCR) products from the founders and their progeny by acrylamide gel electrophoresis showed variability in the ATTCT repeat expansion size. Lane 1 – positive control from plasmid DNA, Lane 2 - wildtype mouse, Lane 3 – Founder 1, Lane 4 – Founder 2, Lane 5 – F1 from founder 1, Lane 6 – F1 from Founder 2, Lane 7 – F2 from Founder 2, Lane 8-F1 from founder 3, Lane 9-14 F2 from founder 3.
D. Real-time RT-PCR analysis revealed that the transgene is widely expressed in the central nervous system (CNS) with the highest level in the hippocampus. Expression was also seen in non-CNS derived tissues with the highest levels present in the heart.
The transgene containing expanded ATTCT repeat was microinjected into fertilized eggs derived from pregnant female FVB/N mice. The microinjected fertilized eggs were then transplanted into the uterus of pseudo-pregnant surrogate mothers to obtain founder transgenic mice. The presence of the transgene and the repeat in the transgenic founder mice was confirmed by both Southern blot and repeat primed PCR (RP-PCR) analyses (Matsuura and Ashizawa 2002; Matsuura et al. 2000).
For southern blot, mouse genomic DNA was digested with enzymes flanking the insertion sequence. DNA was separated on a 0.8% agarose gel and transferred to a Hybond N+ membrane (Amersham). The probe was a synthesized primer containing 12 ATTCT repeats which was random primed with 32P-α-dCTP labeling bead (Amersham) according to manufacturer instructions. The membrane was hybridized in ExpressHyb solution (Clontech) at 65°C overnight. The membrane was washed thoroughly in washing solution (0.04M NaPO4, 1%SDS) twice for 20 minutes each and allowed to air dry before visualizing. Blots were visualized using phosphor imaging screens.
To verify expression of the PrP-driven transgene in the transgenic mice, total RNA isolated from various tissues and regions of the brain was sent to the UTMB RT-PCR Core Lab for quantitative RT-PCR of LacZ transcripts.
Histological, Immunostaining and Fluorescent In Situ Hybridization (FISH)
Transgenic mice at 6 months of age were anesthetized with Avertin and then perfused through the aorta, first rinsing for 15 minutes with PBS and then 60ml of fresh 4% Paraformaldehyde (PFA) in DEPC water. The brain was carefully removed and stored in 4% PFA at 4°C with gentle agitation overnight. Brain tissue was then placed in 30% sucrose overnight and embedded in paraffin for sagittal and coronal sections. Hematoxylin and eosin (H&E) and Periodic Acid-Schiff (PAS) stains were performed using a standard pathological protocol. Neuron numbers in hippocampus CA3 region were counted at high power and averaged from 3 consecutive sections. For immunohistochemical staining, slides were blocked and incubated overnight with a primary antibody against either hnRNP K, PKCδ, COX-IV, or β-galactosidase, which was diluted 1:500 with 1x antibody dilution buffer (10% goat serum and 0.3% triton-X 100 in 1x PBS). Unbound primary antibody was removed by washing with PBS for 10 minutes 3 times each, and then incubated with either goat anti-mouse or goat anti-rabbit secondary antibody for an hour at room temperature. After further washing, confocal microscopy of the slides was performed using Hamamatsu Camera Controller with DP controller software. Immunostaining of GFAP was performed with ABC kit (Vector Laboratories, Burlingame, CA). TUNEL assay was done with In Situ Apoptosis Detection Kit (R&D System, Minneapolis, MN). For PAS staining, the brain sections were deparaffinized, rehydrated and stained using the Sigma-Aldrich Periodic Acid Schiff kit. FISH for detection of AUUCU RNA foci, hnRNP K immunostaining and co-localization of PKCδ and mitochondria were performed as previously described (White et al. 2010).
Locomotive Function Analyses
A battery of analyses were performed to evaluate the behavior of the SCA10 transgenic mice according to established protocols (Brooks and Dunnett 2009). In open field analyses, mice at 6 months of age were placed in a clear, Plexiglas open field apparatus where they were videotaped and analyzed for horizontal and vertical movements, along with total distance, travel speed, rearing and crossings. In footprint studies, the hind limbs of the mice were painted with a non-toxic ink, and mice were encouraged to walk through a square wooden apparatus lined with Whatman paper. The back of the apparatus was closed after the mice were placed on the paper, and the mice were permitted one minute to exit the front of the apparatus. Mice that required more than one minute were re-tested. Step length and gait width of ten consecutive steps were recorded in two trials. For the hindlimb clasping reflex, mice were held by the tail 2-3 inches above a flat surface for a period of 30 seconds and assessed for abnormal hindlimb reflex.
Seizure Susceptibility Analysis
Mice at 12 months of age were given a starting dose of 2.5 mg/kg of pentlylenetetrazol (PTZ) intraperitoneally followed with subsequent increasing doses (Yang and Frankel 2004). Three transgenic mice and three wild-type littermates were studied. Immediately after administration, the animal was placed in the observational area. The time of onset of convulsive behavior, nature and severity of convulsion were recorded using the following scoring system: 0-no response, 1-mild contractions, 2-clonic or myoclonic body jerk, 3-tonic seizure, 4-generalized tonic-clonic seizure. Each mouse was observed for a period of 30 minutes for onset and severity of convulsion. The dose was then escalated to the maximum of 80mg/kg. A mean cumulative score for each dose was calculated for two groups for comparisons and statistical analysis.
Statistical Analysis
The significance of locomotive function studies was conducted with Student’s t -test. Error bars represent standard deviation.
RESULTS
Development of the transgenic mouse lines expressing expanded ATTCT repeats
Transgenic animals were generated with a transgene driven by the prion promoter (PrP) and the ATTCT expansion in the 3′ UTR following the LacZ reporter gene, encoding β-galacatosidase (β-gal) (Figure 1A). Presence of the transgene containing the expanded ATTCT repeat was confirmed by Southern blot analysis (Figure 1B) and repeat-primed PCR (RP-PCR) analysis (Figure 1C) of genomic DNA. The smear phenomenon in the patient lane of the Southern blot is due to somatic mosaicism of the expanded allele. This is commonly seen in SCA 10 patients (Matsuura et al. 2004). Three founders and their F1 progeny had the expanded allele, which were propagated for analysis.
The Transgene is Expressed Widely throughout the Brain of the Transgenic Mice
Expression of the PrP-driven transgene was assessed with quantitative RT-PCR in a variety of tissues at the age of 6 months. Within the brain, analysis of the transgene transcript with the primer set for LacZ showed that the transgene is widely expressed in the central nervous system (CNS) with the highest level of expression in the hippocampus. Expression was also seen in the non-CNS derived tissues with the highest levels present in the heart (Figure 1D). Northern blot analyses of brain tissues were attempted to show direct evidence of the expression of repeats but technical difficulties protected us from yielding meaningful results. However, the FISH study as shown below clearly shows expression of the repeats in the CNS. Immunostaining of β-gal in the brain of 6-month-old transgenic mice showed expression in widespread regions, including cerebral cortex, especially the frontal lobe, pontine nuclei, granular cells of the cerebellum, granule cells and pyramidal cells of the hippocampus, locus ceruleus, and substantia nigra. The Lac Z expression pattern in our transgenic model varies from the typical ATXN10 expression with high expression in pyramidal cells of hippocampus but less expression in Purkinje Cells at the cellular level, most likely due to differences in the use of the PrP promoter in the transgene (Marz et al. 2004) (Table 1).
Table 1.
Comparison of Ataxin-10 Expression to Transgenic LacZ Expression
| Regions of Brain | Human | Mouse | SCA10 Transgenic |
|---|---|---|---|
| ATXN10 | ATXN10 | LacZ | |
| Olfactory Bulb | ND | ++ | ++ |
| Cerebral Cortex | 0/+ | 0/+ | +++ |
| Striatum | 0 | 0/+ | ND |
| Locus Ceruleus | + | + | +++ |
| Substantia Nigra | 0 | 0 | ++ |
| Midbrain Raphe | +/++ | ++ | ND |
| Pontine Nuclei | +/+++ | ++/+++ | +++ |
| Nuclei Cuneatus | +++ | ND | +++ |
| Vestibular Nuclei | ++ | ++ | + |
| Olivary Nuclei | ++ | ++ | ++ |
| Hypoglossal Nuclei | +/++ | ND | +++ |
| Purkinje Cells | +/++ | +/+++ | + |
| Deep Cerebellar Nuclei | ND | +++ | ++ |
| Spinal Motor Neurons | + | + | ND |
| 0 - no observable staining | |||
| + - +++, semi-quantitative staining | |||
| ND- not determined | |||
Modified from Marz et al. (4)
Transgenic mice demonstrate locomotive dysfunction
At 6 months of age, transgenic mice began to exhibit an ataxic gait that primarily involved awkward movements in their hind limbs. Physically, the transgenic mice appeared normal with the exception of patchy hair growth on the back and urine stained fur, suggesting decreased grooming behavior. The mice had normal eye blink, ear twitch, and righting reflexes. All of our transgenic mice at 6 months of age, and less than half of the animals at 3 months of age, displayed an abnormal hindlimb clasping reflex (Figure 2A). Additionally, mice displayed abnormal whisker twitch reflex.
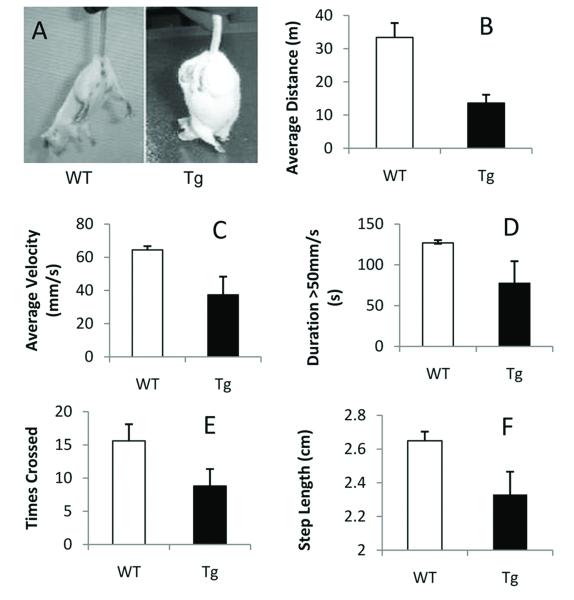
Figure 2.
Transgenic mice show locomotive dysfunction.
A. Hindlimb clasping reflex. A wild-type (WT) mouse extends the hind limbs posteriorly in “V” shape in anticipation of reaching the surface to escape. A transgenic (Tg) mouse displayed an abnormal reflex with hind limbs clasped.
B-E. Open field study. Transgenic mice traveled less distance (B, p=0.011) at a slower speed (C, p=0.013) than their wild-type littermates. Transgenic mice spent less time at a speed of greater than 50mm/s (D, p=0.038) and exhibited significantly fewer crossings (E, p=0.03).
F. Footprint study. Transgenic mice had a significantly shorter step length than their wild-type littermates (F, p<0.05).
The transgenic animals showed abnormalities in open field and footprint analyses. When placed in a new environment, these mice were hesitant to investigate their surroundings. In the open field study, transgenic mice traveled a smaller distance (n=5, p=0.011, Figure 2B) at a slower speed (n=5, p=0.013, Figure 2C) than their wild-type littermates. Detailed analyses revealed that transgenic mice spent less time traveling at a speed greater than 50 mm/s (n=5, p=0.038, Figure 2D). The transgenic mice also exhibited significantly fewer crossings (n=5, p=0.03; Figure 2E). No significant differences were seen in the amount of time spent in the center versus the outside of the arena. In the footprint study, transgenic mice had a significantly shorter step length (n=5, p<0.05, Figure 2F) with a greater variability in the width of steps. However, on fixed-speed and accelerated rotarod analyses, there were no significant differences between transgenic mice and wild-type littermates assessed at 6 months of age (data not shown).
Transgenic mice display seizure behaviors and susceptibility to PTZ-induced seizure
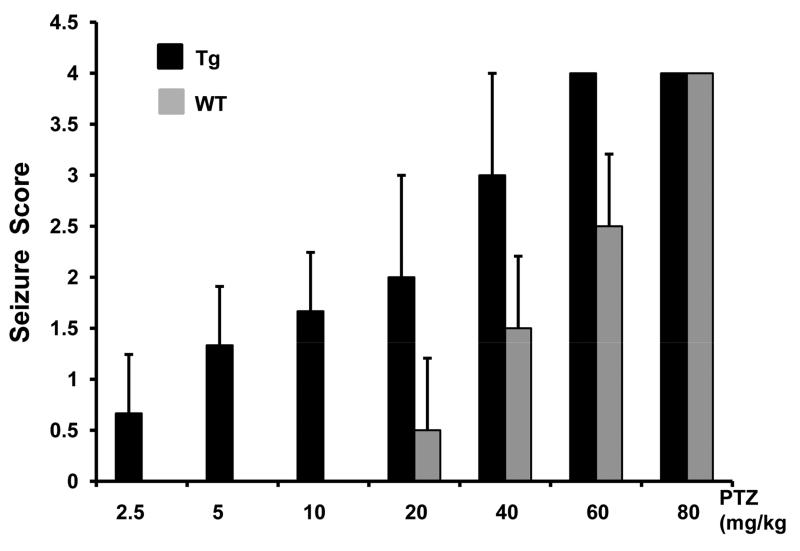
Simply moving the cage elicited an excitable response in transgenic mice, during which they continually run around the cage, a well-known preconvulsant behavior (Bell et al. 1999). To determine their response to induced seizures, we injected transgenic and wild-type littermates with pentylenetetrazol (PTZ) at escalating doses from 2.5mg/kg to 80mg/kg via intraperitoneal injections (Figure 3). Transgenic mice developed seizures at low doses of PTZ while there were no detectable seizures in wild-type mice at the same dose. One transgenic mouse developed full generalized tonic-clonic seizures and died at the dose of 40mg/kg, while the other two died immediately following the dose of 60mg/kg. All wild-type mice survived the 30 minute observation at the escalated dose of 60mg/kg (Figure 3).
Figure 3.
Transgenic mice are susceptible to pentylenetetrazol (PTZ)-induced seizures.
PTZ was given at escalating doses from 2.5mg/kg to 80mg/kg via intraperitoneal injection. Mice were observed for 30 minutes after each dose and scored for the presence and type of seizure. The seizure score was averaged in 3 mice. Transgenic mice developed seizures at a low dose of PTZ while there is no seizure in wild-type mice at the same dose.
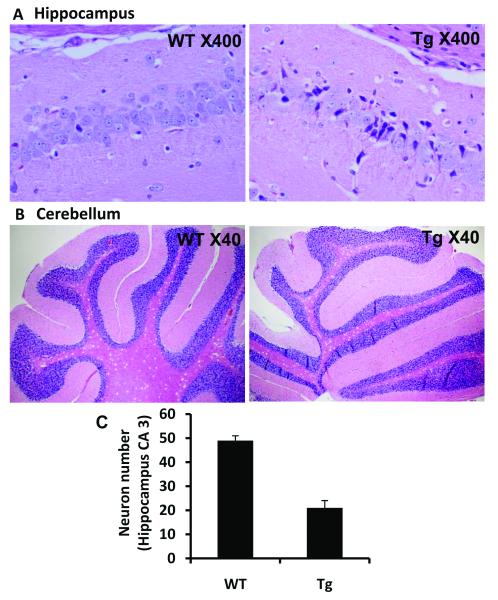
Transgenic brain showed neuronal loss
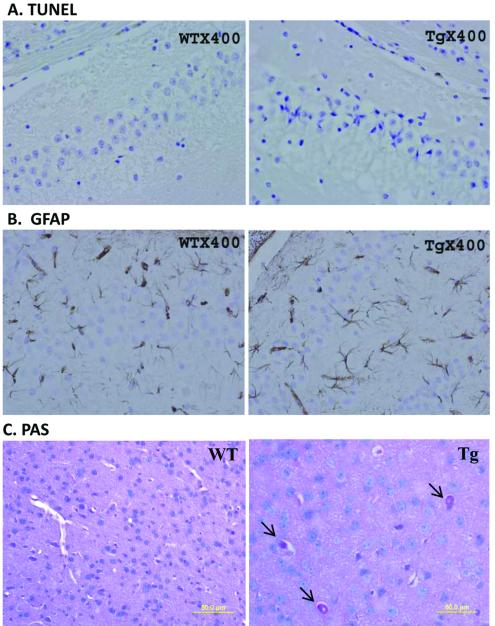
The CA3 region of the hippocampus of transgenic mouse brains showed a conspicuous loss of neurons at 6 months of age. Many neurons were atrophic with small perikaryon, corkscrew-like dendrites and densely basophilic nuclei (Figure 4A). The staining was verified on different mice, showing the same pathological features. Similar neuronal changes were spotted in the pontine nuclei and cerebral cortex of the transgenic mice (data not shown). Unexpectedly, no obvious neuronal loss was observed in the cerebellum (Figure 4B). TUNEL staining showed no evidence of apoptosis in the hippocampus (Figure 5A). The study was repeated in frozen sections and yielded the same result. Serial sections stained for GFAP detected no evidence of gliosis in the area of neuronal loss (Figure 5B). The frontal lobe of the transgenic mice contained neurons that had large vacuoles, which appeared to push the nucleus and cytoplasm to the cell boundary. The vacuoles increased in number and magnitude with increasing age. Brains of 3-month-old animals contained relatively small vacuoles in a small percentage of neurons (~20%) whereas brains of 18-month-old mice contained large vacuoles in nearly 100% of neurons. PAS staining indicated that the vacuoles contained glycogen (Figure 5C), raising the possibility of an autophagic process in neuronal death.
Figure 4.
Pathologic changes in transgenic mouse brain.
A. Neuronal loss in transgenic mouse brain. Many neurons in CA3 region are atrophic with small perikaryon, corkscrew-like dendrites and densely basophilic nuclei.
B. No obvious neuronal loss was observed in the cerebellum.
C. Neuron numbers in hippocampus CA3 region were counted at high power and averaged from 3 consecutive sections. There is dramatic neuron loss at hippocampus CA3 region (WT: 49±2 vs Tg: 21±3).
Figure 5.
Neuronal loss is not associated with apoptosis or gliosis but with glycogen accumulation.
A. Apoptosis was not evident as detected by TUNEL staining in the CA3 region of the hippocampus where neuron loss was conspicuous.
B. Gliosis was not detected by GFAP immunostaining of serial sections in the same region.
C. Large vacuoles were found in neurons in the frontal lobe of transgenic brains. These vacuoles appeared to push the nucleus and cytoplasm to the boundary of the cell and were stained positive for PAS (arrow).
Formation of RNA foci in transgenic brain and interaction with hnRNP K
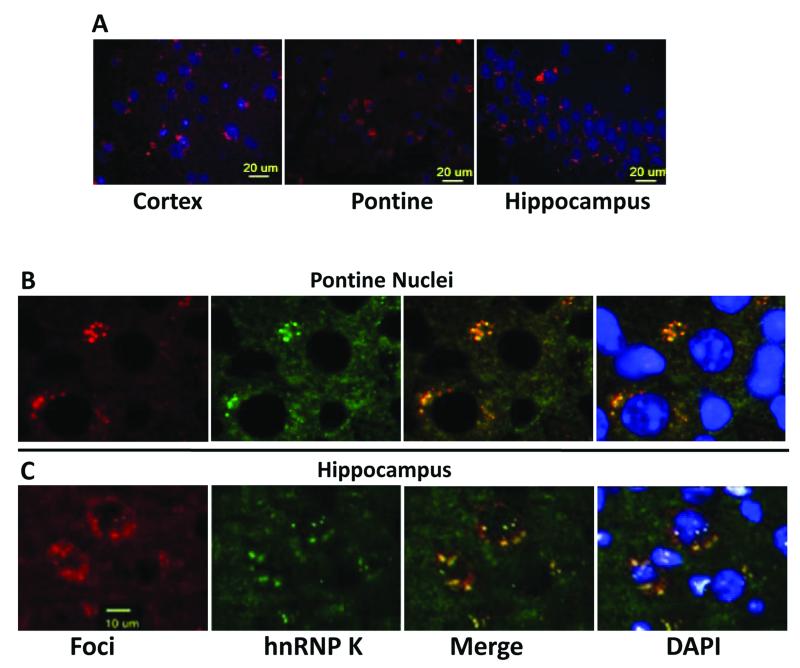
RNA fluorescent in situ hybridization (FISH) showed discrete aggregates of the transgenic transcript containing expanded AUUCU repeats in cortex, pontine nuclei and hippocampal neurons in 6-month-old transgenic mice (Figure 6A). These aggregates were mostly contained within the cytoplasm, but were also found in the nucleus of some cells. RNA aggregates were also found in the brains of 3-month old transgenic mice but not in 1-month-old transgenic mice or age-matched wild-type mice (data not shown). Remarkably, transgenic mice began to demonstrate hindlimb clasp reflex abnormalities at 3 months of age which coincidences with the appearance of RNA foci in the brain. hnRNP K co-localized with the AUUCU repeat containing foci in virtually every part of the brain where RNA foci were detected (Figure 6B), findings that closely resemble those we observed earlier in the intronic AUUCU-repeat transgenic model for SCA10 (White et al. 2010).
Figure 6.
AUUCU repeats form RNA aggregates and sequester hnRNP K in transgenic mouse brain.
A. In sagittal sections of 6 month-old transgenic mouse brains, RNA fluorescent in situ hybridization (FISH) showed distinct RNA aggregates (red) in neurons of the cortex, pontine nuclei and hippocampus. Similar sections of the brain of 6 month-old wild-type littermates showed DAPI stain of nuclei but no red FISH signal (not shown).
B. Co-localization of AUUCU-containing repeats (red) with hnRNPK (green) in the pontine nuclei of transgenic mice.
C. Co-localization of expanded AUUCU repeats (red) and hnRNP K (green) in the hippocampus of transgenic mice.
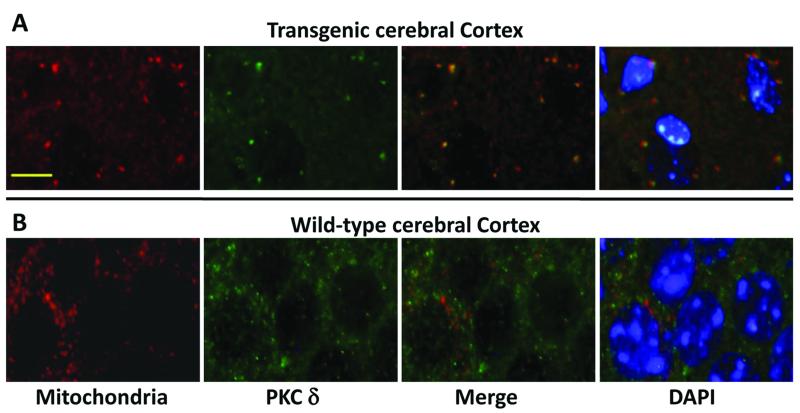
PKCδ accumulated in mitochondria of the transgenic mouse brain
Our previous study showed that PKCδ is translocated to mitochondria in fibroblasts derived from patients with SCA10 and Sy5y cells transfected with the expanded ATTCT repeat (White et al. 2010). Similar to these findings, the brains of our transgenic mice showed increased mitochondrial localization of PKCδ (Figure 7).
Figure 7.
Translocation of PKCδ into mitochondria in transgenic mouse brain
A. Increased localization of PKCδ (green) in mitochondria (COX-IV, red) in cerebral cortex of transgenic mice brain. Co-localization of PKCδ (green) and mitochondria (COX-IV, red) is shown in the merged image as yellow/orange fluorescence in cerebral cortex transgenic mice.
B. PKCδ (green) and mitochondria (COX-IV, red) immunostaining in wild-type littermates do not demonstrate colocalization as shown by a lack of yellow/orange signal in the merged image.
DISCUSSION
This report describes a transgenic mouse model in which expanded ATTCT repeats derived from a SCA10 patient are constructed in the 3′-UTR of the LacZ gene. These mice exhibited locomotive abnormalities and increased seizure susceptibility. However, the motor abnormalities observed in these mice require careful interpretations. Open field tests demonstrated decreased activities of the transgenic mice including reductions in gait speed and crossing; however, while the open field test is sensitive for detecting motor abnormalities, it is not a specific measure. Footprint analysis detected an irregular gait in these mice with awkward movements in their hind limbs suggesting the presence of gait ataxia. Nonetheless, the transgenic mice did not show abnormalities on the rotarod, a more specific measure of motor coordination and balance. Furthermore, as our mice showed no detectable pathologic changes in cerebellar neurons, these motor phenotypes may be attributable to neuronal loss in the cerebral cortex and/or pontine nuclei. Thus, it is premature to conclude that the motor phenotype in our mice recapitulates the cerebellar ataxia of SCA10. Differences in the expression pattern of mutant AUUCU repeats between the transgenic mice, as driven by the PrP promoter, and human SCA10 patients, as driven by the native ATXN10 promoter, as demonstrated in table 1, may partly explain some of these phenotypic differences.
It is postulated that the pathological changes may accumulate with aging, and transgenic mice may have an accelerated declining of locomotive functions. While most of our locomotive analyses were done at the age of 6 months, we found an age-related accumulation of vacuoles, which start appearing at 3 months of age and become robust at 6 months of age, and an increased incidence of the hind limb clasp reflex in transgenic mice. Moreover, the RNA foci containing hnRNP K also start appearing at 3 months of age with a subsequent increase of the number and size at 6 months of age in our transgenic mice. This is consistent with our hypothesis that an RNA-mediated gain of function of the toxic AUUCU repeat RNA is the major pathogenic mechanism of SCA10. In our previous experiments on cell culture models, decreasing the expression of expanded AUUCU repeats and overexpressing hnRNP K rescued the cellular phenotype (White et al. 2010). We expect that similar rescue experiments in our transgenic animals would provide mechanistic evidence for our hypothesis.
The pre-convulsive behavior and susceptibility to PTZ-induced seizures in our transgenic mice suggest that the AUUCU-containing RNA is capable of decreasing the threshold for seizures. Seizure susceptibility may be attributed to neuronal degeneration in the cerebral cortex and hippocampus, neuronal populations which are frequently involved in human epilepsy and mouse models of epilepsy. The seizure susceptibility in our transgenic mice appears to recapitulate the epilepsy phenotype of SCA10. Epilepsy is unique to SCA 10 patients from Mexico but not from Brazil. Our ATTCT construct was amplified from a Mexican patient and was later found to have ATCCT interruptions (Cherng et al. 2011). Repeats from Brazilian patients were found to be homogenous (unreported preliminary data). About 80% of affected members in one family which has interruptions in the expanded allele exhibited seizure, whereas only 25% in another family which has no interrupt suffered from this SCA10-associated phenotype (Grewal et al. 2002; Matsuura et al. 2006). Whether these interruptions render the transgenic mice more susceptible to seizure needs further study. These transgenic mice provide a useful model for further investigations of the mechanism of epilepsy phenotype in SCA10.
In this study, we describe a new transgenic mouse model containing expanded AUUCU repeats within the 3′UTR of a reporter gene and provide evidence that this mouse model recapitulates molecular and cellular abnormalities seen in cells derived from SCA10 patients. Furthermore, we found that the expanded AUUCU repeat forms discrete RNA foci, which co-localize with hnRNP K mainly in the cytoplasm of neurons, leading to translocation of PKCδ to mitochondria in our transgenic mouse brain. The findings duplicate what we observed earlier in another transgenic mouse model expressing expanded ATTCT repeats in the rabbit β-globin intron of the transgene (White et al. 2010). While the intronic expansion model is arguably a more faithful model of SCA10 than the model we examined in this study, the identical molecular and cellular abnormalities in both models confirm that the expanded AUUCU repeat triggers pathogenic RNA gain-of-function events regardless the location of the expanded AUUCU repeat within the transgene. In both models, binding of hnRNP K to the mutant AUUCU repeat may facilitate the trafficking of the RNA-protein complex across the nuclear membrane since hnRNP K is a well-known nuclear shuttle (Michael et al. 1997). We predict that the intronic expansion would cause motor/behavioral abnormalities and histopathological changes similar to the 3′ UTR model. It is interesting to note that both CTG repeat expansion in the 3′UTR of the DMPK gene in myotonic dystrophy type 1 (DM1) and intronic CCTG repeat expansion in the ZFN9 gene in myotonic dystrophy type 2 (DM2) result in an RNA-mediated gain-of-function with sequestration of MBNL1, perturbing the RNA homeostasis and causing closely similar DM phenotypes (Lee and Cooper 2009).
hnRNP K has a wide range of biological functions, including gene expression, DNA methylation, RNA stability and translation (Bomsztyk et al. 2004), and constitutively binds a protein kinase, PKCδ, within the cell (Schullery et al. 1999). However, when hnRNP K binds RNA, PKCδ can no longer bind hnRNP K (Bomsztyk et al. 2004; Schullery et al. 1999). Thus, we postulate that sequestration of hnRNP K in the AUUCU repeat aggregates facilitates the release of PKCδ from hnRNP K, allowing its translocation to the mitochondria and leading to the activation of apoptosis (White et al. 2010).
We found neuronal loss but not active apoptosis in this transgenic model although a previous study of SCA10 cell culture models has shown active apoptosis (White et al. 2010). Similar discrepancies between cell culture and transgenic models have been reported in Huntington’s disease and other neurodegenerative disorders (Stefanis et al. 1997). While the explanation for this difference is elusive, the fast apoptotic process in cell culture models may not faithfully recapitulate the slow degenerative process in vivo. It has been postulated that activation of autophagy pathways in vivo may modulate the acute cell-death process initiated by apoptosis , eventually leading to cell death without features of apoptosis (Lang-Rollin et al. 2008; Pyo et al. 2005). In our transgenic mice, large vacuoles containing glycogen were detected in the frontal lobe, raising the possibility of an autophagic process in neuronal death. The mechanism of neuronal death requires further investigation.
We conclude that our transgenic mouse model expressing an expanded AUUCU repeat in the 3′ untranslated region of the transcript shows motor and seizure susceptibility phenotype and recapitulates the molecular pathophysiology of SCA10.
Acknowledgments
This work was supported by a grant from NINDS (NS041547) to TA.
References
- Bell SM, Schreiner CM, Schultheis PJ, Miller ML, Evans RL, Vorhees CV, Shull GE, Scott WJ. Targeted disruption of the murine Nhe1 locus induces ataxia, growth retardation, and seizures. Am J Physiol. 1999;276(4 Pt 1):C788–795. doi: 10.1152/ajpcell.1999.276.4.C788. [DOI] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26(6):629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- Brooks SP, Dunnett SB. Tests to assess motor phenotype in mice: a user’s guide. Nat Rev Neurosci. 2009;10(7):519–529. doi: 10.1038/nrn2652. [DOI] [PubMed] [Google Scholar]
- Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, Sarkar PS, Ashizawa T, Freudenreich CH, Mirkin SM. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci U S A. 2011;108(7):2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal RP, Achari M, Matsuura T, Durazo A, Tayag E, Zu L, Pulst SM, Ashizawa T. Clinical features and ATTCT repeat expansion in spinocerebellar ataxia type 10. Arch Neurol. 2002;59(8):1285–1290. doi: 10.1001/archneur.59.8.1285. [DOI] [PubMed] [Google Scholar]
- Grewal RP, Tayag E, Figueroa KP, Zu L, Durazo A, Nunez C, Pulst SM. Clinical and genetic analysis of a distinct autosomal dominant spinocerebellar ataxia. Neurology. 1998;51(5):1423–1426. doi: 10.1212/wnl.51.5.1423. [DOI] [PubMed] [Google Scholar]
- Keren B, Jacquette A, Depienne C, Leite P, Durr A, Carpentier W, Benyahia B, Ponsot G, Soubrier F, Brice A, Heron D. Evidence against haploinsuffiency of human ataxin 10 as a cause of spinocerebellar ataxia type 10. Neurogenetics. 2010;11(2):273–274. doi: 10.1007/s10048-009-0227-8. [DOI] [PubMed] [Google Scholar]
- Lang-Rollin I, Dermentzaki G, Vekrellis K, Xilouri M, Rideout HJ, Stefanis L. A novel cell death pathway that is partially caspase dependent, but morphologically non-apoptotic, elicited by proteasomal inhibition of rat sympathetic neurons. J Neurochem. 2008;105(3):653–665. doi: 10.1111/j.1471-4159.2007.05165.x. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37(Pt 6):1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz P, Probst A, Lang S, Schwager M, Rose-John S, Otten U, Ozbek S. Ataxin-10, the spinocerebellar ataxia type 10 neurodegenerative disorder protein, is essential for survival of cerebellar neurons. J Biol Chem. 2004;279(34):35542–35550. doi: 10.1074/jbc.M405865200. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Ashizawa T. Polymerase chain reaction amplification of expanded ATTCT repeat in spinocerebellar ataxia type 10. Ann Neurol. 2002;51(2):271–272. doi: 10.1002/ana.10049. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Fang P, Lin X, Khajavi M, Tsuji K, Rasmussen A, Grewal RP, Achari M, Alonso ME, Pulst SM, Zoghbi HY, Nelson DL, Roa BB, Ashizawa T. Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am J Hum Genet. 2004;74(6):1216–1224. doi: 10.1086/421526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Fang P, Pearson CE, Jayakar P, Ashizawa T, Roa BB, Nelson DL. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am J Hum Genet. 2006;78(1):125–129. doi: 10.1086/498654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, Khajavi M, McCall AE, Davis CF, Zu L, Achari M, Pulst SM, Alonso E, Noebels JL, Nelson DL, Zoghbi HY, Ashizawa T. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26(2):191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. Embo J. 1997;16(12):3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Schullery DS, Ostrowski J, Denisenko ON, Stempka L, Shnyreva M, Suzuki H, Gschwendt M, Bomsztyk K. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J Biol Chem. 1999;274(21):15101–15109. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- Stefanis L, Burke RE, Greene LA. Apoptosis in neurodegenerative disorders. Curr Opin Neurol. 1997;10(4):299–305. doi: 10.1097/00019052-199708000-00004. [DOI] [PubMed] [Google Scholar]
- Wakamiya M, Matsuura T, Liu Y, Schuster GC, Gao R, Xu W, Sarkar PS, Lin X, Ashizawa T. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology. 2006;67(4):607–613. doi: 10.1212/01.wnl.0000231140.26253.eb. [DOI] [PubMed] [Google Scholar]
- Waragai M, Nagamitsu S, Xu W, Li YJ, Lin X, Ashizawa T. Ataxin 10 induces neuritogenesis via interaction with G-protein beta2 subunit. J Neurosci Res. 2006;83(7):1170–1178. doi: 10.1002/jnr.20807. [DOI] [PubMed] [Google Scholar]
- White MC, Gao R, Xu W, Mandal SM, Lim JG, Hazra TK, Wakamiya M, Edwards SF, Raskin S, Teive HA, Zoghbi HY, Sarkar PS, Ashizawa T. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6(6):e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Frankel WN. Genetic approaches to studying mouse models of human seizure disorders. Adv Exp Med Biol. 2004;548:1–11. doi: 10.1007/978-1-4757-6376-8_1. [DOI] [PubMed] [Google Scholar]
- Zu L, Figueroa KP, Grewal R, Pulst SM. Mapping of a new autosomal dominant spinocerebellar ataxia to chromosome 22. Am J Hum Genet. 1999;64(2):594–599. doi: 10.1086/302247. [DOI] [PMC free article] [PubMed] [Google Scholar]