Abstract
Human induced pluripotent stem cells (hiPSC) hold great promise for providing various differentiated cell models for in vitro toxigenicity testing. For Clostridium botulinum neurotoxin (BoNT) detection and mechanistic studies, several cell models currently exist, but none examine toxin function with species-specific relevance while exhibiting high sensitivity. The most sensitive cell models to date are mouse or rat primary cells and neurons derived from mouse embryonic stem cells, both of which require significant technical expertise for culture preparation. This study describes for the first time the use of hiPSC-derived neurons for BoNT detection. The neurons used in this study were differentiated and cryopreserved by Cellular Dynamics International (Madison, WI) and consist of an almost pure pan-neuronal population of predominantly gamma aminoisobutyric acidergic and glutamatergic neurons. Western blot and quantitative PCR data show that these neurons express all the necessary receptors and substrates for BoNT intoxication. BoNT/A intoxication studies demonstrate that the hiPSC-derived neurons reproducibly and quantitatively detect biologically active BoNT/A with high sensitivity (EC50 ∼0.3 U). Additionally, the quantitative detection of BoNT serotypes B, C, E, and BoNT/A complex was demonstrated, and BoNT/A specificity was confirmed through antibody protection studies. A direct comparison of BoNT detection using primary rat spinal cord cells and hiPSC-derived neurons showed equal or increased sensitivity, a steeper dose-response curve and a more complete SNARE protein target cleavage for hiPSC-derived neurons. In summary, these data suggest that neurons derived from hiPSCs provide an ideal and highly sensitive platform for BoNT potency determination, neutralizing antibody detection and for mechanistic studies.
Keywords: botulinum neurotoxin, BoNT, stem cells, induced pluripotent, neurons, cell-based assay
Botulinum neurotoxins (BoNTs), synthesized by the Gram-positive, soil-dwelling bacterium Clostridium botulinum, are the most toxic substances known to humankind and are the causative agents of the neuroparalytic disease botulism (Johnson, 2005; Johnson and Montecucco, 2008). Seven immunologically distinct serotypes of BoNTs designated A through G have been described (Gimenez and Gimenez, 1995). BoNTs are initially synthesized as a single-chain polypeptide of ∼150 kDa, but posttranslational proteolytic cleavage yields distinct heavy and light chains (HC and LC) of ∼100 and ∼50 kDa linked by a disulfide bond. The HC is further functionally divided into the HCC and HCN subdomains. The HCC domain is responsible for recognition and binding to specific neuronal cell surface receptors leading to endocytosis, while the HCN domain is responsible for channel formation in the endocytic vesicle membrane and translocation and internalization of the LC across the endosomal membrane (Fischer and Montal, 2007; Fischer et al., 2009; Montecucco et al., 2004). During translocation, the disulfide bond is cleaved, and the LC is released into the cell cytosol and refolded to the active enzyme component as a zinc-dependent endopeptidase (Fischer and Montal, 2007; Fischer et al., 2009). The LC then specifically targets and cleaves an intracellular SNARE protein at the presynaptic vesicles, which leads to inhibition of neurotransmitter release. Each BoNT serotype has a distinct cleavage target, with BoNT/A and E cleaving SNAP-25 at distinct sites, BoNT/B, D, F, and G cleaving vesicle-associated membrane protein (VAMP)/synaptobrevin at different sites, and BoNT/C cleaving both SNAP-25 and syntaxin (reviewed in Montecucco and Schiavo, 1994).
BoNTs have been classified as a category A Select Agent and present a serious threat as a potential bioterrorism weapon (Arnon et al., 2001; Bossi et al., 2006). Remarkably, BoNT/A, and to a much lesser extent, BoNT/B, are also being used as unique and important pharmaceuticals to treat a variety of neuromuscular disorders and in cosmetics (Chaddock and Acharya, 2011). Cosmetic and clinical applications of BoNTs are increasing, and new formulations of BoNTs for pharmaceutical purposes are being developed necessitating clinical trials, accurate potency determination, and neutralizing antibody screening. Thus, the quantitative and reliable detection of BoNT activity is currently a high priority. Many BoNT detection methods have been published and can be divided into four general categories (reviewed in Cai et al., 2007): (1) in vitro assays that immunologically detect the presence of holotoxin but cannot distinguish between active or inactive states (ELISA); (2) endopeptidase assays that detect the enzymatic activity of the toxin LC but do not distinguish between biologically active holotoxin and the LC only; (3) in vivo assays (mouse bioassay [MBA]); and lastly, (4) in vivo simulation assays such as the hemidiaphragm assay, local injection assays, and cell-based assays using primary or immortalized cells. In order to detect fully active BoNTs, all steps of the intoxication process must be accounted for (i.e., HC binding to cell surface receptors, endocytosis, vesicle channel formation, transduction of the LC into the cell cytosol, and cleavage of SNARE proteins). Only the MBA and the in vivo simulation assays require all these steps to take place.
Although the MBA is quantitative and can monitor all the steps of intoxication, it has a large error rate, is not standardized between laboratories, requires a large number of animals (∼50 per assay), and the corresponding facilities and trained staff. The hemidiaphragm and local injection assays reduce the suffering of animals and are sufficiently sensitive but still require large numbers of animals and skilled staff. The clearly identified shortcomings of these assays have incited a push from regulatory agencies, including the Food and Drug Administration (FDA) and the United States Department of Agriculture, to develop a cell-based model that would provide a specific, sensitive, and quantitative alternative to the MBA (National Institutes of Health, 2008). While continuous cell lines lack the sensitivity to compete with the MBA, primary neurons and neurons derived from mouse embryonic stem cells are significantly more sensitive (Hall et al., 2004; Keller et al., 1999, 2004; Kiris et al., 2011; Lalli et al., 1999; McNutt et al., 2011; Neale et al., 1999; Pellett et al., 2011; Stahl et al., 2007). To our best knowledge, the most sensitive cell type for BoNT detection is the primary rat spinal cord (RSC) cells assay (Pellett et al., 2007), which is more sensitive than and correlates well with the MBA (Pellett et al., 2010). However, the RSC assay still requires the use of some animals and skilled staff for cell preparation and is not easily adaptable to testing standardization.
Here we describe for the first time the use of human induced pluripotent stem cells (hiPSCs)-derived neurons as a novel, highly sensitive, and reproducible platform for BoNT detection. The neurons used in this study were differentiated and cryopreserved by Cellular Dynamics International (Madison, WI) and consist of a purified pan-neuronal population comprised predominantly of gamma aminoisobutyric acid (GABA)ergic and glutamatergic neurons. The data show that these cells express all the necessary receptors and substrates for BoNT intoxication by all BoNT serotypes. In addition, BoNT detection assays demonstrate that the hiPSCs-derived neurons are highly sensitive for quantitative detection of BoNT/A, B, C, and E.
MATERIALS AND METHODS
Neuronal cells.
The cryopreserved hiPSC-derived neurons (iCell Neurons) were supplied by Cellular Dynamics International. The neurons were an ∼98% pure (Tuj1+/Nestin−) pan-neuronal population of GABAergic, glutamatergic, and a very small percentage (< 1%) of dopaminergic neurons. Neurons were thawed according to the provided instructions, and live cells were counted by the Trypan Blue exclusion method. Cells were seeded at a density of 40,000 cells per well into 96-well (Techno Plastic Products [TPP]) plates coated with 0.01% poly-L-ornithine (PLO; Sigma) and 8.3 μg/cm2 Matrigel (BD Biosciences) unless otherwise indicated and incubated in neuronal medium composed of Neurobasal medium supplemented with 2% B-27 Supplement and 2mM Glutamax (all from Invitrogen) at 37°C, 5% CO2 for the indicated maturation times. At 24 h after seeding, the medium was changed completely, and half of the medium was replaced every 2–3 days thereafter.
Primary RSC cells were prepared as previously described (Pellett et al., 2007, 2010) and seeded into Matrigel-coated 96-well plates at a density of 75,000 cells per well.
Botulinum neurotoxin.
Pure BoNT/A, B, C, and E (150 kDa) and BoNT/A complex were prepared from C. botulinum strains Hall A hyper, Okra B, Brazil C, and Beluga E as previously described (Malizio et al., 2000; Prabakaran et al., 2001), with the modification for BoNT/C that 0.2 g/l of Torula yeast RNA (Sigma) was added before acid precipitation. The toxins were dissolved in phosphate buffered saline, pH 7.4, and 40% glycerol and stored at −20°C until use. Activity of the BoNT/A, B, C, and E and BoNT/A complex preparations was determined by the MBA (Hatheway, 1988; Schantz and Kautter, 1978), and specific toxicity was 7 × 107 mouse LD50 U/mg (BoNT/A1), 7.7 × 107 LD50 U/mg (BoNT/A1 complex), 1 × 108 LD50 U/mg (BoNT/B), 1.1 × 107 LD50 U/mg (BoNT/C), and 7.6 × 107 LD50 U/mg (BoNT/E).
Neuronal toxicity assays.
For all neuronal toxicity assays, hiPSC-derived neurons were exposed to the indicated concentrations of BoNTs in 50 μl of neuronal medium unless otherwise indicated. Primary RSC cells were used as control cells in some assays as indicated in the results. All samples were tested in a minimum of triplicate and a negative control without toxin was always included. After the specified exposure time, the toxin solution was removed, and cells were lysed in 50 μl of 1× lithium dodecyl sulfate (LDS) sample buffer (Invitrogen). The cell lysates were analyzed by Western blot for SNAP-25 or VAMP2 cleavage as previously described (Pellett et al., 2007, 2010). Cleaved and uncleaved bands were quantified by densitometry using a Foto/Analyst FX system and TotalLab Quant software (Fotodyne). Data plots and EC50 values were generated using GraphPad PRISM 5 software.
Plating substrate selection.
To select the optimal surface substrate, hiPSC-derived neurons were seeded onto different matrices. The matrices consisted of poly-D-lysine (PDL)–coated plates (BD Biosciences) coated with 1.0 μg/cm2 of laminin (PDL(BD)-laminin) or 8.3 μg/cm2 Matrigel (PDL(BD)-Matrigel), plates coated with 0.01% PLO (Sigma) followed by coating with 1.0 μg/cm2 laminin (PLO(CDI)-laminin) or 8.3 μg/cm2 Matrigel (PLO(CDI)-Matrigel), PLO-laminin–coated plates purchased from BD Biosciences (PLO-laminin(BD)), PDL-coated plates from BD Biosciences (PDL(BD)), or 0.01% PLO-coated plates (PLO(CDI)). In order to reduce cell aggregation, TPP plates were used, which have a more flat surface area and provided less cell aggregation around the well perimeter. Neurons were allowed to remain in culture for 14 days, and sensitivity to BoNT/A was determined by exposing neurons to serial dilutions of the toxin for 48 h. Some of the neurons were maintained for 6 weeks and tested again as above.
Receptor expression analysis.
For the receptor expression analysis, hiPSC-derived neurons were plated onto 24-well plates at a density of 210,000 cells/well in a volume of 0.75 ml. The cells from three wells, respectively, were harvested at 4, 7, 10, 14, and 21 days after plating in 75 μl 1× LDS sample buffer (Invitrogen). Cell lysates were analyzed by Western blot for the expression of SV2A, B, and C isoforms, synaptotagmin I and II, SNAP-25, VAMP using an antibody that recognizes VAMP2 or an antibody that recognizes VAMP1, 2, and 3 isoforms, and syntaxin. Beta-actin was used as a loading control, and primary RSC cells were used as a positive control. The SV2C antibody (Janz and Sudhof, 1999) was generously provided by Roger Janz. All other antibodies were from Synaptic Systems (Göttingen, Germany). All antibodies recognize human proteins.
For mRNA analysis by real-time quantitative PCR (qPCR), three independent cultures of hiPSC-derived neurons were maintained for the indicated times, respectively. Cells were lysed directly on the plate, and RNA was purified using the RNeasy Mini Kit and the RNase-Free DNase Set (Qiagen). For a positive control, human adult total brain RNA was purchased (Agilent Technologies). For all samples, cDNA was generated using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Real-time qPCR amplification was performed on the LightCycler 480 II (Roche) using the TaqMan Gene Expression Master Mix and the following TaqMan Human Gene Expression Assays: SNAP25 (Hs00938962_m1); STX1A (Hs00270282_m1); STX1B (Hs01041315_m1); VAMP1 (Hs00249911_m1); VAMP2 (Hs00360269_m1); VAMP3 (Hs00922166_m1); SV2A (Hs00372069_m1); SV2B (Hs00208178_m1); SV2C (Hs00392676_m1); SYT1 (Hs00194572_m1); SYT2 (Hs00980604_m1); and the Human GAPDH Endogenous Control Primer Set (all from Applied Biosystems). Abs Quant/2nd Derivative analysis was performed on all samples and Cp values were converted to relative fold change via normalization to the Cp for endogenous GAPDH expression. Averages and SD were calculated for each gene within the three biological replicates. Technical quadruplicate PCR reactions were performed for each gene.
Activity-dependent BoNT/A1 uptake assays.
BoNT/A1 was diluted to a concentration of 55 or 275 U per 50 μl of cell stimulation (Invitrogen custom Neurobasal medium containing 2.2mM CaCl and 56mM KCl, supplemented with B27 and Glutamax) or neuronal medium and added to hiPSC-derived neurons cultured for 4 days and RSC cells. The cells were incubated with toxin for 1, 5, 10, and 15 min, respectively. For the negative control, the respective medium without toxin was added to the cells. The toxin was removed, and cells were immediately washed twice with 200 μl of neuronal medium followed by incubation in 200 μl of fresh neuronal medium for 24 h. Samples were collected in replicates of four.
To determine the minimum required toxin concentration for activity-dependent uptake of BoNT/A1 within 5 min, the toxin was diluted to a concentration between 1.72 and 55 U per 50 μl of cell stimulation medium. Four-day–cultured hiPSC-derived neurons were exposed to the toxin dilutions for 5 min, followed by toxin removal and two washes with neuronal medium and incubation in neuronal medium for 24 h. All dilutions were tested in replicates of four.
Antibody protection analysis.
BoNT/A1-specific antibodies were prepared according to (Goodnough et al., 1993). The antibody had a concentration of 0.795 mg/ml. Inhibition of BoNT/A1 activity in hiPSC-derived neurons by neutralizing antibodies was analyzed using two different methods. For the first assay, 55 U of BoNT/A1 was combined with serially diluted antibody in cell stimulation medium and incubated for 1 h at 37°C to allow for antibody-toxin interaction. Four-day–cultured hiPSC-derived neurons were exposed to the toxin-antibody mixture for 5 min, followed by removal of the toxin-antibody mixture and two washing steps with neuronal medium and incubation in neuronal medium for 24 h. For the second assay, 1.5 U of BoNT/A were combined with serial dilutions of antibody in neuronal medium and incubated at 37°C for 1 h. The hiPSC-derived neurons were exposed to the toxin-antibody mixtures for 24 h. For the MBA, serial dilutions of the antibody were preincubated with 5–10 U of BoNT/A1 for 1.5 h at ambient temperatures in a volume of 167 μl. The volume was adjusted to 500 ml and injected into four mice per dilution. The MBA indicated an approximate activity of 25–50 IU/ml.
RESULTS
hiPSC-Derived Neurons Express Receptors Important for BoNT Intoxication
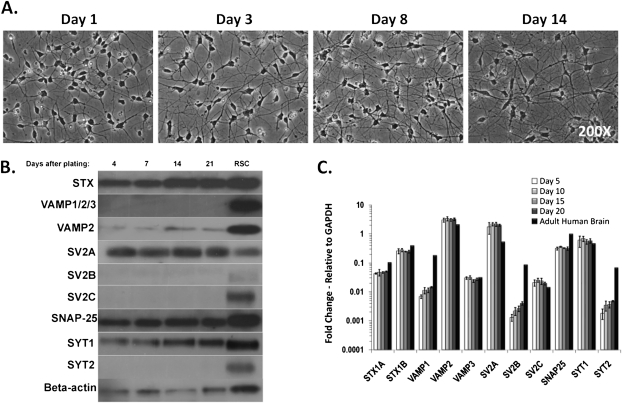
The differentiated and cryopreserved hiPSC-derived neurons (Cellular Dynamics International) formed an increasing network of axons and dendrites over the course of 14 days post plating (Fig. 1A). Neuron-specific staining using anti-beta-III tubulin and MAP2 antibodies confirmed the neuronal phenotype (Supplementary fig. 1). In order to determine whether the hiPSC-derived neurons can be used to detect BoNT activity, expression of the receptors and enzymatic targets necessary for BoNT cell entry and catalytic activity were analyzed by Western blot and qPCR, respectively. The Western blot resulted in signals for SV2A, a faint band for SV2B, synaptotagmin 1, syntaxin, SNAP-25, VAMP2, and beta-actin, which did not change over a time period of 21 days after cell plating (Fig. 1B and Supplementary fig. 2). Analysis of mRNA levels of the same proteins by qPCR indicated expression of all proteins analyzed (Fig. 1C) including SV2B and C isoforms, synaptotagmin 2, and VAMP1 and 3, which were not detected by Western blot. However, the mRNA levels of those isoforms were at least 200-fold lower than those of the isoforms detected by Western blot (SV2A, synaptotagmin 1, and VAMP2). Thus, the qPCR data corroborate the Western blot data and suggest that the hiPSC-derived neurons express primarily SV2A, synaptotagmin 1, and VAMP2 isoforms of these proteins, which is consistent with the neurons possessing a forebrain-like phenotype (Janz and Sudhof, 1999). Expression levels of all proteins did not change throughout the study period, indicating that the neurons could be constructing synaptic networks at their baseline rate by 4 days after plating and remain stable for at least 21 days.
FIG. 1.
BoNT receptor expression in hiPSC-derived neurons. (A) hiPSC-derived neurons were plated on PLO-laminin(BD)–coated six-well plates at a density of 125,000 cells/cm2. Images were acquired at 1, 3, 8, and 14 days after plating at ×200 magnification via an Olympus IX81 microscope. (B) hiPSC-derived neurons were cultured for 4, 7, 14, and 21 days and assayed via Western blot for expression levels of BoNT receptors. A representative Western blot of four replicates is shown. (C) hiPSC-derived neurons were cultured for 5, 10, 15, and 20 days and then used to perform qPCR assays. Adult human brain RNA was used as a comparison sample (STX [syntaxin], SV2 [synaptic vesicle protein], and SYT [synaptotagmin]).
hiPSC-Derived Neurons Sensitively Detect BoNT/A1 and Assay Results Are Not Significantly Influenced by Various Plating Conditions
To determine the optimal cell culture conditions for a BoNT/A1 assay, cells were plated on different matrices and tested for SNAP-25 cleavage. Plates were treated with either PLO or PDL and combinations of PLO or PDL with laminin or matrigel. The neurons were exposed to serial dilutions of BoNT/A after 14 days in culture, and Western blot analysis of cell lysates indicated that SNAP-25 cleavage was nearly identical for all substrates tested (Fig. 2). The limit of detection was 0.05 mouse LD50 units, and cleavage was complete between 1.75 and 3.5 U. The EC50 values ranged from 0.21 to 0.31 U. These data indicate that hiPSC-derived neurons can be plated on any of the tested surface treatments described here (Fig. 2). All subsequent experiments were performed on PLO-Matrigel–coated plates.
FIG. 2.
Comparison of BoNT/A1 sensitivity of hiPSC-derived neurons plated on different substrates. At 2 weeks after plating, the cells were exposed to the indicated concentrations of BoNT/A1 and cell lysates were analyzed for SNAP-25 cleavage by Western blot. Data from three Western blots were quantified by densitometry, and data plots were generated. There were no statistically significant differences in sensitivity of neurons grown on the different substrates. The substrates are shown in the figure legend on the right.
A Cell Culture Time of 4–14 Days Provides an Excellent and Sensitive BoNT/A1 Testing Window
In order to determine whether post thaw culture time affects the BoNT sensitivity of hiPSC-derived neurons, cells were analyzed for SNAP-25 cleavage in parallel at 4 and 7 days post thaw. Primary RSC cells, which are currently the most sensitive in vitro culture system for BoNT detection (Pellett et al., 2007, 2010), were also tested in parallel. The resulting data showed no statistically significant differences in sensitivity of neurons cultured for 4 or 7 days, with EC50 values of ∼0.3 U (Fig. 3A). This is similar to the values observed for day 14 cells (Fig. 2) and for RSC cells. The dose-response curves for the hiPSC-derived neurons were significantly steeper than for RSC cells. In addition, hiPSC-derived neurons reached 100% cleavage with 1.75 U toxin, while 100% cleavage was not achieved in RSC cells at the concentrations tested. This difference in toxin response and cleavage efficiency is likely due to the high purity of the hiPSC-derived neurons compared to RSC cells. It is also noteworthy that the testing of four different lots of hiPSC-derived neurons displayed no major differences in BoNT/A1 sensitivity (data not shown). Thus, hiPSC-derived neurons cultured for 4–14 days provide a reproducible and highly sensitive cell-based model for BoNT/A1 detection and quantification.
FIG. 3.
A) BoNT/A1 sensitivity of hiPSC-derived neurons and RSC cells. hiPSC-derived neurons were plated 4 and 7 days prior to toxin exposure, and RSC cells were cultured for 2 weeks prior to toxin exposure. All cells were exposed to the indicated toxin dilutions for 48 h, and cell lysates were analyzed for SNAP-25 cleavage by Western blot. (B) Time dependence of detecting BoNT/A1 activity in hiPSC-derived neurons. The hiPSC-derived neurons were exposed to serial BoNT/A1 dilutions for 6, 16, 24, and 48 h. The assay time is shown in the figure legend on the right. Data from three Western blots were quantified by densitometry, and data plots and EC50 values were generated. The maturation time and EC50 values are shown in the figure legend below.
Sensitivity of hiPSC-Derived Neurons Increases With Longer Exposure Times
The time dependence of BoNT detection in hiPSC-derived neurons was examined by exposing cells to serial BoNT/A1 dilutions and harvesting samples at 6, 16, 24, and 48 h after toxin addition. The resulting data indicated that a 48-h exposure yielded highest sensitivity, with an approximately threefold increase compared to a 24-h assay and an approximately sixfold increase compared to a 16-h assay (Fig. 3B). A 6-h toxin exposure resulted in an ∼130-fold decrease in sensitivity, with an EC50 of about 40 U.
hiPSC-Derived Neurons Take Up BoNT/A1 Significantly Faster Than RSC Cells in an Activity-Dependent Assay
In order to examine whether hiPSC-derived neurons take up BoNT in an activity-dependent fashion, 4-day cultures of hiPSC-derived neurons and RSC cells were exposed to 55 and 275 U of BoNT/A1 in cell stimulation medium, respectively. The cells were exposed to toxin for 1, 5, 10, or 15 min, followed by complete toxin removal and incubation in neuronal medium for 24 h to allow for SNAP-25 cleavage. Significant SNAP-25 cleavage was observed in hiPSC-derived neurons as early as 1 min after exposure with 55 U of BoNT/A1 and was complete with about 75% SNAP-25 cleavage after 5 min (Fig. 4A). Exposure to 275 U resulted in complete SNAP-25 cleavage at all exposure times tested (Fig. 4A). In contrast, exposure of RSC cells to 55 U of BoNT/A1 did not result in significant SNAP-25 cleavage after exposure times of up to 15 min, and only about 30–40% of SNAP-25 was cleaved after an at least 10 min exposure to 275 U (Fig. 4A). This indicates that hiPSC-derived neurons take up BoNT/A1 in an activity-dependent fashion and that this uptake occurs markedly more efficiently and faster than in RSC cells. Activity dependence was confirmed by exposing hiPSC-derived neurons to 55 U of BoNT/A for 1, 5, or 10 min in cell stimulation medium or neuronal medium. There was significantly more SNAP-25 cleavage in the cells treated with cell stimulation medium, with 50% cleavage of SNAP-25 observed after 1 min and 70% cleavage after 5 min (Fig. 4B). In contrast, only about 20% of SNAP-25 was cleaved after 10 min in neuronal medium (Fig. 4B). Exposure of the hiPSC-derived neurons to 1.7–55 U of BoNT/A1 in cell stimulation medium for 5 min resulted in a concentration-dependent increase in SNAP-25 cleavage, with 50% SNAP-25 cleavage occurring with about 30 U (Fig. 4C).
FIG. 4.
Activity-dependent BoNT/A1 uptake by neurons. (A) hiPSC-derived neurons and RSC cells were exposed to 55 or 275 U of BoNT/A1 in cell stimulation medium for 1, 5, 10, and 15 min, followed by toxin removal and 24 h incubation. Cell lysates were analyzed by Western blot for SNAP-25 cleavage. A representative Western blot of four replicates is shown. (B) hiPSC-derived neurons were exposed to 55 U of BoNT/A1 in both neuronal and cell stimulation medium for 1, 5, and 10 min. Cell lysates were analyzed by Western blot for SNAP-25 cleavage. A representative Western blot of four replicates is shown. (C) Neurons were exposed to 1.7–55 U of BoNT/A1 for 5 min in cell stimulation medium, washed twice with neuronal medium, and incubated for 24 h. Data from four Western blots were quantified by densitometry, and data plots were generated. (D) BoNT/A1 uptake rate of hiPSC-derived neurons compared to RSC cells. The cells were exposed to 82 U of BoNT/A1 for up to 10 h in parallel in either neuronal medium (NM) or cell stimulation medium (CSM). Cell lysates were analyzed for SNAP-25 cleavage by Western blot, and data from three Western blots were quantified by densitometry and data plots were generated. The cell and medium types are shown in the figure legend on the right.
hiPSC-Derived Neurons Have a Faster BoNT/A1 Uptake Rate Than RSC Cells
The BoNT/A1 uptake rate into hiPSC-derived neurons compared to RSC cells was examined by exposing neurons and RSC cells to 82 U of BoNT/A1 in parallel and assessing SNAP-25 cleavage at 2, 4, 6, 8, and 10 h. The hiPSC-derived neurons resulted in significantly earlier and more complete SNAP-25 cleavage than the RSC cells. In hiPSC-derived neurons, 100% of SNAP-25 cleavage was achieved at 8 h and 50% SNAP-25 cleavage at ∼4 h (Fig. 4D). The RSC cells, in contrast, reached only ∼70–80% SNAP-25 cleavage after 10 h, and ∼50% cleavage of SNAP-25 was observed at 6 h. Chemical stimulation of the cells by addition of 56mM KCl and 2.2mM CaCl2 during the assay had no effect on BoNT/A sensitivity, indicating that in the time frame tested, neuronal activity does not affect BoNT uptake into the cells. These data indicate that the hiPSC-derived neurons are significantly more sensitive to BoNT/A than RSC cells and take up the toxin at a faster rate, assuming an equal cleavage rate of human and rat SNAP-25.
BoNT/A1-Specific Antibodies Protect hiPSC-Derived Neurons from SNAP-25 Cleavage by BoNT/A1
Specificity of BoNT/A1 detection in hiPSC-derived neurons was confirmed by an antibody protection assay using two different assay formats. In the first assay, the neurons were exposed to toxin antibody mixtures for 24 h, using the minimal amount of toxin required to achieve nearly complete SNAP-25 cleavage (1.5 U). In the second assay, neurons were exposed to mixtures of 55 U BoNT/A and serially diluted antibody for 5 min in cell stimulation medium, followed by toxin removal and incubation for 24 h. The first assay yielded significantly higher sensitivity in antibody detection. Neurons were fully protected from cleavage of SNAP-25 with as little as 0.0025 μl of antibody. Significant partial protection was observed down to 0.000625 μl of antibody (Fig. 5A). Testing the same antibody dilutions by MBA indicated an ∼10 times greater sensitivity of the cell-based assay than the MBA, as previous data have also indicated for the RSC assay (Pellett et al., 2007). The second (activity dependent) assay was about 10 times less sensitive, requiring 0.016 μl of antibody per 50 μl for full protection, and partial protection was observed with 0.004 μl (Fig. 5B). These results indicate that the hiPSC-derived neurons provide an excellent and highly sensitive assay for neutralizing antibody detection and that a longer exposure with less toxin is more sensitive than an activity-dependent assay, which requires more toxin but a shorter exposure time.
FIG. 5.
Western blot and densitometry data of antibody protection assay in hiPSC-derived neurons. Serial dilutions of antibody and toxin were preincubated for 1 h prior to exposure to neurons, and cell lysates were analyzed for SNAP-25 cleavage by Western blot. The data from three Western blots were quantified by densitometry and average, and SDs are shown in the graph. The amounts of antibody used are indicated. (A) hiPSC-derived neurons were exposed to 1.5 U of toxin and antibody for 24 h. (B) hiPSC-derived neurons were exposed to 55 U of toxin-antibody mixture in cell stimulation medium for 5 min, the mixture was removed, cells were washed twice, and incubated for 24 h.
hiPSC-Derived Neurons Are a Highly Sensitive Cell Model for Detection of BoNT Serotypes B, C, and E
The BoNT receptor analyses indicated that hiPSC-derived neurons express the SNARE proteins and receptors required for cell entry of all BoNT serotypes (Figs. 1B and 1C). To test the sensitivities of neurons to different serotypes, serial dilutions of BoNT/B, C, or E were added to hiPSC-derived neurons or RSC cells for 48 h in parallel, and cell lysates were assayed via Western blot for the cleavage of their respective neuronal substrate. The hiPSC-derived neurons consistently detected all BoNT serotypes with equal or greater sensitivity than RSC cells (Fig. 6). The EC50 values for hiPSC-derived neurons and RSC cells were 15.71 and 29.22 U for BoNT/B (Fig. 6A), 0.4 and 0.36 U for BoNT/C (Fig. 6B), and 1.79 U for BoNT/E in hiPSC-derived neurons (Fig. 6C). Due to the curve shape, an EC50 value for BoNT/E in RSC cells could not be accurately defined with the analysis software but is estimated to be similar to that of hiPSC-derived neurons (Fig. 6C). Interestingly, hiPSC-derived neurons were able to detect BoNT-A complex approximately four times more sensitively than purified BoNT/A (Supplementary fig. 3).
FIG. 6.
Detection of BoNT/B, C, and E activity in hiPSC-derived neurons. Seven-day cultures of hiPSC-derived neurons and RSC cells were exposed to serial dilutions of BoNT/B (A), C (B), and E (C) for 48 h in parallel. Cell lysates were analyzed for the respective SNARE target protein cleavage by Western blot, and data from three Western blots were quantified by densitometry and data plots were generated.
DISCUSSION
In November 2007, two independent groups showed for the first time that human fibroblasts can be reprogrammed into a state of pluripotency by introducing a small set of exogenous factors (Takahashi et al., 2007; Yu et al., 2007). The resulting iPSCs could be continually propagated and cryopreserved as pluripotent cells and opened the opportunity for development of a vast number of applications for hiPSC-derived cell models that resemble fully functioning, differentiated human somatic cells. One such potential application is the potency testing and toxicity screening of pharmaceuticals and neutralizing antibodies against biopharmaceuticals. Although no iPSC-derived differentiated cell models are currently approved for toxigenicity testing, cardiomyocytes, hepatocytes, and neurons have been proposed as desirable cell models for such purposes (reviewed in Anson et al., 2011). With these cell models, the challenge is to reliably and reproducibly produce pure populations of the differentiated cell types in sufficient quantities. Industrial-scale, reproducible differentiation of hiPSCs to nearly pure populations of neurons has now been achieved by Cellular Dynamics International, and the neurons (certified to be over 90% pure) are projected to be commercially available in early 2012.
The data presented here show that hiPSC-derived neurons are exquisitely sensitive to BoNTs (Fig. 3) and express all receptors and substrates necessary for BoNT intoxication (Fig. 1). Compared to an assay using primary RSC cells (Pellett et al., 2007, 2010), the hiPSC-derived neurons show either equal or greater sensitivity to BoNT serotypes and consistently reach 100% substrate cleavage allowing for easy EC50 calculation (Figs. 2 and 6). Even though there were some morphological differences between the neurons grown on some of the different matrices, the matrix did not significantly alter the sensitivity to BoNT/A1, indicating that the surface matrix that is used does not affect the neuronal cell characteristics important for BoNT intoxication (Fig. 2). This allows flexibility for setting up different BoNT assays using these neurons.
Analysis of the optimal post thaw culture time of the hiPSC-derived neurons indicates that culture of 4–14 days yields neurons with excellent and consistent sensitivity to BoNT/A1 (Fig. 3). In contrast, primary RSC cells require at least 2 weeks of maturation (Keller et al., 1999; Neale et al., 1999; Pellett et al., 2007). This allows for an easy and relatively fast set-up of the hiPSC-derived neurons when needed, saving time and expense associated with longer maintenance and culture of cells. The highest sensitivity of hiPSC-derived neurons to BoNT/A1 was reached after a 48-h toxin exposure period with an EC50 of about 0.25–0.3 U (Fig. 3). However, in contrast to primary RSC cells, which require at least 10 times more toxin in a 24-h assay than in a 48-h assay, sensitivity of the hiPSC-derived neurons to BoNT/A1 in a 24-h assay is still equal to the sensitivity of the MBA (EC50 about 0.9 U). Even after an exposure of only 6 h, as little as 17.5 U of BoNT/A1 could be detected, in contrast to 150 U required for detection after a 6-h exposure of RSC cells (Pier et al., 2011).
Activity-dependent assays indicate that hiPSC-derived neurons are able to take up BoNT/A1 within 5 min in a concentration-dependent fashion if chemically stimulated (Figs. 4A–C). This activity-dependent uptake required significantly less toxin than any other activity-dependent cell assay reported thus far (50 U to achieve about 75% SNAP-25 cleavage vs. about 500 U required in primary mouse or RSC cells to achieve 50–60% of SNAP-25 cleavage) (Keller et al., 2004; Pier et al., 2011). The significantly more efficient and faster activity-dependent uptake of BoNT/A1 by hiPSC-derived neurons is likely due to the greater purity of these neurons compared to primary cell preparations, which always contain a mixture of neurons and glial cells. Species specificity may also play a role. Thus, hiPSC-derived neurons provide an excellent cellular model for mechanistic studies of BoNT uptake into neurons as well as for screening BoNT inhibitors.
BoNT/A specificity was confirmed by an antibody protection assay (Fig. 5). Using a 24-h assay format and 1.5 U of BoNT/A, the hiPSC-derived neurons were able to detect antibodies with ∼10 times greater sensitivity than the MBA in a direct comparison. An activity-dependent assay that exposed the hiPSC-derived neurons to the toxin/antibody mixture in cell stimulation medium for only 5 min was about 10 times less sensitive than the 24-h assay. Nevertheless, our data indicate that both styles of antibody protection assays will allow quantitative and sensitive detection of neutralizing antibodies in various samples such as serum samples, antitoxins, etc. Although BoNTs are used effectively in treating a large number of patients for various conditions (reviewed in Dhaked et al., 2010), some will develop neutralizing antibodies, which will prevent the success of further treatments. For example, ∼5% of BoNT/A-treated cervical dystonia patients have been estimated to develop neutralizing antibodies (Kessler et al., 1999). Currently, patients receiving repeated BoNT/A injections are not monitored for development of neutralizing antibodies because a sensitive and quantitative assay is not commercially available (Sesardic et al., 2004). The assay presented here using hiPSC-derived neurons provides the needed testing platform.
BoNTs are expressed in clostridia as a complex with several other proteins (nontoxic nonhemagglutinin protein [NTNH] and hemagglutinins [HA]) (Supplementary fig. 3A) in the case of BoNT/A (reviewed in Johnson and Bradshaw, 2001), which are believed to protect the toxin from the degradative pH of the gastrointestinal tract (Oguma et al., 2000; Sakaguchi et al., 1988). The most commonly used medical preparations of BoNT/A (BOTOX and Dysport) consist of toxin complex, although newer formulations contain only the purified BoNT (Xeomin). The more sensitive detection of BoNT/A complex versus purified BoNT/A could be due to a protective effect of the complex components in the neuronal medium or to increased binding of BoNT/A1 complex to the neuronal cell surface compared to purified BoNT/A1 (Supplementary fig. 3B). Much controversy exists as to which formulation is most beneficial for human use and the precise involvement of the nontoxic protein components (Sesardic et al., 2004), and assays detecting both formulations are crucial for potency determination and research. In addition, hiPSC-derived neurons detected other BoNT serotypes (BoNT/B, C, and E) with equal or greater sensitivity than RSC cells (Fig. 6). Importantly, in all cases, the dose-response curve was significantly steeper and reached 100% cleavage for all serotypes tested. This allows for an easier and more accurate EC50 or IC50 determination. The ability of these neurons to sensitively detect the different BoNT serotypes and complex make them a potentially invaluable tool for BoNT distinction, which is needed for studies such as determination of purity of pharmaceutical preparation of BoNT, basic research studies, and potentially for detection of BoNTs in foods.
In summary, the data presented here demonstrates that hiPSC-derived neurons are a highly sensitive, selective, and species-specific cell model for the detection of BoNTs. Up until the summer of 2011, the in vivo MBA was the only assay approved by the FDA for detection and quantification of BoNT activity of pharmaceutical BoNT preparations. In July 2011, Allergan, who currently produces the vast majority of the pharmaceutically used BoNT as BOTOX (Onabotulinumtoxin A), announced the first approval by the FDA of their in vitro cell-based assay for the detection of BOTOX. No details of this assay have been released. The assay presented here has the potential to largely replace the MBA for BoNT potency determination as well as for antibody detection, screening of inhibitors, and research applications. Assay standardization, optimization, and validation using an appropriate toxin standard will enable the commercial use of this assay platform. Comparative testing of four different lots of hiPSC-derived neurons indicated good lot-to-lot consistency (data not shown), suggesting that standardization will be routine and straightforward. The fact that these neurons are human cells makes for a more species-relevant model system for testing of pharmaceutical BoNT preparations. Also, the neurons are derived from hiPSCs as opposed to cancer or transformed cell lines, which may not be reflective of somatic human neurons. Since hiPSC-derived neurons can now be differentiated and cryopreserved at an industrial scale (Cellular Dynamics International) and only require simple plating and maintenance steps, no expertise in stem cell differentiation techniques is required for this assay. Additionally, this testing platform is amenable to different endpoint measurements including ELISA (Nuss et al., 2010) or intracellularly expressed reporters such as FRET sensors (Dong et al., 2004).
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
University of Wisconsin, Madison; the National Institute of Allergy and Infectious Diseases (R21AI08282); The National Institutes of Allergy and Infectious Diseases (1RO1AI093504-01); subaward (10-01850).
Supplementary Material
Acknowledgments
Antibody specific for SV2C was kindly provided by Roger Janz (University of Texas, Southwestern Medical Center, Dallas, TX). Author Contributions. S.P., E.A.J., W.H.T., and R.C.M.W. conceived of the project and designed and executed the experiments and prepared the manuscript. M.J.S., L.G.C., and C.S. provided the neurons, performed the qPCR experiments, and contributed to data presentation.
References
- Anson BD, Kolaja KL, Kamp TJ. Opportunities for use of human iPS cells in predictive toxicology. Clin. Pharmacol. Ther. 2011;89:754–758. doi: 10.1038/clpt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, et al. Botulinum toxin as a biological weapon: Medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Bossi P, Garin D, Guihot A, Gay F, Crance JM, Debord T, Autran B, Bricaire F. Bioterrorism: Management of major biological agents. Cell Mol. Life Sci. 2006;63:2196–2212. doi: 10.1007/s00018-006-6308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Singh BR, Sharma S. Botulism diagnostics: From clinical symptoms to in vitro assays. Crit. Rev. Microbiol. 2007;33:109–125. doi: 10.1080/10408410701364562. [DOI] [PubMed] [Google Scholar]
- Chaddock JA, Acharya KR. Engineering toxins for 21st century therapies. FEBS J. 2011;278:899–904. doi: 10.1111/j.1742-4658.2011.08013.x. [DOI] [PubMed] [Google Scholar]
- Dhaked RK, Singh MK, Singh P, Gupta P. Botulinum toxin: Bioweapon & magic drug. Indian J. Med. Res. 2010;132:489–503. [PMC free article] [PubMed] [Google Scholar]
- Dong M, Tepp WH, Johnson EA, Chapman ER. Using fluorescent sensors to detect botulinum neurotoxin activity in vitro and in living cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14701–14706. doi: 10.1073/pnas.0404107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Montal M. Crucial role of the disulfide bridge between botulinum neurotoxin light and heavy chains in protease translocation across membranes. J. Biol. Chem. 2007;282:29604–29611. doi: 10.1074/jbc.M703619200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Nakai Y, Eubanks LM, Clancy CM, Tepp WH, Pellett S, Dickerson TJ, Johnson EA, Janda KD, Montal M. Bimodal modulation of the botulinum neurotoxin protein-conducting channel. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1330–1335. doi: 10.1073/pnas.0812839106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez DF, Gimenez JA. The typing of botulinal neurotoxins. Int. J. Food Microbiol. 1995;27:1–9. doi: 10.1016/0168-1605(94)00144-u. [DOI] [PubMed] [Google Scholar]
- Goodnough MC, Hammer B, Sugiyama H, Johnson EA. Colony immunoblot assay of botulinal toxin. Appl. Environ. Microbiol. 1993;59:2339–2342. doi: 10.1128/aem.59.7.2339-2342.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall YH, Chaddock JA, Moulsdale HJ, Kirby ER, Alexander FC, Marks JD, Foster KA. Novel application of an in vitro technique to the detection and quantification of botulinum neurotoxin antibodies. J. Immunol. Methods. 2004;288:55–60. doi: 10.1016/j.jim.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Hatheway CL. Botulism. In: Balows A, Hausler WH, Ohashi M, Turano MA, editors. Laboratory Diagnosis of Infectious Diseases: Principles and Practice. New York, NY: Springer-Verlag; 1988. pp. 111–133. [Google Scholar]
- Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: Anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94:1279–1290. doi: 10.1016/s0306-4522(99)00370-x. [DOI] [PubMed] [Google Scholar]
- Johnson EA. Clostridium botulinum and Clostridium tetani. In: Borriello SP, Murray PR, Funke G, editors. Topley and Wilson's Microbiology and Microbial Infections. 8th ed. London: Hodder Arnold; 2005. pp. 1035–1088. [Google Scholar]
- Johnson EA, Bradshaw M. Clostridium botulinum and its neurotoxins: A metabolic and cellular perspective. Toxicon. 2001;39:1703–1722. doi: 10.1016/s0041-0101(01)00157-x. [DOI] [PubMed] [Google Scholar]
- Johnson EA, Montecucco C. Chapter 11 Botulism. In: Andrew G. Engel, editor. Handbook of Clinical Neurology. Elsevier, Amsterdam, The Netherlands; 2008. pp. 333–368. [DOI] [PubMed] [Google Scholar]
- Keller JE, Cai F, Neale EA. Uptake of botulinum neurotoxin into cultured neurons. Biochemistry. 2004;43:526–532. doi: 10.1021/bi0356698. [DOI] [PubMed] [Google Scholar]
- Keller JE, Neale EA, Oyler G, Adler M. Persistence of botulinum neurotoxin action in cultured spinal cord cells. FEBS Lett. 1999;456:137–142. doi: 10.1016/s0014-5793(99)00948-5. [DOI] [PubMed] [Google Scholar]
- Kessler KR, Skutta M, Benecke R. Long-term treatment of cervical dystonia with botulinum toxin A: Efficacy, safety, and antibody frequency. German Dystonia Study Group. J. Neurol. 1999;246:265–274. doi: 10.1007/s004150050345. [DOI] [PubMed] [Google Scholar]
- Kiris E, Nuss JE, Burnett JC, Kota KP, Koh DC, Wanner LM, Torres-Melendez E, Gussio R, Tessarollo L, Bavari S. Embryonic stem cell-derived motoneurons provide a highly sensitive cell culture model for botulinum neurotoxin studies, with implications for high-throughput drug discovery. Stem Cell Res. 2011;6:195–205. doi: 10.1016/j.scr.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G, Herreros J, Osborne SL, Montecucco C, Rossetto O, Schiavo G. Functional characterisation of tetanus and botulinum neurotoxins binding domains. J. Cell. Sci. 1999;112(Pt 16):2715–2724. doi: 10.1242/jcs.112.16.2715. [DOI] [PubMed] [Google Scholar]
- Malizio CJ, Goodnough MC, Johnson EA. Purification of Clostridium botulinum type A neurotoxin. Methods Mol. Biol. 2000;145:27–39. doi: 10.1385/1-59259-052-7:27. [DOI] [PubMed] [Google Scholar]
- McNutt P, Celver J, Hamilton T, Mesngon M. Embryonic stem cell-derived neurons are a novel, highly sensitive tissue culture platform for botulinum research. Biochem. Biophys. Res. Commun. 2011;405:85–90. doi: 10.1016/j.bbrc.2010.12.132. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Rossetto O, Schiavo G. Presynaptic receptor arrays for clostridial neurotoxins. Trends Microbiol. 2004;12:442–446. doi: 10.1016/j.tim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Schiavo G. Mechanism of action of tetanus and botulinum neurotoxins. Mol. Microbiol. 1994;13:1–8. doi: 10.1111/j.1365-2958.1994.tb00396.x. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. Report on the ICCVAM-NICEATM/ECVAM Scientific Workshop on Alternative Methods to Refine, Reduce or Replace the Mouse LD50 Assay for Botulinum Toxin Testing. 2008. NIH publication number 08-6416. Bethesda, MD. [Google Scholar]
- Neale EA, Bowers LM, Jia M, Bateman KE, Williamson LC. Botulinum neurotoxin A blocks synaptic vesicle exocytosis but not endocytosis at the nerve terminal. J. Cell Biol. 1999;147:1249–1260. doi: 10.1083/jcb.147.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss JE, Ruthel G, Tressler LE, Wanner LM, Torres-Melendez E, Hale ML, Bavari S. Development of cell-based assays to measure botulinum neurotoxin serotype A activity using cleavage-sensitive antibodies. J. Biomol. Screen. 2010;15:42–51. doi: 10.1177/1087057109354779. [DOI] [PubMed] [Google Scholar]
- Oguma K, Fujinaga Y, Inoue K, Yokota K, Watanabe T, Ohyama TK, Inoue K. Microbial Foodborne Diseases: Mechanisms of Pathogenesis and Toxin Synthesis (J. W. Cary, J. E. Linz, and D. Bhatnagar, Eds.) Technomic Pub. Co., Lancaster, PA; 2000. Mechanisms of pathogenesis and toxin synthesis in Clostridium botulinum; pp. 273–293. [Google Scholar]
- Pellett S, Du ZW, Pier CL, Tepp WH, Zhang SC, Johnson EA. Sensitive and quantitative detection of botulinum neurotoxin in neurons derived from mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2011;404:388–392. doi: 10.1016/j.bbrc.2010.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett S, Tepp WH, Clancy CM, Borodic GE, Johnson EA. A neuronal cell-based botulinum neurotoxin assay for highly sensitive and specific detection of neutralizing serum antibodies. FEBS Lett. 2007;581:4803–4808. doi: 10.1016/j.febslet.2007.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett S, Tepp WH, Toth SI, Johnson EA. Comparison of the primary rat spinal cord cell (RSC) assay and the mouse bioassay for botulinum neurotoxin type A potency determination. J. Pharmacol. Toxicol. Methods. 2010;61:304–310. doi: 10.1016/j.vascn.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Pier CL, Chen C, Tepp WH, Lin G, Janda KD, Barbieri JT, Pellett S, Johnson EA. Botulinum neurotoxin subtype A2 enters neuronal cells faster than subtype A1. FEBS Lett. 2011;585:199–206. doi: 10.1016/j.febslet.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Tepp W, DasGupta BR. Botulinum neurotoxin types B and E: Purification, limited proteolysis by endoproteinase Glu-C and pepsin, and comparison of their identified cleaved sites relative to the three-dimensional structure of type A neurotoxin. Toxicon. 2001;39:1515–1531. doi: 10.1016/s0041-0101(01)00124-6. [DOI] [PubMed] [Google Scholar]
- Sakaguchi G, Ohishi I, Koazki S. Bacterial Toxins: Handbook of Natural Toxins, (A.T. Tu, Ed.) Vol. 4. Marcel Dekker Inc., New York, NY; 1988. Botulism—Structure and chemistry of botulinum; pp. 191–216. [Google Scholar]
- Schantz EJ, Kautter DA. Standardized assay for Clostridium botulinum toxins. J. Assoc. Off. Anal. Chem. 1978;61:96–99. [Google Scholar]
- Sesardic D, Jones RG, Leung T, Alsop T, Tierney R. Detection of antibodies against botulinum toxins. Mov. Disord. 2004;19(Suppl. 8):S85–S91. doi: 10.1002/mds.20021. [DOI] [PubMed] [Google Scholar]
- Stahl AM, Ruthel G, Torres-Melendez E, Kenny TA, Panchal RG, Bavari S. Primary cultures of embryonic chicken neurons for sensitive cell-based assay of botulinum neurotoxin: Implications for therapeutic discovery. J. Biomol. Screen. 2007;12:370–377. doi: 10.1177/1087057106299163. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.