Abstract
A shift in toxicity testing from in vivo to in vitro may efficiently prioritize compounds, reveal new mechanisms, and enable predictive modeling. Quantitative high-throughput screening (qHTS) is a major source of data for computational toxicology, and our goal in this study was to aid in the development of predictive in vitro models of chemical-induced toxicity, anchored on interindividual genetic variability. Eighty-one human lymphoblast cell lines from 27 Centre d’Etude du Polymorphisme Humain trios were exposed to 240 chemical substances (12 concentrations, 0.26nM–46.0μM) and evaluated for cytotoxicity and apoptosis. qHTS screening in the genetically defined population produced robust and reproducible results, which allowed for cross-compound, cross-assay, and cross-individual comparisons. Some compounds were cytotoxic to all cell types at similar concentrations, whereas others exhibited interindividual differences in cytotoxicity. Specifically, the qHTS in a population-based human in vitro model system has several unique aspects that are of utility for toxicity testing, chemical prioritization, and high-throughput risk assessment. First, standardized and high-quality concentration-response profiling, with reproducibility confirmed by comparison with previous experiments, enables prioritization of chemicals for variability in interindividual range in cytotoxicity. Second, genome-wide association analysis of cytotoxicity phenotypes allows exploration of the potential genetic determinants of interindividual variability in toxicity. Furthermore, highly significant associations identified through the analysis of population-level correlations between basal gene expression variability and chemical-induced toxicity suggest plausible mode of action hypotheses for follow-up analyses. We conclude that as the improved resolution of genetic profiling can now be matched with high-quality in vitro screening data, the evaluation of the toxicity pathways and the effects of genetic diversity are now feasible through the use of human lymphoblast cell lines.
Keywords: chemical cytotoxicity, apoptosis, HapMap; lymphoblasts, qHTS
The “Registration, Evaluation, Authorisation and Restriction of Chemicals” regulations in Europe and Toxic Substances Control Act reform activities in the United States are creating substantial pressure to develop improved methods for evaluating potential chemical hazards (Plunkett et al., 2010). Current chemical safety evaluation (National Research Council, 2007) relies on in vivo animal testing. In Europe alone, it is expected that 100,000+ chemicals will require new safety data; yet the worldwide capacity to evaluate chemicals for the most animal-intensive in vivo tests is 200–300 chemicals each year (Hartung and Rovida, 2009).
In the United States, the Tox21 program (Collins et al., 2008) is a collaborative initiative of four government agencies. This effort leads the field in its use of a broad spectrum of in vitro assays, many in quantitative high-throughput screening (qHTS) format (Inglese et al., 2006), to screen thousands of environmental chemicals for their potential to affect biological pathways that may result in human disease (Xia et al., 2008). Such data on toxicologically relevant in vitro endpoints can assist in decision making (Reif et al., 2010), serve as predictive surrogates for in vivo toxicity (Martin et al., 2010; Zhu et al., 2008), and generate testable hypotheses on the mechanisms (Xia et al., 2009).
Another important consideration in assessing the potential human health hazard is the degree of interindividual biological variability in the human population (National Research Council, 2008). A comprehensive characterization of human genome sequence variation is important for understanding observed inherited variation in toxicity phenotypes. Indeed, genetic polymorphisms can have a profound influence on disease risk after drug or toxicant exposure (Harrill et al., 2009); yet, these factors are difficult to quantitatively evaluate using current in vivo animal test systems or established cell lines (Rusyn et al., 2010). The availability of genetically diverse genetically defined renewable sources of human cells, such as lymphoblasts from the International HapMap (International HapMap Consortium, 2005) and 1000 Genomes (Durbin et al., 2010) projects, enables in vitro testing at the population scale. As the risk assessment process shifts toward in vitro data, the quantitative assessment of interindividual variability in responses to chemicals as well as an understanding of the underlying genetic causes are needed so that regulatory decisions can be based on data rather than default assumptions.
To demonstrate the feasibility of an in vitro model system to assess interindividual and population-wide variability of chemical-induced toxicity phenotypes, we exposed cells from over 80 Centre d’Etude du Polymorphisme Humain (CEPH) cell lines (O'Shea et al., 2011) to 3 concentrations of 14 environmental chemicals and assessed induction of caspase-3/7, indicative of apoptosis, and cytotoxicity, based on measuring intracellular levels of adenosine triphosphate (ATP) as a surrogate for cell number. This study showed that an in vitro genetics–anchored human model system can be utilized in a population-level screen for chemical toxicity, with the potential to identify candidate genetic susceptibility factors for further study. As a next step, we report here on a larger scale population-based qHTS using hundreds of compounds and covering a more comprehensive range of concentrations. The quantitative assessment of interindividual variability in response at this scale demonstrates the potential of this methodology for toxicity screening, hazard evaluation, and exploration of genetic determinants of susceptibility.
MATERIALS AND METHODS
Experimental Design
Chemicals.
A subset (240 compounds) of the National Toxicology Program's 1408 chemical library (Xia et al., 2008) was used in these experiments. See Supplementary table 1 for a complete list of chemicals used in these experiments. Chemicals were dissolved with dimethyl sulfoxide (DMSO) into 12 different stock concentrations ranging from 56.5nM to 10mM and were aliquoted to 1536-well plate format via pin tool (Kalypsys, San Diego, CA). The final concentration ranges from 0.26nM to 46.08μM in the assay plates. The negative control was DMSO at 0.5% vol/vol; the positive control was staurosporine at the tested concentration range.
Cell lines.
A set of 81 immortalized lymphoblastoid cell lines was acquired from Coriell Cell Repositories (Camden, NJ). The 81 cell lines were from HapMap Consortium's CEPH panel and consisted of 27 trios (father, mother, and a child). Screening was conducted in three batches, and cell lines were randomly divided into batches without regard to family structure. Cells were cultured at 37°C with 5% CO2 in suspension in flasks with upright position in RPMI 1640 media (Gibco, Carlsbad, CA) supplemented with 15% fetal bovine serum (HyClone, South Logan, UT) and 0.1% penicillin-streptomycin (Gibco). Media were changed every 3 days. Cell counts and viability were assessed prior to chemical treatment using Cellometer Auto T4 Plus (Nexcelem Bioscience, Lawrence, MA). Cells were grown to a concentration up to 106 cells/ml, volume of at least 100 ml, and viability of > 85% before treatment. After centrifugation, the cells were resuspended in fresh media. The cell suspension was filtered through a 40-μm nylon cell strainer (BD Biosciences, Durham, NC). Cell stock was diluted with fresh media to final concentrations of 3–4 × 105 cells/ml and plated into a tissue culture–treated 1536-well white/solid bottom assay plates (Greiner Bio-One North America, Monroe, NC) at 2000 cells per 5 μl per well using a flying reagent dispenser (Aurora Discovery, Carlsbad, CA). To increase the robustness of the data and evaluate reproducibility, each cell line was seeded on multiple plates (six plates except for two cell lines where five plates were seeded) so that each compound was screened in each cell line on 2–3 plates (chemicals were randomly divided in half to enable screening of 120 compound × 12 concentrations on each plate).
Cytotoxicity and caspase-3/7 assays.
Two assays were chosen to evaluate cytotoxicity according to the manufacturer’s protocols. CellTiter-Glo Luminescent Cell Viability (Promega Corporation, Madison, WI) assay was used to assess intracellular ATP concentration, a marker for cytotoxicity, 40 h posttreatment. Caspase-Glo 3/7 (Promega) was used to assess activity of caspase-3/7, a marker of apoptosis, 16 h posttreatment. These assays were selected based on their utility for in vitro screening of cytotoxicity in cell type– (Xia et al., 2008) and individual-independent (Choy et al., 2008) manner. Time points were selected based on previous experiments at the National Institutes of Health Chemical Genomics Center (NCGC) (Xia et al., 2008). A ViewLux plate reader (PerkinElmer, Shelton, CT) was used to detect luminescent intensity in each well for both assays. Data are publicly available from PubChem (AIDs: 588812 and 588813).
Data Processing
Response normalization and curve fitting.
Data were normalized relative to the positive/negative controls and corrected as detailed elsewhere (Xia et al., 2008). Concentration-response titration points were fitted to a Hill equation for each chemical. Chemicals were classified into three categories based on their concentration-response curves: active, nonactive, and inconclusive (Huang et al., 2008; Xia et al., 2008). Specifically, in data from cytotoxicity assay, the curve classes −1.1, −1.2, and −2.1 were classified as “active,” any positive curve class as “nonactive,” and others as “inconclusive.” For data from caspase-3/7 assay, curve classes 1.1, 1.2, and 2.1 were classified as active, any negative curve class as nonactive, and others as inconclusive.
Curve P.
To evaluate the cytotoxic potency of each compound, we calculated a “curve P” value for each compound-cell line pair. Curve P is defined as the lowest concentration, which showed a consistent deviation from the baseline response and derived as detailed in Sedykh et al. (2011). It can be regarded as a close approximation for the point of departure. Curve P was derived for all compounds even if little or no toxicity was observed. For the latter compounds, to enable the follow-up statistical analyses, the curve P was assigned to a concentration of 50μM. Batch effects were adjusted using the ComBat method (Johnson et al., 2007).
Data Analysis
Assessing variability across individual, chemical, and assay.
The Pearson correlation coefficient (r) between pairs of replicate plates was used to assess experimental reproducibility. For this analysis, two replicate plates were randomly selected for each chemical and cell line pair (240 chemicals × 81 cell lines = 19,440 total replicate pairs sampled).
Kruskal-Wallis ANOVA (Kruskal and Wallis, 1952) was used to assess the significance of a cell line effect (vs. experimental effect) in curve P for each chemical. The Benjamini-Hochberg false discovery rate (FDR) (Benjamini and Yekutieli, 2001) was used to correct for multiple comparisons. To measure potential confounding with basal metabolic rate, the Spearman (rank) correlation coefficient between curve P and the average ATP level in DMSO-treated cells was computed for each chemical. The Spearman correlation between the average curve P value for the cytotoxicity assay and the average curve P value for the apoptosis assay for each chemical was computed to measure an overall relationship between the two assays. Furthermore, within each chemical, the correlation between the two assays across cell lines (averaged over replications) was computed separately. For both assays, chemical-by-chemical correlation heatmaps were used to identify clusters of chemicals with similar response across cell lines. The order of the chemicals in these heatmaps was determined by complete-linkage distance clustering.
All computations, graphs, and heatmaps used the R programming environment for statistical computing and graphics (2.10.0; R Development Core Team, Vienna, Austria).
Concentration response for populations and individuals.
For the ATP assay data for progesterone, a four-parameter logistic model was fit to the assay versus concentration data for each cell line, using maximum likelihood and the optim routine in R. The model can be written where where (min, max, β0, β1, σ2) are cell line-specific parameter vectors. For a negative concentration-response relationship, EC10 is the concentration for which The variation in the EC10 estimates was used as illustrative of population variation in true EC10 values, although additional sampling variation underlies each EC10 estimate. An overall logistic concentration-response curve was fit to the aggregated data across all individuals.
Assessing heritability and genetic associations.
Heritability calculations were used to determine overall familial effects among the 27 CEPH trios for each chemical, on both assays. Calculations were motivated by the mid-parent regression model where y is the child’s response, ap is the father’s response, am is the mother’s response, and ϵ is an error term. A likelihood ratio significance test is then based on the heritability h2: the variability in response due to shared genetics as a proportion of total variability in response. For this analysis, curve P values for each chemical were quantile normalized to the standard Gaussian distribution.
To measure genotype-toxicity relationships, genome-wide association studies (GWAS) were performed in R using the GenABEL package (Aulchenko et al., 2007). Phase III genotype data, on approximately 1.4 × 106 single-nucleotide polymorphisms (SNPs), were obtained for each cell line from the International HapMap Project (International HapMap Consortium, 2005). GWAS were performed for each chemical on both assays, with quantile-normalized curve P values as the response phenotype. The significance of an association between a given SNP and the response was measured using a likelihood-based score test (Schaid et al., 2002) (qtscore in GenABEL). For our initial screen, the familial trio relationships were not used for the analysis, due to the low evidence for overall heritability, on the grounds that methods such as transmission disequilibrium testing would reduce power and with the intent to follow any significant findings with further testing. LocusZoom (Pruim et al., 2010) was used to visualize the genomic context for suggestive loci determined by GWAS.
RNA-Seq expression versus toxicity assays.
The 42 cell lines in common between Montgomery et al. (2010) and the present study were matched with HapMap IDs, using RNA-Seq tag counts mapped to the genome as previously described for 20,000 genes (Zhou et al., 2011). For computational efficiency, simple read proportions consisting of number of tag counts per gene divided by the mapped library size (Zhou et al., 2011) were used in linear regression as predictors for the cytotoxicity assays. FDR q values were then obtained for the entire set of genes and chemicals, using p.adjust() in R. For the caspase assay, ∼5000 genes were determined to have at least one chemical with q < 0.01, and these genes were retained for clustering. Hierarchical clustering with average linkage was performed directly on the FDR q values using the heatmap function in R.
RESULTS
qHTS in a Population of Human Lymphoblasts Yields Robust and Reproducible Data
Screening was conducted in a 1536-well plate format using a robotic system. The 81 cell lines were randomly subdivided into three batches, and each line was screened against 240 chemical substances (see Supplementary table 1 for a complete list) at 12 concentrations (0.26nM–46.0μM). Each 1536-well plate contained one cell line exposed to 120 chemicals accompanied by concurrent vehicle (DMSO) and positive controls. To increase the robustness of the data, duplicates or triplicates of each plate were run. Assays for intracellular ATP content and caspase-3/7 activity were used based on their utility for in vitro screening of cytotoxicity and apoptosis, respectively, in cell type– and individual-independent manner (Choy et al., 2008; Xia et al., 2008). A combination of the two assays allows for the role of apoptosis in the cytotoxicity response to be evaluated (Shi et al., 2010).
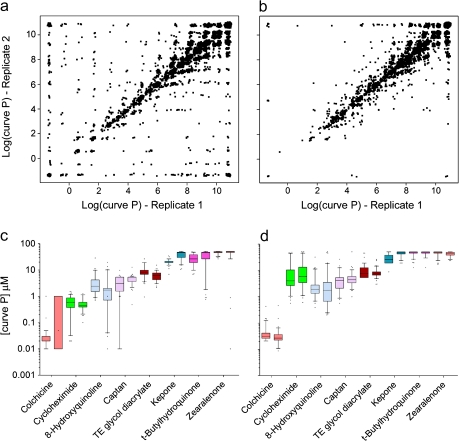
Several metrics were used to evaluate the reproducibility of the toxicity phenotypes. First, the concentration-response curve class (Parham et al., 2009) was identical across replicate plates 95.2% of the time for cytotoxicity and 94.1% for apoptosis. Second, the pair-wise Pearson correlation among replicate plate pairs using log (AC50) values for the compounds with active curve classes for the cytotoxicity and apoptosis assays was r = 0.99 and r = 0.98, respectively. Third, to evaluate the effects correlation for all compounds, we calculated a “curve P” value, the lowest concentration that showed a consistent deviation from the baseline response (Sedykh et al., 2011), which can be regarded as a close approximation for the lowest observed adverse effect level. For chemicals exhibiting no effect across the concentrations tested, the curve P was assigned to 50μM to enable straightforward statistical analyses. The pair-wise correlation among replicate plates of the log (curve P) values was equally high (r[cytotoxicity] = 0.91, r[apoptosis] = 0.95) when all compounds were included (Figs. 1a and b). Finally, there were eight duplicates among the compounds screened. High concordance in median and range of responses for these was observed (Figs. 1c and d).
FIG. 1.
Intraexperimental reproducibility for cytotoxicity (panels a and c) and caspase-3/7 (panels b and d) assays. Panels a and b show log (curve P) values for randomly selected pairs of replicate plates within each chemical and cell line (240 chemicals × 81 cell lines = 19,440 replicate pairs displayed). Panels c and d show side-by-side boxplots for eight duplicate compounds that were tested in two independent wells on each plate.
Range in Cytotoxicity Across the Chemicals
The chemicals selected for screening were a subset of 1408 compounds previously tested in one or more traditional toxicological assays and had been profiled for cytotoxicity and caspase-3/7 induction by the National Toxicology Program and NCGC using qHTS (Xia et al., 2008) in (i) 13 human and rodent cells derived from liver, blood, kidney, nerve, lung, and skin; and in (ii) 26 human lymphoblast cells (data available from PubChem AIDs: 963–989). Of these, 240 compounds that were clearly active in those experiments were selected for the current study (iii).
Comparison of the cytotoxicity average log (curve P) from the current study showed high concordance with that in panels (i) and (ii), see above. Pair-wise correlation analysis for the 240 chemicals across three data sets was highly significant (p < 0.0001). High correlation (r = 0.87; rank correlation = 0.83) was observed between lymphoblast panels (ii) and (iii), whereas the correlations with the diverse panel (i) were moderately high (r = 0.74 or 0.75; rank correlation = 0.72 or 0.75 with (ii) and (iii), respectively). Together, the results indicate high external reproducibility for this measurement of cytotoxicity and, importantly, the potential utility of lymphoblast cell lines as a toll for population-based toxicity screening.
Interindividual Variability in Response Across Cell Lines
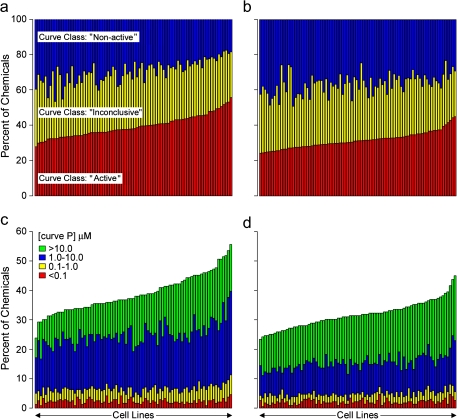
In contrast to the highly invariant reproducible results found within individual cell lines, the chemicals induced a wide range of responses among the lymphoblast lines. The percentage of compounds classified as active in the cytotoxicity assay varied from 28 to 56% (Fig. 2a); an equally broad range of activity (i.e., 24–45%) was seen in the caspase-3/7 assay (Fig. 2b). Among actives, a wide range of potency, assessed from the curve P, was observed for each cell line in both assays (Figs. 2c and d).
FIG. 2.
Distribution of cytotoxicity across chemicals for cytotoxicity (panels a and c) and caspase-3/7 (panels b and d) assays. Panels a and b give the percentage of chemicals classified as “active,” “nonactive,” or “inconclusive” for each cell line. Panels c and d give the range of potency (curve P) for active chemicals in each cell line.
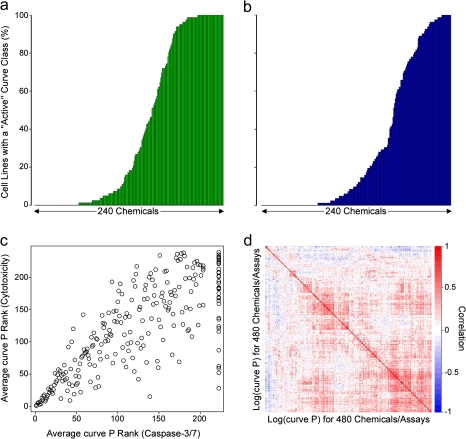
Some chemicals were classified as active for cytotoxicity and caspase-3/7 induction in all of the lymphoblast lines, whereas others were not active for either endpoint (Figs. 3a and b). In both assays, most chemicals were active in some cell lines, whereas not active in others, indicative of interindividual (cell line) variability in response. The significant correlation (rank correlation = 0.77; p = 2.2 × 10−16; all compounds tested) between the chemical’s average curve P for cytotoxicity and caspase-3/7 (Fig. 3c) indicates the primary cause of cell death for these compounds is most likely via apoptosis. A heatmap shows correlations between average log (curve P) for all chemicals in both assays (Fig. 3d). Clusters of chemicals with highly concordant responses across cell lines were evident for cytotoxicity, apoptosis, or both phenotypes. A significant (FDR < 5%) correlation between responses in cytotoxicity and apoptosis assays was observed for most of the compounds screened.
FIG. 3.
The percent of cell lines exhibiting activity for each chemical for cytotoxicity (panel a) and caspase-3/7 (panel b) assays. Panel c displays the rank of the mean ATP curve P value versus the mean caspase curve P value for each chemical. Panel d shows a heatmap of the correlations between log (curve P) values for all chemical-assay combinations.
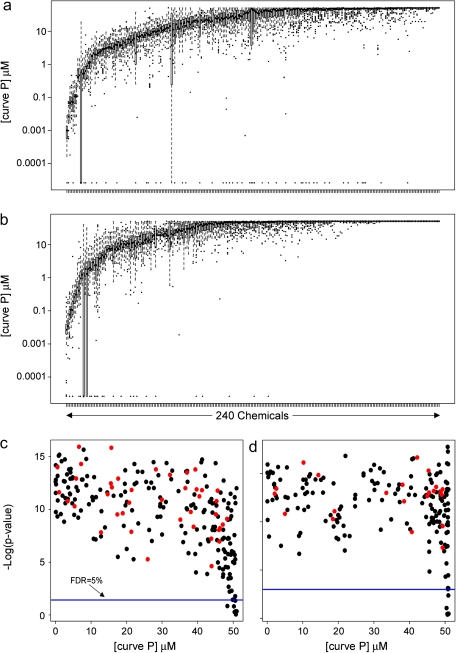
Interindividual variability in cytotoxicity was visualized using boxplots of log (curve P) for each chemical (Figs. 4a and b). Although median cytotoxicity differed between chemicals tested, interindividual variability was observed even for the most active chemicals. Variance components heritability testing for each chemical/assay showed that none of the derived h2 statistics was significant after adjusting for multiple comparisons, an observation which was confirmed using mid-parent assays' values compared with those of the offspring (data not shown).
FIG. 4.
Boxplots of curve P values for each of the 240 chemicals (arranged by mean activity) across the 81 cell lines are shown for cytotoxicity (panel a) and caspase-3/7 (panel b) assays. For cytotoxicity (panel c) and caspase-3/7 (panel d) assays, −log (p values, Kruskal-Wallis test) were plotted against mean curve P (micromolar). The blue line gives a FDR-adjusted significance threshold (FDR = 0.05). Chemicals colored in red had a significant correlation between activity and basal metabolic rate (ATP level in vehicle-treated cells) across the panel of cell lines (Spearman rank correlation; FDR < 0.05).
Interindividual (between cell lines) versus experimental (between replicates) variability for each chemical was evaluated using Kruskal-Wallis ANOVA (Kruskal and Wallis, 1952). Most chemicals show a significant (FDR < 5%) cell line effect (Figs. 4c and d). It has been suggested that differences in chemical’s toxicity among lymphoblast lines could be partly attributed to differences in baseline growth rate and metabolic status (Choy et al., 2008). Correcting for these measurements reduces effect correlation that would otherwise make responses across chemicals appear more similar. We therefore normalized for control levels of intracellular ATP (e.g., metabolic activity) and basal activity of caspase-3/7 as well as for the response of the positive control cytotoxicant. In addition, we directly assessed for each chemical whether the basal metabolic rate, an endpoint which correlates closely with the growth rate (Choy et al., 2008), significantly correlated with cytotoxicity. Approximately 80% and 90% of chemicals (Figs. 4c and d; black dots) exhibited no correlation (FDR > 0.05) between basal metabolic rate (ATP level in vehicle-treated cells) and cytotoxicity or apoptosis, respectively, across the cell panel.
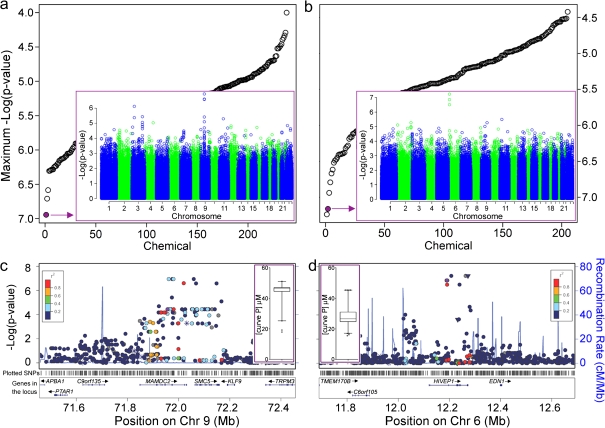
Assessing Relationships Between Cytotoxicity and Genotype
With variability among cells from different individuals demonstrated, we then asked if we could identify genetic loci responsible, utilizing toxicity phenotypes as quantitative traits and publicly available genotypes (International HapMap Consortium, 2005) (Fig. 5). The top two plots in Figure 5 show p values for the most significant SNP associated with cytotoxicity (Fig. 5a) or induction of caspase-3/7 (Fig. 5b) for each chemical. The inset shows a plot of −log10 (p values) for SNP endpoint associations for the selected chemicals. Progesterone had the lowest p value SNPs on chromosome 9, whereas guggulsterones Z (4,17(20)-pregnadiene-3,16-dione, z-isoform) exhibited many suggestive associations on chromosome 6p. Figures 5c and d provide a zoomed-in view of the genomic context for these suggestive regions.
FIG. 5.
Toxicity-genotype relationships were assessed using GWAS analysis for the 240 chemicals on both cytotoxicity (panels a and c) and caspase-3/7 (panels b and d) assays. Panels a and b give p values (-log10 scale) for the most significant SNP associated with toxicity for each chemical. The inset in the diagram gives −log10 (p values) for SNP-toxicity associations across the entire genome, for progesterone (cytotoxicity assay, inset in panel a) and Guggulsterones Z (caspase-3/7 assay, inset in panel b). Panels c and d provide a zoomed-in look at the locus with the most significant p value for each of the two compounds, respectively. Correlation between SNPs is identified with colors. SNP and gene tracks are also shown. Inset: box and whisker plots for each compound’s curve P.
Progesterone was not highly cytotoxic, yet showed an appreciable degree of interindividual variability in curve P values (Fig. 5c inset). A characteristic pattern of SNPs with low p values in linkage disequilibrium is evident in a ∼300 kb region containing two genes, structural maintenance of chromosomes protein 5 (SMC5) and MAM domain containing 2 (MAMDC2). Guggulsterones Z, a bioactive constituent of resinous sap from Commiphora mukul, is a farnesoid X receptor antagonist and is used widely as a nutraceutical. It is known to suppress expression of antiapoptotic genes, promote apoptosis, and inhibit nuclear factor-kappa B (NF-κB) (Shishodia and Aggarwal, 2004). In our study, it was moderately active in inducing caspase-3/7 (Fig. 5d inset) and exhibited interindividual variability. A narrow 100 kb region on chromosome 6p, containing the gene human immunodeficiency virus type I enhancer binding protein 1 (HIVEP1), shows association with the apoptosis phenotype.
Concentration Response for Populations and Individuals
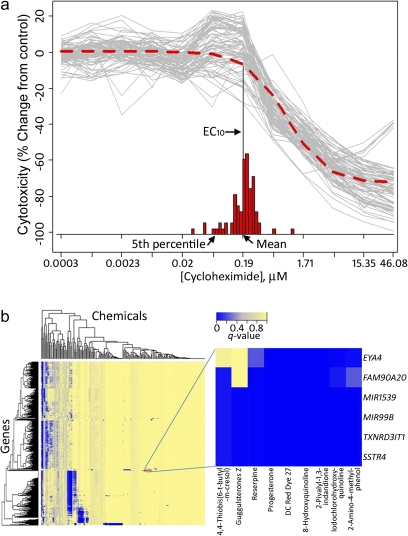
The availability of cytotoxicity screens on 80+ individuals, with the assays performed under controlled conditions, enables sensitive investigation of variation in individual dose-response profiles (National Research Council, 2008). This concept is illustrated in Figure 6a, in which the ATP assay values for cycloheximide are shown in gray for each concentration for all individuals. Separate logistic curve fits were performed, providing for each individual cell line an “effective concentration 10%” (EC10) the estimated concentration at which the response deviates by at least 10% from the control baseline, and these are shown as a histogram. The mean of these EC10 values offers a population-wide summary of the activity (e.g., cytotoxicity, caspase-3/7) of a chemical and is very similar to the EC10 produced when the data are first pooled for all individuals and then fit using a single concentration-response curve (red-dashed curve in Fig. 6a). However, aggregation across the population ignores the variability in toxic susceptibility, and the EC10 estimated fifth percentile may be used to illustrate the concept of a “vulnerable” subpopulation.
FIG. 6.
Panel a, a population concentration response was modeled using in vitro qHTS data using cycloheximide data (cytotoxicity assay) as an example. Logistic dose-response modeling was performed for each individual to the values shown in gray, providing individual 10% effect concentration values (EC10). The EC10 obtained by performing the modeling on average assay values for each concentration (see frequency distribution) are shown in the inset. Panel b, a heatmap of clustered FDRs (q values, see color bar) for association of the data from caspase-3/7 assay with publicly available RNA-Seq expression data on a subset of cell lines. A sample subcluster is shown.
Defining Mode of Action Chemical-Perturbed Pathways
Gene expression data form another rich source of publicly available data, which can be matched with cytotoxicity profiles to provide further evidence of toxicity pathway activity. Many of the HapMap cell lines have been profiled for expression in a number of studies, including highly sensitive RNA-Seq profiling (Montgomery et al., 2010). For the 42 cell lines for which RNA-Seq data are publicly available, expression values for each of ∼20,000 genes were compared with the caspase-3/7 and cytotoxicity assay results, with a number of highly significant associations. A heatmap of clustering performed on FDR q values (Fig. 6b) shows striking patterns of gene-chemical relationships, with much of the structure resolving into distinct sets of genes associated with sets of chemicals. The results for progesterone are shown as a highly specific subgroup, with lymphoblast cytotoxicity for several chemicals being significantly associated with background RNA levels for six transcripts and several microRNAs.
DISCUSSION
New paradigms for the rapid and accurate evaluation of the potential health hazard from environmental chemicals are needed, given the large number of environmental chemicals to be evaluated, and the high cost and low throughput of traditional toxicity testing approaches (Collins et al., 2008). Development of in vitro toxicity tests that can be utilized in a tiered framework is necessary, feasible, and consistent with the needs of scientifically rigorous high-throughput risk assessment (Kavlock et al., 2009). A particular challenge in developing such next generation toxicity testing schemata is the assessment of differential susceptibility among individuals. The results presented here provide proof of principle of such a testing system, demonstrating the feasibility and utility of screening a panel of cells from genetically diverse individuals, whereby both population-wide and individual responses can be evaluated.
The in vitro toxicity–screening paradigm detailed here has focused on a population-based cell culture model, an approach that affords several key benefits compared with collections of unrelated cell lines from different species and tissues (Xia et al., 2008). Our results show that many chemicals exhibit interindividual variation in induction of toxicity, and this information is crucial for chemical-testing prioritization. This screening paradigm also provides quantitative data on population-wide variability in toxicity, which may be used to establish data-driven uncertainty estimates when extrapolating from in vitro data to potential in vivo toxicity (Judson et al., 2011). Even though the data collected herein are on a limited population (81 individuals), it is immediately interpretable for ranking and prioritizing chemicals. For example, a population-based view of dose- or concentration-response is an important concept that directly addresses the issue of subpopulations (National Research Council, 2008); however, actual experimental data-driven implementation has been limited. We reason that the population-based concentration response in vitro qHTS data allows for the development of models to estimate in vitro point of departure and safety/uncertainty factors (Crump et al., 2010) because variation between genetically defined/ diverse cell lines may be treated as reflective of that among individuals. The recognition of underlying genetic causes may further enhance extrapolation and understanding of the shape of the dose-response relationships. In addition, the data may be used to explore potential differences/similarities in modes of action between chemicals on the population-wide level.
By combining toxicity data with publicly available genetic information, such as that provided by the HapMap (International HapMap Consortium, 2005), 1000 Genomes (Durbin et al., 2010), and public RNA–sequencing projects (Montgomery et al., 2011), it is possible to probe the contribution of genomics to toxicity phenotypes. Such an approach represents a substantial savings of cost and time, capitalizing on the extensive prior characterization of these samples. Accordingly, we have begun to explore variation in toxicity susceptibility as a function of genotype as well as the relationship between toxic response and basal expression profiles.
Genotype-phenotype relationships are likely to reflect causal action of underlying physiological variation and are thus of great interest to epidemiologists for understanding the ultimate sources of population variation. However, the effect sizes are typically small, as has been the source of considerable discussion in the genomics community (Manolio et al., 2009). Variation in basal messenger RNA (mRNA) expression, in contrast, may reflect cascades of responses controlled by the underlying genotype and typically involves a smaller multiple testing penalty. Thus, we likely have more power to detect association of expression with toxicity response phenotypes, even though the underlying causality relationships may remain elusive. The highly significant associations identified through the analysis of population-level correlations between basal gene expression variability and chemical-induced toxicity have revealed several reasonable mode of action hypotheses. For example, the in vitro toxicity of 1,3-indandione-containing rodenticides has been shown to occur through the inhibition of the pyrimidine synthetic pathway (Hall et al., 1994), and thioredoxin reductase (e.g., TXNRD3IT1) is required for deoxynucleotide triphosphate pool maintenance during S phase (Koc et al., 2006). Expression of somatostatin receptor 4 correlates with progesterone receptor levels in human breast tumors (Kumar et al., 2005). Thioredoxin reductase affects expression of progesterone receptor–controlled genes in MCF-7 cells (Rao et al., 2009).
Similarly, the quantitative assessment of interindividual genetic variability in responses to environmental agents in vitro demonstrates the potential of this approach to explore the genetic basis for susceptibility through genome-wide association analysis. The genes SMC5 and MAMDC2 implicated in this study as associated with progesterone-induced toxicity are highly plausible and belong to pathways critical for development. The same locus was reported as associated with developmental abnormalities cleft palate and Kabuki syndrome (Kuniba et al., 2009; Marazita et al., 2004), and exposure to progesterone during gestation is known to cause cleft palate in rabbits (Andrew and Staples, 1977). Likewise, the association between guggulsterones Z and polymorphisms in HIVEP1 is highly credible, given the known effects of guggulsterones Z on apoptosis through NF-κB–related signaling (Shishodia and Aggarwal, 2004). HIVEP1 belongs to a family of large zinc finger–containing transcription factors that bind specifically to the NF-κB motif and related sequences (Yu et al., 2009). The alternative splice variant of HIVEP1, the gatekeeper of apoptosis activating proteins (GAAP)-1 protein, can regulate p53 and IRF-1-dependent cell proliferation and apoptosis (Lallemand et al., 2002).
Important limitations to in vitro toxicity profiling using lymphoblasts, as compared with primary cells that may be obtained from other tissues of interest, include inability to assess target organ adverse effects or a potential role of other environmental factors such as lifestyle, diet, or coexposures. In addition, the challenge of assessing the potential toxicity of chemical’s metabolites or the potential lack of the receptor-mediated signaling that may be critical for the downstream adverse molecular events, in lymphoblast cell lines also should be taken into consideration when interpreting the data. Still, whereas lymphocytes do not have the metabolic capacity of the liver or even that of freshly isolated hepatocytes, they do express a number of nuclear receptors, as well as most genes of the phase I and II metabolism, and transporters (Siest et al., 2008). A comparison of the population-wide (250+ individuals of various races, ages, and gender) variability in mRNA levels for several dozen liver-specific thyroid hormone–related genes between human liver (Schadt et al., 2008) and lymphoblast cell lines (Stranger et al., 2007) shows that most of the nuclear receptors and metabolism genes are expressed in lymphoblasts, albeit at 10 to 100 times lower quantity. Importantly, the between subject variability in expression of these genes in either human liver or lymphoblasts is also of appreciable magnitude (4- to 10-fold). To overcome these limitations, both higher concentrations and known metabolites can be tested in vitro because of high throughput. Correcting for the cell growth rate and baseline metabolic rate also reduces effect correlation that may make responses across chemicals appear more similar (Choy et al., 2008).
Based on these results, we reason that a full and sensitive analysis of genomic predictors of toxicity response will be feasible through the joint use of toxicity phenotypes, genotype, and expression information, though considerably larger sample sizes—likely on the order of several hundred or thousands of individual cell lines—will be necessary. Such a population-based in vitro survey would greatly advance our understanding of the genetic underpinnings of susceptibility-related regulatory networks and is ongoing in our laboratories.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This research was supported, in part, by the Intramural Research Programs of the National Toxicology Program, National Institute of Environmental Health Sciences interagency agreement Y2-ES-7020-01 and by grants from the National Institutes of Health (NIH) (R01 ES015241) and U.S. Environmental Protection Agency (U.S. EPA) (RD83382501).
Supplementary Material
Acknowledgments
We thank Srilatha Sakamuru for technical support. The research described in this article has not been subjected to each funding agency’s peer review and policy review and therefore does not necessarily reflect their views and no official endorsement should be inferred. The authors declare no competing financial interests.
References
- Andrew FD, Staples RE. Prenatal toxicity of medroxyprogesterone acetate in rabbits, rats, and mice. Teratology. 1977;15:25–32. doi: 10.1002/tera.1420150104. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, et al. Genetic analysis of human traits in vitro: Drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump KS, Chen C, Louis TA. The future use of in vitro data in risk assessment to set human exposure standards: Challenging problems and familiar solutions. Environ. Health Perspect. 2010;118:1350–1354. doi: 10.1289/ehp.1001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IH, Wong OT, Chi LK, Chen SY. Cytotoxicity and mode of action of substituted indan-1, 3-diones in murine and human tissue cultured cells. Anticancer Res. 1994;14:2053–2058. [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Rovida C. Chemical regulators have overreached. Nature. 2009;460:1080–1081. doi: 10.1038/4601080a. [DOI] [PubMed] [Google Scholar]
- Huang R, Southall N, Cho MH, Xia M, Inglese J, Austin CP. Characterization of diversity in toxicity mechanism using in vitro cytotoxicity assays in quantitative high throughput screening. Chem. Res. Toxicol. 2008;21:659–667. doi: 10.1021/tx700365e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Cohen Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, Dix DJ. Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem. Res. Toxicol. 2011;24:451–462. doi: 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- Kavlock RJ, Austin CP, Tice RR. Toxicity testing in the 21st century: Implications for human health risk assessment. Risk Anal. 2009;29:485–487. doi: 10.1111/j.1539-6924.2008.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Mathews CK, Wheeler LJ, Gross MK, Merrill GF. Thioredoxin is required for deoxyribonucleotide pool maintenance during S phase. J. Biol. Chem. 2006;281:15058–15063. doi: 10.1074/jbc.M601968200. [DOI] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952;47:583–621. [Google Scholar]
- Kumar U, Grigorakis SI, Watt HL, Sasi R, Snell L, Watson P, Chaudhari S. Somatostatin receptors in primary human breast cancer: Quantitative analysis of mRNA for subtypes 1–5 and correlation with receptor protein expression and tumor pathology. Breast Cancer Res. Treat. 2005;92:175–186. doi: 10.1007/s10549-005-2414-0. [DOI] [PubMed] [Google Scholar]
- Kuniba H, Yoshiura K, Kondoh T, Ohashi H, Kurosawa K, Tonoki H, Nagai T, Okamoto N, Kato M, Fukushima Y, et al. Molecular karyotyping in 17 patients and mutation screening in 41 patients with Kabuki syndrome. J. Hum. Genet. 2009;54:304–309. doi: 10.1038/jhg.2009.30. [DOI] [PubMed] [Google Scholar]
- Lallemand C, Plamieri M, Blanchard B, Meritet JF, Tovey MG. GAAP-1: A transcriptional activator of p53 and IRF-1 possesses pro-apoptotic activity. EMBO Rep. 2002;3:153–158. doi: 10.1093/embo-reports/kvf032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Mansilla MA, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am. J. Hum. Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MT, Dix DJ, Judson RS, Kavlock RJ, Reif DM, Richard AM, Rotroff DM, Romanov S, Medvedev A, Poltoratskaya N, et al. Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA's ToxCast program. Chem. Res. Toxicol. 2010;23:578–590. doi: 10.1021/tx900325g. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. [Google Scholar]
- National Research Council. Science and Decisions: Advancing Risk Assessment. Washington, DC: The National Academies Press; 2008. [PubMed] [Google Scholar]
- O'Shea SH, Schwarz J, Kosyk O, Ross PK, Ha MJ, Wright FA, Rusyn I. In vitro screening for population variability in chemical toxicity. Toxicol. Sci. 2011;119:398–407. doi: 10.1093/toxsci/kfq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham F, Austin C, Southall N, Huang R, Tice R, Portier C. Dose-response modeling of high-throughput screening data. J. Biomol. Screen. 2009;14:1216–1227. doi: 10.1177/1087057109349355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett LM, Kaplan AM, Becker RA. An enhanced tiered toxicity testing framework with triggers for assessing hazards and risks of commodity chemicals. Regul. Toxicol. Pharmacol. 2010;58:382–394. doi: 10.1016/j.yrtph.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AK, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. Thioredoxin and thioredoxin reductase influence estrogen receptor alpha-mediated gene expression in human breast cancer cells. J. Mol. Endocrinol. 2009;43:251–261. doi: 10.1677/JME-09-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif DM, Martin MT, Tan SW, Houck KA, Judson RS, Richard AM, Knudsen TB, Dix DJ, Kavlock RJ. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ. Health Perspect. 2010;118:1714–1720. doi: 10.1289/ehp.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: Population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh A, Zhu H, Tang H, Zhang L, Richard A, Rusyn I, Tropsha A. Use of in vitro HTS-derived concentration-response data as biological descriptors improves the accuracy of QSAR models of in vivo toxicity. Environ. Health Perspect. 2011;119:364–370. doi: 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Springer S, Escobar P. Coupling cytotoxicity biomarkers with DNA damage assessment in TK6 human lymphoblast cells. Mutat. Res. 2010;696:167–178. doi: 10.1016/j.mrgentox.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Shishodia S, Aggarwal BB. Guggulsterone inhibits NF-kappaB and IkappaBalpha kinase activation, suppresses expression of anti-apoptotic gene products, and enhances apoptosis. J. Biol. Chem. 2004;279:47148–47158. doi: 10.1074/jbc.M408093200. [DOI] [PubMed] [Google Scholar]
- Siest G, Jeannesson E, Marteau JB, Samara A, Marie B, Pfister M, Visvikis-Siest S. Transcription factor and drug-metabolizing enzyme gene expression in lymphocytes from healthy human subjects. Drug Metab. Dispos. 2008;36:182–189. doi: 10.1124/dmd.107.017228. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Huang R, Sun Y, Semenza GL, Aldred SF, Witt KL, Inglese J, Tice RR, Austin CP. Identification of chemical compounds that induce HIF-1alpha activity. Toxicol. Sci. 2009;112:153–163. doi: 10.1093/toxsci/kfp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Mitchell GA, Richter A. Cirhin up-regulates a canonical NF-kappaB element through strong interaction with Cirip/HIVEP1. Exp. Cell Res. 2009;315:3086–3098. doi: 10.1016/j.yexcr.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Xia K, Wright FA. A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics. 2011;27:2672–2678. doi: 10.1093/bioinformatics/btr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Rusyn I, Richard A, Tropsha A. Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity. Environ. Health Perspect. 2008;116:506–513. doi: 10.1289/ehp.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.