Abstract
Ursodeoxycholic acid (UDCA, ursodiol) is used to prevent damage to the liver in patients with primary biliary cirrhosis. The drug also prevents the progression of colorectal cancer and the recurrence of high-grade colonic dysplasia. However, the molecular mechanism by which UDCA elicits its beneficial effects is not entirely understood. The aim of this study was to determine whether ileal bile acid binding protein (IBABP) has a role in mediating the effects of UDCA. We find that UDCA binds to a single site on IBABP and increases the affinity for major human bile acids at a second binding site. As UDCA occupies one of the bile acid binding sites on IBABP, it reduces the cooperative binding that is often observed for the major human bile acids. Furthermore, IBABP is necessary for the full activation of farnesoid X receptor α (FXRα) by bile acids, including UDCA. These observations suggest that IBABP may have a role in mediating some of the intestinal effects of UDCA.
Keywords: farnesoid X receptor α, colorectal cancer, bile salts
Ursodeoxycholic acid (UDCA) is a bile acid that is abundant in the bile of black bears (Ursudae), and it is the active component of an ancient Chinese remedy for liver disorders (1). In Western society, UDCA is approved as a drug for treating primary biliary cirrhosis (1). It improves the symptoms and decreases the biochemical abnormalities in patients with primary sclerosing cholangitis (PSC) (2). Patients with PSC are at high risk for developing colorectal cancer, and it has been observed that UDCA also mitigates this risk (3, 4). Recently, UDCA was shown to reduce the recurrence of high-grade dysplasia in patients with colorectal cancer (5). Much of the clinical benefit of UDCA has been attributed to a reduction in the hydrophobicity of the systemic bile acid pool (6) and to the ability of UDCA to partially block the transmembrane apical Na+-dependent bile acid transporter (7). Thus far, no intracellular receptor for UDCA has been reported.
The farnesoid X receptor α (FXRα) is a nuclear receptor that binds to and is activated by bile acids (8–10). FXRα forms heterodimers with the retinoid X receptor α (RXRα) and regulates the expression of target genes involved in bile acid homeostasis as well as lipid and glucose metabolism (11). Chenodeoxycholic acid (CDCA) is the best natural ligand of FXRα (8–10). However, deoxycholic acid (DCA), the bile acid most frequently associated with cholestatic diseases and colorectal cancer, also binds to FXRα (8, 9). Given the association of FXRα with the processes underlying cholestatic diseases (12), liver cancer (13, 14), and colorectal cancer (15–17), it would seem to be a likely molecular target for UDCA. However, conflicting reports exist regarding UDCA's ability to bind and activate FXRα (8–10, 18).
Ileal bile acid binding protein (IBABP) is a small cytoplasmic protein mainly expressed in ileal epithelium. Although initially categorized as a member of the fatty acid binding protein family (19), IBABP binds exclusively to bile acids (20). The bile acid selectivity arises from the fact that IBABP has a larger binding pocket than other members of the fatty acid binding protein family (21, 22). The precise biological function of IBABP is not clear, but it is presumed to coordinate with the apical bile acid transporter in the uptake of bile acids into ileocytes. IBABP also associates with FXRα (23) and could potentially help mediate the transcriptional response to bile acids.
IBABP binds two bile acids in a cooperative manner (21, 24). The basis of cooperative binding is the formation of a hydrogen bond between the 3-OH of the bile acid occupying site 1, and either the 7-OH or 12-OH of the bile acid occupying site 2 (supplementary Fig. I). The binding of UDCA to these sites on IBABP has never been examined. Here we show that UDCA binds to IBABP, but unlike major human bile acids, UDCA binds only to a single site. Importantly though, binding of UDCA promotes binding of major human bile acids at the second binding site. This binding mechanism is evident in whole cells, where UDCA potentiates the activation of FXRα by major human bile acids. These findings explain how UDCA can promote the transcriptional response of FXRα without binding to it.
EXPERIMENTAL PROCEDURES
Reagents
All free, glycine-, and taurine-conjugated cholic acid (CA), chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), and ursodeoxycholic acid (UDCA) in sodium salt form were purchased from Sigma and Calbiochem. 15N-labeled bile acids were synthesized in-house via coupling of 15N-labeled glycine (Cambridge Isotope Laboratories) to unconjugated bile salts by the method of Momose et al. (25). Rabbit antiserum to human IBABP was raised by our laboratory using recombinant protein as antigen. Oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. siRNA SMARTpools were purchased from Dharmacon. Lipofectamine 2000 was ordered from Invitrogen. The molecular cloning reagents were purchased from Promega and New England Biolabs, unless otherwise indicated.
Plasmid constructs
The IBABP promoter containing a FXRα binding element was amplified from human genomic DNA as previously described (26) and inserted into pGL3-Basic vector (Promega) to produce the FXRE-Luc reporter gene for monitoring FXRα activity. The pCDNA3.1-hFXRα2 (27) plasmid was a gift from Dr. Peter A. Edwards (University of California at Los Angeles). To construct the mammalian expression vector for human IBABP, the open reading frame (ORF) of IBABP was amplified from IMAGE clone 1916019 (Research Genetics) using pfu DNA polymerase (Stratagene), and then inserted into pcDNA6/His C (Invitrogen) between BamH I and Xho I sites (pcDNA6/His-IBABP). To construct the recombinant IBABP expression vector, the ORF of IBABP was inserted into the prokaryotic expression vector pGEX-KG-1 between EcoR I and Xho I sites (pGEX-KG-1/IBABP). The ORF was fused in-frame in N terminus with glutathione S-transferase (GST) coding sequence separated by a thrombin site and a glycine linker.
Protein expression and purification
pGEX-KG-1/IBABP was transformed into E. coli strain BL21(DE3) (Stratagene). The recombinant IBABP was expressed in LB medium with induction of 25 µM isopropyl-β-D-thiogalactopyranoside (IPTG) at 28°C for 4 h. Recombinant IBABP was purified in mild buffer by affinity chromatography on glutathione agarose, coupling with on-column cleavage by thrombin protease to remove the GST tag. Briefly, 5 g of cells were suspended in 30 ml of ice-cold PBS (pH = 7.4) with 1.5 ml of bacterial protease inhibitor cocktail (Sigma) and lysed by the French Pressure Cell Press (Aminco). After brief sonication to break the host DNA, the crude bacterial extract was centrifuged at 13,000 rpm for 30 min at 4°C, and the supernatant was adjusted with 1 M DTT to a final concentration of 5 mM, and loaded onto PBS-equilibrated 2 × 5 ml GSTrap HP columns (Amersham) at 0.5 ml/min overnight in cold room. The columns were then washed with 180 ml PBS at 1 ml/min, injected with 12 ml thrombin protease solution at 20 U/ml (Amersham), and incubated at room temperature for 20 h. A PBS-equilibrated 1 ml HiTrap Benzamidine FF (high sub) column (Amersham) was connected after GSTrap column to remove thrombin protease, and the recombinant IBABP was eluted using PBS at 0.5 ml/min. The protein preparations were then delipidated by passing through hydroxyalkoxypropyl-dextran (type VI; Sigma) column preequilibrated with PBS at 37°C. 15N-labeled IBABP was expressed and purified similarly, except that the M9 minimal medium supplemented with 15NH4Cl was used. The purity of IBABP was estimated as >98% by SDS-PAGE gel and analytical gel-filtration chromatography. The protein was correctly folded as indicated by the sharp melting curve in differential scanning calorimetry (DSC) assay. Protein concentration was determined by BCA protein assay (Pierce).
Tryptophan fluorescence spectroscopy
Tryptophan fluorescence was measured in volts at 20°C with 450 volt input using MOS 250 fast UV/Vis spectrometer (Bio-Logic). IBABP (250–270 µl of 10–20 µM) in PBS was titrated stepwise at 1–2 µl increments with 2.5–5.0 mM of the different bile acids and UDCA in the same buffer. The detailed concentration of ligand and protein, and the titration volume and step are specified in the legend of each figure. After each titration, the protein and ligand mixture was incubated for 5 min to allow the binding to reach equilibrium. Emission spectra were recorded in triplicate from 310 to 400 nm at a rate of 125 nm/s, with excitation at 280 nm. Both excitation and emission slits were 10 nm. Fluorescence gain (ΔF) at 336 nm was calculated by subtracting the fluorescence intensity of apo-protein from that of the holo-protein. The binding data were analyzed with two independent approaches. The Hill equation, ΔF/ΔFmax=[BA]HN/(KDHN+[BA]HN), was used to obtain binding affinity, and the Hill coefficient from a plot of the normalized fluorescence change ΔF/ΔFmax (specific binding) was plotted against bile acid concentration [BA]. In a second analysis, the Scatchard plot of ΔF/ΔFmax /[BA] versus ΔF/ΔFmax was used to identify binding cooperativity. In these plots, convex downward curvature indicates cooperativity. The value of the ordinate at the maximum abscissa value on these curves can also be used to calculate the Hill coefficient, HN (HN=1/(1-ΔF/ΔFmax).
NMR spectroscopy
Protein-observed NMR experiments were performed on 0.03−0.1 mM uniformly 15N-labeled human IBABP samples in the presence or absence of bile acids in 20 mM potassium phosphate, pH 7.2, at 303 K. NMR spectra were acquired on 500 MHz 5 mm TXI Bruker Avance and 600 MHz Bruker Avance Spectrometer with 5 mm TCI cryoprobe. Bile acids were dissolved in NMR buffer as 2.0–10.0 mM stock solutions prior to NMR titration experiments. Saturation with bile acid ligands was monitored up to 10-fold molar excess. Ligand-observed NMR experiments were performed on 0.6 mM uniformly 15N-labeled GUDCA or GCA samples in the presence or absence of 0.2 mM unlabeled human IBABP at 298 K. 2D 1H, 15N HSQC spectra were recorded over an experimental time of 6 to 24 h per spectrum with 512 indirect 15N data points, at 30 ppm 15N sweep width, 119.5 ppm carrier position, 4,096 direct 1H increments, 3s recycling delay, and 16–64 scans. Spectral processing was conducted with NMRPipe, NMRDraw, in-house scripts, and NMRView. Before Fourier transformation, the data were multiplied with a squared sine-bell window function, phase corrected, and zero-filled to 2,048 data points for indirect 15N sampling.
Cell culture
Caco-2 cells were grown in complete Dulbecco's modified Eagle's media (DMEM; Mediatech) supplemented with antibiotics (Omega Scientific) and 10% fetal bovine serum (Invitrogen). Cells were maintained in 100 mm standard cell culture dishes (BD Biosciences) and grown at 37°C under a humidified 5% CO2 atmosphere. Cells were split twice a week.
Overexpression of IBABP in cells and bile acid treatment
All transfections were performed with 12,000 cells per well in 24-well plate (for luciferase assay) or scaled up in 6-well plate (for Western blot) using Lipofectamine 2000 (Invitrogen). The day before transfection, proliferating Caco-2 cells were seeded in antibiotics-free DMEM with 10% FBS. pcDNA6/His-IBABP was introduced into cells with FXRE-Luc reporter and pRL-CMV using Lipofectamine 2000 according to manufacturer's protocol. The plasmid load was 0.9 µg per well in a 24-well plate and 4.0 µg per well in a 6-well plate. The cells were incubated in OptiMEM (Invitrogen) containing the DNA-lipid complexes for 24 h, and then treated with 125 µM UDCA or control medium for 5 h, followed by addition of 25 µM CDCA or DCA alone or plus 125 µM UDCA for an additional 24 h. In bile acid treatment experiments, cells were fed with phenol red-free DMEM (Mediatech) supplemented with 10% charcoal-stripped fetal bovine serum (Hyclone).
RNA interference and bile acid treatment
Two days before siRNA transfection, Caco-2 cells were seeded in antibiotics-free DMEM with 10% FBS. pCDNA3.1-hFXRα2 was introduced into Caco-2 cells. After 24 h incubation in OptiMEM, IBABP-specific siRNAs were transfected into Caco-2 cells using Lipofectamine 2000 according to manufacturer's protocol; the final siRNA concentration was 40 nM. After 24 h incubation in OptiMEM, cells were transfected with FXRE-Luc reporter and pRL-CMV, and incubated for 24 h. The cells were then treated with 125 µM UDCA or control medium for 5 h, followed by addition of 25 µM CDCA alone or plus 125 µM UDCA for an additional 24 h in phenol red-free DMEM supplemented with 10% charcoal-stripped fetal bovine serum.
Determination of FXRα activity with a luciferase reporter construct
The activation of FXRα in cells was measured by luciferase reporter construct FXRE-Luc that contains coding sequence for firefly luciferase regulated by IBABP promoter. Renilla luciferase under CMV promoter control (pRL-CMV) was also included for normalization of transfection efficiency. After 24 h bile acid treatment, cells were harvested in 1× Passive Lysis Buffer (Promega), and firefly and Renilla luciferase activities were measured using the Dual-Luciferase Assay System with Veritas Microplate Luminometer (Promega) according to the manufacturer's manual.
Western blotting and quantitative PCR
The cellular proteins were extracted with CelLytic buffer (Sigma), and 50 µg of total proteins were separated in 10–20% Criterion SDS-PAGE gel (Bio-Rad). The protein level of IBABP was determined by Western blot with standard procedures. The protein level of β-actin was used a loading control. Cellular RNA was isolated using the RNeasy Mini Kit (Qiagen) according to manufacturer instruction. The mRNA expression of organic solute transporter α (OSTα) was measured by qPCR using Mx3000P qPCR system (Stratagene) and normalized to housekeeping gene ARPP0 as described previously (28). The qPCR primers for OSTα are 5′-TTG CTT GTT CGC CTC CCT ATT CCT C-3′ and 5′-GTC TTT CCT TCG GTA GTA CAT TCG TG-3′.
Statistical analysis
All values are reported as mean ± SEM. Data from multiple treatment groups were compared using two-way ANOVA with Bonferroni posttest. Data from two treatment groups were compared using unpaired t-test. A probability value of P < 0.05 was considered statistically significant.
RESULTS
UDCA binds to a single site on IBABP
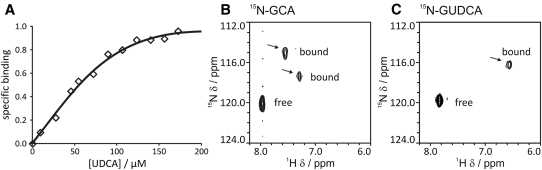
The ability of IBABP to bind bile acids found in humans is well established, but its binding to UDCA has never been tested. Binding between IBABP and UDCA was measured with tryptophan fluorescence spectroscopy. IBABP contains a single tryptophan, making interpretation of the binding data straightforward. Purified IBABP was titrated with UDCA in a stepwise manner, yielding a typical hyperbolic binding curve with Hill coefficient of 1, indicative of a single class of binding site(s) (Fig. 1A). The affinity of UDCA for IBABP is 63 µM. The glyco (G) and tauro (T) conjugates of UDCA, which are observed in humans treated with UDCA, have similar affinity (81 µM for GUDCA, and 56 µM for TUDCA). We also used ligand-observed 1H, 15N HSQC NMR spectroscopy to probe the number of binding sites for UDCA on IBABP. Changes to the 2D 1H, 15N correlation spectra of 15N-bile acids were monitored in response to addition of IBABP (Fig. 1B, C). Using the glyco conjugates of bile acids, two peaks of bound 15N-GCA are observed in the presence of IBABP (Fig. 1B), whereas only a single bound peak of 15N-GUDCA is detected (Fig. 1C). In conjunction with the experiments from tryptophan fluorescence, this finding suggests that IBABP contains a single binding site for UDCA.
Fig. 1.
UDCA binds a single site in IBABP. (A) Tryptophan fluorescence spectroscopy was used to quantify the binding affinity of UDCA to IBABP. IBABP (250 µl at 10 µM) was titrated in a stepwise manner with 1 µl increments of UDCA (5 mM). The specific binding measured by normalized changes in emission fluorescence is plotted against UDCA concentration. The hyperbolic binding curve indicates IBABP can bind UDCA in a single binding site. (B, C) The binding sites of UDCA and CA to IBABP were determined by ligand-observed 1H, 15N HSQC NMR spectroscopy. The contour plots of NMR spectra of 15N-GCA (B) or 15N-GUDCA (C) bound to IBABP at a three-to-one molar ratio are shown. Arrows indicate peaks corresponding to bound bile acids.
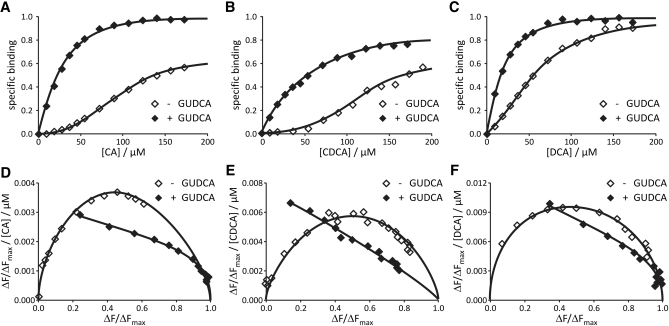
IBABP contains two binding sites for major human bile acids and binds them in a cooperative manner (21). Because UDCA appeared to bind only a single site on IBABP, we tested its effect on binding to major human bile acids. The binding experiments were conducted by saturating IBABP with GUDCA, and then titrating with other bile acids. GUDCA was used as the ligand because it is considered to be the active form of UDCA in humans (29). GUDCA shifted the binding curve between IBABP and other bile acids to the left (Fig. 2A–C), increasing the affinity of IBABP for major human bile acids by a factor of 2- to 5-fold (Table 1). In addition, when calculated with the Hill equation, we observe a 50–60% decrease in binding cooperativity of major human bile acids in the presence of UDCA (Table 2). This is consistent with the fact that UDCA already occupies one binding site on IBABP. To confirm the loss of cooperative binding, the data were also plotted according to the method of Scatchard (Fig. 2D–F). These plots show a convex downward curve in the absence of UDCA, with the ordinate of the apex ∼0.5, which equates to a Hill coefficient of 2, a value very similar to that obtained with the Hill equation (Table 2). However, in the presence of UDCA, the Scatchard plots for other bile acids lack convex curvature, indicating that binding is far less cooperative. The affinities and cooperativity of the major human bile acids for IBABP measured in these experiments are entirely consistent with values reported from published reports using different detection methods and kinetic models (20, 22).
Fig. 2.
UDCA increases the binding affinity and lowers the binding cooperativity of IBABP for major human bile acids. Tryptophan fluorescence spectroscopy was used to assess the effect of UDCA on binding affinity and cooperativity of major human bile acids. (A–C) IBABP (270 µl at 20 µM) containing GUDCA (200 µM) was titrated in a stepwise manner with CA, CDCA, or DCA (2.5 mM). The left-shift of the binding curve (closed diamonds) indicates that GUDCA increases the affinity for other bile acids. (D–F) Scatchard plot was used to identify binding cooperativity. ΔF/ΔFmax / [BA] was plotted against ΔF/ΔFmax. It yields a straight line for noncooperative binding, whereas cooperativity results in high curvature of the plot.
TABLE 1.
UDCA and its conjugates increase the binding affinity between IBABP and major human bile acids
| KD / µM | Control | + UDCA | + GUDCA | + TUDCA |
| CA | 177.1 | 23.3 | 28.0 | 28.9 |
| CDCA | 125.8 | 30.0 | 22.6 | 24.7 |
| DCA | 56.3 | 28.5 | 22.7 | 25.7 |
| GCA | 43.7 | 13.6 | 19.0 | 24.1 |
| GCDCA | 49.6 | 22.0 | 24.2 | 25.3 |
| GDCA | 38.9 | 18.6 | 23.2 | 20.0 |
| TCA | 31.5 | 11.7 | 12.1 | 13.8 |
| TCDCA | 25.3 | 18.6 | 17.3 | 20.9 |
| TDCA | 29.4 | 11.8 | 13.0 | 12.8 |
The binding affinity KD of CA, CDCA, DCA and their glycine (G) and taurine (T) conjugates with IBABP was calculated using the Hill equation.
TABLE 2.
UDCA and its conjugates decrease the binding cooperativity between IBABP and major human bile acids
| HN | Control | + UDCA | + GUDCA | + TUDCA |
| CA | 1.96 | 1.28 | 1.32 | 1.07 |
| CDCA | 1.99 | 1.46 | 1.31 | 1.36 |
| DCA | 1.93 | 1.43 | 1.52 | 1.30 |
| GCA | 2.02 | 1.56 | 1.26 | 1.13 |
| GCDCA | 1.89 | 1.41 | 1.43 | 1.33 |
| GDCA | 1.69 | 1.28 | 1.43 | 1.39 |
| TCA | 2.28 | 1.42 | 1.36 | 1.40 |
| TCDCA | 2.16 | 1.61 | 1.37 | 1.32 |
| TDCA | 2.04 | 1.45 | 1.31 | 1.26 |
The binding cooperativity HN of CA, CDCA, DCA and their glycine (G) and taurine (T) conjugates with IBABP was calculated using the Hill equation.
UDCA induces unique conformational changes in IBABP
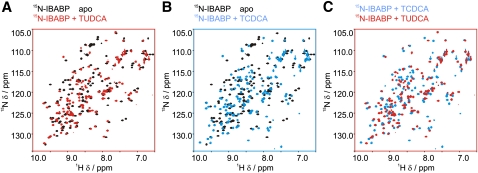
We used protein-observed 1H, 15N HSQC NMR spectroscopy to monitor structural changes of IBABP induced by the stereoisomers TUDCA and TCDCA. The 7-hydroxyl group in TCDCA is in the α-conformation, but in TUDCA, it is in the β-conformation. The HSQC spectra of IBABP were obtained in its apo state when bound to TUDCA and when bound to TCDCA. The spectra show that both bile acids induce substantial conformational changes in IBABP (Fig. 3A, B) and that the conformation of IBABP is significantly different when bound to TCDCA compared with TUDCA (Fig. 3C).
Fig. 3.
UDCA induces unique conformational changes in IBABP. Changes to the chemical shift of IBABP upon binding by bile acids were compared using protein-observed 1H, 15N HSQC NMR spectra. All complexes are formed by adding bile acids in 10-fold molar excess of [U-15N]IBABP (100 µM). (A) Overlay of apo-IBABP (black) and TUDCA-bound IBABP (red). (B) Overlay of apo-IBABP (black) and TCDCA-bound IBABP (blue). (C) Overlay of TCDCA-bound IBABP (blue) and TUDCA-bound IBABP (red).
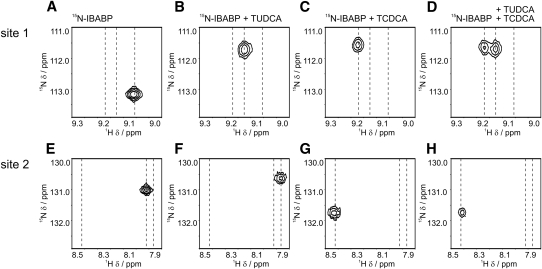
From the same HSQC spectra, we focused on resonances for an amino acid indicative of site 1 of IBABP (Gly66), which was previously reported to exhibit strong changes to the chemical shift upon binding of major human bile acids (30). A significant shift is observed upon binding of TUDCA (compare Fig. 4A, B), and TCDCA (compare Fig. 4A, C). These findings are consistent with the idea that this site (site 1) can bind either TUDCA or TCDCA. In contrast, we observed that an amino acid indicative of site 2 (Val37) undergoes only a minor chemical shift perturbation in response to TUDCA (compare 4E, F); however, it is perturbed upon binding of TCDCA (compare Fig. 4E, G). These findings are consistent with the idea that major human bile acids, but not UDCA, bind to site 2 on IBABP.
Fig. 4.
UDCA induces strong changes to the chemical shift at bile acid binding site 1. The chemical shifts of residues Gly66, reporting on site 1 (top panels), and Val37, reporting on site 2, (bottom panels) of IBABP were examined in detail in protein-observed 1H, 15N HSQC NMR spectra of different bile acid binding. The positions of these residues are shown in apo state (A, E), when bound to TUDCA (B, F), when bound to TCDCA (C, G), and when bound to TUDCA and TCDCA (D, H).
We also performed an experiment with NMR to determine if TUDCA could be displaced from IBABP by TCDCA. Gly66, the indicator of occupancy at binding at site 1, exhibits two resonances when both of these bile acids are present (compare Fig. 4A, D). These resonances are in equal portion, consistent with the conclusion that half the IBABP population has TUDCA at site 1 and the other half contains TCDCA. This displacement is independent of the order of the ligand addition because both experiments resulted in the same end state. In contrast, the resonance of Val37 is the same in the presence of TCDCA versus TCDCA and TUDCA (compare Fig. 4G, H). Taken together, these results show that, at these concentrations of TUDCA and TCDCA, two populations of IBABP are possible. One population contains TUDCA at site 1 and TCDCA at site 2. The other population contains TCDCA at both sites.
Activation of FXRα by major human bile acids is potentiated by UDCA
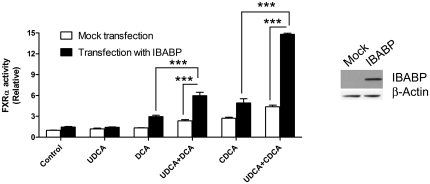
As we show that UDCA binds to IBABP and a recent study showed that IBABP interacts with FXRα (23), we hypothesized that IBABP may have a role in mediating the activation of FXRα by UDCA. This idea was tested in Caco-2 cells, which are commonly used as model colonocytes. CDCA was chosen as a representative bile acid for two reasons. One, it is the best bile acid agonist of FXRα (8–10). Two, it is the most abundant bile acid in human colon tissue and colon polyps (31, 32). In these experiments, we chose to preincubate the cells with UDCA because it is lipophobic and has a much slower diffusion rate than CDCA (33, 34). In our view, this preincubation is more likely to recapitulate the human clinical condition in which UDCA is administered daily to maintain its concentration at a relatively steady-state. These experiments were performed on both Caco-2 cells where IBABP cannot be detected by Western blot and on Caco-2 cells transfected with an expression vector encoding IBABP. UDCA increased the effect of DCA and CDCA on activation of FXRα in Caco-2 cells (Fig. 5, white bars). However, this effect was far more pronounced when IBABP was overexpressed (Fig. 5, black bars).
Fig. 5.
UDCA promotes activation of FXRα by bile acids when IBABP is present. Caco-2 cells were transfected with IBABP and treated with UDCA (125 µM) for 5 h. The cells were then treated with CDCA or DCA (25 µM) for 24 h. The relative FXRα activity was determined using a Luciferase reporter assay. Values are the average ± SEM of three independent experiments. ***P < 0.001. The expression levels of the transfected IBABP are shown by Western blot using β-actin as a loading control.
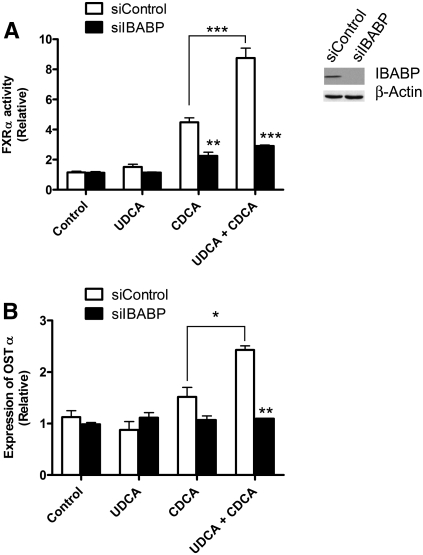
To confirm that IBABP is necessary for the enhanced activation of FXRα by UDCA, the expression of endogenous IBABP was suppressed with RNAi. Caco-2 cells have no detectable IBABP by Western blot (Fig. 5). Because IBABP is a target gene of FXRα, the cells were transfected with an FXRα construct to increase the endogenous expression of IBABP (Fig. 6). The cells were then treated with IBABP-specific siRNA to knock down the induced IBABP. This knockdown eliminated the ability of UDCA to potentiate the activation of FXRα by CDCA. These effects are indicated by changes in the activity of the Luciferase reporter (Fig. 6A), and by changes to the expression of the OSTα, a target gene of FXRα (Fig. 6B). The expression levels of FXRα and RXRα remained constant in these studies (not shown), proving that the primary effect on activation results from a reduction in IBABP.
Fig. 6.
IBABP is necessary for the full activation of FXRα by CDCA in Caco-2 cells. Caco-2 cells were treated for 24 h with siRNA targeting IBABP (siIBABP) or scrambled siRNA (siControl). Cells were then treated with UDCA (125 µM) for 5 h followed by incubation with CDCA (25 µM) for 24 h. (A) Activation of FXRα was measured by Luciferase reporter assay. (B) Expression level of the FXRα target gene OSTα was determined by qPCR. Values are the average ± SEM of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. The knockdown of endogenous IBABP is shown by Western blot using β-actin as a loading control.
DISCUSSION
The results of the study support the following conclusions: i) UDCA binds to a single site on IBABP; ii) occupation of this site by UDCA increases the affinity of a second binding site for major human bile acids; iii) UDCA augments the activation of FXRα by major human bile acids; and iv) this augmentation requires IBABP.
The primary finding of this study is that UDCA binds to a single site on IBABP. In a series of papers published by Cistola's group, IBABP has been shown to have two bile acid binding sites. The selectivity and cooperativity of these sites are governed by the hydroxylation pattern on steroid rings of major human bile acids (21, 22, 24, 35). Here, we found that UDCA binds only one of the bile acid binding sites in IBABP. This conclusion is strongly supported by the binding isotherms evident in tryptophan fluorescence studies, which show that UDCA binds to a single class of sites. The conclusion is further substantiated by the ligand-observed NMR spectrum from 15N-GUDCA bound to IBABP, which shows a single peak corresponding to bound GUDCA. Additional support for the presence of only a single binding site for UDCA can be taken from the fact that TUDCA has little influence on protein resonance shift perturbation of an amino acid that is an indicator of occupancy at site 2. Because CDCA induced shifts in site 2, the lack of resonance shifts upon UDCA binding are consistent with the conclusion that it binds at only one site on IBABP. Finally, the fact that TUDCA fails to displace TCDCA bound to site 2 also supports the idea that UDCA cannot bind to site 2.
The second important finding of this study is that occupation of site 1 by UDCA increases the affinity of IBABP at the second binding site for the major human bile acids by 2- to 5-fold. In patients treated with UDCA, the concentration of this bile acid in fecal water is ∼50 μM (36), which is near the affinity we report here. Therefore, in patients treated with UDCA, one would expect a substantial fraction of the IBABP to have UDCA at site 1. Although we observed displacement of TUDCA from site 1 by TCDCA, those experiments were conducted at equimolar concentrations of each bile acid and at concentrations above their KD. In patients treated with UDCA, its levels in fecal water are 2-fold higher than DCA levels (37), which has a similar affinity for the protein. Consequently, in patients treated with UDCA, it is likely that much of the IBABP has UDCA at site 1 and another bile acid at site 2.
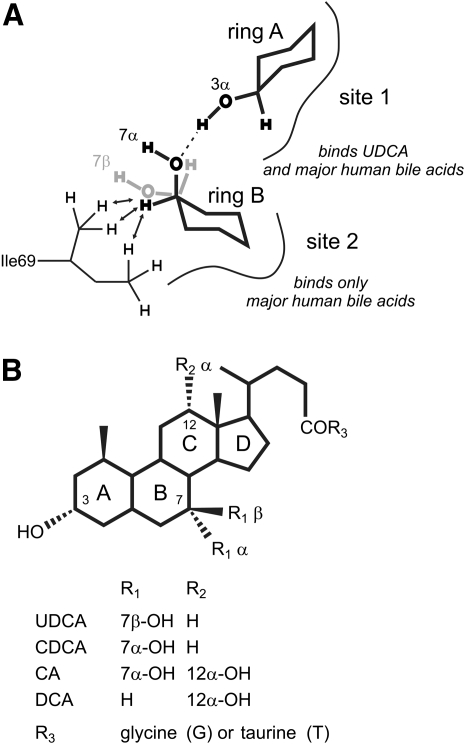
We constructed a model of IBABP bound to CDCA (Fig. 7A) based on the structure of chicken liver bile acid binding protein (LBABP), bound to two cholic acids (38), to provide structural basis for the selectivity of UDCA for binding site 1. The model shows that the 7α-OH of the steroid ring of CDCA, occupying binding site 2, makes two important contacts. One contact is a hydrogen bond with the 3-OH of CDCA at site 1. The second contact includes Van der Waals interactions between the 7β-H and the two methyl groups of Ile69 in IBABP. If CDCA at site 2 is replaced with UDCA, the 7-OH group is in the β rather than the α conformation, so neither of these key contacts can be made. In addition, Cistola's group showed that bile acids lacking a 12-OH preferentially bind to site 1 (35). As UDCA lacks a 12-OH, it falls into this category.
Fig. 7.
Model of CDCA bound to IBABP. (A) Model of the binding of two CDCA molecules to human IBABP based on structures of chicken LBABP in complex with two bile acids (1TW4.pdb, 2JU3.pdb). For clarity, we only show the steroid ring A of CDCA at site 1 and ring B of CDCA at site 2. The 7α-OH group of CDCA at site 2 forms a hydrogen bond (dashed line) to the 3α-OH group of the bile acid at site 1. In addition, the ring B 7β-H of CDCA at site 2 interacts with Ile69 in IBABP by Van der Waals forces (arrows). The stereoisomer, UDCA with 7β-OH (gray), cannot form these two interactions. (B) Chemical structure of UDCA, CDCA, CA, and DCA.
Importantly though, when UDCA is bound at site 1, the 3-OH group of its steroid ring is in the same conformation as major human bile acids. Therefore, UDCA can still engage in cooperative binding with major human bile acids at site 2. In this case, a hydrogen bond can form between the 3-OH of UDCA and the 7-OH or 12-OH group of the bile acid at site 2. These hydrogen bonds are illustrated in Fig. 7 and supplementary Fig. I. This conclusion is also consistent with the work of Cistola's group, which showed that binding cooperativity is governed by patterns of hydroxylation in the steroid B- and C-rings of bile acids (21).
Interestingly, binding cooperativity is also determined by two amino acid residues, 99 and 101, located in the inner cavity of IBABP (39). Chicken IBABP (H99/A101) has a rigid H-bond pattern and disfavors conformational flexibility needed for coupling between the two sites, and indeed, chicken IBABP shows noncooperative binding of bile acids (39). On the other hand, human IBABP (A99/S101) has an extended H-bond network, which allows cooperative bile acid binding. Although the H-bond network does not appear to affect the bile acid selectivity, the presence of UDCA in site 1 is likely be communicated through this H-bond network.
The third important observation of this study is that UDCA augments the activation of FXRα by major human bile acids. Our results show that IBABP mediates UDCA's enhancement of activation of FXRα by other bile acids in cells. However, the mechanism by which IBABP bridges UDCA and FXRα is not entirely clear. A prior report shows that IBABP directly interacts with FXRα (23), so this binding could enable bile acid transfer from IBABP to FXRα in the nucleus. Other proteins from the lipid binding protein family appear to act in this manner. For example, cellular retinoic acid binding protein II (CRABP II) transfers retinoic acid to the retinoic acid receptor and directly binds to this nuclear receptor (40). Similarly, liver fatty acid binding protein (LFABP) increases the intracellular levels of fatty acids and directly binds to peroxisome proliferator-activated receptor (PPAR) (41, 42).
The findings presented here are counter to the report of Campana et al. (43), who concluded that UDCA could compete for the binding of CDCA to FXRα and thereby block the transcriptional response of FXRα to CDCA. There are several potential explanations for the discrepancy in the two studies. One, the conclusion that FXRα binds to UDCA comes from binding studies performed on cell lysates rather than purified protein, and it runs counter to three other reports (8–10). Two, we interpret the binding isotherms of Campana et al. (43) to indicate that CDCA actually promotes binding of UDCA to proteins in the lysate, rather than inhibiting binding. If this is the case and the binding in the lysate is attributed to IBABP, then this result would be consistent with our observations. Three, and most significantly, Campana et al. used different concentrations of bile acids and different treatment times when measuring the effect of UDCA on the transcriptional response of FXRα. This is an important point, and it may have clinical relevance. It is entirely conceivable that the effects of UDCA on cells are dose-dependent and that there may be multiple effects. Low concentrations of UDCA, which are not toxic and do not appreciably change the hydrophobicity of the extracellular milieu, are likely to potentiate the stimulation of FXRα by other bile acids (this study). At higher concentrations, UDCA may have altogether different effects, like those observed by Campana et al. (43). Consequently, it may ultimately prove useful to monitor the FXRα response in patients treated with UDCA to determine whether there is a more appropriate dosing regimen.
The discrepancy between our conclusions and those of Campana et al. (43) also underscore the conflicting views on the ability of UDCA to bind directly to FXRα. Only one study has reported direct binding between UDCA and FXRα (18). In that report, the affinity was measured by an in vitro scintillation proximity assay. Weak binding between UDCA and FXRα was detected (affinity of ∼185 μM). Importantly though, in the same study, the affinity of CDCA for FXRα was reported to be 7-fold higher than that reported in other publications (44), so scintillation proximity appears to overestimate affinity. Furthermore, UDCA also fails to recruit coactivators for FXRα (10, 18), and it fails to activate FXRα in cultured cells (8–10). Altogether, the overwhelming body of evidence indicates that UDCA is unlikely to directly bind to and activate FXRα and supports our conclusion that IBABP is a necessary bridge between these two molecules.
To piece together a precise mechanism of action for IBABP, we will need to know the intracellular concentrations and movement of bile acids, especially in the presence of clinically relevant levels of UDCA. However, technical challenges currently preclude the measurement of intracellular bile acid concentrations and the tracking of bile acid movement in cells, especially combinations of bile acids. Although fluorescently labeled bile acids have been synthesized, their value as intracellular probes is uncertain because they are transported differently than natural bile acids and because their choleretic properties differ (45–47). To our best knowledge, there is no report on the measurement of intracellular bile acid concentration in intestinal cells. Consequently, these measures remain at the forefront of key hurdles to overcome to enable a complete understanding of the cellular function of IBABP.
Given the findings of this study, any interpretation of the therapeutic effects of UDCA should take IBABP into account. As outlined above, the concentration of UDCA in fecal water is near its KD for IBABP, so a significant fraction of IBABP is occupied by this bile acid at its therapeutic concentration. On the basis of the observations presented here, UDCA bound to IBABP is likely to modulate the activity of FXRα in ileocytes and thus enhance the excretion of major human bile acids (7, 48). In addition, UDCA may increase the buffering capacity of IBABP in colonocytes. Due to the efficient absorption at distal small intestine, colon bile acids are present at low concentration (49), and IBABP is not fully bound by major human bile acids due to their low affinity. UDCA increases the binding affinity of major human bile acids, thus reducing the levels of free bile acids in colonocytes. This would reduce cytotoxic stress placed on the gastrointestinal system. In colorectal cancer, this effect would protect against bile acid-induced mutations in the genome (50) and the acquisition of bile acid resistance, a property that is essential for disease progression (50). Although the clinical effects of UDCA have been attributed to its ability to reduce the hydrophobicity and, therefore, cytotoxicity of the systemic bile acid pool (6), the stimulation of FXRα could be another reason for its clinical benefit.
Supplementary Material
Acknowledgments
The authors thank Dr. Russell Dahl at our Medicinal Chemistry facility for synthesizing 15N labeled bile acids. The authors also thank Christina Niemeyer for editorial assistance with this manuscript.
Footnotes
Abbreviations:
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- DCA
- deoxycholic acid
- FXRα
- farnesoid X receptor α
- GUDCA
- glyco conjugate of UDCA
- IBABP
- ileal bile acid binding protein
- OSTα
- organic solute transporter α
- RXRα
- retinoid X receptor α
- TUDCA
- tauro conjugate of UDCA
- UDCA
- ursodeoxycholic acid
This work was supported by National Institutes of Health grants R21 CA-116329, R01 CA-108959, and R01 CA-135300. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Beuers U. 2006. Drug insight: mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3: 318–328 [DOI] [PubMed] [Google Scholar]
- 2.LaRusso N. F., Shneider B. L., Black D., Gores G. J., James S. P., Doo E., Hoofnagle J. H. 2006. Primary sclerosing cholangitis: summary of a workshop. Hepatology. 44: 746–764 [DOI] [PubMed] [Google Scholar]
- 3.Tung B. Y., Emond M. J., Haggitt R. C., Bronner M. P., Kimmey M. B., Kowdley K. V., Brentnall T. A. 2001. Ursodiol use is associated with lower prevalence of colonic neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis. Ann. Intern. Med. 134: 89–95 [DOI] [PubMed] [Google Scholar]
- 4.Pardi D. S., Loftus E. V. J., Kremers W. K., Keach J., Lindor K. D. 2003. Ursodeoxycholic acid as a chemopreventive agent in patients with ulcerative colitis and primary sclerosing cholangitis. Gastroenterology. 124: 889–893 [DOI] [PubMed] [Google Scholar]
- 5.Alberts D. S., Martinez M. E., Hess L. M., Einspahr J. G., Green S. B., Bhattacharyya A. K., Guillen J., Krutzsch M., Batta A. K., Salen G., et al. 2005. Phase III trial of ursodeoxycholic acid to prevent colorectal adenoma recurrence. J. Natl. Cancer Inst. 97: 846–853 [DOI] [PubMed] [Google Scholar]
- 6.Hofmann A. F. 2007. Biliary secretion and excretion in health and disease: current concepts. Ann. Hepatol. 6: 15–27 [PubMed] [Google Scholar]
- 7.Hofmann A. F. 2003. Inappropriate ileal conservation of bile acids in cholestatic liver disease: homeostasis gone awry. Gut. 52: 1239–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553 [DOI] [PubMed] [Google Scholar]
- 9.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365 [DOI] [PubMed] [Google Scholar]
- 10.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368 [DOI] [PubMed] [Google Scholar]
- 11.Lee F. Y., Lee H., Hubbert M. L., Edwards P. A., Zhang Y. 2006. FXR, a multipurpose nuclear receptor. Trends Biochem. Sci. 31: 572–580 [DOI] [PubMed] [Google Scholar]
- 12.Trauner M. 2004. The nuclear bile acid receptor FXR as a novel therapeutic target in cholestatic liver diseases: hype or hope? Hepatology. 40: 260–263 [DOI] [PubMed] [Google Scholar]
- 13.Yang F., Huang X., Yi T., Yen Y., Moore D. D., Huang W. 2007. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67: 863–867 [DOI] [PubMed] [Google Scholar]
- 14.Kim I., Morimura K., Shah Y., Yang Q., Ward J. M., Gonzalez F. J. 2007. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 28: 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gottardi A., Touri F., Maurer C. A., Perez A., Maurhofer O., Ventre G., Bentzen C. L., Bentzen E. J., Dufour J-F. 2004. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig. Dis. Sci. 49: 982–989 [DOI] [PubMed] [Google Scholar]
- 16.Modica S., Murzilli S., Salvatore L., Schmidt D. R., Moschetta A. 2008. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 68: 9589–9594 [DOI] [PubMed] [Google Scholar]
- 17.Maran R. R. M., Thomas A., Roth M., Sheng Z., Esterly N., Pinson D., Gao X., Zhang Y., Ganapathy V., Gonzalez F. J., et al. 2009. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 328: 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lew J-L., Zhao A., Yu J., Huang L., de Pedro N., Pelaez F., Wright S. D., Cui J. 2004. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 279: 8856–8861 [DOI] [PubMed] [Google Scholar]
- 19.Zimmerman A. W., Veerkamp J. H. 2002. New insights into the structure and function of fatty acid-binding proteins. Cell. Mol. Life Sci. 59: 1096–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman A. W., van Moerkerk H. T., Veerkamp J. H. 2001. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 33: 865–876 [DOI] [PubMed] [Google Scholar]
- 21.Tochtrop G. P., Bruns J. L., Tang C., Covey D. F., Cistola D. P. 2003. Steroid ring hydroxylation patterns govern cooperativity in human bile acid binding protein. Biochemistry. 42: 11561–11567 [DOI] [PubMed] [Google Scholar]
- 22.Toke O., Monsey J. D., DeKoster G. T., Tochtrop G. P., Tang C., Cistola D. P. 2006. Determinants of cooperativity and site selectivity in human ileal bile acid binding protein. Biochemistry. 45: 727–737 [DOI] [PubMed] [Google Scholar]
- 23.Nakahara M., Furuya N., Takagaki K., Sugaya T., Hirota K., Fukamizu A., Kanda T., Fujii H., Sato R. 2005. Ileal bile acid-binding protein, functionally associated with the farnesoid X receptor or the ileal bile acid transporter, regulates bile acid activity in the small intestine. J. Biol. Chem. 280: 42283–42289 [DOI] [PubMed] [Google Scholar]
- 24.Tochtrop G. P., Richter K., Tang C., Toner J. J., Covey D. F., Cistola D. P. 2002. Energetics by NMR: site-specific binding in a positively cooperative system. Proc. Natl. Acad. Sci. USA. 99: 1847–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momose T., Tsubaki T., Iida T., Nambara T. 1997. An improved synthesis of taurine- and glycine-conjugated bile acids. Lipids. 32: 775–778 [DOI] [PubMed] [Google Scholar]
- 26.Barley N. F., Taylor V., Shaw-Smith C. J., Chakravarty P., Howard A., Legon S., Walters J. R. F. 2003. Human ileal bile acid-binding protein promoter and the effects of CDX2. Biochim. Biophys. Acta. 1630: 138–143 [DOI] [PubMed] [Google Scholar]
- 27.Lee H., Zhang Y., Lee F. Y., Nelson S. F., Gonzalez F. J., Edwards P. A. 2006. FXR regulates organic solute transporters α and β in the adrenal gland, kidney, and intestine. J. Lipid Res. 47: 201–214 [DOI] [PubMed] [Google Scholar]
- 28.Fang C., Dean J., Smith J. W. 2007. A novel variant of ileal bile acid binding protein is up-regulated through NF-κB activation in colorectal adenocarcinoma. Cancer Res. 67: 9039–9046 [DOI] [PubMed] [Google Scholar]
- 29.Paumgartner G., Beuers U. 2002. Ursodeoxycholic acid in cholestatic liver disease: mechanisms of action and therapeutic use revisited. Hepatology. 36: 525–531 [DOI] [PubMed] [Google Scholar]
- 30.Kurz M., Brachvogel V., Matter H., Stengelin S., Thüring H., Kramer W. 2003. Insights into the bile acid transportation system: the human ileal lipid-binding protein-cholyltaurine complex and its comparison with homologous structures. Proteins. 50: 312–328 [DOI] [PubMed] [Google Scholar]
- 31.Nakashima T., Seto Y., Nakajima T., Shima T., Sakamoto Y., Sano A., Takino T. 1989. Distribution of tissue bile acids in the human alimentary tract and colon polyps. Jpn. J. Med. 28: 25–29 [DOI] [PubMed] [Google Scholar]
- 32.Gelb A. M., McSherry C. K., Sadowsky J. R., Mosbach E. H. 1982. Tissue bile acids in patients with colon cancer and colonic polyps. Am. J. Gastroenterol. 77: 314–317 [PubMed] [Google Scholar]
- 33.Hofmann M., Zgouras D., Samaras P., Schumann C., Henzel K., Zimmer G., Leuschner U. 1999. Small and large unilamellar vesicle membranes as model system for bile acid diffusion in hepatocytes. Arch. Biochem. Biophys. 368: 198–206 [DOI] [PubMed] [Google Scholar]
- 34.Aldini R., Roda A., Montagnani M., Cerre C., Pellicciari R., Roda E. 1996. Relationship between structure and intestinal absorption of bile acids with a steroid or side-chain modification. Steroids. 61: 590–597 [DOI] [PubMed] [Google Scholar]
- 35.Tochtrop G. P., DeKoster G. T., Covey D. F., Cistola D. P. 2004. A single hydroxyl group governs ligand site selectivity in human ileal bile acid binding protein. J. Am. Chem. Soc. 126: 11024–11029 [DOI] [PubMed] [Google Scholar]
- 36.van Gorkom B. A., van der Meer R., Boersma-van Ek W., Termont D. S., de Vries E. G., Kleibeuker J. H. 2002. Changes in bile acid composition and effect on cytolytic activity of fecal water by ursodeoxycholic acid administration: a placebo-controlled cross-over intervention trial in healthy volunteers. Scand. J. Gastroenterol. 37: 965–971 [DOI] [PubMed] [Google Scholar]
- 37.Hess L. M., Krutzsch M. F., Guillen J., Chow H-H. S., Einspahr J., Batta A. K., Salen G., Reid M. E., Earnest D. L., Alberts D. S. 2004. Results of a phase I multiple-dose clinical study of ursodeoxycholic acid. Cancer Epidemiol. Biomarkers Prev. 13: 861–867 [PubMed] [Google Scholar]
- 38.Nichesola D., Perduca M., Capaldi S., Carrizo M. E., Righetti P. G., Monaco H. L. 2004. Crystal structure of chicken liver basic fatty acid-binding protein complexed with cholic acid. Biochemistry. 43: 14072–14079 [DOI] [PubMed] [Google Scholar]
- 39.Zanzoni S., Assfalg M., Giorgetti A., D'Onofrio M., Molinari H. 2011. Structural requirements for cooperativity in ileal bile acid-binding proteins. J. Biol. Chem. 286: 39307–39317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong D., Ruuska S. E., Levinthal D. J., Noy N. 1999. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 274: 23695–23698 [DOI] [PubMed] [Google Scholar]
- 41.Huang H., Starodub O., McIntosh A., Kier A. B., Schroeder F. 2002. Liver fatty acid-binding protein targets fatty acids to the nucleus. Real time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 277: 29139–29151 [DOI] [PubMed] [Google Scholar]
- 42.Huang H., Starodub O., McIntosh A., Atshaves B. P., Woldegiorgis G., Kier A. B., Schroeder F. 2004. Liver fatty acid-binding protein colocalizes with peroxisome proliferator activated receptor α and enhances ligand distribution to nuclei of living cells. Biochemistry. 43: 2484–2500 [DOI] [PubMed] [Google Scholar]
- 43.Campana G., Pasini P., Roda A., Spampinato S. 2005. Regulation of ileal bile acid-binding protein expression in Caco-2 cells by ursodeoxycholic acid: role of the farnesoid X receptor. Biochem. Pharmacol. 69: 1755–1763 [DOI] [PubMed] [Google Scholar]
- 44.Gardès C., Blum D., Bleicher K., Chaput E., Ebeling M., Hartman P., Handschin C., Richter H., Benson G. M. 2011. Studies in mice, hamsters, and rats demonstrate that repression of hepatic apoA-I expression by taurocholic acid in mice is not mediated by the farnesoid-X-receptor. J. Lipid Res. 52: 1188–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holzinger F., Schteingart C. D., Ton-Nu H., Eming S. A., Monte M. J., Hagey L. R., Hofmann A. F. 1997. Fluorescent bile acid derivatives: relationship between chemical structure and hepatic and intestinal transport in the rat. Hepatology. 26: 1263–1271 [DOI] [PubMed] [Google Scholar]
- 46.Holzinger F., Schteingart C. D., Ton-Nu H-T., Cerrè C., Steinbach J. H., Yeh H-Z., Hofmann A. F. 1998. Transport of fluorescent bile acids by the isolated perfused rat liver: kinetics, sequestration, and mobilization. Hepatology. 28: 510–520 [DOI] [PubMed] [Google Scholar]
- 47.Monte M. J., Rosales R., Macias R. I. R., Iannota V., Martinez-Fernandez A., Romero M. R., Hofmann A. F., Marin J. J. G. 2008. Cytosol-nucleus traffic and colocalization with FXR of conjugated bile acids in rat hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G54–G62 [DOI] [PubMed] [Google Scholar]
- 48.Lanzini A., De Tavonatti M. G., Panarotto B., Scalia S., Mora A., Benini F., Baisini O., Lanzarotto F. 2003. Intestinal absorption of the bile acid analogue 75Se-homocholic acid-taurine is increased in primary biliary cirrhosis, and reverts to normal during ursodeoxycholic acid administration. Gut. 52: 1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton J. P., Xie G., Raufman J-P., Hogan S., Griffin T. L., Packard C. A., Chatfield D. A., Hagey L. R., Steinbach J. H., Hofmann A. F. 2007. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G256–G263 [DOI] [PubMed] [Google Scholar]
- 50.Bernstein H., Bernstein C., Payne C. M., Dvorakova K., Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589: 47–65 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.