Abstract
Kr-pok (kidney cancer-related POZ domain and Krüppel-like protein) is a new proto-oncogenic POZ-domain transcription factor. Fatty acid synthase gene (FASN) encodes one of the key enzymes in fatty acids synthesis and is the only enzyme that synthesizes fatty acids in cancer cells. Sp1 and SREBP-1c are the two major transcription activators of FASN. We investigated whether Kr-pok modulates transcription of the FASN. FASN expression is significantly decreased in Kr-pok knockout murine embryonic fibroblasts. Coimmunoprecipitation, GST fusion protein pull-down, and immunocytochemistry assays show that the zinc-finger domain of Kr-pok interacts directly with the bZIP DNA binding domain of SREBP-1. Electrophoretic mobility shift assay, oligonucleotide pull-down, and chromatin immunoprecipitation assays showed that Kr-pok changes the transcription factor binding dynamics of Sp1 and SREBP-1c to the SRE/E-box elements of the proximal promoter. We found that Kr-pok expression increased during 3T3-L1 preadipocyte differentiation and that FASN expression is decreased by the knockdown of Kr-pok. Kr-pok facilitates the SREBP-1c-mediated preadipocyte differentiation and/or fatty acid synthesis. Kr-pok may act as an important regulator of fatty acid synthesis and may induce rapid cancer cell proliferation by increasing palmitate synthesis.
Keywords: kidney cancer-related POZ domain and Krüppel-like protein, sterol responsive element binding protein-1c, specificity protein 1, fatty acid synthase, transcription, oncoprotein, lipid, fatty acid
Fatty acid synthase (FASN), a 250- to 270-kDa cytosolic protein and one of the main lipogenic enzymes in mammals, catalyzes reactions that contribute to the conversion of acetyl-CoA and malonyl-CoA to palmitate (1–4). FASN gene transcription is under tight nutritional and hormonal control in lipogenic tissues, such as the liver and white adipose tissue. The transcription factors for this gene, namely specificity proteins 1 and 3 (Sp1 and Sp3), nuclear factor Y (NF-Y), and sterol regulatory element binding protein-1 (SREBP-1), have cognate binding sites on the proximal promoter of the FASN (5–7). Additionally, tumor-associated FASN not only functions as a key component of the anabolic energy-storage pathway but also confers growth and survival advantages to most human cancers, such as prostate, breast, ovarian, endometrial, colorectal, lung, stomach, and skin cancers (8–10). FASN plays an important role in cancer cell proliferation by providing a cell membrane lipid component that is needed for rapid cell growth. Some anti-cancer drugs target FASN, aiming to repress FASN expression or inhibit FASN enzyme activity. An increase in FASN expression is part of the overall genetic reprogramming of cancer cells, as evidenced by the concomitant increase in other SREBP-1c-regulated enzymes of the lipogenic pathways (4).
SREBPs are a family of basic helix-loop-helix leucine zipper transcription factors that are synthesized as inactive precursor proteins and are anchored to the ER (endoplasmic reticulum) membrane (11–13). SREBPs interact with SCAP (SREBP cleavage-activating protein), which is retained in the ER by Insig protein (14). The SCAP-SREBP-Insig complex is stabilized by cholesterol. When sterol levels are low, the SCAP-SREBP complex is released from Insig and moves to the Golgi, where the N-terminus of SREBP is cleaved by proteolysis and translocated to the nucleus. Activated SREBPs, by binding to the SRE elements, increase the transcription of many genes involved in cholesterol and fatty acid synthesis. There are three isoforms of SREBPs: SREBP-1a, SREBP-1c, and SREBP-2. SREBP-1a and SREBP-1c are transcribed from the same SREBP-1 gene, but each is driven by a distinct promoter. SREBP-2 is encoded by a separate gene, SREBP-2, which encodes a single mRNA (13, 15 and references therein).
The POZ domain-containing proteins play various cellular regulatory roles by interacting with regulatory proteins or by controlling transcription of target gene expression (16–18). In particular, interactions between some of the POK family proteins and co-regulators are major determinants in differentiation, development, hematopoiesis, tumor suppression, and oncogenesis (19–31). FBI-1, one of the POK family transcription factors, was recently identified as a potential determinant of adipocyte differentiation (25–27). Kr-pok, a recently characterized proto-oncoprotein, contains a POZ-domain and four Krüppel-like ZFs that are similar to those of FBI-1 in two key functional domains: the POZ-domain (81% similarity) and the four Krüppel-like ZFs (88% similarity) (supplementary Fig. I) (28, 29). Accordingly, Kr-pok is expected to have some properties comparable to those of FBI-1, such as adipocyte differentiation and cell proliferation.
In this study, we found that Kr-pok and SREBP-1c interact to synergistically activate FASN expression. Kr-pok changes the binding dynamics of SREBP-1c and Sp1 at the core regulatory elements of the FASN promoter, which results in the transcriptional up-regulation of FASN. Kr-pok may be one of the key regulators of fatty acid synthesis and cancer cell proliferation.
MATERIALS AND METHODS
Cell culture
Stable HEK293T-Rex-Kr-pok cells, which are inducible by doxycycline, were prepared by transfecting mammalian Flp-InTM T-RExTM host HEK293 cells with pOG44 and pcDNA5/FRT/TO©-Kr-pok plasmids and selecting with hygromycin and blasticidin (Invitrogen, Carlsbad, CA). To prepare Kr-pok+/+ and Kr-pok−/− mouse embryonic fibroblasts (MEFs), pregnant female Kr-pok+/− mice were mated with Kr-pok+/− male mice and euthanized at 13.5 days post coitum. The embryos were homogenized, treated with a trypsin-EDTA solution, and placed in a CO2 incubator for 6 h. Fresh DMEM was added to the culture media, and embryo homogenates were continuously incubated to obtain MEFs. HEK293A, HCT116, and LNcaP cells were cultured in DMEM (Gibco-BRL, Gaithersburg, MD) supplemented with 10% FBS (Gibco-BRL).
Plasmid preparation
Various fatty acid synthase promoter-luciferase reporter gene plasmids were provided by Dr. Kyung-Sup Kim of the Yonsei University School of Medicine (Seoul, Korea) and Dr. Timothy F. Osborne of the Sanford/Burnham Medical Research Institute (Orlando, FL). pcDNA3.1-Kr-pok-Myc-His and pcDNA3.0-FLAG-Kr-pok plasmids were prepared by cloning mouse brain cDNA fragments into pcDNA3.1 or pcDNA3.0 plasmids (Invitrogen). pcDNA3.1-Myc-SREBP-1c was prepared by cloning pCMV-SREBP-1c into the pcDNA3.1 plasmid (Invitrogen). To prepare recombinant GST-POZ Kr-pok, GST-ZF Kr-pok, and GST-bZIP SREBP-1 proteins, cDNA fragments encoding Kr-pok POZ, Kr-pok zinc fingers, and the SREBP-1 bZIP domain, respectively, were cloned into pGEX4T3 (Amersham Biosciences, Piscataway, NJ). All plasmid constructs were verified by sequencing.
Total RNA isolation and RT-PCR or quantitative PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen). cDNA was synthesized using 5 μg of total RNA, random hexamers (10 pmol), and superscript reverse transcriptase II (200 units) in a total volume of 20 μl using a reverse transcription kit (Invitrogen). PCR was performed using the following amplification conditions: 94°C denaturation for 3 min, 30 cycles of amplification reaction (94°C for 30 s, 55°C for 30 s, 72°C for 30 s), and a final extension reaction at 72°C for 5 min. Quantitative PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) and ABI PRISM 7300 RT-PCR System (Applied Biosystems). GAPDH mRNA was used as a control.
Western blot analysis
Cells were harvested and lysed in RIPA buffer. Cell extracts (30 µg) were separated using 12% SDS-PAGE, transferred to an Immun-BlotTM PVDF Membrane (Bio-Rad, Hercules, CA), and blocked in 5% skim milk (BD Biosciences, Sparks, MD). Blotted membranes were incubated with antibodies against FLAG-tag (Abcam, Cambridge, UK), GAPDH (Chemicon, Temecula, CA), SREBP-1, Sp1, and Myc-Tag (SantaCruz Biotech, Santa Cruz, CA) and then incubated with HRP-conjugated mouse, rabbit, or goat IgGs (Vector Laboratory, Burlingame, CA). A rabbit polyclonal antibody against Kr-pok by us as reported elsewhere (29). Protein bands were visualized with an ECL solution (PerkinElmer, Waltham, MA).
Determination of FASN enzyme activity
Cells (HCT116, LNcaP) infected with the recombinant adenovirus overexpressing Kr-pok, the shRNA Kr-pok knock-down, and the control adenovirus were cultured for 2 days, harvested, and lysed in RIPA buffer. FASN enzyme activity was analyzed by spectrophotometrically measuring the oxidation of NADPH by the conversion of Malonyl-CoA to palmitate. The reaction was initiated by adding 100 μg of cell lysates to the assay mixture (0.1 M potassium phosphate buffer [pH 7.0], 30 µM Acetyl-CoA, 100 µM Malonyl-CoA, 0.1 mM NADPH, and 1 mM EDTA). The decrease in absorbance at 340 nm was monitored over 30 min. The oxidation of cell lysates was background corrected for NADPH oxidation in the presence of only Malonyl-CoA and Acetyl-CoA.
Immunoprecipitation assays
Cells were washed, pelleted, and resuspended in a lysis buffer that was supplemented with protease inhibitors. The cell lysate was precleared, and the supernatant was incubated with antibodies on a rotating platform at 4°C overnight, followed by incubation with protein A-Sepharose Fast Flow beads. The beads were collected, washed, and resuspended in equal volumes of 5× SDS loading buffer. Immunoprecipitated proteins were separated with 12% SDS-PAGE. The Western blot assay was performed as described above.
GST fusion protein purification, in vitro transcription and translation, and GST fusion protein pull-down assays
Recombinant GST, GST-POZ Kr-pok, GST-ZF Kr-pok, and GST-bZIP SREBP-1 fusion proteins were prepared from Escherichia coli BL21 (DE3) cells grown overnight at 18°C in medium containing 0.2 mM IPTG. The E. coli were lysed and purified using glutathione-agarose 4 bead affinity chromatography (Peptron, Taejeon, Korea). The purified proteins were then resolved with 12% SDS-PAGE to quantitate and assess purity. Kr-pok and SREBP-1c polypeptides were prepared using the TNT extract in the presence of [35S]methionine (Promega, Madison, WI). GST fusion protein-agarose bead complexes were incubated with in vitro translated [35S]methionine (1175.0 Ci/mol) labeled Kr-pok or SREBP-1c polypeptides at 4°C for 4 h in HEMG buffer. The reaction mixtures were centrifuged, the pellets were rinsed, and the bound proteins were separated using 12% SDS-PAGE. The gels were then exposed to X-ray film (Kodak, Rochester, NY).
Immunostaining and cellular localization of Kr-pok and SREBP-1c
HEK293A cells were grown on coverslips placed in a culture dish. The cells were then transfected with pcDNA3.0-FLAG-Kr-pok and pcDNA3.1-SREBP-1c-Myc plasmids. After 24 h, the cells were washed with cold PBS and fixed in 97:3 cold methanol:formaldehyde for 20 min at −20°C. The cells were permeabilized in 0.2% Triton X-100 and washed with PBS. Next, the cells were incubated in 5% normal horse serum and then incubated with mouse anti-FLAG primary antibody for 2 h at room temperature. The cells were washed and incubated with FITC-conjugated anti-mouse IgG secondary antibody (Invitrogen). For double staining, the cells were washed and incubated with rabbit anti-Myc antibody and then with Rhodamine-conjugated anti-rabbit IgG secondary antibody (Invitrogen). After DAPI staining, washing, and mounting, the immunostained cells were examined on a Carl Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Germany).
Transcriptional analysis of 6xSRE and various FASN gene promoters
HEK293A cells were transiently cotransfected with the SREBP-1c expression plasmid, increasing amounts of the Kr-pok expression plasmid, and the reporter plasmid (pGL2-6xSRE-Luc, pGL2-FAS1, 2, 3-Luc, and pGL3-FASN Wt or Mt-Luc reporter fusion plasmids) using Lipofectamine Plus reagent (Invitrogen). After 24 h of incubation, the cells were harvested and analyzed for luciferase activity. The reporter activity was normalized with cotransfected β-galactosidase activity or protein concentration for transfection efficiency.
Electrophoretic mobility shift assay
The oligonucleotide probes were annealed by heating at 93°C for 5 min and then slowly cooled to room temperature. The annealed oligonucleotides were prepared by labeling with [α-32P]ATP and Klenow enzyme by incubating for 30 min at 37°C (Roche, Nutley, NJ). 32P-labeled, double-stranded oligonucleotide probes were purified with SephadexTM G-50 (Amersham Biosciences, Uppsala, Sweden). The sequences of the oligonucleotides used in the EMSA are as follows (only the top strand is listed): FASN SRE/E-box: 5′-GATCGTCCAGCCCATGTGGCGTGGC-3′. The remaining procedure was performed as reported previously (27).
Oligonucleotide pull-down assays
HEK293A cells transfected with pcDNA3.0-FLAG-Kr-pok and/or pcDNA3.0-SREBP-1c-Myc were lysed in HKMG buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, and 0.5% Nonidet P-40). The cellular extracts were incubated with 1 μg of biotinylated double-stranded oligonucleotides. The sequence of the SRE/E-box oligonucleotide was 5′-GTCCAGCCCATGTGGCGTGGC-3′ (only the top strand is listed). The mixtures were incubated with Streptavidin-agarose beads for 2 h to collect the DNA-protein complex and then spun, and the pellets were washed with HKMG buffer. The precipitates were resolved by 10% SDS-PAGE and analyzed using Western blot.
Chromatin immunoprecipitation assays
Using chromatin immunoprecipitation (ChIP) assays, we investigated the molecular interaction between Sp1 or SREBP-1c and the SREBP-1c binding site on the FASN promoter in the presence of Kr-pok. Subconfluent HEK293A cells on a culture dish were transfected with pcDNA3.0-FLAG Kr-pok and/or pcDNA3.1-SREBP-1c using Lipofectamine Plus and grown for an additional 48 h. The cells were fixed with formaldehyde (final concentration, 1%) to cross-link Kr-pok, SREBP-1c, and Sp1 onto the FASN promoter. The remaining ChIP procedures were performed as reported previously (27). PCR reactions of immunoprecipitated DNA were carried out using oligonucleotide primer sets designed to amplify the FASN gene region. FASN WT or MT primers (forward: 5′-CAGGCGCGTTCCCGCGCAGG-3′; reverse: 5′-GAGAGCGAGGCTGGAGCGCG-3′) and FAS2 or FAS3 primers (forward: 5′-TCCAAACTCATCAATGTA-3′; reverse: 5′-AAAGCAATTGTTCCAGGAACCAGGG-3′) were used for the ChIP assays.
Preparation of recombinant adenovirus overexpressing Kr-pok and shRNA against Kr-pok mRNA
The Kr-pok cDNA was cloned into an adenovirus E1 shuttle vector pCA14 (Microbix, Mississauga, Ontario, Canada) to generate pCA14-Kr-pok. To prepare a recombinant adenovirus expressing shRNA against Kr-pok, annealed shRNA DNA sequences (sense: 5′-GATCCCTCCAGTGCATCGTGAATGTTTTTCAAGAGA(loop)-ACATTCACGATGCACTGGATTTTTTTGGAA(loop)-A-3′; antisense: 5′-AGCTTTTCCAAAAA(loop)-AATCCAGTGCATCGTGAATGTTCTCTTGAA(loop)-AAACATTCACGATGCACTGGAGG-3′) were cloned into pSilencer 2.0-U6 (Ambion, Austin, TX) and subcloned into the pΔE1sp1A vector. The pCA14-Kr-pok shuttle vector or pΔE1sp1A-U6-shKr-pok vector and the adenovirus vector vmdl324Bst were linearized by restriction enzyme digestion for homologous recombination into E. coli BJ518. The homologous recombinant adenoviral plasmid was digested and transfected into HEK293 cells to generate the adenovirus expressing Kr-pok (dl324-Kr-pok) or shRNA against Kr-pok (dl324-shKr-pok).
Knock-down of Srebp-1c mRNA by siRNA
siRNAs against Srebp-1c mRNA were designed and purchased from Bioneer (Seoul, Korea): siSrebp-1c, 5′-CCACGGAGCCAUGGAUUGCACAUUUdTdT-3′, 5′-AAAUGUGCAAUCCAUGGCUCCGUGGdTdT-3′. The siRNAs (50 pmol) were transfected into 3T3-L1 cells using LipofectamineTM RNAiMAX (Invitrogen).
Differentiation of 3T3L1 preadipocyte
3T3-L1 preadipocytes were maintained at low passage and grown to confluence in DMEM supplemented with 10% calf serum (Gibco-BRL). Differentiation was induced by placing 2 day postconfluent cultures in DMEM supplemented with 10% calf serum for up to 8 days. The medium for differentiating 3T3-L1 preadipocytes was supplemented with 0.525 mM methylisobutylxanthine (Sigma, MO), 1 μM dexamethasone (Sigma), and 0.167 μM insulin (Roche). Forty-eight hours later, this medium was replaced with medium supplemented only with 0.167 μM insulin (Roche).
RESULTS
Kr-pok activates transcription of the FASN gene
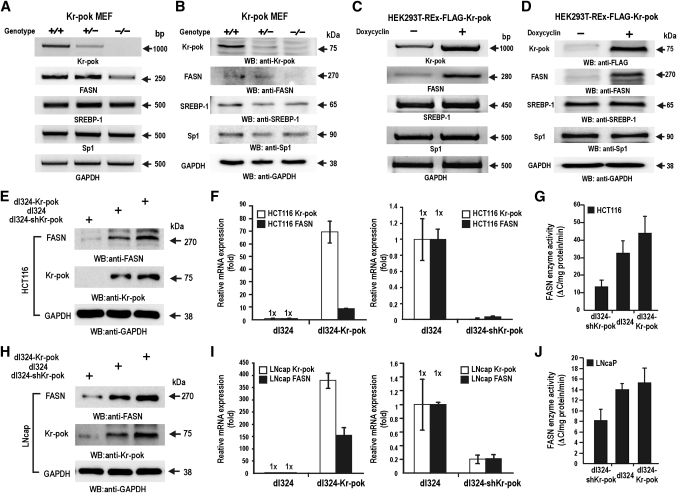
Kr-pok, recently characterized as a proto-oncoprotein expressed abundantly in most kidney cells, is similar to FBI-1 at the amino acid sequence level in two key functional domains (Supplementary Fig. I) (28, 29). FBI-1 expression has been shown to increase during the 6- to 48-h time period of human preadipocyte differentiation, and it was suggested that FBI-1 may play an important role in adipogenesis and rapid cancer cell growth (26, 27). In this investigation, we tested whether Kr-pok regulates FASN gene expression. RT-PCR of mRNAs and Western blot analysis of total cell lysates prepared from MEF cells revealed that the knockout of the Kr-pok gene decreased FASN gene expression (Fig. 1A, B). Kr-pok induction of stable HEK293T-REx-Kr-pok cells by doxycyclin increased FASN gene expression (Fig. 1C, D).
Fig. 1.
Kr-pok activates FASN gene expression. A, B: RT-PCR and Western blot analysis of Kr-pok+/+ and Kr-pok−/− MEF cells. In comparison to FASN expression in Kr-pok+/+ MEF cells, FASN expression decreased in Kr-pok−/− MEF cells. C, D: RT-PCR and Western blot analysis of HEK293T-REx-FLAG-Kr-pok cells after induction by doxycycline. Upon induction of Kr-pok expression, FASN mRNA and protein increased. E–J: Western blot, RT-qPCR, and FASN enzyme activity assays of HCT116 (E–G) and LNcaP (H–J). The cells were infected with recombinant dl324-shKr-pok, dl324, and dl324-Kr-pok adenovirus. The cell lysates were analyzed for FASN expression of protein and mRNA. Additionally, FASN enzyme activities were analyzed by spectrophotometry. GAPDH, control.
We investigated whether Kr-pok can regulate transcription of FASN gene in cancer cells. RT-qPCR and Western blot analysis of the cell lysates prepared from HCT116 (colon cancer) and LNcaP (prostate cancer) cells infected with recombinant dl324-Kr-pok adenovirus revealed that Kr-pok increases transcription of the FASN gene and FASN expression in cancer cells (Fig. 1E, F, H, I). We also analyzed lipogenic activity at the enzyme level by monitoring the oxidation of NADPH to NADP+ caused by the conversion of Malonyl-CoA and Acetyl-CoA to palmitate. Whereas ectopic Kr-pok increased FASN enzyme activity (ΔC/mg protein/min.; ΔC = ΔA/E; ΔA = change in absorbance; E = extinction coefficient of NADPH [E340nm = 6.22 mM−1 cm−1]), the knock-down of Kr-pok expression clearly decreased FASN enzyme activity in the two cancer cell lysates tested (Fig. 1G, J). These data show that the transcription of the FASN gene, a major player in the synthesis of the phospholipids of the cell membrane, was potently activated by Kr-pok.
The zinc-finger domain of Kr-pok interacts with the bZIP DNA binding domain of SREBP-1c
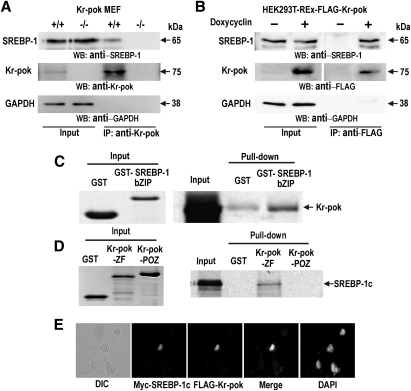
SREBPs are major transcription regulators that control the expression of enzymes involved in cholesterol and fatty acid synthesis, including FASN. Above, we showed that Kr-pok increased FASN expression. Accordingly, we tested whether SREBP-1 and Kr-pok could interact to increase FASN gene transcription. Coimmunoprecipitation and Western blot analysis of the cell lysates prepared from Kr-pok+/+, Kr-pok−/− MEF, and doxycyclin-induced HEK293T-REx-Kr-pok cells showed that SREBP-1 interacts with Kr-pok and forms a protein complex (Fig. 2A, B). GST fusion protein pull-down assays showed that the zinc-finger domain of Kr-pok directly interacts with the bZIP domain of SREBP-1 (Fig. 2C, D). Immunocytochemical analysis of the HEK293A cells that were cotransfected with the FLAG-Kr-pok and Myc-SREBP-1c expression vectors showed that the two proteins colocalize in the nucleus (Fig. 2E). These data suggest that the interaction is direct and occurs through the DNA binding domains of each protein.
Fig. 2.
The zinc-finger domain of Kr-pok interacts with the bZIP DNA binding domain of SREBP-1c. A, B: Coimmunoprecipitation and Western blot analysis of Kr-pok and SREBP-1c. Lysates prepared from Kr-pok+/+, Kr-pok−/− MEF, and doxycycline-induced HEK293T-REx-Kr-pok cells were immunoprecipitated using the anti-Kr-pok antibody and analyzed by Western blotting with the anti-SREBP-1 antibody. C, D: In vitro GST fusion protein pull-down assays. Recombinant GST, GST-ZF Kr-pok, GST-POZ Kr-pok, or GST-bZIP SREBP-1 proteins were incubated with [35S]methionine-labeled SREBP-1c or Kr-pok and were pulled down and resolved with 12% SDS-PAGE. The gel was then exposed to X-ray film. Input: 10% of Kr-pok was added to the binding reactions. E: Immunocytochemistry and cellular colocalization of Kr-pok and SREBP-1c. The HEK293A cells transfected with the expression vectors of FLAG-Kr-pok and Myc-tagged SREBP-1c were analyzed by immunocytochemical staining using mouse anti-FLAG antibody and rabbit anti-Myc antibody. FITC-conjugated anti-mouse IgG or Rhodamine-conjugated anti-rabbit IgG antibody were used as secondary antibodies. GAPDH, control.
Kr-pok increases transcriptional activation of the FASN gene by interacting with SREBP-1c
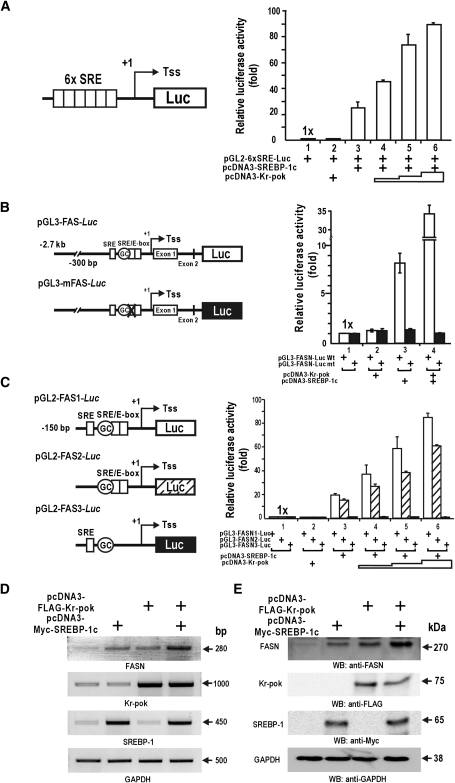
We have shown that Kr-pok increases transcription of the FASN gene and interacts with SREBP-1. We analyzed the functional significance of the protein interaction between Kr-pok and SREBP-1 on the SREBP-1 target genes, including FASN. First, SREBP-1c alone is able to activate transcription on the artificial 6×(SRE)-Luc promoter. Although Kr-pok is not able to activate transcription alone, Kr-pok increases transcription in the presence of SREBP-1c in HEK293A cells (Fig. 3A). These data suggest that the interaction between Kr-pok and SREBP-1c is important for synergistic transcriptional activation of SREBP-1c target genes with SRE.
Fig. 3.
Kr-pok further increases transcriptional activation of the FASN gene by SREBP-1c. A–C: Kr-pok enhances transcriptional activation of the artificial pGL2-6×[SRE]Luc or FASN promoter-Luc with SREBP-1c in HEK293A cells cotransfected with expression vectors of SREBP-1c (25 ng) and Kr-pok (25, 125, and 500 ng). Reporter activity was normalized with cotransfected β-galactosidase activity or protein concentration for transfection efficiency. The data presented are the average of three independent assays. Bars represent standard deviation. D, E: RT-PCR and Western blot analysis. HEK293A cells were cotransfected with the expression vector of FLAG-Kr-pok and/or Myc-SREBP-1c. The cell extracts were analyzed for FASN expression. Tss (+1), transcription start site; GAPDH, control.
We also tested how Kr-pok and SREBP-1c regulated transcription of the human FASN gene promoter with proximal SREs and long upstream regulatory sequences (−2.7 kb from Tss +1) in HEK293A cells. Kr-pok alone showed no effect on the transcription of the pGL3-FASN-Luc promoter. SREBP-1c increased transcription 8-fold, and Kr-pok augmented this increase to 35-fold. To investigate the functional significance of the SRE elements and to determine if Kr-pok affects the molecular events between SREBP-1c and proximal SRE/E-box binding sites (−65 to 45 bp) of the FASN gene, we prepared the pGL3-mFASN-Luc reporter plasmid. Transient transfection and transcriptional analysis of the mutated plasmid revealed that neither Kr-pok nor SREBP-1c has any effect on transcription. This finding suggests that the SRE elements are important in regulating the transcriptional activation of the FASN promoter by SREBP-1c and Kr-pok (Fig. 3B).
To map which regulatory element is important for transcriptional activation by SREBP-1c and Kr-pok, we prepared three different promoter reporter gene fusion constructs that differed in the proximal regulatory element (Fig. 3C). The three FASN constructs were distinguished by the inclusion of SRE, Sp1 binding GC-box, and SRE/E-box elements. In HEK293A cells, SREBP-1c activated transcription of only the promoter constructs with the proximal SRE/E-box. Although Kr-pok alone had little effect on transcription of any of the promoter constructs, Kr-pok significantly increased SREBP-1c's transcription of FASN1 and FASN2 promoter constructs. Kr-pok was unable to activate transcription of the FASN3 promoter construct, which lacked the SRE/E-box (Fig. 3C). The data suggest that, although Kr-pok does not show any effect on transcription by itself, it potently increases transcription of the FASN promoter, most likely by modulating the molecular interaction between SREBP-1c and the SRE/E-box.
We tested whether the protein interaction between Kr-pok and SREBP-1c influences gene transcription of endogenous FASN using RT-PCR analysis of the mRNA. SREBP-1c alone increased FASN mRNA transcription significantly, and Kr-pok also increased FASN expression, likely with the help of endogenous SREBP-1. Cotransfection of Kr-pok and SREBP-1c synergistically activated FASN gene transcription (Fig. 3D). Western blot analyses also showed that Kr-pok and SREBP-1c increased FASN protein expression (Fig. 3E).
The modulation of dna binding activity of SREBP-1c and Sp1 at the SRE/E-box of FASN proximal promoter by the zinc-finger DNA binding domain of Kr-pok
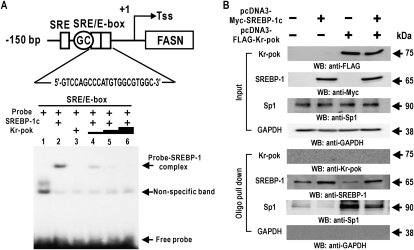
Kr-pok synergistically increased transcriptional activation of the FASN gene via SREBP-1, likely by modulating the molecular interaction of SREBP-1c at the SRE/E-box. We investigated whether Kr-pok influences the DNA binding activity of SREBP-1 using EMSA. EMSA showed that in vitro translated Kr-pok did not show DNA binding activity and that SREBP-1 binds to the SRE/E-box probe. Initially, we expected that Kr-pok may increase the DNA binding activity of SREBP-1 because Kr-pok and SREBP-1c synergistically activated FASN gene transcription. Unexpectedly, Kr-pok decreased the DNA binding activity of SREBP-1 (Fig. 4A).
Fig. 4.
Kr-pok decreases binding of the SREBP-1 to the SRE/E-box. A: The 32P-labeled SRE/E-box binding element probe of the FASN gene was incubated with His-SREBP-1 (200 ng) and/or in vitro translated Kr-pok (75–675 ng) and separated by 4% nondenaturing PAGE. The gel was exposed to X-ray film. B: Oligonucleotide pull-down assays of Kr-pok, SREBP-1c, and Sp1 binding to the SRE/E-box elements of the FASN gene promoter. HEK293A cell extracts were incubated with biotinylated double-stranded oligonucleotides. The mixtures were further incubated with Streptavidin-agarose beads and precipitated by centrifugation. The precipitate was analyzed by Western blot assays using antibodies against Kr-pok, SREBP-1, and Sp1. SRE/E-box, downstream SREBP-1 binding element; Tss, transcription start point (+1); GAPDH, control.
The transcriptional regulation of the FASN gene was previously characterized, and Sp1 and SREBP-1c were shown to synergistically activate transcription by acting on SRE, Sp1 binding GC-box, and SRE/E-box (32). We investigated whether Kr-pok influenced DNA binding activity of SREBP-1c and Sp1. An oligonucleotide pull-down assay of DNA-protein binding events on the SRE/E-box showed that, although Kr-pok does not bind to the SRE/E-box, ectopic Kr-pok increases Sp1 binding but decreases SREBP-1 binding. Ectopic SREBP-1c decreases Sp1 binding but increases its own binding. When Kr-pok and SREBP-1c were coexpressed, binding of SREBP-1c and Sp1 increased in comparison to the binding by endogenous SREBP-1c and Sp1, a condition in which FASN can be synergistically activated by the two factors (Fig. 4B). These data suggest that transcriptional regulation of the FASN gene by Kr-pok and SREBP-1c involves the Sp1 transcription factor acting at the SRE/E-box.
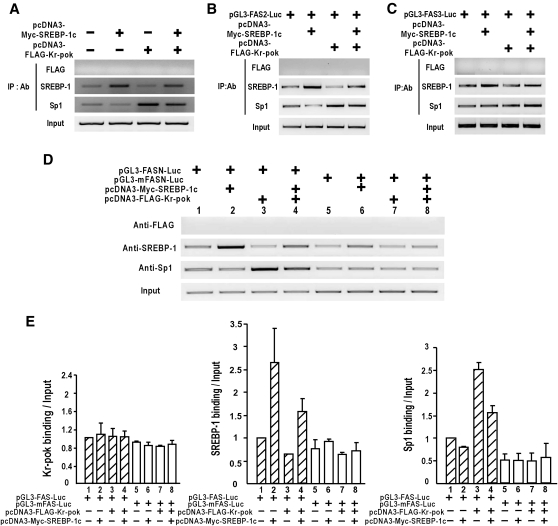
ChIP analysis of DNA-protein binding events on the endogenous FASN proximal promoter showed that Kr-pok decreases SREBP-1 binding and increases Sp1 binding without direct DNA binding. When Kr-pok and SREBP-1c were coexpressed, binding of SREBP-1c and Sp1 increased relative to the binding of endogenous SREBP-1c and Sp1 (Fig. 5A). To investigate the mechanistic role of the SRE/E-box in the transcriptional activation of FASN by Kr-pok and SREBP-1, we performed ChIP assays using the pGL3-FASN2, pGL3-FASN-3, pGL3-FASN WT (2.7 kb), and pGL3-mFASN MT (2.7 kb) reporter plasmids. When the SRE/E-box was present, such as in FASN2 and FASN WT plasmids, the SREBP-1c and Sp1 binding patterns were similar to that of the endogenous FASN promoter (Fig. 5B, D). However, with an SRE/E-box mutation or deletion, as with the FASN3 and mFASN MT plasmids, the DNA binding of SREBP-1c and Sp1 was not altered by Kr-pok (Fig. 5C, D). Our data potentially explain why Kr-pok does not show any effect on the transcriptional regulation of FASN3 or mFASN MT reporter gene by SREBP-1 and Sp1 (Fig. 3B, C). The change in transcription factor binding initiated by Kr-pok may be important for the synergistic transcriptional activation of the FASN gene.
Fig. 5.
The ChIP assay reveals the modulatory effect of Kr-pok on the DNA binding of SREBP-1c and Sp1 to the proximal promoter of FASN. A: ChIP assays of the DNA binding activity of FLAG-Kr-pok, SREBP-1c, and Sp1 on the endogenous FASN gene in HEK293A cells transfected with the expression vector of FLAG-Kr-pok or/and SREBP-1c. B, C: ChIP assays of the DNA binding activity of FLAG-Kr-pok, SREBP-1c, and Sp1 on the FASN2 or FASN3 promoter construct that was transfected into the HEK293A cells. D: ChIP assays of the DNA binding activity of FLAG-Kr-pok, SREBP-1c, and Sp1 on the pGL3-FASN-Luc WT or MT (−2.7 kb) plasmid transfected into HEK293A cells. Sheared chromatin was immunoprecipitated using the antibodies indicated. PCR amplification primers were designed to bind to the plasmid vector sequences and not to the endogenous FASN promoter for FASN2, FASN3, Wt, and Mt. E: Histograms of ChIP assay binding band intensities of Kr-pok, SREBP-1c, and Sp1 were divided by input band intensities. Histogram showing the average of all independent ChIP assays shown in (D).
Kr-pok expression is increased during adipocyte differentiation and facilitates the differentiation of 3T3-L1 preadipocytes
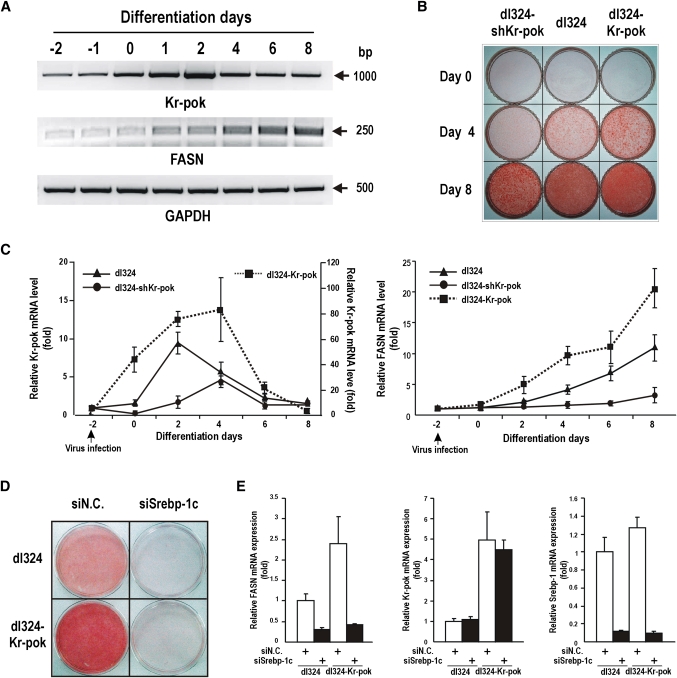
Our data showed that Kr-pok and SREBP-1c interact with each other and that this molecular interaction is important for the transcriptional activation of the FASN gene. SREBP-1c is known to promote adipocyte differentiation and is involved in the insulin-mediated regulation of the FASN gene. We investigated whether Kr-pok participates in the differentiation of adipocytes. First, we examined the expression profile of Kr-pok mRNA during adipocyte differentiation. 3T3-L1 preadipocytes, which represent a well characterized in vitro model of adipocyte differentiation, were differentiated into mature adipocytes upon exposure to a mixture of hormones (MDI, methylisobutylxanthine, dexamethasone, and insulin). During differentiation, the expression of endogenous Kr-pok mRNA increased from induction day 0 until it reached a peak on day 2; it gradually declined until day 8. FASN mRNA began to rise on day 1 of differentiation and continued to increase until day 8 of differentiation (Fig. 6A, C).
Fig. 6.
Kr-pok mRNA expression increases during early adipocyte differentiation and increases FASN mRNA expression and fatty acid synthesis. A: RT-PCR analysis of Kr-pok and FASN mRNA expression during 3T3-L1 preadipocyte differentiation. The cells were treated with a differentiation-inducing MDI mixture and harvested at the indicated times. B, C: 3T3-L1 preadipocytes were infected with recombinant adenovirus dl324-Kr-pok or dl324-shKr-pok at −2 days of differentiation induction and allowed to differentiate for 8 days. The cells were stained with Oil Red O at the indicated times (B). The cells were harvested at the indicated times and analyzed for the expression of Kr-pok and FASN mRNA by RT-qPCR (C). D, E: Roles of Kr-pok and Srebp-1c in 3T3-L1 preadipocytes differentiation and fatty acid synthesis. The preadipocytes were transfected with siSrebp-1c RNA at −3 days and infected with recombinant adenovirus dl324-Kr-pok at −2 days of differentiation induction. The cells were allowed to differentiate for 4 days and stained with Oil Red O (D). The cells were harvested at 4 days after differentiation induction and analyzed for the expression of Kr-pok, Srebp-1, and FASN mRNA by RT-qPCR (E). GAPDH, control.
To investigate whether Kr-pok participated in adipocyte differentiation, 3T3-L1 preadipocytes were infected with each of the recombinant dl324, dl324-Kr-pok, and dl324-shKr-pok adenoviruses 2 days before differentiation induction. Then, adipocyte differentiation was induced by incubating the cells with fresh medium containing MDI at day 0 of differentiation induction. The 3T3-L1 preadipocytes infected with dl324-Kr-pok adenovirus show intense staining with Oil Red O. Cells infected with dl324-shKr-pok adenovirus show much weaker staining. These data suggested that Kr-pok increases adipocyte differentiation and/or fatty acid synthesis in the cells (Fig. 6B). Kr-pok increases transcription of the FASN gene in adenovirus-infected 3T3-L1 preadipocytes and produces more lipid droplets. The dl324-shKr-pok adenovirus, which knocked down Kr-pok mRNA, decreases transcription of the FASN gene in 3T3-L1 preadipocytes and significantly decreases lipid accumulation (Fig. 6B, C).
We tested whether an increase in lipid synthesis or adipocyte differentiation by Kr-pok is dependent on the presence of SREBP-1c. The 3T3-L1 preadipocytes were transfected with negative control siRNA (siN.C.) or siSrebp-1c RNA 3 days before differentiation induction. The cells were further infected with the recombinant dl324 or dl324-Kr-pok adenovirus 2 days before differentiation induction. The 3T3-L1 cells infected with recombinant dl324-Kr-pok adenovirus show intense staining with Oil Red O. The 3T3-L1 cells transfected with siSrebp-1c RNA show no staining with Oil Red O, which suggested that adipocyte differentiation was blocked. Alternatively, when the 3T3-L1 cells were transfected with siSrebp-1c RNA and infected with recombinant dl324-Kr-pok adenovirus, the cells did not differentiate into adipocytes. These results suggested that SREBP-1c is critical in adipocyte differentiation and that coexpression of Kr-pok and SREBP-1c increases fatty acid synthesis and/or adipocyte differentiation (Fig. 6D). Additionally, RT-qPCR analysis of the cells showed that Kr-pok does not increase transcription of the FASN gene in the absence of Srebp-1c and that Srebp-1c does not affect Kr-pok expression (Fig. 6E).
DISCUSSION
Proto-oncogene FBI-1, a member of the POZ zinc finger protein family, has been suggested to play roles in the differentiation of adipocytes, fatty acid synthesis in cancer cells, and oncogenesis (25–27). Kr-pok is a member of the POK family of proteins and is most closely related to FBI-1 in secondary protein structure of key functional domains and perhaps also in some cellular functions. Recently, we have shown that Kr-pok promoted cell proliferation by repressing transcription of a negative regulator of cell cycle progression, CDKN1A, by competitively binding with MIZ-1 and/or by interacting with p53 (28, 29). We suspected that Kr-pok might increase the expression of some lipogenic genes because it increased cell proliferation, a cellular process that requires rapid lipid synthesis.
It has become apparent that FASN-driven lipogenesis is important in cancer cell proliferation (1–4, 8, 9). FASN is critical for cancer cell proliferation because it is the only enzyme involved in the synthesis of palmitate, a critical component of the cell membrane of cancer cells. Therefore, we investigated whether Kr-pok regulated FASN expression in Kr-pok+/+, Kr-pok−/− MEF, HEK293T-REx-Kr-pok, and two cancer cell lines. The knock-out of Kr-pok expression in MEF cells decreased FASN expression, and ectopic Kr-pok expression increased FASN expression in various cell lines (Fig. 1).
Transcriptional regulation of the FASN gene depends largely on the protein-protein interactions of the transcription factors that bind to the proximal promoter of the FASN gene (5–7). SREBP-1c plays a major role in lipogenic gene expression, including FASN. Kr-pok increases FASN gene expression at the transcriptional level in HEK293A and some other cancer cells. We have demonstrated that SREBP-1c alone activates transcription and that Kr-pok synergistically enhances this transcription. In contrast, mutation or deletion of the SRE/E-box element of the FASN promoter abolishes the effects of Kr-pok and SREBP-1c on the transcriptional regulation of the FASN gene. These data suggest that the SRE/E-box element is important for the synergistic transcriptional activation of the FASN promoter by Kr-pok and SREBP-1c (Fig. 3). In addition, we were able to show that Kr-pok and SREBP-1c interact directly through their DNA binding domains and that this interaction is critical for the synergistic activation of FASN gene transcription (Fig. 2).
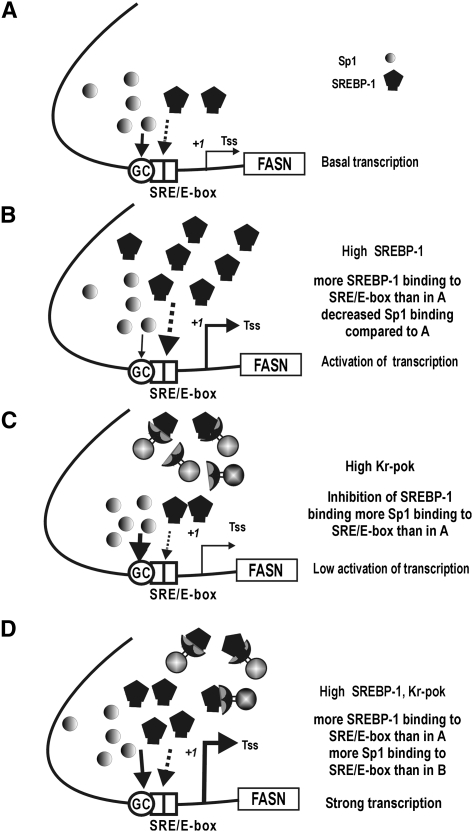
To study the importance of protein-protein interactions between Kr-pok, SREBP-1, and Sp1 in the transcriptional regulation of FASN at the FASN proximal promoter, we performed EMSA, oligonucleotide pull-down, and ChIP assays. The SRE/E-box element of the FASN gene is a SREBP-1c binding element, but it can be bound by Sp1 as well (27, 32). Accordingly, we investigated the transcription factor binding dynamics of Kr-pok, SREBP-1c, and Sp1 on the proximal promoter of the FASN gene by oligonucleotide pull-down and in vivo ChIP assays. Ectopic SREBP-1c decreases Sp1 binding on the proximal promoter of the FASN gene that contains the SRE/E-box element (Figs. 4B, 5A), and SREBP-1c activates transcription of the FASN gene (Fig. 3). Although ectopic Kr-pok increases Sp1 binding on the proximal promoter, Kr-pok does not significantly affect transcription of the FASN gene in the reporter gene constructs (Figs. 3, 5A). Kr-pok also slightly decreases SREBP-1c binding. When the expression of Kr-pok and SREBP-1c is high, as is the case in some cancer tissues, SREBP-1c and Sp1 binding is maintained at a level that allows the synergistic activation of FASN gene transcription (Figs. 3, 5). This situation resulted in a 2- to 3-fold increase in FASN gene transcription compared with that observed when only SREBP-1c was overexpressed. For these molecular events, the SRE/E-box of the proximal promoter of FASN appears to be the center of transcriptional activation because a FASN MT (−2.7 kb) construct with a mutation at the SRE/E-box resulted in no transcriptional activation by Kr-pok and SREBP-1c (hypothetical model in Fig. 7).
Fig. 7.
Hypothetical model: binding dynamics of three transcription factors (Kr-pok, SREBP-1c, and Sp1) on the SRE/E-box of the FASN gene promoter. A: Sp1 is abundant, whereas SREBP-1c was not induced and is scarce. The promoter elements are mainly bound by Sp1, and transcription is at a basal level. B: Upon induction of SREBP-1c, Sp1 binding is reduced and SREBP-1c binding is increased, resulting in potent transcriptional activation. C: Upon induction of Kr-pok, Sp1 binding is induced and SREBP-1c binding is reduced. Kr-pok did not directly bind to the promoter, and Kr-pok had little effect on transcription. D: Synergistic activation of FASN gene transcription by Kr-pok and SREBP-1c. In the presence of high levels of Kr-pok and SREBP-1c, binding of SREBP-1c decreased compared with that of (B), and Sp1 binding decreased compared with that of (C). However, unlike (B) or (C), SREBP-1c and Sp1 binding are maintained at certain level that allows for the synergistic activation of FASN gene transcription. Arrows represent DNA-protein interactions. The thickness of the arrow indicates relative binding activity. Tss (+1), transcription start site.
Finally, we found that Kr-pok gene transcription was induced by treatment of 3T3-L1 cells with hormone mixtures that are known to induce adipocyte differentiation. The induction of Kr-pok gene transcription precedes the induction of FASN gene transcription. Kr-pok overexpression or knock-down experiments, using recombinant adenovirus dl324-Kr-pok or dl324-shKr-pok, showed that Kr-pok activates FASN gene expression (Fig. 6). Additionally, these assays raised the possibility that Kr-pok might play a role in adipocyte differentiation. It appears that although Kr-pok plays important roles in fatty acid synthesis and/or adipocyte differentiation, SREBP-1c is critically required in the processes (Fig. 6D, E).
In summary, our study revealed novel roles for the proto-oncoprotein Kr-pok in FASN gene expression. We found that the molecular events that occur among the proximal SRE/E-box, Kr-pok, SREBP-1c, and Sp1 are important for transcriptional regulation of FASN gene expression. FASN, a key multifunctional enzyme of de novo fatty acid synthesis, is highly expressed in most human carcinomas. FASN and fatty acid metabolism in cancer have become a focus for the diagnosis and treatment of cancer. The molecular mechanism revealed by this study provides critical information on how the proto-oncoprotein Kr-pok uses the cellular regulatory system of the FASN promoter to provide cellular membrane components necessary for rapid cancer cell growth and proliferation. Furthermore, these molecular mechanisms may provide a novel target for the development of future anti-cancer drugs.
Supplementary Material
Footnotes
Abbreviations:
- BTB/POZ
- bric-à-brac, tramtrack, and broad complex and pox virus zinc finger
- ChIP
- chromatin immunoprecipitation
- CMV
- cytomegalovirus
- DAPI
- 4’,6-diamidino-2-phenylindole
- EMSA
- electrophoretic mobility shift assay
- FASN
- fatty acid synthase
- FBI-1
- factor that binds to the inducer of short transcripts of human immunodeficiency virus-1
- GC-box
- Sp1 binding GC-rich box
- GST
- glutathione S-transferase
- IP
- immunoprecipitation
- IPTG
- isopropyl-1-thio-β-D-galactopyranoside
- MEF
- murine embryonic fibroblast
- POK
- POZ domain and Krüppel like
- Sp1
- specificity protein 1
- SRE
- sterol responsive element
- SREBP
- sterol responsive element binding protein
- ZF
- zinc finger
This work was supported by a Mid-career Researcher Program Grant (2009-0081294) (M.-W. H.), a Do-Yak Program Grant (2011-0028817) (M.-W. H.), an Atomic Energy Research Grant (2008-2001735) (M.-W. H.), and a Medical Research Center Grant (2011-0030708) (M.-W. H.) from the National Research Foundation of the Korean Ministry of Education, Science and Technology.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and two figures.
REFERENCES
- 1.Swinnen J. V., Van Veldhoven P. P., Timmermans L., De Schrijver E., Brusselmans K., Vanderhoydonc F., Van de Sande T., Heemers H., Heyns W., Verhoeven G. 2003. Fatty acid synthase drives the synthesis of phospholipids partitioning into detergent-resistant membrane microdomains. Biochem. Biophys. Res. Commun. 302: 898–903 [DOI] [PubMed] [Google Scholar]
- 2.Chirala S. S., Wakil S. J. 2004. Structure and function of animal fatty acid synthase. Lipids. 39: 1045–1053 [DOI] [PubMed] [Google Scholar]
- 3.Costello L. C., Franklin R. B. 2005. ‘Why do tumour cells glycolyse?’: from glycolysis through citrate to lipogenesis. Mol. Cell. Biochem. 280: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menendez J. A., Lupu R. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7: 763–777 [DOI] [PubMed] [Google Scholar]
- 5.Magaña M. M., Osborne T. F. 1996. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J. Biol. Chem. 271: 32689–32694 [DOI] [PubMed] [Google Scholar]
- 6.Kim J. B., Sarraf P., Wright M., Yao K. M., Mueller E., Solanes G., Lowell B. B., Spiegelman B. M. 1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Invest. 101: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf S. S., Roder K. H., Schweizer M. 1998. The general transcription factor Sp1 plays an important role in the regulation of fatty acid synthase. Biochem. Soc. Trans. 26: S95. [DOI] [PubMed] [Google Scholar]
- 8.Kusakabe T., Nashimoto A., Honma K., Suzuki T. 2002. Fatty acid synthase is highly expressed in carcinoma, adenoma and in regenerative epithelium and intestinal metaplasia of the stomach. Histopathology. 40: 71–79 [DOI] [PubMed] [Google Scholar]
- 9.Innocenzi D., Alo P. L., Balzani A., Sebastiani V., Silipo V., La Torre G., Ricciardi G., Bosman C., Calvieri S. 2003. Fatty acid synthase expression in melanoma. J. Cutan. Pathol. 30: 23–28 [DOI] [PubMed] [Google Scholar]
- 10.Sebastiani V., Visca P., Botti C., Santeusanio G., Galati G. M., Piccini V., Capezzone de Joannon B., Di Tondo U., Alo P. L. 2004. Fatty acid synthase is a marker of increased risk of recurrence in endometrial carcinoma. Gynecol. Oncol. 92: 101–105 [DOI] [PubMed] [Google Scholar]
- 11.Tontonoz P., Kim J. B., Graves R. A., Spiegelman B. M. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13: 4753–4759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborne T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275: 32379–32382 [DOI] [PubMed] [Google Scholar]
- 13.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500 [DOI] [PubMed] [Google Scholar]
- 15.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardwell V. J., Treisman R. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8: 1664–1677 [DOI] [PubMed] [Google Scholar]
- 17.Costoya J. A. 2007. Functional analysis of the role of POK transcriptional repressors. Brief. Funct. Genomics Proteomics. 6: 8–18 [DOI] [PubMed] [Google Scholar]
- 18.Kelly K. F., Daniel J. M. 2006. POZ for effect-POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 16: 578–587 [DOI] [PubMed] [Google Scholar]
- 19.Barna M., Hawe N., Niswander L., Pandolfi P. P. 2000. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25: 166–172 [DOI] [PubMed] [Google Scholar]
- 20.Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J., Pandolfi P. P. 2004. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36: 653–659 [DOI] [PubMed] [Google Scholar]
- 21.Issa J. P., Zehnbauer B. A., Kaufmann S. H., Biel M. A., Baylin S. B. 1997. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 57: 1678–1681 [PubMed] [Google Scholar]
- 22.Chen W., Cooper T. K., Zahnow C. A., Overholtzer M., Zhao Z., Ladanyi M., Karp J. E., Gokgoz N., Wunder J. S., Andrulis I. L., et al. 2004. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 6: 387–398 [DOI] [PubMed] [Google Scholar]
- 23.Phan R. T., Dalla-Favera R. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 432: 635–639 [DOI] [PubMed] [Google Scholar]
- 24.Phan R. T., Saito M., Basso K., Niu H., Dalla-Favera R. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6: 1054–1060 [DOI] [PubMed] [Google Scholar]
- 25.Maeda T., Hobbs R. M., Merghoub T., Guernah I., Zelent A., Cordon-Cardo C., Teruya-Feldstein J., Pandolfi P. P. 2005. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 433: 278–285 [DOI] [PubMed] [Google Scholar]
- 26.Laudes M., Christodoulides C., Sewter C., Rochford J. J., Considine R. V., Sethi J. K., Vidal-Puig A., O'Rahilly S. 2004. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J. Biol. Chem. 279: 11711–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi W. I., Jeon B. N., Park H., Yoo J. Y., Kim Y. S., Koh D. I., Kim M. H., Kim Y. R., Lee C. E., Kim K. S., et al. 2008. Proto-oncogene FBI-1 (Pokemon) and SREBP-1 synergistically activate transcription of fatty-acid synthase gene (FASN). J. Biol. Chem. 283: 29341–29354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K. M., Choi W. I., Koh D. I., Kim Y. J., Jeon B. N., Yoon J. H., Lee C. E., Kim S. H., Oh J., Hur M. W. 2011. The proto-oncoprotein KR-POK represses transcriptional activation of CDKN1A by MIZ-1 through competitive binding. Oncogene. In press [DOI] [PubMed] [Google Scholar]
- 29.Jeon B. N., Kim M. K., Choi W. I., Koh D. I., Hong S. Y., Kim K. S., Kim M., Yun C. O., Yoon J., Choi K. Y., et al. Kenneth, and M. W. Hur. 2011. KR-POK interacts with p53 and represses CDKN1A transcriptional activation by p53. Cancer Res. In press [DOI] [PubMed] [Google Scholar]
- 30.He X., Dave V. P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B. A., Kappes D. J. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433: 826–833 [DOI] [PubMed] [Google Scholar]
- 31.Sun G., Liu X., Mercado P., Jenkinson S. R., Kypriotou M., Feigenbaum L., Galéra P., Bosselut R. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6: 373–381 [DOI] [PubMed] [Google Scholar]
- 32.Athanikar J. N., Sanchez H. B., Osborne T. F. 1997. Promoter selective transcriptional synergy mediated by sterol regulatory element binding protein and Sp1: a critical role for the Btd domain of Sp1. Mol. Cell. Biol. 17: 5193–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.