Abstract
We previously reported that adenosine monophosphate-activated protein kinase (AMPK) activity is lower in adipose tissue of morbidly obese individuals who are insulin resistant than in comparably obese people who are insulin sensitive. However, the number of patients and parameters studied were small. Here, we compared abdominal subcutaneous, epiploic, and omental fat from 16 morbidly obese individuals classified as insulin sensitive or insulin resistant based on the homeostatic model assessment of insulin resistance. We confirmed that AMPK activity is diminished in the insulin resistant group. A custom PCR array revealed increases in mRNA levels of a wide variety of genes associated with inflammation and decreases in PGC-1α and Nampt in omental fat of the insulin resistant group. In contrast, subcutaneous abdominal fat of the same patients showed increases in PTP-1b, VEGFa, IFNγ, PAI-1, and NOS-2 not observed in omental fat. Only angiotensinogen and CD4+ mRNA levels were increased in both depots. Surprisingly, TNFα was only increased in epiploic fat, which otherwise showed very few changes. Protein carbonyl levels, a measure of oxidative stress, were increased in all depots. Thus, adipose tissues of markedly obese insulin resistant individuals uniformly show decreased AMPK activity and increased oxidative stress compared with insulin sensitive patients. However, most changes in gene expression appear to be depot-specific.
Keywords: insulin sensitivity, adenosine monophosphate-activated protein kinase, inflammation
Obesity is a risk factor for disorders such as type 2 diabetes, atherosclerotic cardiovascular disease, nonalcoholic fatty liver disease, hypertension, certain cancers, and even dementia (1). Insulin resistance is thought to play a significant role in the pathogenesis of these disorders. However, not all obese individuals are insulin resistant (2, 3). A subgroup of such patients has been shown to be insulin sensitive and to have lower levels of ectopic fat in liver and skeletal muscle and decreased intra-abdominal fat compared with their insulin resistant counterparts (4, 5). It has also been noted that they are less predisposed to atherosclerotic cardiovascular disease (3). Approximately 25% of morbidly obese individuals [defined as body mass index (BMI) > 35 kg/m2] are estimated to be insulin sensitive (6, 7). At a molecular level, what distinguishes them from their insulin resistant counterparts is unclear.
In a very small study, we recently reported that AMP kinase (AMPK) activity is lower in morbidly obese humans who are insulin resistant [homeostatic model assessment of insulin resistance (HOMA-IR) > 2.3] than in comparably obese individuals who are insulin sensitive (8). AMPK is a fuel-sensing enzyme that has been implicated in the regulation of glucose and lipid homeostasis and insulin sensitivity (9–12). Moreover, diminished AMPK activity or impairment of its activation has been observed in skeletal muscle of individuals with obesity and diabetes in some (13, 14) but not other (15) studies, and in visceral adipose tissue of centrally obese humans with Cushings syndrome, a disorder associated with insulin resistance (16).
Obesity is also frequently associated with chronic low-grade inflammation, and a causal relationship between such inflammation and the development of insulin resistance has been described (17). Likewise, increased inflammation has been found in adipose tissue of insulin resistant compared with insulin sensitive obese patients (1, 18), suggesting that it could be a key factor that distinguishes the two populations.
Yet another abnormality observed in obese humans is an increase in oxidative stress. Thus, obese individuals are reported to have higher serum levels of isoprostanes (19), malondialdehyde (20), protein carbonyls (21), and urinary isoprostanes (22). In addition, oxidative stress has been implicated in the pathogenesis of both insulin resistance (23–25) and inflammation (26). Whether differences in oxidative stress distinguish adipose tissue of insulin sensitive and insulin resistant obese individuals and, if so, whether this correlates with inflammation and/or diminished AMPK activity, has not been determined, however.
In the present study, we assessed these questions in abdominal subcutaneous, omental, and epiploic adipose tissue of morbidly obese individuals undergoing bariatric surgery. The results indicate that decreases in AMPK activity and increases in oxidative stress distinguish insulin resistant from insulin sensitive patients in all three fat depots. In contrast, although the expression of genes linked to inflammation was increased in the insulin resistant group, specific changes in the three depots were quite different.
MATERIALS AND METHODS
Patients
Sixteen patients (7 men and 9 women) with a mean age of 42.4 ± 15 years and a BMI of 44.6 ± 7.0 kg/m2 were studied (Table 1). Limited results from eight of these patients were reported previously (8). The study was approved by the Boston University Medical Center Institutional Review Board. All participants were approved for surgical intervention via laparoscopic roux-en-y gastric bypass surgery and had signed an informed consent form prior to their enrollment. Subjects were divided into insulin resistant and insulin sensitive subgroups based on their HOMA-IR. A value of 2.3 was considered the cut off point as described by Matthews et al. (27). All measurements were carried out on blood taken after an overnight fast. Biopsies of abdominal subcutaneous, epiploic (mesenteric adipose tissue taken from the epiploica of the transverse colon), and omental fat were obtained at the time of surgery. The tissues were immediately frozen in liquid nitrogen and stored at −80°C until further processing.
TABLE 1.
Baseline characteristics of the study population
| Parameter | Insulin sensitive (n = 8) | Insulin resistant (n = 8) |
| Age, years | 42.4 ± 5.4 | 42.5 ± 5.3 |
| Sex, female/male | 5/3 | 4/4 |
| HOMA | 1.6 ± 0.2 | 5.1 ± 0.8 ** |
| Weight, kg | 174.2 ± 12.6 | 167.8 ± 15.4 |
| Height, m | 1.7 ± 0.04 | 1.6 ± 0.02 |
| BMI, kg/m2 | 41.9 ± 1.7 | 47.4 ± 2.8 |
| Waist circumference, cm | 130 ± 0.7 | 130 ± 0.7 |
| Hip circumference, m | 1.4 ± 0.04 | 1.4 ± 0.04 |
| Heart rate, bpm | 65.6 ± 3.1 | 72.4 ± 5.1 |
| Plasma insulin, μIU/ml | 6.1 ± 1.2 | 19.1 ± 4.5 ** |
| Glucose, mg/dl | 118.3 ± 21.7 | 114.1 ± 7.7 |
| HbA1C, % | 6.3 ± 0.4 | 6.7 ± 0.3 |
| Total cholesterol, mg/dl | 180.3 ± 10.6 | 175.5 ± 9.2 |
| HDL, mg/dl | 44.1 ± 4.0 | 43.6 ± 2.5 |
| Systolic BP, mm Hg | 127.4 ± 4.3 | 132.1 ± 5.1 |
| Diastolic BP, mm Hg | 70.5 ± 1.9 | 75.8 ± 4.5 |
| Triacylglycerol, mg/dl | 91.5 ± 8.7 | 165.9 ± 34.4 |
| hsCRP, mg/L | 3.9 ± 1.0 | 6.4 ± 1.7 |
| Metformin | 2 | 7 |
| Metabolic syndrome | 6 | 8 |
| Diabetes | 3 | 7 |
| Antidepressants | 3 | 5 |
Data are means ± S.E. **p < 0.01 compared with insulin sensitive patients.
Abbreviations: HbA1c, glycosylated hemoglobin A1C; hsCRP, high-sensitivity C-reactive protein.
RNA isolation and real-time quantitative PCR array
Total RNA from each fat depot was extracted using the RNeasy lipid tissue mini kit (Qiagen, Valencia, CA). Two hundred nanograms of total RNA from each sample were reverse transcribed into cDNA using the RT2 first strand kit (Qiagen). An RT2 Profiler Custom PCR Array (Qiagen) was used to examine the mRNA levels of 43 genes (complete list shown in Table 2). Both 18S and cyclophilin A were used as house-keeping genes. Of the two, 18S gave the most consistent result and was therefore used for normalization analysis. Several negative controls were included in each run. All PCR experiments were conducted with a StepOne Real Time PCR system (Applied Biosystems, Carlsbad, CA). The data analysis was performed using the ΔΔCt based calculations (28).
TABLE 2.
Relative gene expression level in different fat depots of insulin resistant versus sensitive patients
Western blot analysis
Total proteins were isolated from subcutaneous, epiploic, and omental adipose tissue that had been homogenized in cell lysis buffer (Cell Signaling Technology, Danvers, MA) supplemented with protease (Roche, Mannheim, Germany) and phosphatase (Sigma-Aldrich, Saint Louis, MO) inhibitors. Homogenates were centrifuged at 14,000 g for 15 min at 4°C. The protein concentration of the supernatant was determined using the bicinchoninic acid assay (Thermo Scientific, Rockford, IL) with BSA as a standard. Twenty micrograms of protein lysate were loaded onto each lane of a 4-15% polyacrylamide gradient gel (Bio-Rad, Hercules, CA) and separated by electrophoresis. The separated proteins were transferred to a PVDF membrane (Millipore, Billerica, MA), blocked with 5% nonfat dry milk in tris-buffered saline supplemented with Tween-20 for 1 h and then incubated overnight with primary antibodies against phospho-AMPK (Thr172), total AMPK (Cell Signaling Technology), Nampt (Bethyl Laboratories, Montgomery, TX), and β-actin (Sigma-Aldrich). Bound antibodies were detected with the appropriate horseradish peroxidase-linked secondary antibodies (Cell Signaling Technology). Proteins were visualized by enhanced chemiluminescence (Thermo Scientific), and bands were quantified with Scion Image Software (National Institutes of Health). The size of each antibody-bound protein was verified using standard molecular mass markers (Bio-Rad).
Protein carbonylation assay
Protein carbonylation was determined with an OxyBlot protein oxidation detection kit (Millipore, Billerica, MA) to provide a measure of oxidative stress. In brief, 10 μg of protein lysate was derivatized with 4-dinitrophenylhydrazine (DNPH) and then neutralized according to the manufacturer's instructions. The neutralized samples were next fractionated by SDS-PAGE and the carbonylated proteins detected by Western blotting with an anti-DNPH antibody. A negative control was included in each blot. Total carbonylation was visualized by enhanced chemiluminescence (Thermo Scientific), and the bands quantified with Scion Image Software.
Statistical analysis
Data are expressed as means ± SE. GraphPad Prism software (La Jolla, CA) was used for all analyses. The comparison was performed using Student's t-test or one-way ANOVA. Minimal level of significance was set at p < 0.05.
RESULTS
Baseline patient characteristics
The clinical characteristics of insulin sensitive and resistant patients are presented in Table 1. The two groups were similar with respect to weight, height, BMI, waist and hip circumference, heart rate, blood pressure, and fasting plasma glucose. Fasting plasma insulin levels were significantly higher in the insulin resistant patients. In addition, the insulin resistant subjects showed higher levels of triacylglycerol, hsCRP, and HbA1c than those in the insulin sensitive group; although the differences did not achieve statistical significance. A greater number of insulin resistant patients were on metformin and antidepressants. All eight patients in the insulin resistant group and six of the insulin sensitive subjects had the metabolic syndrome based on ATP III criteria (29). Based on HbA1C levels of 5.7% or greater and known diagnoses, six patients in the insulin resistant group and three in the insulin sensitive group were either diabetic or prediabetic.
Decreased AMPK activity (phosphorylation) in insulin resistant compared with insulin sensitive patients
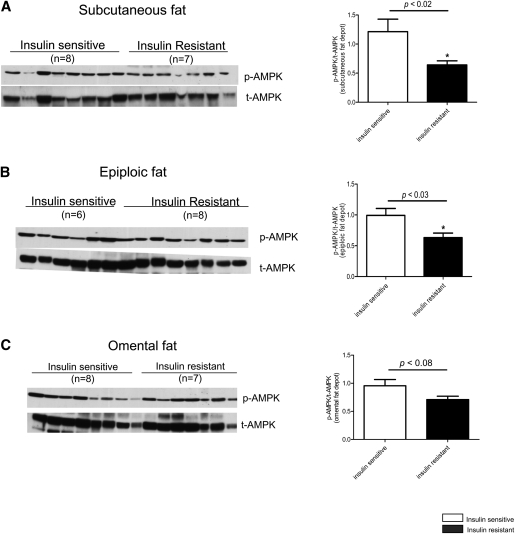
Western blot analysis was carried out to assess AMPK phosphorylation at threonine 172, an indicator of its activation (30). As shown in Fig. 1, the abundance of phospho-AMPK was 47% and 36% lower, respectively, in abdominal subcutaneous and epiploic fat of the insulin resistant compared with the insulin sensitive patients (Fig. 1A, B). In omental fat, phospho-AMPK was 26% lower in the insulin resistant group; however, the difference (p = 0.08) did not reach statistical significance (Fig. 1C).
Fig. 1.
Quantification of AMPK phosphorylation on threonine 172 (activation) in abdominal subcutaneous, epiploic, and omental adipose tissue of insulin sensitive and insulin resistant humans (n = 6–8 per group). Relative protein levels were normalized against total AMPK. * p < 0.05 relative to insulin sensitive group.
Differences in gene expression profiles in adipose tissue
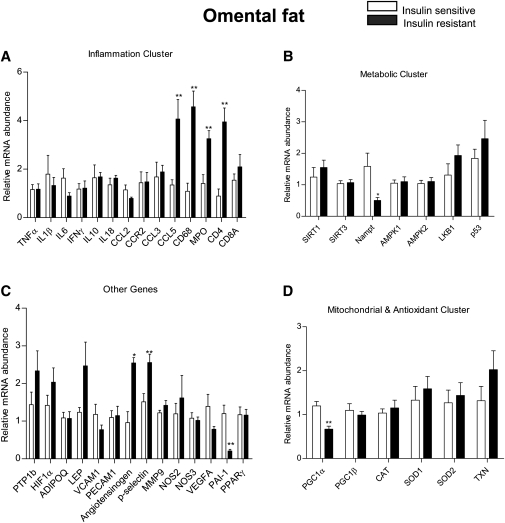
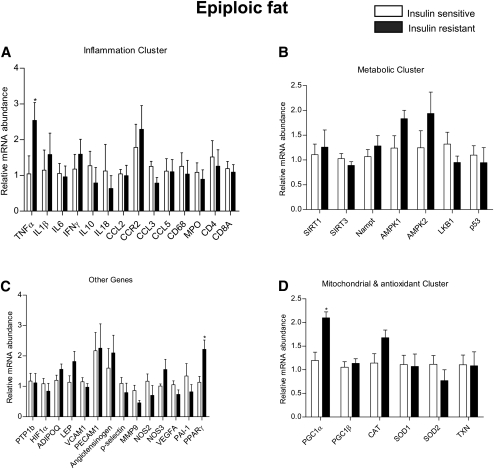
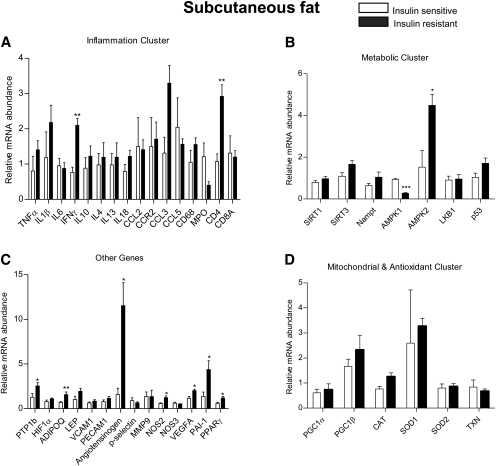
We utilized a Profiler Custom PCR array approach to measure mRNA levels of 43 genes. These genes were grouped into subclasses based on their potential link to the pathogenesis of insulin resistance. PCR array was the method of choice because it enabled us to use as little as 200 ng of RNA to carry out amplification reactions for the 43 genes. A complete list of the genes examined is shown in supplementary Table II. The data for each depot are presented according to gene subclass (inflammatory, metabolic, mitochondrial, and anti-oxidant and others) in Figs. 2–4. In addition, the key differences between the two groups in each depot are shown in Table 2 and supplementary Table I.
Fig. 2.
Relative mRNA expression of genes by RT2 profiler PCR array in human abdominal subcutaneous fat. Results are means ± SEM (n = 6–8 per group). *p < 0.05, **p < 0.01 versus insulin sensitive group.
Fig. 4.
Relative mRNA expression of genes by RT2 profiler PCR array in human omental fat. Results are means ± SEM (n = 6–8 per group). *p < 0.05, **p < 0.01 versus insulin sensitive group.
In omental fat (Fig. 2), the expression of many genes involved in the immune response was increased in the insulin resistant group. They include the chemokine CCL5 (also known as RANTES), macrophage marker CD68, neutrophil marker MPO, and the T cell marker CD4+ (Fig. 2A). Levels of angiotensinogen [a gene associated with insulin resistance (31)] and p-selectin (32), an adhesion molecule important for the recruitment of leukocytes into adipose tissue, were also higher in the insulin resistant group (Fig. 2C). In contrast, Nampt, a molecule that activates SIRT1, and is itself activated by AMPK (33), showed a 50% decrease in its mRNA expression in the insulin resistant group, as did the mitochondrial biogenesis marker PGC1α (Fig. 2B, D). Surprisingly, the mRNA for PAI-1, a molecule generally increased in the insulin resistant state, was decreased in the insulin resistant group (Fig. 2C).
In the abdominal subcutaneous depot, mRNA levels of CD4+ and IFNγ, a T cell inflammatory cytokine that has been linked to insulin resistance (34), were upregulated in the insulin resistant compared with the insulin sensitive group (Fig. 3A). In addition, we found higher mRNA levels of PTP-1b, angiotensinogen, NOS2 (also known as iNOS), VEGFa, PAI-1, and PPARγ in the insulin resistant group (Fig. 3C). Unexpectedly, the mRNA for the anti-inflammatory adipokine adiponectin was higher in the insulin resistant group (Fig. 3C). In the ‘metabolic cluster’, the mRNA of AMPKα1 [the major isoform expressed in adipose tissue (35)] was downregulated, whereas that of AMPK isoform α2 was upregulated in the insulin resistant group (Fig. 3B). No differences in gene expression between the two groups were detected in the mitochondrial and antioxidant subsets (Fig. 3D).
Fig. 3.
Relative mRNA expression of genes by RT2 profiler PCR array in human epiploic fat. Results are means ± SEM (n = 6–8 per group). *p < 0.05, **p < 0.01, ***p < 0.001 versus insulin sensitive group.
In epiploic fat, we found increased mRNA levels for TNFα, PPARγ, and PGC1α in the insulin resistant group (Fig. 4). The changes in TNFα and PGC1α were unique to epiploic fat, whereas a similar increase in PPARγ occurred in subcutaneous fat and a decrease in PGC1α in omental fat. Interestingly, in the insulin sensitive patients, the mRNA levels of CCL5, CD4+, CD8+, and angiotensinogen were comparable in epiploic and omental fat and were substantially greater than those in subcutaneous fat (supplementary Table I).
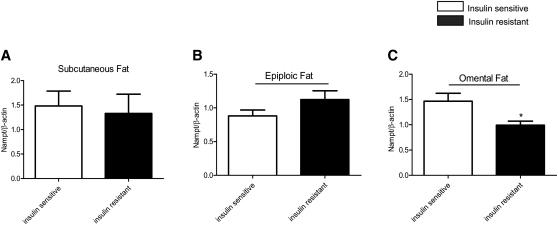
Nampt protein expression is decreased in omental fat of insulin resistant patients
Nampt is a rate-limiting enzyme involved in NAD+ biosynthesis and it positively regulates the activity of the histone/protein deacetylase SIRT1 and other sirtuins (36). Western blot analysis indicated reduced Nampt protein abundance in omental fat of the insulin resistant group in keeping with the decreased expression of its mRNA (Fig. 5A–C). No change in SIRT1 mRNA (Figs. 2–4) or protein (data not shown) was detected in any of the depots.
Fig. 5.
Nampt protein expression in adipose tissue of insulin sensitive and insulin resistant humans (n = 6–8 per group). Relative protein levels were normalized against β-actin. * p < 0.05 relative to insulin sensitive group.
Oxidative stress in insulin sensitive and insulin resistant patients
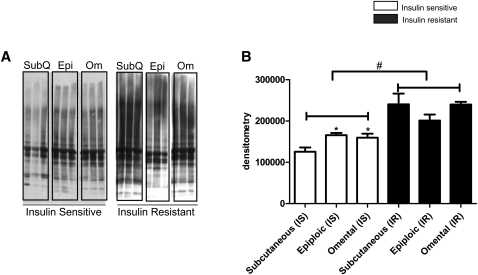
Protein carbonylation, a marker of oxidative stress, was assessed in all three depots. A representative image is shown in Fig. 6A. Quantification of signal intensities by densitometry indicated that within the insulin sensitive group, carbonylation of proteins was greater in both epiploic and omental than in abdominal subcutaneous fat (Fig. 6B). As a group, the insulin resistant patients had significantly higher levels of oxidative damage in all three depots, with the greatest increase (vs. insulin sensitive group) found in the subcutaneous abdominal fat (Fig. 6B).
Fig. 6.
Carbonylation of proteins in adipose tissue from insulin sensitive and insulin resistant patients. A: Protein lysate (10 μg) from adipose tissue was derivatized with DNPH (Oxyblot, Millipore) and resolved by SDS-PAGE. Representative images are shown on the left. B: Relative quantification of carbonylation was determined by densitometry. (n = 5 per group, *, p < 0.05 compare with subcutaneous depot of insulin sensitive group; #, p < 0.0001 compare with insulin resistant group).
DISCUSSION
The overall objective of this study was to identify factors in adipose tissue that distinguish massively obese humans who are insulin sensitive from comparably obese individuals who are insulin resistant. Measurements were performed on abdominal subcutaneous, epiploic, and omental fat obtained during bariatric surgery. The key findings include the following: 1) higher levels of inflammatory genes were present in all three depots of the insulin resistant group; however, the specific genes that were increased varied among depots; 2) genes involved in cell signaling, metabolism, and mitochondrial function also showed distinct changes in their expression pattern; and 3) in contrast, increases in protein carbonylation (an indicator of oxidative stress) and decreases in AMPK activity were observed in all three depots of the insulin resistant patients.
It has long been appreciated that visceral and subcutaneous fat differ with respect to both morphology and function (37). For instance, omental fat appears to be more sensitive to the action of lipolytic agents (38), whereas subcutaneous fat has a greater capacity for lipogenesis and expansion (39). In addition, visceral fat is more vascular and typically contains more immune cells (37). In the present study, we found striking differences in the gene profile of omental and subcutaneous abdominal fat when insulin sensitive and insulin resistant patients were compared. For instance, omental fat from the insulin resistant group showed increases in genes associated with inflammation (CCL5, CD68, MPO, CD4+, p-selectin), and decreases in PGC1α and Nampt compared with insulin sensitive subjects (Fig. 2). Although somewhat overlapping changes in gene expression were observed in subcutaneous fat, CD4+T and angiotensinogen were the only genes that showed consistently higher expression in the insulin resistant group in both depots (Figs. 2, 3). The mRNA levels of IFNγ (an inflammatory cytokine produced by CD4+ T cells), PTP-1b, PPARγ, VEGFa, PAI-1, and NOS2, none of which was significantly increased in omental fat of the insulin resistant patients, were all upregulated in subcutaneous fat (Fig. 3). Increases in most of these molecules have been reported to associate with insulin resistance (31, 40–42).
As already noted, in the insulin sensitive group, epiploic and omental fat showed a greater abundance of CCL5, CD4+, CD8+, and angiotensinogen mRNA than did subcutaneous fat (supplementary Table I), suggesting that they both had the changes in gene expression that distinguish visceral from subcutaneous fat. However, in contrast to omental fat, none of these mRNAs were upregulated in the epiploic depot of the insulin resistant patients (Fig. 4). Indeed, of the inflammatory cytokines studied, only TNFα mRNA was increased in epiploic fat of the insulin resistant group, and it was not similarly elevated in either omental or subcutaneous fat. The reason for such a finding remains to be determined.
The PCR array generated a few other unexpected results. For example, increased PAI-1 and decreased adiponectin levels have been causally linked to the development of obesity and insulin resistance (43, 44), yet we found decreased PAI-1 (omental) (Fig. 2C) and increased adiponectin (subcutaneous) (Fig. 3C) in the insulin resistant group. Moreover, decreased AMPKα1 and increased AMPKα2 mRNA levels were detected in the subcutaneous fat depot of the insulin resistant population, but not in the visceral depots (Figs. 2–4). Such a finding is puzzling because most cells types found in adipose tissue (i.e., adipocytes, macrophages, endothelial cells) predominantly express the AMPKα1 isoform (45–47). To the best of our knowledge, the only cell type in adipose tissue that expresses AMPKα2 is the fibroblast (48). Future studies involving the separation of adipocytes from various stromal fractions may help discern the source of the elevated AMPKα2. In a recent report by Kloting et al. (1), SIRT1 mRNA in omental and, to a lesser extent, subcutaneous fat were lower and IL-6 and Nampt (see below) mRNA higher in obese insulin resistant than insulin sensitive patients. We did not observe these changes. Such discrepancies could be attributable to differences in methods (i.e., the use of a custom PCR array in which all primers were designed by Qiagen vs. conventional real-time quantitative PCR) or study subject selection. With respect to the latter, we relied on HOMA-IR to distinguish insulin sensitive from insulin resistant patients and Kloting et al. utilized the euglycemic hyperinsulinemic clamp for this purpose. In addition, patients with diabetes or a family history of diabetes were excluded in the Kloting study, whereas six of the eight patients in the insulin resistant group in the present study were diabetic or prediabetic as were three of the eight insulin sensitive patients. Also of note, more of the insulin resistant than insulin sensitive patients enrolled in our study were on metformin therapy. However, the use of this drug has been shown to increase AMPK activity in adipose tissue (49). Thus, if anything, this would have made for higher rather than lower AMPK activity in the insulin resistant subjects. Finally, it should be noted that a potential limitation of this study is that it is based on 16 patients and has limited power for analyses that involve multiple comparisons. Nonetheless, using separate Bonferroni and Hochberg adjustments by depot and gene subclass to control the overall false discovery rate, adjusted p values showed that differences between the insulin sensitive and insulin resistant groups remained significant or trended to significance (p < 0.1) in almost all instances.
Excessive production of reactive oxygen species, like inflammation, has been implicated in the pathogenesis of insulin resistance (23–25). In humans, protein carbonylation, an indicator of chronic oxidative stress (50), has been reported to correlate directly with adiposity and may contribute to the development of insulin resistance (51). Here, we found that protein carbonylation was greater in adipose tissue of the insulin resistant patients than their insulin sensitive counterparts (Fig. 6). Furthermore, in contrast to the differing changes in gene expression, the increase in protein carbonylation was detected in all three depots. Interestingly, subcutaneous abdominal fat had less protein carbonylation than the two visceral depots in the insulin sensitive patients and underwent the largest increase in protein carbonyl levels in the insulin resistant population. Whether this contributed to the increased expression of specific genes in this depot remains to be determined. Finally, it has been demonstrated that in severely obese women, bariatric surgery leads to parallel and progressive improvements in insulin sensitivity and markers of oxidative stress in plasma (dROMs test) over 2 years (52).
The results confirmed our previous finding that AMPK activity is diminished in adipose tissue (subcutaneous and epiploic depots) of insulin resistant compared with insulin sensitive patients (8). A similar trend was observed in omental fat, although the difference was not statistically significant (p < 0.08) (Fig. 1C). We also found decreases in Nampt mRNA and protein abundance in omental fat of insulin resistant patients (Figs. 2, 5). Nampt has been identified as a stress- and nutrient-responsive enzyme that increases cellular NAD+ and promotes survival during genotoxic stress (53). Given the role of Nampt in conferring cellular resistance to oxidative stress and fostering immune cells to survive under inflammatory conditions (54), the reduced Nampt expression found here could have contributed to the elevated oxidative stress as well as heightened inflammation in omental fat of the insulin resistant patients. It has also been suggested, based on studies in skeletal muscle, that Nampt can be activated by AMPK and that such regulation could account for the ability of AMPK to activate SIRT1 (33, 55, 56). Whether the observed decrease in Nampt expression would affect SIRT1 activity in adipose tissue remains to be determined. Downregulation of SIRT1 mRNA expression has been reported in visceral fat of morbidly obese patients with severe hepatic steatosis (57) and, as already noted, in omental fat of morbidly obese individuals who are insulin resistant (1). In the latter study, however, the decrease in SIRT1 mRNA was associated with an increase in Nampt gene expression. Clearly, measurements of SIRT1 activity are required to address the possible discrepancy between these findings and those reported here.
The association between decreased AMPK activity and the increases in oxidative stress and inflammatory genes observed in the present study are in keeping with the relationship of these factors in cultured adipocytes and other cells. Thus, both pharmacologically (58) and genetically (Gauthier et al., unpublished observation) induced decreases in AMPK activity have been reported to increase oxidative stress in 3T3-L1 adipocytes when lipolysis is stimulated. Likewise, genetically-induced decreases in AMPK activity have been shown to increase the inflammatory effects of palmitate and other molecules (TNFα, LPS) in cultured endothelial cells (59) and macrophages (60). Such associations raise the possibility that decreased AMPK could be responsible for the increases in oxidative stress and inflammation observed here in adipose tissue of obese insulin resistant patients. On the other hand, it has also been demonstrated that inflammation caused by TNFα (61) and palmitate (62) decreases AMPK activity in rodent skeletal muscle and aorta, respectively, and that a lipid infusion has a similar effect on mouse adipose tissue (46). Likewise, oxidative stress, as reflected by an increase in the lipid peroxidation by-product hydroxynonenal (51), has been reported to downregulate AMPK upstream kinase LKB1. Collectively, these findings raise the intriguing possibility that diminished AMPK activity and increased oxidative stress and inflammation could be causal factors for each other. If so, whether one of them is the initiating event that leads to adipose tissue and systemic insulin resistance in most obese humans remains to be determined. Also requiring consideration in such a discussion is ER stress, which has been linked to all of these factors, as well as to insulin resistance (63). Longitudinal studies of adipose tissue in people before, during, and after weight gain as well as before and after bariatric surgery could clarify this issue. Yet another intriguing model might be patients with Cushing's syndrome, who are insulin resistant and have decreased AMPK activity in their visceral fat (16).
Supplementary Material
Acknowledgments
The authors thank Christopher Kohberger, Nga Tong, and Anupriya Mundra for their assistance in preparing the manuscript and Drs. Barbara Nikolajczyk, Vishwajeet Puri, and Walter Pories for constructive criticism during its preparation. Finally, we are very grateful to Drs. Lisa Sullivan and Timothy Heeren at the Boston University School of Public Health for their help and advice with the statistical analyses.
Footnotes
Abbreviations:
- AMPK
- AMP kinase
- BMI
- body mass index
- HOMA-IR
- homeostatic model assessment of insulin resistance
This work was supported by National Institutes of Health grants R01-DK19514 and P01-HL068758 (to N.B.R.), a mentor-based Fellowship Grant from the American Diabetes Association ADA-7-11-MN-43 (to N.B.R), and National Institutes of Health grants R01HL084213 and P01 HL 081587 (to N.G.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. M-S.G. was supported by a postdoctoral research fellowship from Fonds de la Recherche en Santé du Québec and is presently a Canadian Diabetes Association fellow. M.F. and R.V. were supported by T32 HL07224 training grant from the American Heart Association. None of the authors has a conflict of interest relevant to this manuscript.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables.
REFERENCES
- 1.Klöting N., Fasshauer M., Dietrich A., Kovacs P., Schon M. R., Kern M., Sutumvoll M., Blüher M. 2010. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 299: E506–E515 [DOI] [PubMed] [Google Scholar]
- 2.Sims E. A. 2001. Are there persons who are obese, but metabolically healthy? Metabolism. 50: 1499–1504 [DOI] [PubMed] [Google Scholar]
- 3.Reaven G. 2005. All obese individuals are not created equal: insulin resistance is the major determinant of cardiovascular disease in overweight/obese individuals. Diab. Vasc. Dis. Res. 2: 105–112 [DOI] [PubMed] [Google Scholar]
- 4.Stefan N., Kantartzis K., Machann J., Schick F., Thamer C., Rittig K., Balletshofer B., Machicao F., Fritsche A., Häring H. U. 2008. Identification and characterization of metabolically benign obesity in humans. Arch. Intern. Med. 168: 1609–1616 [DOI] [PubMed] [Google Scholar]
- 5.Gauthier M. S., Ruderman N. B. 2010. Adipose tissue inflammation and insulin resistance: all obese humans are not created equal. Biochem. J. 430: e1–e4 [DOI] [PubMed] [Google Scholar]
- 6.Ferrannini E., Natali A., Bell P., Cavallo-Perin P., Lalic N., Ningrone G. 1997. Insulin resistance and hypersecretion in obesity. European Group for the Study of Insulin Resistance (EGIR). J. Clin. Invest. 100: 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogardus C., Lillioja S., Mott D. M., Hollenbeck C., Reaven G. 1985. Relationship between degree of obesity and in vivo insulin action in man. Am. J. Physiol. 248: E286–E291 [DOI] [PubMed] [Google Scholar]
- 8.Gauthier M-S., O'Brien E. L., Bigornia S., Mott M., Cacicedo J. M., Xu X. J., Gokce N., Apovian C., Ruderman N. 2011. Decreased AMP-activated protein kinase activity is associated with increased inflammation in visceral adipose tissue and with whole-body insulin resistance in morbidly obese humans. Biochem. Biophys. Res. Commun. 404: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergeron R., Previs S. F., Cline G. W., Perret P., Russell R. R., 3rd, Young L. H., Shulman G. I. 2001. Effect of 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside infusion on in vivo glucose and lipid metabolism in lean and obese Zucker rats. Diabetes. 50: 1076–1082 [DOI] [PubMed] [Google Scholar]
- 10.Fisher J. S., Gao J., Han D. H., Holloszy J. O., Nolte L. A. 2002. Activation of AMP kinase enhances sensitivity of muscle glucose transport to insulin. Am. J. Physiol. Endocrinol. Metab. 282: E18–E23 [DOI] [PubMed] [Google Scholar]
- 11.Iglesias M. A., Ye J. M., Frangioudakis G., Saha A. K., Tomas E., Ruderman N. B., Cooney G. J., Kraegen E. W. 2002. AICAR administration causes an apparent enhancement of muscle and liver insulin action in insulin-resistant high-fat-fed rats. Diabetes. 51: 2886–2894 [DOI] [PubMed] [Google Scholar]
- 12.Ruderman N., Prentki M. 2004. AMP kinase and malonyl-CoA: targets for therapy of the metabolic syndrome. Nat. Rev. Drug Discov. 3: 340–351 [DOI] [PubMed] [Google Scholar]
- 13.Bandyopadhyay G. K., Yu J. G., Ofrecio J., Olefsky J. M. 2006. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis. Diabetes. 55: 2277–2285 [DOI] [PubMed] [Google Scholar]
- 14.Sriwijitkamol A., Coletta D. K., Wajcberg E., Balbontin G. B., Reyna S. M, Barrientes J., Eagan P. A., Jenkinson C. P., Cersosimo E., DeFronzo R. A., Sakamoto K., Musi N. 2007. Effect of acute exercise on AMPK signaling in skeletal muscle of subjects with type 2 diabetes: a time-course and dose-response study. Diabetes. 56: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Højlund K., Mustard K. J., Staehr P., Hardie D. G., Beck-Nielsen H., Richter E. A., Wojtaszewski J. F. 2004. AMPK activity and isoform protein expression are similar in muscle of obese subjects with and without type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 286: E239–E244 [DOI] [PubMed] [Google Scholar]
- 16.Kola B., Christ-Crain M., Lolli F., Arnaldi G., Giacchetti G., Boscaro M., Grossman A. B., Korbontis M. 2008. Changes in adenosine 5′-monophosphate-activated protein kinase as a mechanism of visceral obesity in Cushing's syndrome. J. Clin. Endocrinol. Metab. 93: 4969–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellen K. E., Hotamisligil G. S. 2005. Inflammation, stress, and diabetes. J. Clin. Invest. 115: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbarroja N., Lopez-Pedrera R., Mayas M. D., Garcia-Fuentes E., Garrido-Sánchez L., Macías-González M., El Bekay R., Vidal-Puig A., Tinahones F. J. 2010. The obese healthy paradox: is inflammation the answer? Biochem. J. 430: 141–149 [DOI] [PubMed] [Google Scholar]
- 19.Block G., Dietrich M., Norkus E. P., Morrow J. D., Hudes M., Caan B., Packer L. 2002. Factors associated with oxidative stress in human populations. Am. J. Epidemiol. 156: 274–285 [DOI] [PubMed] [Google Scholar]
- 20.Olusi S. O. 2002. Obesity is an independent risk factor for plasma lipid peroxidation and depletion of erythrocyte cytoprotectic enzymes in humans. Int. J. Obes.Relat. Metab. Disord. 26: 1159–1164 [DOI] [PubMed] [Google Scholar]
- 21.Uzun H., Konukoglu D., Gelisgen R., Zemgin K., Mustafa T. 2007. Plasma protein carbonyl and thiol stress before and after laparascopic gastric banding in morbidly obese patients. Obes. Surg. 17: 1367–1373 [DOI] [PubMed] [Google Scholar]
- 22.Tadros T. M., Massaro J. M., Rosito G. A., Hoffmann U., et al. 2010. Pericardial fat volume correlates with inflammatory markers: the Framingham Heart Study. Obesity (Silver Spring). 18: 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., Nakayama O., Makishima M., Matsuda M., Shimomura I. 2004. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 114: 1752–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceriello A., Motz E. 2004. Is oxidative stress the pathogenic mechanism underlying insulin resistance, diabetes, and cardiovascular disease? The common soil hypothesis revisited. Arterioscler. Thromb. Vasc. Biol. 24: 816–823 [DOI] [PubMed] [Google Scholar]
- 25.Houstis N., Rosen E. D., Lander E. S. 2006. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 440: 944–948 [DOI] [PubMed] [Google Scholar]
- 26.Brownlee M. 2001. Biochemistry and molecular cell biology of diabetic complications. Nature. 414: 813–820 [DOI] [PubMed] [Google Scholar]
- 27.Matthews D. R., Hosker J. P., Rudenski A. S., Naylor B. A., Treacher D. F., Turner R. C. 1985. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 28: 412–419 [DOI] [PubMed] [Google Scholar]
- 28.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 29.Grundy S. M., Cleeman J. I., Daniels S. R., et al. 2005. Diagnosis and management of the metabolic syndrome: an AHA/NHLBI scientific statement. Circulation. 112: 2735–2752 [DOI] [PubMed] [Google Scholar]
- 30.Hawley S. A., Davison M., Woods A., Davies S. P., Beri R. K., Carling D., Hardie D. G. 1996. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271: 27879–27887 [DOI] [PubMed] [Google Scholar]
- 31.Cooper R., Forrester T., Ogunbiyi O., Muffinda J. J. 1998. Angiotensinogen levels and obesity in four black populations. ICSHIB Investigators. J. Hypertens. 16: 571–575 [DOI] [PubMed] [Google Scholar]
- 32.Burns A. R., Bowden R. A., Abe Y., Walker D. C., Simon S. I., et al. 1999. P-selectin mediates neutrophil adhesion to endothelial cell borders. J. Leukoc. Biol. 65: 299–306 [DOI] [PubMed] [Google Scholar]
- 33.Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 458: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGillicuddy F. C., Chiguoine E. H., Hinkle C. C., Kim R. J., Shah R., Roche H. M., Smyth E. M., Reilly M. P. 2009. Interferon gamma attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J. Biol. Chem. 284: 31936–31944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kola B., Grossman A. B., Korbonits M. 2008. The role of AMP-activated protein kinase in obesity. Front. Horm. Res. 36: 198–211 [DOI] [PubMed] [Google Scholar]
- 36.Yang H., Lavu S., Sinclair D. A. 2006. Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity? Exp. Gerontol. 41: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wajchenberg B. L. 2000. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr. Rev. 21: 697–738 [DOI] [PubMed] [Google Scholar]
- 38.Hellmér J., Marcus C., Sonnenfeld T., Arner P. 1992. Mechanisms for differences in lipolysis between human subcutaneous and omental fat cells. J. Clin. Endocrinol. Metab. 75: 15–20 [DOI] [PubMed] [Google Scholar]
- 39.Joe A. W. B., Yi L., Even Y., Vogl A. W., Rossi F. M. V. 2009. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 27: 2563–2570 [DOI] [PubMed] [Google Scholar]
- 40.Vidal-Puig A., Jimenez-Linan M., Lowell B. B., Hamann A., Hu E., Spiegelman B., Flier J. S., Moller D. E. 1996. Regulation of PPARg Gene Expression by Nutrition and Obesity in Rodents. J. Clin. Invest. 97: 2553–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferder L., Inserra F., Martinez-Maldonado M. 2006. Inflammation and the metabolic syndrome: role of angiotensin II and oxidative stress. Curr. Hypertens. Rep. 8: 191–198 [DOI] [PubMed] [Google Scholar]
- 42.Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., Loy A. L., Normandin D., Cheng A., Himms-Hagen J., Chan C. C., et al. 1999. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 283: 1544–1548 [DOI] [PubMed] [Google Scholar]
- 43.Tschritter O., Fritsche A., Thamer C., Haap M., Thamer C., Bachmann O., Dahl D., Maerker E., Teigeler A., Machicao F., et al. 2003. Plasma adiponectin concentrations predict insulin sensitivity of both glucose and lipid metabolism. Diabetes. 52: 239–243 [DOI] [PubMed] [Google Scholar]
- 44.Alessi M. C., Poggi M., Juhan-Vague I. 2007. Plasminogen activator inhibitor-1, adipose tissue and insulin resistance. Curr. Opin. Lipidol. 18: 240–245 [DOI] [PubMed] [Google Scholar]
- 45.Daval M., Diot-Dupuy F., Bazin R., Hainault I., Viollet B., Vaulont S., Hajduch E., Ferré P., Foufelle F. 2005. Anti-lipolytic action of AMP-activated protein kinase in rodent adipocytes. J. Biol. Chem. 280: 25250–25257 [DOI] [PubMed] [Google Scholar]
- 46.Yang Z., Kahn B. B., Shi H., Xue B. Z. 2010. Macrophage alph1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J. Biol. Chem. 285: 19051–19059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colombo S. L., Moncada S. 2009. AMPKα1 regulates the antioxidant status of vascular endothelial cells. Biochem. J. 421: 163–169 [DOI] [PubMed] [Google Scholar]
- 48.Morizane Y., Thanos A., Takeuchi K., Murakami Y., Kayama M., Trichonas G., Miller J., Foretz M., Viollet B., Vavvas D. G. 2011. AMP-activated protein kinase suppresses matrix metalloproteinase-9 expression in mouse embryonic fibroblasts. J. Biol. Chem. 286: 16030–16038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle J. G., Logan P. J., Jones G. C., Small M., Sattar N., Connell J. M., Cleland S. J., Salt I. P. 2011. AMP-activated protein kinase is activated in adipose tissue of individuals with type 2 diabetes treated with metformin: a randomised glycaemia-controlled crossover study. Diabetologia. 54: 1799–1809 [DOI] [PubMed] [Google Scholar]
- 50.Davies K. J. 1987. Protein damage and degradation by oxygen radicals. J. Biol. Chem. 262: 9895–9901 [PubMed] [Google Scholar]
- 51.Frohnert B., Sinaiko A. R., Serrot F. J., Foncea R. E., Moran A., Ikramuddin S., Choudry U., Bernlohr D. A. 2011. Increased adipose protein carbonylation in human obesity. Obesity (Silver Spring). 19: 1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gletsu-Miller N., Hansen J. M., Jones D. P., Go Y-M., Torres W. E., Ziegler T. R., Lin E. 2009. Loss of total and visceral adipose tissue mass predicts decreases in oxidative stress after weight-loss surgery. Obesity (Silver Spring). 17: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., et al. 2007. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 130: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rongvaux A., Galli M., Denanglaire S., Van Gool F., Dreze P. L., Szpirer C., Bureau F., Andris F., Leo O. 2008. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J. Immunol. 181: 4685–4695 [DOI] [PubMed] [Google Scholar]
- 55.Ruderman N. B., Xu J. X., Nelson L. E., Cacicedo J. M., Saha A. K., Lan F., Ido Y. 2010. AMPK and SIRT1: a long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 298: E751–E760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. 2008. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell. 14: 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa C. S., Hammes T. O., Rohden F., Margis R., Bortolotto J. W., Padoin A. V., Mottin C. C., Guaragna R. M. 2010. SIRT1 transcription is decreased in visceral adipose tissue of morbidly obese patients with severe hepatic steatosis. Obes. Surg. 20: 633–639 [DOI] [PubMed] [Google Scholar]
- 58.Gauthier M. S., Miyoshi H., Souza S. C., Cacicedo J. M., Saha A. K., Greenberg A. S., Ruderman N. B. 2008. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: potential mechanism and physiological relevance. J. Biol. Chem. 283: 16514–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cacicedo J. M., Yagihashi N., Keaney J. F., Jr, Ruderman N. B., Ido Y. 2004. AMPK inhibits fatty acid-induced increases in NF-kB transactivation in cultured human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 324: 1204–1209 [DOI] [PubMed] [Google Scholar]
- 60.Sag D., Carling D., Stout R. D., Suttles J. 2008. Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 181: 8633–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steinberg G. R., Michell B. J., van Denderen B. J. W., Watt M. J., Carey A. L., Fam B. C., Andrikopoulos S., Proietto J., Görgün C. Z., Carling D., et al. 2006. Tumor necrosis factor a-induced skeletal muscle insulin resistance involves suppression of AMP-kinase signaling. Cell Metab. 4: 465–474 [DOI] [PubMed] [Google Scholar]
- 62.Wu Y., Song P., Xu J., Zhang M., Zou M-H. 2007. Activation of protein phosphatase 2a by palmitate inhibits AMP-activated protein kinase. J. Biol. Chem. 282: 9777–9788 [DOI] [PubMed] [Google Scholar]
- 63.Hummasti S., Hotamisligil G. S. 2010. Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ. Res. 107: 579–591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.