Abstract
Phosphoinositides (PI) play important regulatory roles in cell physiology. Localization and quantitation of PIs within the cell is necessary to understand their precise function. Currently, ectopic expression of green fluorescent protein (GFP)-fused PI-binding domains is used to visualize PIs localized to the cell membrane. However, ectopically expressed PI-binding domains may compete with endogenous binding proteins, thus altering the physiological functions of the PIs. Here, we establish a novel method for quantification and visualization of PIs in cells and tissue samples using PI-binding domains labeled with quantum dots (Qdot) as specific probes. This method allowed us to simultaneously quantify three distinct PIs, phosphatidylinositol 3,4,5-triphosphatase [PtdIns(3,4,5)P3), PtdIns(3,4)P2, and PtdIns(4,5)P2, in crude acidic lipids extracted from insulin-stimulated cells. In addition, the method allowed the PIs to be visualized within fixed cells and tissues. Sequential and spatial changes in PI production and distribution were detected in platelet-derived growth factor (PDGF)-stimulated NRK49F cells. We also observed accumulation of PtdIns(3,4)P2 at the dorsal ruffle in PDGF-stimulated NIH3T3 cells. Finally, we found PtdIns(3,4,5)P3 was enriched in lung cancer tissues, which also showed high levels of phosphorylated Akt. Our new method to quantify and visualize PIs is expected to provide further insight into the role of lipid signaling in a wide range of cellular events.
Keywords: pleckstrin homology domain, measurement, membrane ruffle, phosphoinositide 3-kinase
Phosphoinositides (PI) play important regulatory roles in a variety of cellular events, including signal transduction, cytoskeletal regulation, and membrane trafficking (1–3). The formation and breakdown of PIs are catalyzed by families of PI kinases and phosphatases. Recent studies have revealed that the improper functioning of these enzymes is associated with disorders such as diabetes and cancer, which suggests that cellular concentrations of each PI species is usually under strict control (4, 5).
A number of PI-binding domains have been identified, including pleckstrin homology (PH); Phox (PX); Fab1, YOTB, Vac1p, and EEA1 (FYVE); Epsin N-terminal homology (ENTH); and Glucosyltransferase, Rab-like GTPase activator, and myotubularin (GRAM) domains (6, 7). Although most PH domains show little specificity in the PIs they bind, several show relatively high specificity; these include the PH domains of phospholipase Cδ1 (PLCδ1) for PtdIns(4,5)P2 and the general receptor for phosphoinositides-1 (GRP1) for PtdIns(3,4,5)P3 (8, 9). In addition, the tandem-PH domain-containing protein-1 (TAPP1) and the four phosphate-adaptor protein-1 (FAPP1) PH domains specifically bind PtdIns(3,4)P2 and PtdIns4P, respectively (10, 11). Other domains that bind specific PIs have also been described. The FYVE domains of hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and early endosome antigen-1 (EEA1) specifically recognize PtdIns3P, which is primarily found in endosomes, multivesicular bodies, and phagosomes (12). The GRAM domains of myotubularin and myotubularin-related protein 2 (MTMR2) are involved in endosomal and vacuolar targeting through interaction with PtdIns(3,5)P2 (13). Moreover, some PI-binding domains have functions beyond simply recruiting host proteins to the membrane surface. Such domains, including ENTH, Bin Amphiphysin Rvs (BAR), and FCH and BAR (F-BAR), bind PIs with little specificity and participate in endocytosis, cytokinesis, and other processes through their intrinsic membrane-bending abilities (14).
The intracellular localization of each PI species has been studied by transfection of cultured cells with green fluorescent protein (GFP)-fused PH domains (15). However, this method may not accurately detect intracellular PIs. Balla et al. reported that the GFP-PLCδ1-PH domain was localized only to the plasma membrane despite some of its target phosphoinositide, PtdIns(4,5)P2, being present in the Golgi membrane and secretory vesicles as well (16). In addition, simultaneously visualizing distinct PI species is difficult by using this conventional method, and overexpression of PI-binding domains could interfere with the normal function of PIs by competing with their target proteins. Indeed, Varnai et al. demonstrated that overexpression of isolated PH domains could inhibit PtdIns(3,4,5)P3-regulated cellular pathways (17). Finally, the major drawback of the GFP-PH domain method is that it cannot be applied to tissues or clinical samples.
In this study, we developed a new method for quantifying and visualizing multiple PIs using quantum dot (Qdot)–labeled PI-binding domains as specific probes. With this method, we succeeded in simultaneously quantifying the levels of PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 in crude lipid samples. We also visualized each PI in fixed cells or tissues, and we demonstrated their differential membrane distribution, both spatially and temporally.
MATERIALS AND METHODS
Materials
Phosphoinositides (diC16; dipalmitoyl) were purchased from CellSignals Inc. (Columbus, OH). Phosphatidylethanolamine (PE) and phosphatidylcholine (PC) were purchased from Avanti Polar Lipids (Alabaster, AL).
Constructs
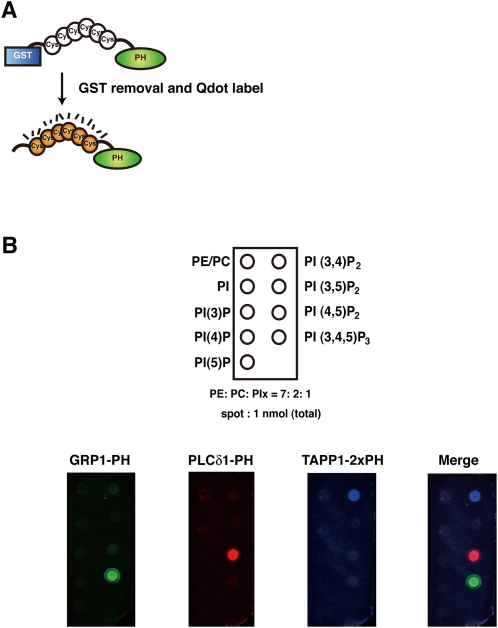
To construct the GST fusion 6Cys proteins, tgctgctgctgctgctgc was inserted into the BamHI site of the pGEX6P-1 vector (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The sequences of the GRP1-PH, PLCδ1-PH, and TAPP1-2×PH domains were inserted into the appropriate sites of the vector. Site-specific mutagenesis of GRP1-PH and TAPP1-2×PH was performed by PCR using mutated primers. GST fusion proteins were purified from E. coli BL21 with glutathione-sepharose (GE Healthcare) according to the manufacturer's protocol, and the GST tag was removed by incubation with PreScission protease at 4°C overnight. Each purified 6Cys-PH domain was labeled with the Qdot antibody conjugation kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Dot blot assay
One nanomole of each liposome (PE: PC: PIx = 7: 2: 1, where PIx indicates the appropriate PI species) was spotted onto a nitrocellulose membrane. The membrane was dried and blocked with 5% skim milk, 1% BSA (BSA), and 0.05% Tween 20 in phosphate buffered saline (PBS). The membrane was then coincubated with Qdot-labeled 6Cys-PLCδ1-PH, 6Cys-GRP1-PH, and 6Cys-TAPP1-2×PH domains for 2 h at room temperature. Following incubation, this membrane was washed several times with PBS containing 0.05% Tween 20, and then scanned with a FLA-8000 fluorescent image analyzer (Fujifilm, Tokyo, Japan).
Phosphoinositide extraction from cells
Cells were seeded into 60 mm dishes and starved with serum-free medium before stimulation with insulin, as described below. Lipids were extracted from cell lysates as described previously (18). The lipids were spotted onto nitrocellulose membranes and quantified with Qdot-labeled 6Cys-PH domains, as described above.
Cell culture, transfection, and fluorescence microscopy
NIH3T3 cells were cultured in DMEM (Sigma-Aldrich, St. Louis, MO) containing 10% calf serum. Chinese hamster ovary (CHO) cells stably expressing the wild-type human insulin receptor (CHO-IR) were cultured in Ham's F-12 medium (Invitrogen) containing 10% fetal calf serum (FCS). NRK49F cells were maintained in DMEM with 10% FCS. NIH3T3 and NRK49F cells were starved with DMEM for 8 h before stimulation with 20 ng/ml PDGF-BB. CHO-IR cells were starved with Ham's F-12 medium for 8 h before stimulation with 100 nM insulin.
Transient transfection of CHO-IR cells was performed using Lipofectamine 2000 reagents (Invitrogen). After 24 h incubation, cells were starved with Ham's F-12 medium and stimulated for varying times with 100 nM of insulin.
Cells were fixed and permeabilized at 37°C for 1 h with 3.7% (w/v) formaldehyde, 0.1% (w/v) glutaraldehyde, and 1.5 mg/ml saponin in buffer (5 mM KCl, 137 mM NaCl, 4 mM NaHCO3, 0.4 mM KH2PO4, 1.1 mM Na2HPO4, 2 mM MgCl2, 5 mM PIPES, pH 7.2, plus 2 mM EGTA and 5.5 mM glucose), as described previously (19). The cells were stained with Qdot-6Cys-GRP1-PH, Qdot-6Cys-PLCδ1-PH, and Qdot-6Cys-TAPP1-2×PH domains and observed with an FV-1000 confocal microscope (Olympus, Tokyo, Japan).
Wound healing
NIH3T3 cells were grown on glass cover slips in 12-well plates. Confluent cells were ruptured using a P1000 pipette tip. These cells were then incubated in fresh culture medium, fixed as described above, and stained with Qdot-6Cys-GRP1-PH, Qdot-6Cys-PLCδ1-PH, and Qdot-6Cys-TAPP1-2×PH domains.
Immunohistochemistry
Arrays of frozen human cancer tissues were purchased from ISU ABXIS (Seoul, South Korea). Tissues were fixed as described above and incubated with anti-pAkt Ser473 antibody (Cell Signaling Technology, Beverley, MA) and Qdot-6Cys-GRP1-PH domains. TO-PRO-3 iodide was used for nuclear staining. Fluorescence images were obtained using an FV-1000 confocal microscope.
RESULTS
Construction of PI-specific probes
To determine whether ectopically expressed PI-binding domains influence cell morphology, we transfected CHO-IR cells with various PH domain constructs. Cells overexpressing GFP were used as a control and developed membrane ruffling upon insulin stimulation (supplementary Fig. I). In contrast, cells overexpressing any of the individual PH domains, except PLCδ1, showed poor membrane ruffling, consistent with previous observations (17).
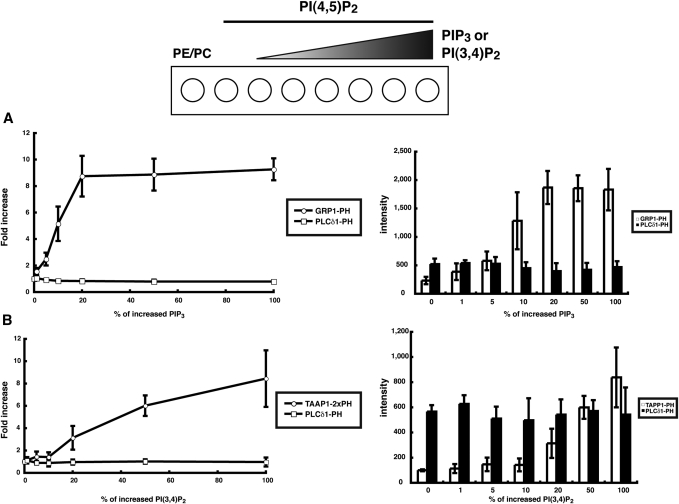
To overcome the limitations associated with the GFP-PH domain constructs, we established a new method using PI-binding domains as specific probes. For this, GRP1-PH, PLCδ1-PH, and TAPP1-2×PH were selected as specific probes for PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2, respectively. We used TAPP1-2×PH for the detection of PtdIns(3,4)P2 because this probe possesses two PH domains in tandem, owing to which the binding affinity of this probe is approximately 10-fold higher than that with a single PH domain (6). Initially, we labeled the PH domain by covalently modifying the lysine residues with Qdots. However, these PH domains showed very low affinities for the PIs, possibly due to electrostatic repulsion between the positively charged residues in the PH domains and the negatively charged phosphates in the PIs (data not shown). Thus, six tandem cysteine residues were introduced at the amino-termini of the PH domains to allow labeling of thiols with amine-derivatized, polyethylene glycol (PEG)-coated Qdot nanocrystals, using the thiol crosslinker succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxy-(6-amidocaproate) (SMCC) (Fig. 1A). Newly designed Qdot probes (hereafter referred to as Qdot-6Cys-PH) retained the lipid specificities and sensitivities of the parent PH domains, as shown by overlay assays. Thus, Qdot-6Cys-GRP1-PH, Qdot-6Cys-PLCδ1-PH, and Qdot-6Cys TAPP1-2×PH specifically bound to PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2, respectively, whereas proteins containing point mutations failed to do so (Fig. 1B and supplementary Fig. II). Each probe recognized PIs in a concentration-dependent manner with a sensitivity of less than 5 pmol (Fig. 2). PtdIns(4,5)P2 was present in cell membranes at significantly higher concentrations than PtdIns(3,4,5)P3 or PtdIns(3,4)P2, and thus might interfere with their recognition by the probes. Therefore, we tested the probe specificities by mixing increasing concentrations of PtdIns(3,4,5)P3 or PtdIns(3,4)P2 with a constant and high level of PtdIns(4,5)P2 in the same area of the membrane. We found that both Qdot-6Cys-GRP1-PH and Qdot-6Cys-TAAP1-2xPH probes specifically recognized PtdIns(3,4,5)P3 and PtdIns(3,4)P2, respectively, even in the presence of high levels of PtdIns(4,5)P2 (Fig. 3), indicating that these probes can be used to detect PIs in crude lipid samples extracted from cultured cells or tissues.
Fig. 1.
Construction of specific PI binding probes by using PH domains. (A) Scheme for building PI-specific binding PH domains. PI-binding PH domains were expressed and purified as GST fusion proteins. GST was removed by incubation with PreScission protease, and PH domains were then covalently coupled to Qdots. (B) Binding specificity of probes in a dot blot assay. One nanomole of liposome (PE: PC: PIx = 7: 2: 1) was spotted onto a nitrocellulose membrane. The membrane was incubated with Qdot-6Cys-GRP1-PH, Qdot-6Cys-PLCδ1-PH, and Qdot-6Cys-TAPP1-2×PH domains.
Fig. 2.
Sensitivity of Qdot-labeled probes for each PI. The indicated amount of liposomes consisting of (PE: PC: PIx = 7: 2: 1) were spotted onto a nitrocellulose membrane and incubated with Qdot-labeled probes.
Fig. 3.
Specificity of Qdot-labeled probes. Liposomes were formed consisting of PE: PC = 7: 2, a constant amount of PtdIns(4,5)P2, and 0, 1, 5, 10, 20, 50, or 100 pmol of either PtdIns(3,4,5)P3 (A) or PtdIns(3,4)P2 (B). Liposomes were spotted on the membrane, which was then incubated with either Qdot-6Cys-GRP1-PH and Qdot-6Cys-PLCδ1-PH (A) or Qdot-6Cys-TAPP1-2×PH and Qdot-6Cys-PLCδ1-PH (B). The circles in the left graphs indicate the increase in GRP1-PH signals (A) or TAPP1-2×PH signals (B), and the squares show the corresponding increases in the PLCδ1-PH signals (A and B). Histograms on the right show the absolute intensity values of the indicated probes. Results are the mean ± SD of three independent experiments.
Quantification of PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 in cells
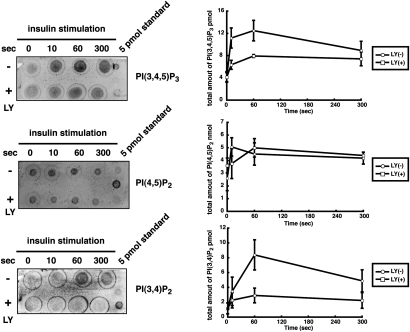
We next measured the amount of PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 in insulin-stimulated CHO-IR cells. Acidic lipids were extracted from cell lysates, spotted onto a nitrocellulose membrane, and the levels of all three PIs were quantified simultaneously with the Qdot-labeled probes. PtdIns(3,4,5)P3 was found to be elevated within 10 s of insulin stimulation and then decreased over the next 5 min (Fig. 4). In contrast, PtdIns(3,4)P2 levels increased more slowly and reached a maximum 60 s after stimulation. This result supports previous observations that PtdIns(3,4)P2 is produced mainly from the dephosphorylation of PtdIns(3,4,5)P3 by PI 5-phosphatase (20). In contrast, PtdIns(4,5)P2 levels were largely unchanged during the 5 min incubation following addition of insulin (Fig. 4). As expected, the PI 3-kinase (PI3K) inhibitor LY294002 significantly blocked the production of both PtdIns(3,4,5)P3 and PtdIns(3,4)P2. Collectively, these results show that the simultaneous measurement of PIs in a crude lipid mixture is feasible using our method, which does not rely on thin layer chromatography (TLC) or high performance lipid chromatography (HPLC) separation procedures.
Fig. 4.
Quantification of PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 in insulin-stimulated CHO-IR cells. Before stimulation, cells were incubated for 15 min with 25 μM of LY294002 [LY(+) or LY(−)]. For quantification of PtdIns(4,5)P2, lipids were diluted 100-fold before spotting. A standard curve was used for quantification. Data are mean ± SD of three independent experiments.
Visualization of PIs in insulin-stimulated CHO-IR cells
Next, we determined if the Qdot probes could be used to visualize PIs in cultured cells. First, various concentrations of saponin for permeabilization were tested with a slight modification as described previously (19). As shown in supplementary Fig. III, saponin concentration of 1.5 mg/ml was sufficient for the Qdot probes to visualize plasma membrane, although lower concentration appeared less efficient. We also carried out the same staining using a single Qdot probe to exclude the possibility that the localizations of multiple probes were affected by each other. Data shown in supplementary Fig. IV clearly demonstrate that there is virtually no difference between single- and multiple-stained samples (compare supplementary Fig. IV with Fig. 5), confirming that Qdot probes can be used not only to detect single PI but also to simultaneously detect three distinct PIs in cultured cells. In addition, we stained cells that expressed GFP-lipid binding domain with Qdot-labeled probe. GFP-PLCδ1 PH domain was localized throughout plasma membranes and, correspondingly, Qdot-labeled PLCδ1 PH probe also stained plasma membranes (supplementary Fig. V).
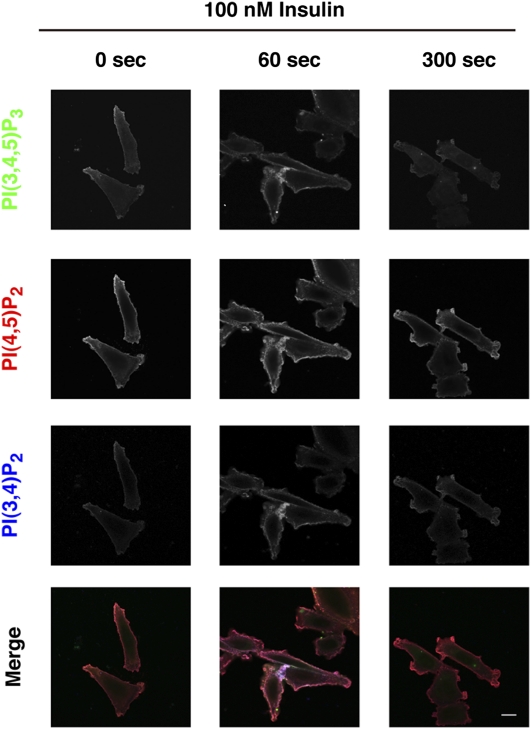
Fig. 5.
Costaining of PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 in insulin-stimulated CHO-IR cells. PtdIns(3,4,5)P3 and PtdIns(3,4)P2 were produced at plasma membranes in response to insulin stimulation. Cells were incubated with 100 nM insulin for the indicated times, fixed, and stained with lipid binding probes. Bar = 10 μm.
In quiescent CHO-IR cells, PtdIns(3,4,5)P3 and PtdIns(3,4)P2 were only weakly detectable, whereas PtdIns(4,5)P2 was present constitutively at the plasma membrane (Fig. 5). Following insulin stimulation, the levels of all three PIs increased, particularly at the lamellipodia, which are actin-rich plasma membrane protrusions that rapidly form in response to activation of PI3K. The observed increase in PtdIns(4,5)P2 could result from activation of type I phosphatidylinositol-4-phosphate-5-kinases (PIP5KI), which are known to generate PtdIns(4,5)P2 and affect organization of the actin cytoskeleton (21). LY294002 blocked the insulin-stimulated increases in PtdIns(3,4,5)P3 and PtdIns(3,4)P2, as well as lamellipodia formation (supplementary Fig. VI). Importantly, the insulin-stimulated increase in PtdIns(3,4,5)P3 and PtdIns(3,4)P2 was not detected in cells expressing mutated PH domains (supplementary Fig. VII).
Distinct localization of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 in PDGF-stimulated NRK49F cells
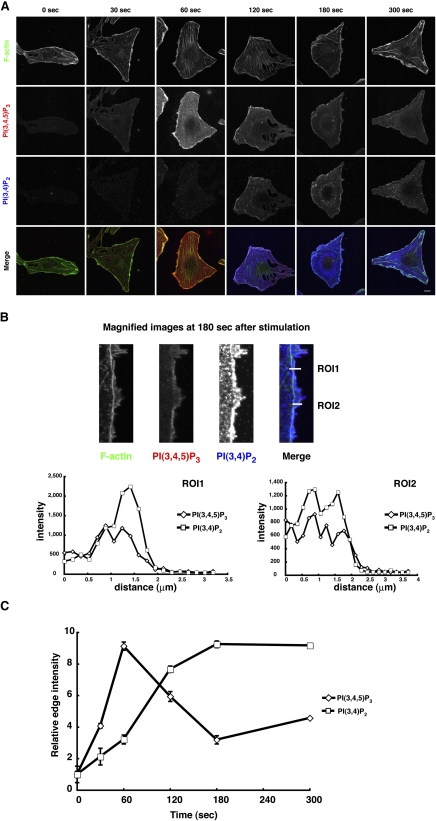
We next used confocal microscopy of the Qdot-PH domain probes to detect the kinetics of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 production in NRK49F cells. Accumulation of PtdIns(3,4,5)P3 at the plasma membrane was detected 60 s after cell stimulation with PDGF and then it was gradually decreased, whereas production of PtdIns(3,4)P2 was delayed and became detectable 120 s following stimulation (Fig. 6A, C). High-resolution analysis was performed to show the difference in localization of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 at 180 s poststimulation (Fig. 6B). In both regions of interest (ROI), we observed high PtdIns(3,4)P2 and low PtdIns(3,4,5)P3 levels (ROI1 and ROI2 in Fig. 6B), indicating that PtdIns(3,4)P2 was likely produced from PtdIns(3,4,5)P3. These results show that our probes can simultaneously detect the spatial differences in PI localization.
Fig. 6.
Distinct localization of PtdIns(3,4,5)P3 and PtdIns(3,4)P2 in PDGF-stimulated NRK49F cells. (A) NRK49F cells were stimulated with PDGF for the indicated times. The cells were fixed and then stained with probes for PtdIns(3,4,5)P3 and PtdIns(3,4)P2. Bar = 10 μm. (B) Magnified images of the cell edge in PDGF-stimulated cells after 180 s. The graphs show the intensity of PtdIns(3,4,5)P3 (diamonds) and PtdIns(3,4)P2 (squares) in ROI1 and ROI2. (C) Fluorescence intensity of Qdot-6Cys-GRP1-PH and Qdot-6Cys-TAPP1-2×PH staining at the cell edge was quantified. Data represents mean ± SD (n = 3), with the signals at time 0 taken as the standard.
PtdIns(3,4)P2 enrichment in PDGF-induced dorsal ruffles
The PI3K-driven signaling pathway induces rapid reorganization of the actin cytoskeleton, which gives rise to not only lamellipodia but also peripheral and dorsal membrane ruffles (22, 23). These transient and dynamic changes in the actin cytoskeleton are initiated by the production of PIs at discrete regions of the plasma membrane. We examined whether there was a difference in the localization of the PIs in PDGF-stimulated NIH3T3 fibroblasts. We found that PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4)P2 were all localized at peripheral membrane ruffles in response to PDGF stimulation (Fig. 7A). Interestingly, PtdIns(3,4)P2 was more concentrated at dorsal ruffles, consistent with previous reports (24, 25) (Fig. 7B). As expected, no increase in PtdIns(3,4,5)P3 and PtdIns(3,4)P2 was detected at peripheral or dorsal ruffles using the PH domain mutants (supplementary Fig. VIII).
Fig. 7.
Localization of PtdIns(3,4)P2 in PDGF-induced dorsal ruffles of NIH3T3 cells. (A) Both PtdIns(3,4,5)P3 and PtdIns(3,4)P2 were present in peripheral membrane ruffles of NIH3T3 cells after PDGF stimulation. (B) PtdIns(3,4)P2 became concentrated in dorsal ruffles in response to PDGF stimulation. Arrow indicates the enrichment of PtdIns(3,4)P2 at the dorsal ruffle. The bar graph indicates the intensity profile in ROI1. Magnified images are derived from the regions shown by boxes. Bars = 10 μm in (A) and 20 μm in (B).
Localization of PIs in polarized cells
The morphological changes that occur during cell migration are associated with a polarized distribution of PIs. PtdIns(3,4,5)P3 is known to mediate the effect of growth factors on the formation of peripheral ruffles implicated in cell movement (26). Consistently, we noted that all three PIs were localized at the leading edge of the cell membrane in migrating cells; however, only PtdIns(4,5)P2 was also found at the opposing side of the plasma membrane (Fig. 8A–C). This finding confirms that the PIs display polarized distribution to the front and back of the cell during movement.
Fig. 8.
Localization of PIs in migrating cells. (A) NIH3T3 cells were stained with Qdot-6Cys-GRP1-PH, Qdot-6Cys-PLCδ1-PH, and Qdot-6Cys-TAPP1-2×PH after wound induction. Magnified images are derived from the solid and dashed boxes. An image of wound edge area is shown. (B) Graph shows intensity profiles of green, red, and blue channels in the ROI. (C) Graph shows mean ± SD (n = 3) of the ratio of front-to-rear fluorescence intensity of each channel. Bars = 20 μm.
Colocalized elevation of Akt phosphorylation and PtdIns(3,4,5)P3 in lung cancer tissue
Many human cancers exhibit elevated PI3K activity or reduced PtdIns(3,4,5)P3 3-phosphatase (PTEN) activity, each of which results in the accumulation of PtdIns(3,4,5)P3 (27, 28). We stained various cancer tissues, including thyroid, stomach, liver, lung, colon, and breast cancers, with our Qdot probes to detect regions of high PI concentration. We found clear staining of lung cancer tissue with the Qdot-6Cys-GRP1-PH probe (Fig. 9A), which indicates a significant accumulation of PtdIns(3,4,5)P3. Importantly, staining of phosphorylated Akt (pAkt) colocalized with that of PtdIns(3,4,5)P3 (Fig. 9A). In contrast, neither pAkt nor PtdIns(3,4,5)P3 was detected in control nonneoplastic lung tissue (Fig. 9B).
Fig. 9.
Staining of PtdIns(3,4,5)P3 in lung tissue. Lung cancer tissue (squamous cell carcinoma) (A) and lung nonneoplastic tissue (B) were stained with pAkt (Ser473) antibody and Qdot-6Cys-GRP1-PH domains. Arrow indicates colocalization of PtdIns(3,4,5)P3 and phosphorylated Akt. TO-PRO-3 iodide was used for nuclear staining. Bars = 10 μm.
DISCUSSION
In this study, we describe a new method that allows quantification of cellular PIs and their noninvasive visualization in cultured cells and tissue samples. Our method enables sensitive and simultaneous quantification of three PIs in small amounts of crude acidic lipids more easily than conventional methods. PI metabolism has been studied using radioisotope labeling combined with TLC or HPLC techniques, which are technically demanding and complex (29, 30). In addition, variations in metabolic labeling efficiency cannot theoretically be excluded. Although a new nonradioisotope method has been developed to quantify PtdIns(3,4,5)P3 (31), specialized HPLC-mass spectrometry equipment is required. In contrast, our method permits quantification and visualization of PIs more easily and in a more direct manner. Moreover, PtdIns(3,4,5)P3 and PtdIns(3,4)P2, which are present in very low amounts in the cell, were readily detected by the Qdot-labeled PH domain probes.
There are currently two methods to visualize phosphoinositides in cells: one is to express lipid binding domains as GFP-fused proteins in intact cells, and the second is to use commercially available anti-phosphoinositide antibodies or lipid binding domains in fixed cells. Although GFP-fusion PH domain probes can be used for live cell imaging, their expression allows limited resolution and may alter cell function, either by sequestering lipid or by promoting and/or interfering with lipid-protein interactions. Indeed, Varnai et al. showed that overexpression of isolated PH domains could inhibit PtdIns(3,4,5)P3-regulated pathways (17). In addition, because the expressed PH domains were distributed throughout the plasma membrane, this approach lacks the ability to spatially resolve two or three PIs at the plasma membrane.
Electron microscopic techniques have been used to map PI distribution at high resolution, using lipid binding domains and antibodies. Chemical fixation of lipids is an essential methodological consideration when measuring PIs. Downes et al. have shown that aldehyde-based fixation methods more efficiently immobilize membrane proteins than membrane lipids (32). Fixation of lipids requires the inclusion of glutaraldehyde, either alone or in combination with formaldehyde (33, 34). By using this method, PtdIns(3,4,5)P3 was shown to localize at both plasma and nuclear membranes of agonist-induced cells, whereas PtdIns(3,4)P2 localized at the plasma membrane and intracellular organelles (35, 36). More recently, Fujita et al. developed an electron microscopic method that used freeze-fracture replicas for the detection of PtdIns(4,5)P2 without chemical fixation (37, 38). Their method is superior in resolution but requires more sophisticated techniques.
Light microscopy imaging techniques have also been used to detect PIs in fixed cells, including detection by anti-phosphoinositide antibodies (19, 34, 39). For this method, glutaraldehyde was used to preserve plasma membrane structures. Triton, which is commonly used in immunocytochemical experiments, induces clustering of PtdIns(4,5)P2 detectable by fluorescence resonance energy transfer (FRET), with visible clustering observed at 0.005% Triton (40). Therefore, saponin was used for membrane permeabilization. By doing so, PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(4)P at the plasma membrane were visualized in fixed cells. However, this method allows only one species of PI to be stained (19, 34, 39), and it is important to analyze a number of PIs simultaneously because cellular events are coordinately controlled by different PI species. Our method allows visualization of three different PIs in stimulated cells, and the spatial resolution is much improved compared with the method using GFP-fusion PH domains.
With the new method, we observed an enrichment of PtdIns(3,4)P2 in dorsal ruffles of PDGF-stimulated NIH3T3 cells, and PtdIns(3,4,5)P3 was present in peripheral ruffles (Fig. 7). TAPP1, a PtdIns(3,4)P2-binding protein, is recruited to dorsal ruffles, which suggests that PtdIns(3,4)P2 may be involved in dorsal ruffle formation (24). In another study, we reported that PtdIns(3,4,5)P3-binding SH3YL1 regulates dorsal ruffle formation by recruiting SHIP2, which dephosphorylates PtdIns(3,4,5)P3 to PtdIns(3,4)P2 at dorsal ruffles (25). Collectively, these findings indicate that PtdIns(3,4)P2 is a key molecule in the formation of circular F-actin–enriched structures, and it will be of interest to investigate this further.
The number of cellular processes known to be directly or indirectly controlled by PIs has expanded dramatically. Thus, the ability to detect and visualize multiple species of PIs will play a critical role in understanding the many functions of these signaling lipids.
Supplementary Material
Footnotes
Abbreviations:
- CHO-IR
- Chinese hamster ovary cells expressing the wild-type human insulin receptor
- CHO-IR
- GFP, green fluorescent protein
- GRP1
- general receptor for phosphoinositides-1
- PC
- phosphatidylcholine
- PDGF
- platelet-derived growth factor
- PE
- phosphatidylethanolamine
- PH
- pleckstrin homology
- PI
- phosphoinositide
- PI3K
- PI 3-kinase
- PLCδ1
- phospholipase Cδ1
- Qdot
- quantum dot
- ROI
- region of interest
- TAPP-1
- tandem-PH domain-containing protein-1
This work was supported in part by a Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Creative Scientific Research.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures.
REFERENCES
- 1.Janmey P. A., Lindberg U. 2004. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 5: 658–666 [DOI] [PubMed] [Google Scholar]
- 2.Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443: 651–657 [DOI] [PubMed] [Google Scholar]
- 3.Vicinanza M., D'angelo G., Di Campli A., De Matteis M. A. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27: 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazar D. F., Saltiel A. R. 2006. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat. Rev. Drug Discov. 5: 333–342 [DOI] [PubMed] [Google Scholar]
- 5.Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419: 29–49 [DOI] [PubMed] [Google Scholar]
- 6.Furutani M., Tsujita K., Itoh T., Ijuin T., Takenawa T. 2006. Application of phosphoinositide-binding domains for the detection and quantification of specific phosphoinositides. Anal. Biochem. 355: 8–18 [DOI] [PubMed] [Google Scholar]
- 7.Lemmon M. A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9: 99–111 [DOI] [PubMed] [Google Scholar]
- 8.Várnai P., Balla T. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray A., Van Der Kaay J., Downes C. P. 1999. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem. J. 344: 929–936 [PMC free article] [PubMed] [Google Scholar]
- 10.Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6: 393–404 [DOI] [PubMed] [Google Scholar]
- 12.Raiborg C., Bremnes B., Mehlum A., Gillooly D. J., D'Arrigo A., Stang E., Stenmark H. 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114: 2255–2263 [DOI] [PubMed] [Google Scholar]
- 13.Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. 2004. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 279: 13817–13824 [DOI] [PubMed] [Google Scholar]
- 14.Itoh T., De Camilli P. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 1761: 897–912 [DOI] [PubMed] [Google Scholar]
- 15.Várnai P., Balla T. 2006. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta. 1761: 957–967 [DOI] [PubMed] [Google Scholar]
- 16.Balla T., Bondeva T., Várnai P. 2000. How accurately can we image inositol lipids in living cells? Trends Pharmacol. Sci. 21: 238–241 [DOI] [PubMed] [Google Scholar]
- 17.Várnai P., Bondeva T., Tamas P., Toth B., Buday L., Wen L. 2005. Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns (3, 4, 5)P3 suggest their interaction with protein binding partners. J. Cell Sci. 118: 4879–4888 [DOI] [PubMed] [Google Scholar]
- 18.Gray A., Olsson H., Batty I. H., Priganica L., Peter Downes C. 2003. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 313: 234–245 [DOI] [PubMed] [Google Scholar]
- 19.Yip S. C., Eddy R. J., Branch A. M., Pang H., Wu H., Yan Y., Drees B. E., Neilsen P. O., Condeelis J., Backer J. M. 2008. Quantification of PtdIns(3,4,5)P(3) dynamics in EGF-stimulated carcinoma cells: a comparison of PH-domain-mediated methods with immunological methods. Biochem. J. 411: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke T. F., Kaplan D. R., Cantley L. C., Toker A. 1997. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 275: 665–668 [DOI] [PubMed] [Google Scholar]
- 21.Kisseleva M., Feng Y., Ward M., Song C., Anderson R. A., Longmore G. D. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25: 3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovceva E., Larsen M. R., Waterfield M. D., Baum B., Timms J. F. 2007. Dynamic cofilin phosphorylation in the control of lamellipodial actin homeostasis. J. Cell Sci. 120: 1888–1897 [DOI] [PubMed] [Google Scholar]
- 23.Weiger M. C., Wang C. C., Krajcovic M., Melvin A. T., Rhoden J. J., Haugh J. M. 2009. Spontaneous phosphoinositide 3-kinase signaling dynamics drive spreading and random migration of fibroblasts. J. Cell Sci. 122: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan A., Yakubchyk Y., Chabot J., Obagi C., Daher E., Maekawa K., Gee S. H. 2004. The phosphoinositol 3,4-bisphosphate-binding protein TAPP1 interacts with syntrophins and regulates actin cytoskeletal organization. J. Biol. Chem. 279: 53717–53724 [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa J., Tokuda E., Tenno T., Tsujita K., Sawai H., Hiroaki H., Takenawa T., Itoh T. 2011. SH3YL1 regulates dorsal ruffle formation by a novel phosphoinositide-binding domain. J. Cell Biol. 193: 901–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oikawa T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., Takenawa T. 2004. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 6: 420–426 [DOI] [PubMed] [Google Scholar]
- 27.Marone R., Cmiljanovic V., Giese B., Wymann M. P. 2008. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim. Biophys. Acta. 1784: 159–185 [DOI] [PubMed] [Google Scholar]
- 28.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., et al. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science. 304: 554. [DOI] [PubMed] [Google Scholar]
- 29.Blank M. L., Cress E. A., Lee P., Stephens N., Piantadosi C., Snyder F. 1983. Quantitative analysis of ether-linked lipids as alkyl- and alk-1-enyl-glycerol benzoates by high-performance liquid chromatography. Anal. Biochem. 133: 430–436 [DOI] [PubMed] [Google Scholar]
- 30.Wenk M. R., Pellegrini L., Klenchin V. A., Di Paolo G., Chang S., Daniell L., Arioka M., Martin T. F., De Camilli P. 2001. PIP kinase Igamma is the major PI(4,5)P(2) synthesizing enzyme at the synapse. Neuron. 32: 79–88 [DOI] [PubMed] [Google Scholar]
- 31.Clark J., Anderson K. E., Juvin V., Smith T. S., Karpe F., Wakelam M. J., Stephens L. R., Hawkins P. T. 2011. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 8: 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Downes C. P., Gray A., Watt S. A., Lucocq J. M. 2003. Advances in procedures for the detection and localization of inositol phospholipid signals in cells, tissues, and enzyme assays. Methods Enzymol. 366: 64–84 [DOI] [PubMed] [Google Scholar]
- 33.Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. 2002. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 363: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammond G. R., Schiavo G., Irvine R. F. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 422: 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watt S. A., Kimber W. A., Fleming I. N., Leslie N. R., Downes C. P., Lucocq J. M. 2004. Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy. Biochem. J. 377: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsay Y., McCoull D., Davidson L., Leslie N. R., Fairservice A., Gray A., Lucocq J., Downes C. P. 2006. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 119: 5160–5168 [DOI] [PubMed] [Google Scholar]
- 37.Fujita A., Cheng J., Tauchi-Sato K., Takenawa T., Fujimoto T. 2009. A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA. 106: 9256–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita A., Cheng J., Fujimoto T. 2010. Quantitative electron microscopy for the nanoscale analysis of membrane lipid distribution. Nat. Protoc. 5: 661–669 [DOI] [PubMed] [Google Scholar]
- 39.Hammond G. R., Dove S. K., Nicol A., Pinxteren J. A., Zicha D., Schiavo G. 2006. Elimination of plasma membrane phosphatidylinositol (4,5)-bisphosphate is required for exocytosis from mast cells. J. Cell Sci. 119: 2084–2094 [DOI] [PubMed] [Google Scholar]
- 40.van Rheenen J., Achame E. M., Janssen H., Calafat J., Jalink K. 2005. PIP2 signaling in lipid domains: a critical re-evaluation. EMBO J. 24: 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.