Rather than directly impairing 26S proteasomes, misfolded huntingtin may disrupt cellular proteostasis and lead to competition for limited 26S proteasome capacity.
Abstract
Pathognomonic accumulation of ubiquitin (Ub) conjugates in human neurodegenerative diseases, such as Huntington’s disease, suggests that highly aggregated proteins interfere with 26S proteasome activity. In this paper, we examine possible mechanisms by which an N-terminal fragment of mutant huntingtin (htt; N-htt) inhibits 26S function. We show that ubiquitinated N-htt—whether aggregated or not—did not choke or clog the proteasome. Both Ub-dependent and Ub-independent proteasome reporters accumulated when the concentration of mutant N-htt exceeded a solubility threshold, indicating that stabilization of 26S substrates is not linked to impaired Ub conjugation. Above this solubility threshold, mutant N-htt was rapidly recruited to cytoplasmic inclusions that were initially devoid of Ub. Although synthetically polyubiquitinated N-htt competed with other Ub conjugates for access to the proteasome, the vast majority of mutant N-htt in cells was not Ub conjugated. Our data confirm that proteasomes are not directly impaired by aggregated N-terminal fragments of htt; instead, our data suggest that Ub accumulation is linked to impaired function of the cellular proteostasis network.
Introduction
Ubiquitin (Ub) has been recognized for >20 yr to be a prominent and invariant feature of intracellular inclusion bodies (IBs) in the vast majority of neurodegenerative diseases, and Ub immunohistochemistry is considered to be the definitive marker in postmortem neuropathological diagnosis of these disorders (Lehman, 2009). In the absence of disease, Ub is diffusely distributed in brain tissue, whereas in diseased brain and in cell culture models thereof, cellular Ub becomes visibly concentrated together with aggregated proteins within IBs (Mayer et al., 1989), and markedly elevated levels of poly-Ub conjugates can be detected by mass spectrometry (MS) in tissue lysates (Bennett et al., 2007). Although the linkage of disrupted Ub homeostasis and protein aggregation to disease is undisputed, the functional significance of these correlations remains unresolved, and the mechanistic implications for pathogenesis remain controversial.
One prominent hypothesis holds that aggregated proteins inhibit protein degradation by the 26S proteasome (26S; Taylor et al., 2002; Ciechanover and Brundin, 2003; Valera et al., 2005; Ortega et al., 2007). Poly-Ub chains are best known for their role in targeting proteins for degradation by 26S, and enzymatic disassembly of these chains by 26S-associated deubiquitinating enzymes is essential to allow entry of substrates into the 20S proteolytic chamber and to “recycle” Ub to maintain cellular-free Ub levels (Finley, 2009). Experimental impairment of proteasome function leads to accumulation of Ub conjugates and to broad dysregulation of cellular Ub homeostasis, and mice harboring loss-of-function mutations in genes required for maintenance of Ub homeostasis develop neurodegenerative disorders (Saigoh et al., 1999; Wilson et al., 2002; Ryu et al., 2008). However, despite the well-documented accumulation of Ub conjugates in the neurodegenerative disease brain, there is no consistent evidence documenting an accompanying decrease in levels of free Ub or disruption of Ub pool dynamics in neurodegenerative disease or models thereof, and Ub conjugate accumulation may be a benign but robust indication of an underlying lesion in 26S function. In addition to degrading aggregation-prone, folding-defective proteins, proteasomes control the levels of critical cellular regulators, and impaired 26S function has severe and pleiotropic consequences for cell function and survival (Kawazoe et al., 1998).
The hypothesis that protein aggregates impair 26S function is supported by the demonstration that endogenous short-lived 26S substrates accumulate in models of Huntington’s disease (HD; Jana et al., 2001; Zemskov and Nukina, 2003) and by studies using synthetic reporters consisting of GFP fused to destabilizing degrons (Dantuma et al., 2000; Bence et al., 2001; Bennett et al., 2005). In cells expressing these reporters, steady-state levels of GFP fluorescence should—assuming constant synthesis rates—provide a readout of Ub–proteasome system (UPS) functional capacity. Multiple studies have confirmed that the levels of these reporters, when expressed in mammalian cell culture, increase significantly upon coexpression of the aggregation-prone mutant but not wild-type, nonaggregating variants of neurodegenerative disease-linked proteins (Bence et al., 2001; Illing et al., 2002; Petrucelli et al., 2002; Mandrusiak et al., 2003; Bennett et al., 2005; van Tijn et al., 2007; Deriziotis and Tabrizi, 2008). Although these studies are consistent with the hypothesis that protein aggregates can inhibit UPS function, direct measurement of proteasome function in extracts from brains of patients or animal models of neurodegenerative disease has yielded equivocal results, with decreased (Kabashi et al., 2004), increased (Bett et al., 2006), or unchanged (Diaz-Hernández et al., 2003; Gillardon et al., 2007) levels of proteasome activity being reported in different diseases and model systems.
In the present study, we have investigated the temporal and mechanistic relationship between aggregation of an N-terminal exon 1 fragment of huntingtin (htt; N-htt) and UPS function using a quantitative flow cytometry assay and live-cell time-lapse imaging to simultaneously interrogate N-htt expression, Ub distribution, and UPS function in single cells. Our data establish that N-htt—whether aggregated or not—does not choke or clog 26S and lead us to propose a model in which Ub accumulation is the consequence of simple competition for limited 26S capacity by cellular proteins that become diverted to the UPS in the face of disrupted protein folding homeostasis, which occurs when one or more components of the cell’s proteostasis network becomes overwhelmed by folding-impaired, aggregation-prone N-htt.
Results
UPS substrates accumulate above a critical concentration threshold of mutant htt expression
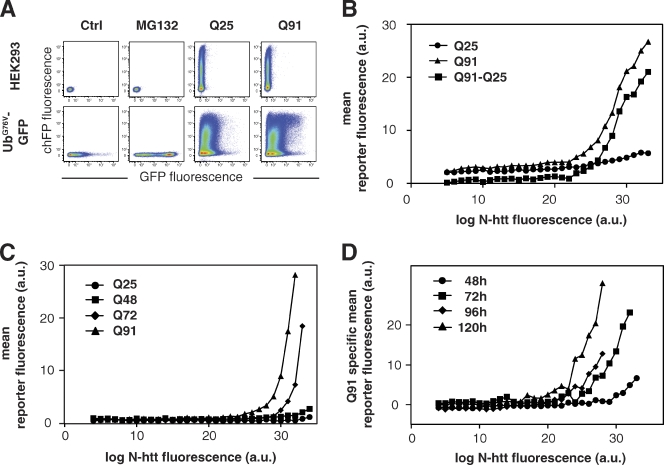
To precisely assess the quantitative relationship between the amount of N-htt expressed in cells and accumulation of a UPS reporter, we devised a two-color flow cytometry–based assay to simultaneously analyze UPS function using an unstable green fluorescent UPS reporter, UbG76V-GFP (Dantuma et al., 2000), and N-htt expression using a red (mCherry) fluorescent N-htt fragment, N-htt(Qn)–chFP (Fig. 1). Control experiments established that the presence of chFP does not alter the aggregation properties of N-htt(Qn) (Fig. S1 A) or its impact on the UPS (Fig. S1 B). Because N-htt(Q25)–chFP or N-htt(Q91)–chFP differ only in the length of the polyglutamine tract, the red fluorescence signal can be used to directly compare the expression levels of the two proteins. Expression of N-htt(Q25)–chFP in cells stably expressing UbG76V-GFP resulted in the appearance of a population of doubly labeled cells in which the distribution of green fluorescence was not substantially different from that of untransfected reporter cells (Fig. 1 A, bottom). In contrast, expression of N-htt(Q91)–chFP resulted in the appearance of a distinct subpopulation of cells displaying high levels of both green and red fluorescence. Transformation of the data in Fig. 1 A to plot the concentration of N-htt(Qn)–chFP versus UbG76V-GFP fluorescence (Fig. S2) revealed that although N-htt(Q25)–chFP had only a minimal effect on the level of green fluorescence, N-htt(Q91)–chFP induced accumulation of the UPS reporter at high expression levels (Fig. 1 B).
Figure 1.
UPS impairment occurs above a critical, polyglutamine length-dependent concentration of mutant htt. (A) Flow cytometry analysis of UPS function in HEK293 cells. Two-color scatter plots of HEK293 cells (top row) or HEK293 cells stably expressing UbG76V-GFP (bottom row). Cells were analyzed 72 h after transient transfection of N-htt(Q25)–chFP or N-htt(Q91)–chFP as indicated. Control experiments included cells incubated overnight with 10 µM MG132 or left untreated (control [Ctrl]). Graphs are presented as pseudocolor density plots with “hot” colors (i.e., red) indicating high frequency areas and “cold” colors (i.e., blue) indicating rare events. (B) UbG76V-GFP accumulates above a critical concentration threshold of mutant htt. Relationship between UbG76V-GFP fluorescence (vertical axis) and the fluorescence of N-htt(Q25)–chFP, N-htt(Q91)–chFP, or the difference between the Q91 and Q25 curves derived from flow cytometry as in A. a.u., arbitrary unit. Details of data transformation are presented in Fig. S2. (C) Effect of polyglutamine length on dose response of chFP-CL1 fluorescence to N-htt(Qn)–GFP expression, in which n = 25 (•), 48 (■), 72 (◆), and 91 (▴). (D) Effect of equivalent levels of expression of mutant N-htt on UbG76V-GFP accumulation becomes more severe with increasing expression time. HEK293 cells stably expressing UbG76V-GFP were transfected with N-htt(Q91)–chFP or N-htt(Q25)–chFP and analyzed by flow cytometry. The Q91-specific mean reporter fluorescence was calculated as in B at 48 (•), 72 (■), 96 (◆), or 120 (▴) h after transfection. The data shown are from single representative experiments out of at least two independent repeats.
The Q91-specific mean reporter fluorescence, determined by subtracting green fluorescence in cells expressing N-htt(Q25)–chFP from that of cells expressing N-htt(Q91)–chFP, shows that UbG76V-GFP accumulates significantly only in cells expressing N-htt(Q91)–chFP above a threshold (∼22 arbitrary units [a.u.]; Fig. 1 B). This threshold concentration of N-htt(Qn)–GFP at which the UPS reporter chFP-CL1 accumulated was inversely related to polyglutamine length (Fig. 1 C), suggesting a strict relationship between N-htt aggregation and UPS impairment. We further found that the concentration of N-htt(Q91)–chFP at which we observed stabilization of UbG76V-GFP was strongly affected by the length of time that the cells had been expressing mutant N-htt (Fig. 1 D). These data suggest that chronic expression of mutant N-htt decreases the intrinsic capacity of cells to degrade UPS substrates.
htt aggregation is associated with accumulation of diverse 26S substrates
Stabilization of UbG76V-GFP by high levels of mutant N-htt could be caused by interference by aggregated or oligomeric protein with 26S function, with some aspect of the Ub conjugation cascade, or both. To discriminate between these possibilities, we generated HEK293 cell lines stably expressing GFP molecules that are destabilized by degrons that target proteins to 26S by different Ub-dependent mechanisms. These reporters include, in addition to UbG76V-GFP (Dantuma et al., 2000), a substrate of the Ub fusion degradation pathway that recognizes proteins with an uncleavable N-terminal Ub (Johnson et al., 1995), Ub-Arg-GFP (Dantuma et al., 2000), a substrate of the N-terminal end rule pathway that recognizes proteins with destabilizing N-terminal amino acids (Varshavsky, 1997), and GFP-CL1 (Bence et al., 2001), which is destabilized by a short amphipathic motif that, in yeast, targets proteins for ubiquitination by a distinct set of E2 and E3 enzymes (Gilon et al., 2000). In addition, we examined the effect of N-htt(Qn)–chFP expression on the levels of cODC-GFP, a 26S substrate that is destabilized by a Ub-independent degron consisting of the 37 C-terminal amino acids from ornithine decarboxylase (ODC; Zhang et al., 2003). All four reporters, stably expressed in clonal HEK293 cell lines, exhibit low steady-state fluorescence in the absence of stress and time-dependent increases in diffuse cytoplasmic fluorescence after challenge with MG132 (Fig. S3, A and B). Control experiments using ts20 Balb/C 3T3 fibroblasts harboring a thermolabile E1 (Kulka et al., 1988) verified that the cODC-GFP fusion protein degradation is indeed subject to Ub-independent degradation by 26S (Fig. S3 C).
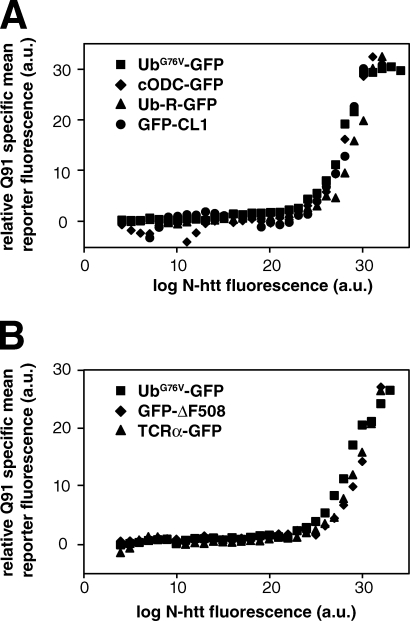
Expression of N-htt(Q91)–chFP, but not N-htt(Q25)–chFP, resulted in an N-htt(Q91)–chFP dose-dependent increase in the fluorescence of all four reporters (Fig. 2 A), strongly suggesting that expression of N-htt(Q91)–chFP impairs the UPS downstream of any Ub-dependent targeting steps, most likely at the level of 26S itself. To further test this hypothesis, we assessed the effect of N-htt(Q91)–chFP expression on the degradation of substrates of the ER-associated degradation (ERAD), a quality control pathway that delivers misfolded or unassembled proteins in the early secretory pathway to the UPS (Vembar and Brodsky, 2008). We found that N-htt(Q91)-chFP, but not N-htt(Q25)-chFP, expression resulted in the accumulation of a folding-defective mutant cystic fibrosis transmembrane conductance regulator, GFP-ΔF508 (Ward et al., 1995), and of unassembled TCR-α–GFP, a subunit of the T cell receptor (Fig. 2 B; Yu et al., 1997). Two additional ERAD substrates, an amyloidogenic mutant of the nonglycosylated protein transthyretin (Sekijima et al., 2005), and GluR1, an unassembled subunit of the n-methyl-d-aspartate receptor, exhibited similar behavior (unpublished data). Remarkably, all of these ERAD substrates accumulate at precisely the same threshold of N-htt(Q91)–chFP expression as found for the four cytosolic UPS reporters (Fig. 2, A and B). It is possible that the elevated fluorescence of all of these unstable GFP conjugates could be caused by increased transcription of the reporter cDNAs in response to N-htt(Q91)–chFP expression (Bowman et al., 2005; Alvarez-Castelao et al., 2009). However, we found no evidence of increased levels of GFP in response to the highest levels of N-htt(Q91)–chFP expression (Fig. S3 D). Because GFP is a very stable protein in mammalian cells (Bence et al., 2001), small changes in its rate of synthesis are integrated over time to generate robust changes in steady-state levels. The observation that all of the unstable reporters accumulate above the same level of N-htt(Q91)–chFP expression strongly suggests that mutant N-htt expression above a critical concentration threshold has pleiotropic effects on the activity or capacity of 26S. The finding that a Ub-independent 26S substrate, cODC-GFP, along with substrates that depend on different Ub conjugation pathways, accumulates in response to expression of N-htt(Q91)-chFP above a critical concentration threshold suggests that this pleiotropic effect is not caused by an impaired Ub metabolism.
Figure 2.
All 26S substrates are stabilized at the same critical concentration threshold of mutant N-htt expression. (A) All cytoplasmic UPS reporters accumulate above the same critical concentration threshold of mutant N-htt expression. Effect of N-htt(Q91)–chFP expression (x axis) on the fluorescence levels of different unstable GFP-degron constructs (y axis): Ub fusion degradation substrate UbG76V-GFP, Ub-independent substrate cODC-GFP, N-terminal end rule substrate Ub-R-GFP, and CL1-dependent substrate GFP-CL1. (B) ERAD substrates accumulate above the same critical concentration threshold of mutant N-htt expression. Relationship between N-htt(Q91)–chFP expression (x axis) plotted against the fluorescence levels of GFP-tagged ERAD substrates (y axis): GFP-ΔF508 and TCR-α–GFP. UbG76V-GFP was included for comparison. The data shown are from single representative experiments out of two independent repeats.
Ubiquitinated polyglutamine aggregates do not choke or clog the 26S proteasome
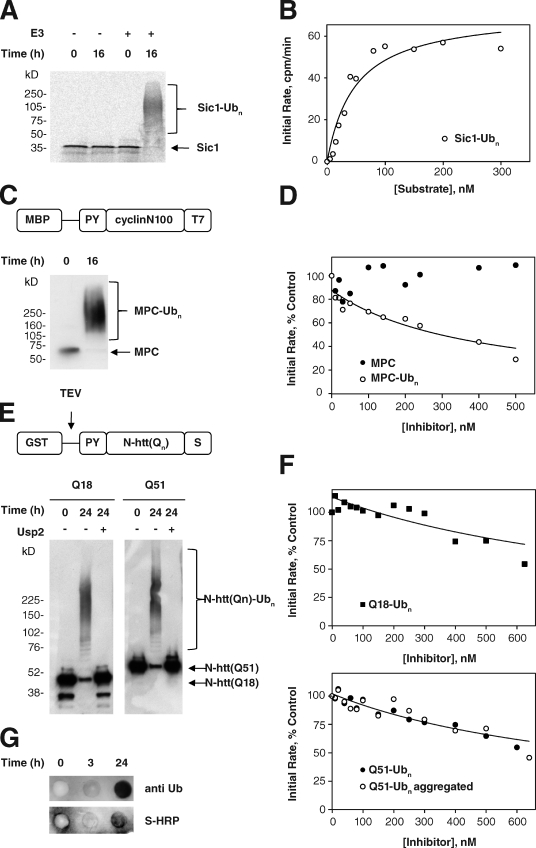
The most parsimonious model to explain why all UPS substrates accumulate in response to N-htt aggregation would predict that aggregated N-htt interacts directly with and inhibits 26S activity. Although we have previously shown that neither soluble nor aggregated polyglutamine peptides nor N-htt fragments inhibit 20S or 26S proteasomes in vitro (Bennett et al., 2005), it is conceivable that if polyubiquitinated, these aggregates could antagonize 26S proteasomes by virtue of their affinity for Ub receptors, but resistance to ATP-dependent unfolding associated with the 19S ATPases. To directly test the possibility that 26S proteasomes are inhibited in such a manner, we established an in vitro assay for 26S proteasome activity to measure the effect of polyubiquitination on the ability of aggregated polyglutamine-containing proteins to interfere with substrate degradation in trans. A purified 35S-labeled substrate (PY-Sic1) was efficiently and quantitatively converted to a high molecular weight polyubiquitinated PY-Sic1(Ubn) form in vitro by the E3 Ub ligase Rsp5 (Fig. 3 A; Saeki et al., 2005). Addition of purified proteasomes to this preparation resulted in efficient degradation that was dependent on ATP (unpublished data) and inhibited by MG132 and by pretreatment of the PY-Sic1(Ubn) substrate with the catalytic core of the deubiquitinating enzyme Usp2 (USP2-cc; Fig. S4, A and B; Catanzariti et al., 2004). Michaelis–Menten analysis of 35S–PY-Sic1(Ubn) degradation yielded a Michaelis constant (Km) of 46.9 ± 3.5 nM (Fig. 3 B), in good agreement with a previous study with other 26S proteasome substrates in vitro (Thrower et al., 2000) and confirming that this model substrate can be used to quantify the activity of 26S in vitro. To assess inhibition of 26S activity in vitro, we used the same enzymatic strategy to generate high molecular weight polyubiquitinated forms of an unlabeled chimeric protein (MPC) consisting of an N-terminal maltose binding protein fused to a fragment of Xenopus laevis cyclin (N100; Chen and Fang, 2001) and a T7 epitope tag (Fig. 3 C). The initial rate of 35S–PY-Sic1(Ubn) degradation by purified 26S rat liver proteasomes was strongly inhibited (inhibition constant [Ki] of 150 nM) by MPC(Ubn) but not by nonubiquitinated (Fig. 3 D) or deubiquitinated (not depicted) MPC, confirming that ubiquitinated proteins can inhibit 26S function and establishing the utility of this assay for the evaluation of proteasome inhibition by ubiquitinated N-htt aggregates.
Figure 3.
Ubiquitinated polyglutamine aggregates do not choke the 26S proteasome in vitro. (A) In vitro ubiquitination of radiolabeled Sic1 substrate. 35S–PY-Sic1 purified from E. coli was ubiquitinated in vitro as described in Materials and methods. Aliquots of the reaction were removed at the indicated times, separated on a 4–20% gradient gel, and visualized by autoradiography. Mobilities of unmodified Sic1 and polyubiquitinated Sic1 are indicated. (B) Kinetic analysis of Sic1(Ubn) degradation. 100 nM 35S-Sic1(Ubn) was incubated in the presence of 10 nM 26S proteasomes, and degradation kinetics were assessed by SDS-PAGE (Fig. S4 A) or release of TCA-soluble 35S radioactivity (Fig. S4 B). Initial rates of substrate degradation were determined from the kinetics of 35S radiolabel release (Fig. S4 B) and fitted to the Michaels–Menten equation by least-squares analysis assuming a Km of 50 nM. The data shown are from a single representative experiment out of three independent repeats. (C) Preparation of ubiquitinated MPC. MPC, consisting of an N-terminal maltose binding protein (MBP) fused to a fragment of Xenopus cyclin (cyclin N100) and a T7 epitope tag, was purified from E. coli and was ubiquitinated in vitro as described in Materials and methods. Aliquots of the reaction were removed at the indicated times, separated on a 4–20% gradient gel, and visualized by immunoblotting with an anti-T7 HRP conjugate. (D) Ubiquitinated MPC is a competitive inhibitor of Sic1(Ubn) degradation. Dependence of the initial rate of 35S-Sic1(Ubn) degradation on the concentration of MPC or ubiquitinated MPC (MPC(Ubn)). 10 nM proteasomes were incubated with 100 nM substrate, and initial rates were determined by a linear fit to soluble TCA radioactivity as in Fig. S4 B. Initial rates are expressed as a percentage of the control reaction without MPC or MPC(Ubn). The data were fit by least-squares analysis as described in Materials and methods. The data shown are from a single representative experiment out of two independent repeats. (E) Preparation of ubiquitinated N-htt fragments. GST–PY-N-htt(Qn) containing a C-terminal S tag was purified from E. coli and ubiquitinated in vitro as described in Materials and methods. Aliquots of the reaction were removed at the indicated times, treated as indicated with the deubiquitinating enzyme Usp2-cc, separated on a 4–20% gradient gel, and visualized by immunoblotting with an anti-T7 horse HRP conjugate. (F) Competitive inhibition of Sic1(Ubn) degradation by ubiquitinated N-htt is independent of polyglutamine length or aggregation state. Dependence of the initial rate of 35S-Sic1(Ubn) degradation on the concentration of N-htt(Q18)(Ubn) (top) or N-htt(Q51)(Ubn) (bottom). Analysis was performed with nonaggregated (uncleaved) N-htt(Q51)(Ubn) (closed circles) or after TEV cleavage and aggregation of the ubiquitinated N-htt(Q51)(Ubn) (open circles). 10 nM proteasomes were incubated with 100 nM substrate, and initial rates were determined by a linear fit of soluble TCA radioactivity as in Fig. S4 B. Initial rates are expressed as a percentage of the control reaction without inhibitor present. The data were fit by least-squares analysis as described in Materials and methods. The data shown are from a single representative experiment out of two independent repeats. (G) N-htt(Q51)(Ubn) aggregates are insoluble. N-htt(Q51)(Ubn) was aggregated and filtered through a 0.2-µm cellulose acetate filter as described in Materials and methods. The blot was probed with anti-Ub (FK2) monoclonal antibody (top) or S protein–HRP to detect polyubiquitinated trapped N-htt aggregates.
To assess the ability of ubiquitinated protein aggregates to inhibit 26S activity, we used a similar approach to append poly-Ub chains on GST–PY-N-htt(Qn) purified from Escherichia coli. A previous study has established that the presence of GST retards the aggregation of N-htt fusions containing intermediate (∼Q50) length polyglutamine repeats and that aggregation of Q > 40 fusions can be efficiently induced upon cleavage of GST with a site-specific protease (Scherzinger et al., 1997). Both GST–PY-N-htt(Q18) and GST–PY-N-htt(Q51) were efficiently converted to high molecular weight Ub-conjugated species. Treatment with Usp2-cc resolved both fusion proteins to their native, unmodified molecular weights, indicating that their reduced mobility was caused by Ub conjugation rather than to aggregation (Fig. 3 E). Hydrolysis of the GST moiety with tobacco etch virus (TEV) protease rapidly converted GST–PY-N-htt(Q51) into high molecular weight aggregates that did not migrate beyond the stacking gel on SDS-PAGE and were efficiently retained on a 0.2-µm cellulose acetate filter trap (Fig. S4 C; Wanker et al., 1999), confirming that the presence or absence of GST can be used to switch this fusion protein from a soluble nonaggregated protein into an aggregated state. We found that GST–N-htt(Q18)(Ubn) inhibited degradation of 35S–PY-Sic1(Ubn) by 26S in a dose-dependent manner (Fig. 3 F), with a Ki of ∼250 nM, similar to the value obtained for inhibition by MPC(Ubn). Importantly, soluble GST–N-htt(Q51)(Ubn) and aggregated PY-N-htt(Q51)(Ubn) (but not nonubiquitinated forms; not depicted; Bennett et al., 2005) also inhibited 35S–PY-Sic1(Ubn) degradation with inhibitory constants of 150–215 nM (Fig. 3 F), demonstrating that 26S inhibition is associated with the presence of a poly-Ub chain on these proteins but is independent of their aggregation state. TEV-cleaved GST–N-htt(Q51)(Ubn) aggregates, retained on a 0.2-µm cellulose acetate filter, were immunoreactive with antibodies to Ub and S protein, confirming the presence of polyubiquitinated N-htt aggregates (Fig. 3 G). The similar Ki values for MPC(Ubn), GST–PY-N-htt(Q51)(Ubn), and aggregated PY-N-htt(Q51)(Ubn) support the conclusion that neither the presence of a polyglutamine tract nor the aggregation state influences the ability of a polyubiquitinated protein to inhibit the degradation of other ubiquitinated proteins by 26S in vitro. Therefore, our data lead to the conclusion that, at least in vitro, proteasomes are neither choked nor clogged by N-htt but leave open the possibility that ubiquitinated N-htt aggregates can compete with other ubiquitinated substrates for 26S if these species accumulate to sufficient levels relative to other 26S substrates.
Stabilization of UPS reporters is not caused by direct competition for 26S proteasome by ubiquitinated N-htt
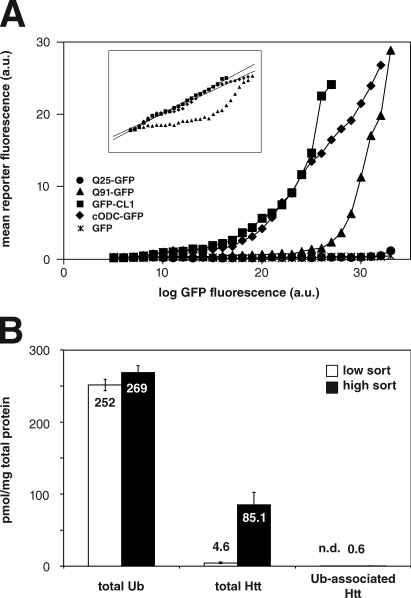
It is possible that ubiquitinated forms of N-htt could accumulate to sufficiently high concentrations in cells expressing polyglutamine-expanded N-htt(Qn)–chFP to be able to outcompete other poly-Ub–tagged substrates, including highly expressed UPS reporters. However, if ubiquitinated N-htt were to act as a simple competitor for binding of other ubiquitinated proteins to proteasomes, the concentration dependence of UbG76V-GFP stabilization would not be expected to exhibit such a steep threshold as is observed in Fig. 1. To test this, we examined the effect of expression of one short-lived fluorescent proteasome substrate on the accumulation of another (Fig. 4 A). We found that expression of GFP-CL1 or cODC-GFP resulted in accumulation of chFP-CL1 in a concentration-dependent manner but exhibited no sharp threshold, as reflected by the nearly linear relationship of the fluorescence profiles of the two proteins when represented on a double logarithmic plot (Fig. 4 A, inset). This suggests that these two UPS substrates must compete with each other for one or more limiting UPS component. Because the two CL1-tagged proteins share the same degron, they might compete with each other for limiting Ub conjugation machinery, but the observation that a Ub-independent proteasome substrate (cODC-GFP) also competes for degradation of a Ub-dependent substrate (chFP-CL1) strongly suggests that the competition between these two reporters is at the level of proteasome binding, as the ODC degron functions as a molecular mimic that competes directly with Ub chains for binding to the 19S cap (Zhang et al., 2003). Therefore, competition for limited substrate binding capacity is a potential mechanism by which 26S activity might be reduced by a highly expressed protein, such as N-htt(Qn)–chFP, only to the extent that such a protein was modified with poly-Ub chains or a Ub mimetic, such as the ODC degron, and whether this Ub-conjugated species was present at sufficient levels relative to other 26S substrates to be effective.
Figure 4.
Stabilization of proteasome reporters by aggregation-prone N-htt is not caused by direct competition for 26S. (A) Short-lived proteasome substrates, but not N-htt, compete for degradation of chFP-CL1 degradation. HEK293 cells stably expressing chFP-CL1 were transfected with GFP-CL1, cODC-GFP, GFP, N-htt(Q25)–GFP, or N-htt(Q91)–GFP and analyzed by two-color flow cytometry. (inset) Double logarithmic plot shows that cODC-GFP and GFP-CL1 data fit well with a linear least-squares regression (r2 > 0.98), whereas N-htt(Q91)–GFP does not. The data shown are from a single representative experiment out of two independent repeats. (B) Concentration of Ub, N-htt, and Ub-associated N-htt in HEK293 cells sorted on the basis of N-htt(Q91)–chFP fluorescence intensity. HEK293 cells stably expressing UbG76V-GFP and transfected with N-htt(Q91)–chFP were sorted based on high and low N-htt(Q91)–chFP fluorescence intensity followed by Ub AQUA-MS analysis to determine the amount of each species in the two sorted populations. All data are represented as means ± SEM (n = 3).
We therefore used absolute quantitative (AQUA)–MS (Gerber et al., 2003; Kirkpatrick et al., 2005) to determine the extent to which N-htt is ubiquitinated and the fraction of cellular poly-Ub conjugated to N-htt in FACS-sorted cells expressing N-htt(Q91)–chFP at levels above (high sort [HS]) or below (low sort [LS]) the threshold at which we observe accumulation of UbG76V-GFP (Fig. S5 A). Total N-htt levels in lysates of these cells reflected this enrichment, increasing nearly 20-fold from 4.64 ± 1.04 pmol/mg in the LS population to 85.14 ± 17.59 pmol/mg in HS cells (Fig. 4 B). Poly-Ub conjugates were isolated by hP2UBA affinity capture (Bennett et al., 2007) from the same lysates and analyzed by AQUA-MS to determine the abundance of N-htt(Q91)–chFP present in the poly-Ub–associated population (Fig. 4 B). N-htt was undetectable in the poly-Ub fraction from LS cells and represented <6% of total protein in the poly-Ub fraction from HS cells (unpublished data). Because this technique does not distinguish between the amount of covalently polyubiquitinated N-htt and N-htt that is noncovalently (i.e., indirectly associated with other polyubiquitinated proteins) associated with poly-Ub chains, this value necessarily overestimates the actual amount of N-htt–Ub conjugates. Indeed, we have never detected any N-htt peptides containing a diglycine covalent Ub signature in these experiments or in MS/MS analysis in cell lines and R6/2 transgenic mice (unpublished data). Therefore, only a minor fraction of total poly-Ub chains are covalently linked to N-htt in the cells expressing the highest levels of N-htt in which we observe elevated levels of Ub chains and the UbG76V-GFP reporter (Fig. S5 A), making it unlikely that polyubiquitinated N-htt represents a sufficient fraction of total poly-Ub conjugates to effectively outcompete other substrates.
We also used the AQUA-MS approach to assess the fraction of the total soluble N-htt pool that is associated with Ub, which, as in the previous paragraph, overestimates the actual fraction of covalent N-htt–Ub conjugates. This analysis revealed that of the 85.14 ± 17.59 pmol/mg total N-htt, at most 0.57± 0.08 pmol/mg (0.7%) was Ub associated (Fig. 4 B), indicating that soluble N-htt, in cells sorted to express the highest levels of this protein, is not ubiquitinated to a significant extent.
N-htt and Ub are recruited to IBs independently
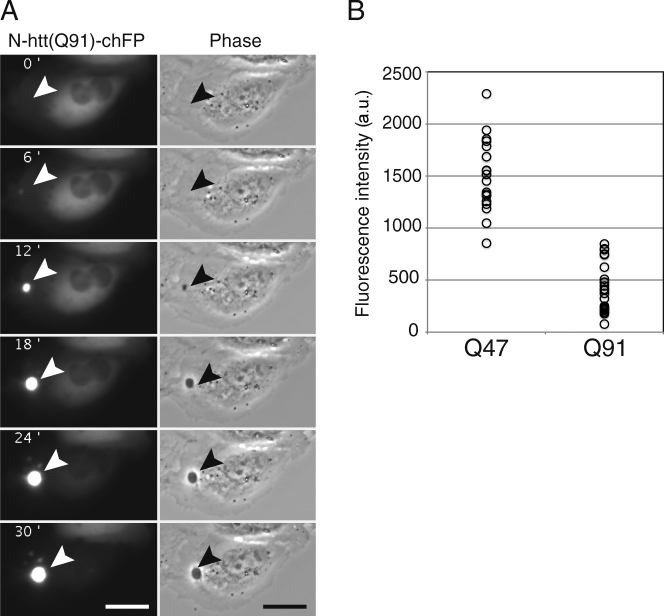
To reconcile the persistent presence of elevated poly-Ub chains associated with N-htt inclusions in this and previous studies (Bence et al., 2001; Bennett et al., 2005), with the AQUA-MS data indicating little or no covalent Ub modification of N-htt, we used real-time fluorescence video microscopy to assess the temporal and spatial relationship between N-htt aggregation and Ub recruitment to IBs. Because HEK293 cells are not optimal for live-cell imaging experiments, we performed these experiments in U2OS cells. Control experiments in U2OS cells stably expressing UbG76V-GFP verified that the relationship between UPS reporter accumulation and N-htt expression in these cells is similar to that observed in HEK293 cells (Fig. S5 B). N-htt(Q91)–chFP fluorescence was initially diffusely distributed throughout the cytoplasm, and the overall fluorescence intensity increased linearly after transfection (Fig. 5 A and Video 1). The IB appeared as an initially dim spot of chFP fluorescence detectable above the background of diffuse fluorescence. Once it appeared, the diameter and intensity of this spot increased rapidly, and within 25 min, the vast majority of diffuse chFP fluorescence had coalesced into it. These IBs could also be visualized as phase-dense spots that became evident in parallel with recruitment of fluorescent N-htt. Quantification of total cellular chFP fluorescence in individual N-htt(Q91)–chFP–expressing cells at the time point immediately before the first appearance of detectable fluorescent puncta revealed that puncta formation occurred when total N-htt(Q91)–chFP concentration exceeded a threshold of total abundance of 431 ± 49 (mean [a.u.] ± SEM; Fig. 5 B). In cells expressing N-htt(Q47)-chFP, a similar threshold behavior was observed; but the threshold occurred at significantly (P < 0.001) higher total N-htt concentration (1,510 ± 81 a.u.), consistent with the relationship between N-htt concentration and aggregation determined with pure N-htt fragments in vitro. IBs were never observed in cells expressing N-htt(Q25)–chFP, which is below the glutamine length threshold for N-htt aggregation in vitro and in vivo, even though total chFP fluorescence in these cells often increased to levels comparable with that observed in the N-htt(Q47)–chFP–expressing cells (unpublished data). These data suggest that IB formation in cells expressing mutant N-htt is a two-step process, punctuated by the initial formation of a focus of insoluble N-htt and followed by rapid recruitment of the remaining diffuse N-htt into the nascent IB. The kinetics and glutamine length dependence of this process strongly suggest that it is driven by the self-association properties intrinsic to polyglutamine.
Figure 5.
IBs form rapidly in a concentration- and repeat length–dependent manner. (A) U2OS cells were transiently transfected with N-htt(Q91)–chFP, and formation of IBs was followed by time-lapse fluorescence video microscopy at 6-min intervals. Note the absence of diffuse mCherry fluorescence outside of the IB at the 30-min time point (Video 1). The arrowheads indicate the site of a forming IB. Bars, 25 µm. (B) Threshold for IB formation. Cells transiently transfected with N-htt(Q91)–chFP or N-htt(Q47)–chFP were monitored by time-lapse fluorescence video microscopy as in A. Total chFP fluorescence intensity in individual cells in the frame just before the first detectable IB was quantified and plotted. The data shown are from a single representative experiment out of two independent repeats.
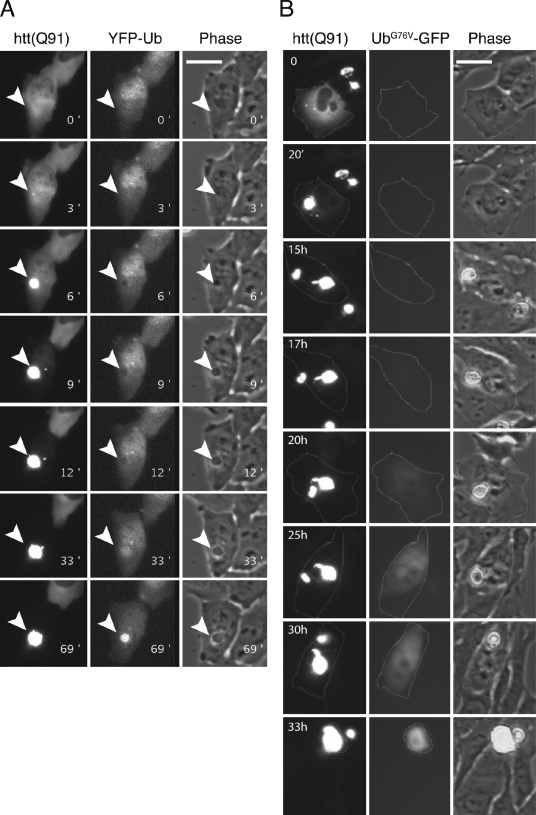
To evaluate the role of Ub conjugation in recruitment of N-htt to IBs, we expressed N-htt(Q91)–chFP in cells stably expressing YFP-Ub to monitor changes in Ub distribution during the process of N-htt IB formation (Fig. 6 A and Video 2). Ub molecules bearing N-terminal epitope tags or protein fusions can functionally complement deletion of chromosomal Ub in yeast (Ellison and Hochstrasser, 1991), and in mammalian cells, GFP-Ub fusions have been used to monitor the localization and trafficking of intracellular Ub conjugates (Qian et al., 2002; Dantuma et al., 2006). We observed that YFP fluorescence was diffusely distributed in cells before IB formation but, strikingly, was distinctly excluded from the initial N-htt(Q91)–chFP puncta (Fig. 6 A, 6- and 9-min time points). YFP-Ub became recruited to the nascent IBs only after a delay (average time of 43 min)—a time at which the IB had reached its mature size. These data demonstrate the N-htt molecules that are recruited into IBs are not conjugated to Ub and that Ub conjugation is not required for IB formation.
Figure 6.
Ub recruitment and UPS substrate stabilization are delayed after IB formation. (A) U2OS cells stably expressing YFP-Ub were transiently transfected with N-htt(Q91)–chFP and followed by time-lapse two-color fluorescence video microscopy (Video 2). Sequential frames show the formation of the N-htt IB (left hand column) and the complete recruitment of chFP fluorescence by 33 min. Note that during the initial formation of the IB (6- and 9-min time points), YFP fluorescence is excluded from the IB and is recruited there only after a ∼40-min delay. The arrowheads indicate the site of the forming IB. (B) U2OS cells stably expressing UbG76V-GFP were transiently transfected with N-htt(Q91)–chFP and monitored by time-lapse two-color fluorescence video microscopy at 20-min intervals. Selected frames of the recorded image series (Video 3) are presented. The outlined cell formed an IB at t = 0, and UbG76V-GFP became detectably stabilized in the cell after an ∼17-h delay. Bars, 25 µm.
UPS reporter accumulation is delayed relative to IB formation
To assess the spatial and temporal relationship between IB formation and stabilization of UPS reporters, we used real-time single-cell analysis in a U2OS cell line stably expressing UbG76V-GFP. These cells were transiently transfected with N-htt(Q91)–chFP, and double-positive cells were continuously monitored at 20-min intervals for >72 h (Table S1). UbG76V-GFP stabilization was never observed in cells lacking IBs. However, we observed UbG76V-GFP stabilization in 8/33 (24%) cells that formed IBs (Fig. 6 B, Video 3, and Table S1). This stabilization occurred on average 22.8 ± 4 h after IB formation, revealing a significant delay between IB formation and UPS substrate stabilization. The cells that never exhibited UbG76V-GFP stabilization died, on average, within 12 h after IB formation, suggesting that the low frequency of cells that accumulate UbG76V-GFP is a reflection of the poor survival of cells burdened with N-htt above the critical concentration at which IBs form. Thus, stabilization of UbG76V-GFP and Ub-R-GFP (unpublished data) is a late event after IB formation in cells expressing high levels of aggregated N-htt; most cells experiencing a high burden of N-htt aggregation die before accumulating this reporter.
Discussion
Proteasomes are not directly impaired by aggregated N-htt
The data in the present study establish that polyglutamine fragments with covalently attached poly-Ub can inhibit 26S activity in vitro. However, because the Ki for N-htt fragments is independent of polyglutamine length and aggregation state and is similar to the Ki of an unrelated polyubiquitinated protein, MPC, these data suggest that ubiquitinated N-htt, whether aggregated or not, does not choke or otherwise noncompetitively inhibit 26S.
Thus, N-htt expression can directly inhibit proteasome activity only to the extent that ubiquitinated forms of the protein are able to accumulate to concentrations approaching or in excess of other polyubiquitinated species with which they compete for binding to 26S. Although some N-htt exon 1 can be recovered from metal affinity chromatography of extracts of cultured HeLa cells or striatal neurons transfected with His6-tagged Ub, the fraction of N-htt that was Ub associated (which does not necessarily reflect covalent ubiquitination of N-htt) was exceedingly low (Steffan et al., 2004), suggesting that the vast majority of N-htt exon 1 expressed in cells is not modified with poly-Ub, a finding that is confirmed by our AQUA-MS results and by the distinctly different kinetics with which N-htt and Ub are recruited to IBs and the complete exclusion of Ub during IB formation. We conclude that N-htt is neither a direct competitive nor a noncompetitive inhibitor of 26S.
Why do Ub conjugates accumulate in neurodegenerative disease?
The molecular genetics of dominantly inherited neurodegenerative diseases like HD strongly support a gain-of-function mechanism in which the disease-causing mutation—for example, expansion of the Gln repeat in N-htt—causes the disease-encoded protein to adopt one or more nonnative conformations that are toxic to cells (Hatters, 2008; Finkbeiner, 2011). In our experiments, the sharp transition between diffuse localization of cytoplasmic N-htt(Qn)–chFP and its rapid recruitment to IBs is observed when total N-htt(Qn)–chFP accumulates above a critical concentration threshold. Dependence of this critical concentration upon the length of the Gln tract argues strongly that this transition is caused by the intrinsic capacity of this protein to form organized, fibrillar aggregates. However, because molecular chaperones, including Hsp70/Hsp40, which constitute part of the cell’s proteostasis machinery, are able to suppress the aggregation of polyglutamine-expanded proteins in vitro and in vivo (Warrick et al., 1999; Chan et al., 2000; Jana et al., 2000; Kazemi-Esfarjani and Benzer, 2000; Howarth et al., 2007), the critical concentration for N-htt(Qn)–chFP aggregation in any given cell must be determined by both the absolute concentration of the aggregation-prone species and the capacity of the complement of molecular chaperones to resist this thermodynamically driven process. Therefore, the point at which diffusely localized N-htt transitions to IBs must represent the point at which the production of aggregation-prone N-htt species can no longer be offset by the cellular proteostasis network. At this point, preoccupation of one or more critical chaperones by the ever-increasing burden of N-htt production erodes the capacity of the cellular machinery to sustain the folding of its normal complement of cellular substrates. This proteostatic collapse is observed in Caenorhabditis elegans, in which expression of aggregation-prone, disease-associated proteins, including polyglutamine-expanded N-htt fragments (Gidalevitz et al., 2006) or mutant Cu and Zn superoxide dismutase (SOD1) linked to amyotrophic lateral sclerosis (Gidalevitz et al., 2009), leads to pleiotropic disruption of the protein-folding environment, indicated by the emergence of temperature-sensitive mutant phenotypes in unrelated genes at permissive temperatures. Conversely, the presence of temperature-sensitive polymorphic variants of cellular genes dramatically enhances the aggregation and toxicity of N-htt and SOD1 mutants in worms (Gidalevitz et al., 2006, 2009).
Therefore, proteins that depend on the factors that become limiting as a result of the burden of maintaining N-htt solubility will be delayed in their ability to fold, and some of them will be targeted to the UPS for degradation. We propose that the increased flux of normal cellular chaperone clients that are diverted to the UPS eventually will lead to a “queue” of ubiquitinated proteins waiting their turn to be degraded by 26S, and it is these proteins, collectively, that compete with each other and with artificial fluorescent UPS reporters for degradation. Some of the excess undegraded Ub-conjugated proteins would be recruited to IBs, composed of a Ub-free core of aggregated N-htt. This model provides a plausible explanation for why the ability of cells to degrade unstable GFP reporters declines with increasing time of exposure to aggregation-prone mutant N-htt and why IBs are enriched in so many different unrelated proteins, including Ub, transcription factors, cytoskeleton proteins, nuclear pore complex components, kinases, enzymes, and cell cycle regulators (Suhr et al., 2001; Leverenz et al., 2007; Xia et al., 2008).
Our model is consistent with previous findings from a longitudinal study of primary striatal neurons in culture, which reveal that cell survival is negatively correlated with the level of diffuse N-htt and positively correlated with IB formation (Arrasate et al., 2004). Our observation that IBs form abruptly, recruiting diffuse htt from a preexisting pool and excluding soluble Ub, suggests the possibility that IB formation may transiently “liberate” proteostasis machinery that would be otherwise preoccupied with maintaining monomeric or oligomeric N-htt in a soluble state. Our finding that stabilization of UbG76V-GFP is a late event relative to IB formation and YFP-Ub recruitment is consistent with the finding of Mitra et al. (2009), who observed that neurons that formed IBs had significantly smaller increases in UPS reporter fluorescence, indicating that less UPS impairment occurs in cells after IB formation than in cells that did not form IBs. It is not entirely clear why we did not observe increased UbG76V-GFP before IB formation, as was observed in the Mitra et al. (2009) study. We speculate that variation in the expression profiles of different components of the proteostasis machinery could contribute to the tissue- and cell type–specific effects observed in models of HD and other conformational diseases. This variability could also partially explain the apparent discrepancy between in vitro models, which consistently show accumulation of GFP UPS reporters in cell culture models of HD and mouse models of HD (Maynard et al., 2009; Ortega et al., 2010) and spinocerebellar ataxia type 1 (Bowman et al., 2005), in which these reporters have not been observed to accumulate or accumulate only transiently, although it is likely that technical details, such as expression and signal to noise levels or the sensitivity of these reporters in the context of intact tissue, could also impact the in vivo observations.
An outstanding question raised by our findings is the extent to which activation of cellular stress response pathways is able to compensate for the increasing burden imposed by chronic expression of mutant N-htt. Preliminary data indicate that cells that express the highest levels of N-htt(Q91)–chFP fail to activate HSF1, the principal transcription activator of the cytoplasmic heat shock response (unpublished data). This lack of heat shock activation in response to mutant N-htt may not be unique to our artificial cellular model; levels of several heat shock proteins are observed to decline by >50% in brains from transgenic (Hay et al., 2004) or HdhQ150/Q150 knockin (Woodman et al., 2007) HD model mice without apparent change in the corresponding mRNA levels. Further studies will be necessary to determine why chronic expression of polyglutamine-expanded N-htt or other aggregation-prone proteins (Olzscha et al., 2011) fail to activate the heat shock response while at the same time leading to pleiotropic changes to the protein-folding environment in model organisms (Gidalevitz et al., 2006; Ben-Zvi et al., 2009) and, more recently, to the misfolding of a metastable, luciferase-based proteostasis sensor (Gupta et al., 2011).
Materials and methods
Plasmids
The plasmids expressing GFP-CL1, N-htt–GFP, and N-htt exon 1 were described previously (Bence et al., 2001). In brief, the GFP-CL1 plasmid was created by ligating an oligonucleotide encoding ACKNWFSSLSHFVIHL into the GFP-C1 plasmid (Takara Bio Inc.). N-htt–GFP encodes htt exon 1 and contains mixed CAG/CAA repeats fused to a C-terminal GFP tag in pcDNA3.1 (Invitrogen). N-htt exon 1 was created by removing GFP from N-htt–GFP and ligating a linker between the BamHI and XbaI sites. Plasmids expressing Ub-R-GFP and UbG76V-GFP were gifts from N. Dantuma (Karolinska Institutet, Stockholm, Sweden). The cODC-GFP–expressing plasmid was created by E. Bennett (University of California, San Diego, San Diego, CA) by amplification of the C-terminal 37 amino acids of ODC by PCR and cloned into pEGFP1 (Takara Bio Inc.) using the HindIII and BamHI sites. Plasmids expressing N-htt(Q25)–chFP and N-htt(Q91)–chFP were created by inserting chFP (a gift from R. Tsien, University of California, San Diego, La Jolla, CA) into the BamHI site of the N-htt exon 1 plasmids described in this paragraph. The chFP-CL1 plasmid was created by insertion of chFP and the CL1 sequence into pcDNA3.1 (Invitrogen). pET3a-Ub and pET15b-Ubc4 were provided by C. Pickart (Johns Hopkins University, Baltimore, MD) and D. Rotin (The Hospital for Sick Children, Toronto, Canada), respectively. pHUE and pHUsp2-cc were gifts from R. Baker (Australian National University, Canberra, Australia). Plasmids for GST-Rsp5 and GST-ΔC2Rsp5 were obtained from J. Huibregtse (University of Texas, Austin, TX). The ΔC2Rsp5 coding sequence was PCR amplified and cloned into a pET28a vector (EMD) to yield a His6-tagged construct (pET28a-ΔC2Rsp5). The plasmid for expressing His6–PY-Sic1 was obtained from R. Deshaies (Howard Hughes Medical Institute, California Institute of Technology, Pasadena, CA). The PY-Sic1 coding sequence was PCR amplified and cloned into the pHUE vector as previously described (Catanzariti et al., 2004) to create the pHUE–PY-Sic1 plasmid. A plasmid containing the cyclin B N100 fragment (Chen and Fang, 2001) was obtained from G. Fang (Stanford University, Stanford, CA). The cyclin B N100 coding sequence was PCR amplified and cloned into the pMAL-c2X plasmid (New England Biolabs, Inc.). Site-directed mutagenesis was used to introduce a Pro-Pro-Pro-Tyr sequence (PY motif) in front of the cyclin N100 sequence and a T7 tag at the end of cyclin N100 to create pMAL-PY-cyclinN100-T7 (MPC). Plasmids expressing the original GST–N-htt fusion constructs containing either Q18 or Q51 were obtained from E. Wanker (Max Delbrück Center, Berlin, Germany). These constructs were modified by the addition of a TEV protease cleavage site between the GST and htt exon 1 coding regions (GST–N-htt(Qn)-ΔS) or by the addition of a TEV protease cleavage site between the GST and htt exon 1 coding regions and the addition of an S tag C terminal to the htt exon 1 coding region (GST–N-htt(Qn); Bennett et al., 2005). Within these constructs, site-directed mutagenesis was used to insert a PY motif immediately after the TEV protease site to create plasmids encoding for GST–PY-N-htt(Q18)-S and GST–PY-N-htt(Q51)-S.
Cell lines
The GFP-CL1 (Bence et al., 2001), CFTRΔF508-GFP (Johnston et al., 1998), and TCR-α–GFP (DeLaBarre et al., 2006) cell lines were previously described. These cell lines were prepared by transfection of HEK293 cells followed after 48 h by selection of transformed cells by growth in G418. Also, the other stable HEK293 cell lines expressing the constructs described in this study were created by transfection, selected with G418, and cloned by limiting dilution. The temperature-sensitive ts20 Balb/C 3T3 clone A31 fibroblast cell line (Kulka et al., 1988) was a gift from D.T. Madden (The Buck Institute for Research on Aging, Novato, CA). All cells were grown in DME with 10% animal serum complex, l-glutamine, and antibiotics.
Flow cytometry and cell sorting
Unless indicated, cells were harvested 72 h after transfection and analyzed with a flow cytometer (LSR II; BD) with a 488- and 535-nm laser (BD). To ensure a sufficient number of cells with elevated levels of the transfected protein, >200,000 cells were analyzed per condition in a typical experiment. To plot the level of the reporter protein versus the level of the transfected protein (described in Fig. S2), a set of 41 gates of equal width (on a logarithmic scale) was set up in the channel for the transfected protein. The mean compensated fluorescence of the reporter protein in each of these gates was calculated and plotted on the ordinate with the gate number (corresponding to the log of fluorescence intensity of the transfected protein) plotted on the abscissa. Each construct and condition in singly transfected HEK293 cells was used as a single-color control to compensate the spillover between chFP and GFP individually for each gate. Gates with <100 events were not included in the analysis. The data shown in Figs. 1, 2, and 4 are from single representative experiments out of a minimum of two independent repeats. Raw flow cytometry data were analyzed using FlowJo (version 8.8.6; Tree Star) software.
To isolate high and low N-htt(Q91)–GFP–expressing populations, cells were harvested 72 h after transient transfection and sorted according to N-htt(Q91)–chFP intensity using a cytometer (Digital Vantage; BD) with a 80-µm nozzle and 570–595-nm tunable laser. To define the LS and HS populations, the photomultiplier tube voltage was set to center the nonexpressing population over the 102-a.u. intensity mark, and the LS gate was defined to include cells in the 102–103-a.u. interval, whereas the HS gate included cells with >103 a.u. 1–2 × 106 cells were sorted from each gate, and pellets were flash frozen in liquid N2.
Protein purification
Recombinant human E1 enzyme was purchased from Boston Biochem. Recombinant Ub was expressed and purified as previously described (Kaiser et al., 2011). In brief, Ub was expressed from the pET3a-Ub expression vector in BL21 (DE3) pLysS RIL cells upon induction with IPTG. Bacterial cells were harvested, resuspended in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% [vol/vol] glycerol, 1% [vol/vol] Triton X-100, 1 mM EDTA, and protease inhibitor cocktail [Complete; Roche]), lysed by french press, and clarified by centrifugation (SS-34 rotor; Sorvall; 18,500 rpm for 60 min). Glacial acetic acid was added dropwise with mixing on ice until the solution reached pH 4. The precipitate was pelleted by centrifugation (SS-34 rotor; 13,000 rpm for 20 min), and the supernatant was dialyzed against 25 mM sodium acetate, pH 4.5. The dialysate was purified using a column (HiTrap SP XL; GE Healthcare) and eluted with a 0–500 mM NaCl linear gradient. Elution fractions containing Ub were pooled and further purified by gel filtration chromatography on a column (Sephacryl S-200; GE Healthcare) with 50 mM Tris, pH 7.5, 500 mM NaCl, and 1 mM DTT. Eluted fractions were dialyzed into 25 mM Hepes, pH 7.5, aliquoted, and stored at −80°C.
Recombinant Ubc4 was expressed and purified as previously described (Kaiser et al., 2011). In brief, Ubc4 was expressed from the pET15b-Ubc4 plasmid in BL21 (DE3) cells upon induction with IPTG. Bacterial cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 0.2% Triton X-100, 1 mM PMSF, and EDTA-free protease inhibitor cocktail [Complete]), and lysed by french press. The lysate was clarified by centrifugation, and the supernatant was batch bound to Ni-nitrilotriacetic acid resin for 2 h. The resin was washed five times with 10 bed vol lysis buffer, and Ubc4 was eluted with lysis buffer supplemented with 250 mM imidazole. The eluate was dialyzed into 50 mM Tris, pH 7.6, 150 mM NaCl, 10% glycerol, and 1 mM DTT, aliquoted, and stored at −80°C.
Recombinant Usp2-cc was expressed from the pHUsp2-cc plasmid in BL21 (DE3) as an N-terminal His6-tagged fusion protein. Expression was induced with 0.4 mM IPTG for 4 h at 37°C. Cells were harvested by centrifugation, resuspended in buffer A (50 mM Hepes, pH 7.5, 300 mM NaCl, 30% glycerol, 0.1% Triton X-100, and 2 mM β-mercaptoethanol), and lysed by french press. The lysate was clarified by centrifugation, and the supernatant was bound to Ni-nitrilotriacetic acid beads. The beads were washed with buffer A without Triton X-100 and eluted with 400 mM imidazole. The eluted protein was dialyzed into 50 mM Hepes, pH 7.5, 150 mM NaCl, 30% glycerol, and 5 mM DTT, aliquoted, and stored at −80°C.
The His6-tagged ΔC2Rsp5 protein was expressed from the pET28a-ΔC2Rsp5 plasmid in E. coli and purified by using metal affinity chromatography (TALON; Takara Bio Inc.). In brief, cell pellets were resuspended in 40 vol of 50 mM Tris-HCl, pH 7.6, 300 mM NaCl, 5 mM β-mercaptoethanol, and 1 mM PMSF and lysed by sonication. The lysate was clarified at 10,000 g for 30 min before being loaded onto a gravity column packed with 3 ml of TALON beads. The column was washed with 20 column vol resuspension buffer without β-mercaptoethanol and eluted with 3 column vol of 50 mM Tris, pH 7.6, 300 mM NaCl, 200 mM imidazole, and 10% glycerol. The eluted protein was dialyzed into 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM DTT, and 10% glycerol. GST-Rsp5, GST–N-htt(Q51)-ΔS, GST–PY-N-htt(Q18)-S, and GST–PY-N-htt(Q51)-S were expressed in BL21 (DE3) cells and affinity purified with glutathione–Sepharose 4b resin (GE Healthcare) as recommended by the manufacturer. PY-Sic1 was expressed as an N-terminal His6-tagged Ub fusion protein using the pHUE–PY-Sic1 plasmid and metabolically labeled in BL21 (DE3) cells grown in minimal media supplemented with [35S]methionine (Zhang et al., 2003). In brief, a 100-ml culture of E. coli was grown at 37°C in M9 minimal media to OD600 = 0.5, washed once with M9 media, and resuspended in 80 ml M9 media containing 0.4% glucose and 0.063% methionine assay media. After further incubation at 37°C for 30 min, protein expression was induced with 1 mM IPTG. After a 1-h incubation, 1 mCi [35S]methionine was added. After 10 min, 1 mM of unlabeled methionine was added to the culture and grown for an additional 10 min. Cells were harvested by centrifugation and lysed by 3× freeze/thaw steps in the presence of 1 mg/ml lysozyme and DNase I (Sigma-Aldrich). Clarified lysate was then bound to TALON metal affinity resin for 1 h at RT. Resin-bound protein was incubated with His-tagged Usp2-cc. PY-Sic1 was released from the TALON beads upon cleavage with Usp2-cc, collected in the unbound fraction, and supplemented with 0.2 mg/ml BSA as a carrier. The final protein concentration was measured using a bicinchoninic acid protein assay (Thermo Fisher Scientific) relative to a BSA standard. MPC was expressed in BL21 (DE3) cells and affinity purified with amylose resin according to the manufacturer’s instructions (New England Biolabs, Inc.). 26S proteasomes were purified from Sprague-Dawley rats as previously described (Zhang et al., 2003). In brief, frozen rat livers were resuspended in 3 ml/g of tissue with buffer A (20 mM Tris, pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 50 mM NaCl, and 20% [vol/vol] glycerol), dounce homogenized, and clarified by centrifugation (SS-34 rotor; 9,300 rpm for 80 min). From the clarified supernatant, S100 extracts were prepared by centrifugation (Ti45 rotor; Beckman Coulter; 30,000 rpm for 58 min). The 26S proteasome within the S100 extract was enriched by sedimentation centrifugation (Ti45 rotor; 35,000 rpm for 18 h). The resulting pellet was resuspended in buffer A without glycerol and fractionated by centrifugation using a 15–40% glycerol gradient (SW27 rotor; Beckman Coulter; 25,000 rpm for 16 h). Gradient fractions containing peptidase activity were further purified using a column (Mono Q HR 5/50; GE Healthcare) equilibrated with 20 mM Tris, pH 7.5, 1 mM DTT, 50 mM NaCl, and 10% (vol/vol) glycerol. Proteasomes were eluted using a 50–800-mM NaCl linear gradient. Active fractions were pooled, enriched by sedimentation centrifugation as described in this paragraph, and finally resuspended in 20 mM Tris, pH 7.5, 1 mM DTT, 50 mM NaCl, and 10% (vol/vol) glycerol. The concentration of 26S proteasomes was measured using a Bradford assay relative to a BSA standard curve.
Substrate ubiquitination
0.05–0.2 mg/ml 35S–PY-Sic1 and MPC were ubiquitinated for 16 h in the presence of 50 nM E1 (Boston Biochem), 2.4 µM E2 (His6-Ubc4), 2 µM E3 (GST-Rsp5), 300 µM Ub, and ATP buffer (40 mM Tris-HCl, pH 7.2, 2 mM ATP, 5 mM MgCl2, and 1 mM DTT). The reaction was then mixed with 0.5 vol glutathione–Sepharose beads for 3 h at RT to remove contaminating autoubiquitinated GST-Rsp5. Similarly, 0.05–0.2 mg/ml GST–PY-N-htt(Qn)-S was ubiquitinated, except that 2 µM E3 (ΔC2Rsp5) was used, and 1 vol TALON metal affinity resin was needed to remove any autoubiquitinated E3 enzyme. 35S–PY-Sic1 ubiquitination (35S-Sic1(Ubn)) was monitored by SDS-PAGE and autoradiography. The ubiquitination of MPC (MPC(Ubn)) was monitored by SDS-PAGE and detected with T7-HRP conjugate (Thermo Fisher Scientific) followed by chemiluminescence. The ubiquitination of GST–PY-N-htt(Qn)-S was monitored by separation on SDS-PAGE and detection with S-HRP conjugate (EMD) followed by chemiluminescence.
In vitro Sic1 degradation assay
The substrate degradation reaction was performed at RT in 50–100 µl of total volume containing 10 nM 26S, ATP buffer, and 100 nM 35S-Sic1(Ubn). 5-µl reaction aliquots were removed at various time points and separated on a 4–20% gradient gel followed by autoradiography. Alternately, aliquots were added to 10% TCA or double-distilled H20 on ice for 30 min and 2 mg/ml BSA as a carrier. Samples were centrifuged at 12,000 g for 30 min, and the supernatant was removed for analysis by scintillation spectrometry. The percentage of degradation is determined by dividing the measured counts after TCA precipitation by the total counts measured in double-distilled H20. To assess the Ub dependence of degradation, 35S-Sic1(Ubn) was pretreated with 10 nM Usp2-cc (Baker et al., 2005) for 30 min at 37°C before the addition of proteasomes. To inhibit substrate degradation, 26S proteasomes were pretreated with 10 µM MG132 (Enzo Life Sciences) before the addition of substrate. The initial rate of degradation was determined by least-squares linear regression analyses of 5-, 10-, and 20-min time points using SigmaPlot (Systat Software, Inc.). The dependence of the initial rate of degradation on substrate concentration was plotted, and the data were fit to the Michaelis–Menten equation using SigmaPlot.
Proteasome inhibition assay
The proteasome degradation inhibition assay was set up in 50 µl total volume containing 10 nM 26S proteasomes, 100 nM 35S-Sic1(Ubn), and 0–500 nM MPC(Ubn) in ATP buffer. 5-µl aliquots were removed at 5, 10, and 20 min for TCA precipitation, and scintillation counting was performed as described in the previous paragraph. The initial rate of degradation as a percentage of the control reaction is reported as the percent ratio of the initial rate of degradation of 35S-Sic1(Ubn) in the presence of varying concentrations of MPC(Ubn) and the initial rate of degradation for 35S-Sic1(Ubn) in a control reaction with no MPC(Ubn). The dependence of the initial rate of degradation of 35S-Sic1(Ubn) on the concentration of MPC(Ubn) was plotted, and the data fit to the following equation for competitive inhibition using SigmaPlot: v0 = (viK0.5)/(K0.5 + [Ubn]), in which K0.5 = (1 + [S]/KM)Ki, Ubn is the concentration of ubiquitinated inhibitor, [S] is the concentration of 35S-Sic1(Ubn), and vi is the initial rate in the presence of inhibitor (Thrower et al., 2000).
In vitro formation of ubiquitinated N-htt aggregates
The TEV protease-induced cleavage and aggregation of GST–N-httEx1(Q51)-S and filter trap analysis were performed as previously described (Bennett et al., 2005). Aggregation of 0.15 mg/ml ubiquitinated GST–PY-N-htt(Q51)-S was initiated by the addition of 1:1 (wt/wt) TEV protease and 1:1 (wt/wt) GST–N-htt(Q51)-ΔS in aggregation buffer (40 mM Tris-HCl, pH 7.2, 1 mM DTT, and 0.5 mM EDTA). The aggregation reaction was incubated at 37°C and considered complete after 24-h filter trap analysis of ubiquitinated aggregates was performed using both the anti-Ub antibody (clone FK2; Enzo Life Sciences) and an S tag HRP conjugate followed by chemiluminescence. The degradation of 100 nM 35S-Sic1(Ubn) in the presence of 0–625 nM ubiquitinated aggregates was assessed, and the Ki was determined as described in the previous paragraph.
Microscopy
HEK293 cells were transfected with the indicated N-htt–chFP plasmid and grown on poly-d-lysine–coated coverslips. 72 h after transfection, cells were fixed with 4% paraformaldehyde. Cells were imaged by epifluorescence on a microscope (Axiovert 200M; Carl Zeiss) with a 100× oil lens, NA 1.4 (Carl Zeiss). Digital (12 bit) images were acquired with a cooled charge-coupled device (CoolSNAP HQ; Photometrics) and processed using MetaMorph software (Universal Imaging).
For live-cell imaging of transiently transfected cells U2OS cells, cells were plated in DME with 10% FCS and 5% CO2 on glass-bottom 35-mm plates (MatTek) or on 8-well cover glass-bottom plates. Cells were transfected the next day with N-htt(Q91)–chFP or N-htt(Q47)–chFP with FuGENE 6 (Roche) according to the manufacturer’s instructions. Imaging of cells was started 16 h after transfection when most of the cells expressed the fluorescent protein at low but detectable levels.
For imaging of cells expressing YFP-Ub and UbG76V-GFP, cells were stably cotransfected with pEF-YFP-Ub or UbG76V-GFP with a vector that confers G418 resistance. Drug-resistant colonies were pooled, and expressing cells were sorted by FACS and further propagated. These cells were then transiently transfected with N-htt(Q91)–chFP as described in the previous paragraph.
Live-cell imaging was performed on an inverted microscope (Axiovert 200M) encased in a perspex chamber that was heated to 37°C. Plates were placed in a smaller internal chamber that was continuously perfused with humidified 5% CO2. The set up included a motorized stage (before) that enabled multiple simultaneous imaging of multiple fields. Digital (12 bit) images were acquired with a cooled charge-coupled device camera (CoolSNAP HQ) with an exacte light source (X-Cite; Lumen Dynamics), filter cubes for visualizing mCherry, YFP, or GFP, and phase contrast with a 20× NA 0.8 air objective. The entire set up was controlled by MetaMorph software, and ImageJ (National Institutes of Health) was used for image processing, analysis, and assembly.
Poly-Ub affinity capture and MS
For poly-Ub affinity capture, HEK293 cells stably expressing UbG76V-GFP were transfected with N-htt(Q91)–chFP. Cells were then sorted into low (102–103 a.u.) and high (>103 a.u.) N-htt(Q91)–chFP fluorescence as described in Flow cytometry and cell sorting. Cells were lysed in 20 mM Hepes, pH 7.2, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 5 mg/ml N-ethylmaleimide, 2% SDS, and EDTA-free protease inhibitors (Complete) followed by 3× sonication for 10–15 s (Kaiser et al., 2011). Extracts were centrifuged at 20,000 g in a microcentrifuge (Centrifuge 5424; Eppendorf) for 10 min, and supernatants were quantified by bicinchoninic acid before enrichment of Ub species by affinity capture with human P2UBA resin (Ub association domain from human ubiquilin 2, UBQLN2, also known as PLIC-2; 1:1 slurry).
MS was performed as previously described (Bennett et al., 2007; Riley et al., 2011). In brief, trypsin-digested samples were analyzed on a liquid chromatography–MS system that consisted of a capillary HPLC system (Agilent Technologies) coupled to a mass spectrometer (mitrOTOF system [Bruker Daltonics]; LTQ Orbitrap XL [Thermo Fisher Scientific]). Peptide mixtures were separated on a C-18 reversed-phase column using a linear gradient of acetonitrile (0–45%). Relative intensities of the isotope-labeled standards and analyses were obtained by measuring the intensities of the monoisotopic molecular ions after averaging ∼10 scans across the elution profile of the standard. Absolute quantification was performed using the ion intensities of the tracked endogenous peptides relative to the spiked labeled peptides. Ub ions measured included LIFAGK-GG-QLEDGR (UbK48) isopeptide unlabeled (mass/charge [m/z] = 487.60) and heavy isotope labeled (m/z = 489.94), ESTLHLVLR (ESTL) peptide unlabeled (m/z = 356.55) and heavy isotope labeled (m/z = 358.88), TLTGK-(GG)-TITLEVEPSDTIENVK (UbK11) isopeptide unlabeled (m/z = 801.43) and heavy isotope labeled (m/z = 803.43), and TLSDYNIQK-(GG)-ESTLHLVLR (UbK63) isopeptide unlabeled (m/z = 561.81) and heavy isotope labeled (m/z = 563.56). In addition, ions were measured for chFP tryptic peptide LSFPEGFK (m/z = 462.74) and its heavy isotope-labeled standard (m/z = 466.25) and for N-htt tryptic peptide AFESLK (m/z = 347.69) and its heavy leucine-labeled standard (m/z = 351.20). The chFP peptide is unique to chFP and is not found in GFP. To calculate the percentage of Ub-associated N-htt, the amount of P2UBA-associated N-htt(Q91)–-chFP (as measured by chFP) was divided by the total amount of N-htt(Q91)–chFP (as measured by chFP) using Orbitrap analysis. The chFP and N-htt peptides gave similar results.
Online supplemental material
Fig. S1 shows that the chFP tag does not alter the aggregation or UPS impairment phenotypes of htt. Fig. S2 explains the method of transformation of FACS data used to generate the plots in Figs. 1, 2, and 4. Fig. S3 shows experiments further characterizing the reporter lines used in this study. Fig. S4 accompanies Fig. 3 and shows the kinetics of radiolabeled Sic1 degradation by 26S proteasomes and controls for the production of ubiquitinated and aggregated htt(Q51) in vitro. Fig. S5 shows MS data accompanying Fig. 4 B and characterizes the cell line used in Figs. 5 and 6. Video 1 shows aggregation of N-htt(Q91)–chFP in U2OS cells. Video 2 shows that N-htt(Q91)–chFP aggregates recruit Ub. Video 3 shows that N-htt(Q91)–chFP aggregates form many hours before impaired proteasomal degradation. Table S1 shows the event log for time-lapse video microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110093/DC1.
Acknowledgments
We are grateful to C. Crumpton, D. Parks, and L. Herzenberg and the Stanford shared flow cytometry facility for help with flow cytometry and data analysis. We thank E. Bennett for reagents, sharing of unpublished results, and discussions. We thank B. Chen, E. Sofos, and R. Trevino for help with plasmid construction, J. Olzmann, D. Dowlatshahi, and M. Pearce for critical reading of the manuscript, and the members of the Kopito laboratory for helpful discussion.
This work was supported by grants to R.R. Kopito from the National Institutes of Neurological Diseases and Stroke, the Huntington’s Disease Society of America, and the Cure Huntington’s Disease Initiative. B.E. Riley was supported in part by a postdoctoral fellowship from the Hereditary Disease Foundation. M.S. Hipp was supported in part by a fellowship from the Deutsche Forschungsgemeinschaft. K. Bersuker was supported by a predoctoral training grant from the National Institute of General Medical Sciences.
Footnotes
Abbreviations used in this paper:
- AQUA
- absolute quantitative
- a.u.
- arbitrary unit
- ERAD
- ER-associated degradation
- IB
- inclusion body
- HD
- Huntington’s disease
- HS
- high sort
- htt
- huntingtin
- LS
- low sort
- MS
- mass spectrometry
- ODC
- ornithine decarboxylase
- TEV
- tobacco etch virus
- Ub
- ubiquitin
- UPS
- Ub-proteasome system
References
- Alvarez-Castelao B., Martín-Guerrero I., García-Orad A., Castaño J.G. 2009. Cytomegalovirus promoter up-regulation is the major cause of increased protein levels of unstable reporter proteins after treatment of living cells with proteasome inhibitors. J. Biol. Chem. 284:28253–28262 10.1074/jbc.M109.004101 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Baker R.T., Catanzariti A.M., Karunasekara Y., Soboleva T.A., Sharwood R., Whitney S., Board P.G. 2005. Using deubiquitylating enzymes as research tools. Methods Enzymol. 398:540–554 10.1016/S0076-6879(05)98044-0 [DOI] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Bence N.F., Jayakumar R., Kopito R.R. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 17:351–365 10.1016/j.molcel.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Shaler T.A., Woodman B., Ryu K.Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., Kopito R.R. 2007. Global changes to the ubiquitin system in Huntington’s disease. Nature. 448:704–708 10.1038/nature06022 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A., Miller E.A., Morimoto R.I. 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA. 106:14914–14919 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bett J.S., Goellner G.M., Woodman B., Pratt G., Rechsteiner M., Bates G.P. 2006. Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington’s disease mice: exclusion of proteasome activator REGgamma as a therapeutic target. Hum. Mol. Genet. 15:33–44 10.1093/hmg/ddi423 [DOI] [PubMed] [Google Scholar]
- Bowman A.B., Yoo S.Y., Dantuma N.P., Zoghbi H.Y. 2005. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 14:679–691 10.1093/hmg/ddi064 [DOI] [PubMed] [Google Scholar]
- Catanzariti A.M., Soboleva T.A., Jans D.A., Board P.G., Baker R.T. 2004. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 13:1331–1339 10.1110/ps.04618904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.Y., Warrick J.M., Gray-Board G.L., Paulson H.L., Bonini N.M. 2000. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 9:2811–2820 10.1093/hmg/9.19.2811 [DOI] [PubMed] [Google Scholar]
- Chen J., Fang G. 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15:1765–1770 10.1101/gad.898701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Brundin P. 2003. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 40:427–446 10.1016/S0896-6273(03)00606-8 [DOI] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., Masucci M.G. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543 10.1038/75406 [DOI] [PubMed] [Google Scholar]
- Dantuma N.P., Groothuis T.A., Salomons F.A., Neefjes J. 2006. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173:19–26 10.1083/jcb.200510071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B., Christianson J.C., Kopito R.R., Brunger A.T. 2006. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 22:451–462 10.1016/j.molcel.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Deriziotis P., Tabrizi S.J. 2008. Prions and the proteasome. Biochim. Biophys. Acta. 1782:713–722 [DOI] [PubMed] [Google Scholar]
- Díaz-Hernández M., Hernández F., Martín-Aparicio E., Gómez-Ramos P., Morán M.A., Castaño J.G., Ferrer I., Avila J., Lucas J.J. 2003. Neuronal induction of the immunoproteasome in Huntington’s disease. J. Neurosci. 23:11653–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M.J., Hochstrasser M. 1991. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 266:21150–21157 [PubMed] [Google Scholar]
- Finkbeiner S. 2011. Huntington’s Disease. Cold Spring Harb. Perspect. Biol. 3:a007476 10.1101/cshperspect.a007476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P. 2003. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 100:6940–6945 10.1073/pnas.0832254100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Krupinski T., Garcia S., Morimoto R.I. 2009. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 5:e1000399 10.1371/journal.pgen.1000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillardon F., Kloss A., Berg M., Neumann M., Mechtler K., Hengerer B., Dahlmann B. 2007. The 20S proteasome isolated from Alzheimer’s disease brain shows post-translational modifications but unchanged proteolytic activity. J. Neurochem. 101:1483–1490 10.1111/j.1471-4159.2006.04438.x [DOI] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., Kulka R.G. 2000. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol. Cell. Biol. 20:7214–7219 10.1128/MCB.20.19.7214-7219.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F.U., Raychaudhuri S. 2011. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods. 8:879–884 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- Hatters D.M. 2008. Protein misfolding inside cells: the case of huntingtin and Huntington’s disease. IUBMB Life. 60:724–728 10.1002/iub.111 [DOI] [PubMed] [Google Scholar]
- Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., Mahal A., Smith D.L., Woodman B., Bates G.P. 2004. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 13:1389–1405 10.1093/hmg/ddh144 [DOI] [PubMed] [Google Scholar]
- Howarth J.L., Kelly S., Keasey M.P., Glover C.P., Lee Y.B., Mitrophanous K., Chapple J.P., Gallo J.M., Cheetham M.E., Uney J.B. 2007. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol. Ther. 15:1100–1105 [DOI] [PubMed] [Google Scholar]
- Illing M.E., Rajan R.S., Bence N.F., Kopito R.R. 2002. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J. Biol. Chem. 277:34150–34160 10.1074/jbc.M204955200 [DOI] [PubMed] [Google Scholar]
- Jana N.R., Tanaka M., Wang G., Nukina N. 2000. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum. Mol. Genet. 9:2009–2018 10.1093/hmg/9.13.2009 [DOI] [PubMed] [Google Scholar]
- Jana N.R., Zemskov E.A., Gh Wang, Nukina N. 2001. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 10:1049–1059 10.1093/hmg/10.10.1049 [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Ma P.C., Ota I.M., Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456 10.1074/jbc.270.29.17442 [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E., Agar J.N., Taylor D.M., Minotti S., Durham H.D. 2004. Focal dysfunction of the proteasome: a pathogenic factor in a mouse model of amyotrophic lateral sclerosis. J. Neurochem. 89:1325–1335 10.1111/j.1471-4159.2004.02453.x [DOI] [PubMed] [Google Scholar]
- Kaiser S.E., Riley B.E., Shaler T.A., Trevino R.S., Becker C.H., Schulman H., Kopito R.R. 2011. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods. 8:691–696 10.1038/nmeth.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe Y., Nakai A., Tanabe M., Nagata K. 1998. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur. J. Biochem. 255:356–362 10.1046/j.1432-1327.1998.2550356.x [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P., Benzer S. 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 287:1837–1840 10.1126/science.287.5459.1837 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick D.S., Gerber S.A., Gygi S.P. 2005. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 35:265–273 10.1016/j.ymeth.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Kulka R.G., Raboy B., Schuster R., Parag H.A., Diamond G., Ciechanover A., Marcus M. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726–15731 [PubMed] [Google Scholar]
- Lehman N.L. 2009. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 118:329–347 10.1007/s00401-009-0560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz J.B., Umar I., Wang Q., Montine T.J., McMillan P.J., Tsuang D.W., Jin J., Pan C., Shin J., Zhu D., Zhang J. 2007. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol. 17:139–145 10.1111/j.1750-3639.2007.00048.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrusiak L.M., Beitel L.K., Wang X., Scanlon T.C., Chevalier-Larsen E., Merry D.E., Trifiro M.A. 2003. Transglutaminase potentiates ligand-dependent proteasome dysfunction induced by polyglutamine-expanded androgen receptor. Hum. Mol. Genet. 12:1497–1506 10.1093/hmg/ddg161 [DOI] [PubMed] [Google Scholar]
- Mayer R.J., Lowe J., Lennox G., Doherty F., Landon M. 1989. Intermediate filaments and ubiquitin: a new thread in the understanding of chronic neurodegenerative diseases. Prog. Clin. Biol. Res. 317:809–818 [PubMed] [Google Scholar]
- Maynard C.J., Böttcher C., Ortega Z., Smith R., Florea B.I., Díaz-Hernández M., Brundin P., Overkleeft H.S., Li J.Y., Lucas J.J., Dantuma N.P. 2009. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl. Acad. Sci. USA. 106:13986–13991 10.1073/pnas.0906463106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Tsvetkov A.S., Finkbeiner S. 2009. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in huntington disease. J. Biol. Chem. 284:4398–4403 10.1074/jbc.M806269200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., Vendruscolo M., Hayer-Hartl M., Hartl F.U., Vabulas R.M. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 144:67–78 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Ortega Z., Díaz-Hernández M., Lucas J.J. 2007. Is the ubiquitin-proteasome system impaired in Huntington’s disease? Cell. Mol. Life Sci. 64:2245–2257 10.1007/s00018-007-7222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z., Díaz-Hernández M., Maynard C.J., Hernández F., Dantuma N.P., Lucas J.J. 2010. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J. Neurosci. 30:3675–3688 10.1523/JNEUROSCI.5673-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli L., O’Farrell C., Lockhart P.J., Baptista M., Kehoe K., Vink L., Choi P., Wolozin B., Farrer M., Hardy J., Cookson M.R. 2002. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 36:1007–1019 10.1016/S0896-6273(02)01125-X [DOI] [PubMed] [Google Scholar]
- Qian S.B., Ott D.E., Schubert U., Bennink J.R., Yewdell J.W. 2002. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J. Biol. Chem. 277:38818–38826 10.1074/jbc.M205547200 [DOI] [PubMed] [Google Scholar]
- Riley B.E., Kaiser S.E., Kopito R.R. 2011. Autophagy inhibition engages Nrf2-p62 Ub-associated signaling. Autophagy. 7:338–340 10.4161/auto.7.3.14780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K.Y., Garza J.C., Lu X.Y., Barsh G.S., Kopito R.R. 2008. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc. Natl. Acad. Sci. USA. 105:4016–4021 10.1073/pnas.0800096105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y., Isono E., Toh-E A. 2005. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399:215–227 10.1016/S0076-6879(05)99014-9 [DOI] [PubMed] [Google Scholar]
- Saigoh K., Wang Y.L., Suh J.G., Yamanishi T., Sakai Y., Kiyosawa H., Harada T., Ichihara N., Wakana S., Kikuchi T., Wada K. 1999. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat. Genet. 23:47–51 [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., Wanker E.E. 1997. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 90:549–558 10.1016/S0092-8674(00)80514-0 [DOI] [PubMed] [Google Scholar]
- Sekijima Y., Wiseman R.L., Matteson J., Hammarström P., Miller S.R., Sawkar A.R., Balch W.E., Kelly J.W. 2005. The biological and chemical basis for tissue-selective amyloid disease. Cell. 121:73–85 10.1016/j.cell.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. 2004. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 304:100–104 10.1126/science.1092194 [DOI] [PubMed] [Google Scholar]
- Suhr S.T., Senut M.C., Whitelegge J.P., Faull K.F., Cuizon D.B., Gage F.H. 2001. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J. Cell Biol. 153:283–294 10.1083/jcb.153.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.P., Hardy J., Fischbeck K.H. 2002. Toxic proteins in neurodegenerative disease. Science. 296:1991–1995 10.1126/science.1067122 [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman L., Rechsteiner M., Pickart C.M. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94–102 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera A.G., Díaz-Hernández M., Hernández F., Ortega Z., Lucas J.J. 2005. The ubiquitin-proteasome system in Huntington’s disease. Neuroscientist. 11:583–594 10.1177/1073858405280639 [DOI] [PubMed] [Google Scholar]
- van Tijn P., de Vrij F.M., Schuurman K.G., Dantuma N.P., Fischer D.F., van Leeuwen F.W., Hol E.M. 2007. Dose-dependent inhibition of proteasome activity by a mutant ubiquitin associated with neurodegenerative disease. J. Cell Sci. 120:1615–1623 10.1242/jcs.03438 [DOI] [PubMed] [Google Scholar]
- Varshavsky A. 1997. The N-end rule pathway of protein degradation. Genes Cells. 2:13–28 10.1046/j.1365-2443.1997.1020301.x [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker E.E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. 1999. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 309:375–386 10.1016/S0076-6879(99)09026-6 [DOI] [PubMed] [Google Scholar]
- Ward C.L., Omura S., Kopito R.R. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 83:121–127 10.1016/0092-8674(95)90240-6 [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Chan H.Y., Gray-Board G.L., Chai Y., Paulson H.L., Bonini N.M. 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23:425–428 10.1038/70532 [DOI] [PubMed] [Google Scholar]
- Wilson S.M., Bhattacharyya B., Rachel R.A., Coppola V., Tessarollo L., Householder D.B., Fletcher C.F., Miller R.J., Copeland N.G., Jenkins N.A. 2002. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 32:420–425 10.1038/ng1006 [DOI] [PubMed] [Google Scholar]
- Woodman B., Butler R., Landles C., Lupton M.K., Tse J., Hockly E., Moffitt H., Sathasivam K., Bates G.P. 2007. The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res. Bull. 72:83–97 10.1016/j.brainresbull.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Xia Q., Liao L., Cheng D., Duong D.M., Gearing M., Lah J.J., Levey A.I., Peng J. 2008. Proteomic identification of novel proteins associated with Lewy bodies. Front. Biosci. 13:3850–3856 10.2741/2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Kaung G., Kobayashi S., Kopito R.R. 1997. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J. Biol. Chem. 272:20800–20804 10.1074/jbc.272.33.20800 [DOI] [PubMed] [Google Scholar]
- Zemskov E.A., Nukina N. 2003. Impaired degradation of PKCalpha by proteasome in a cellular model of Huntington’s disease. Neuroreport. 14:1435–1438 10.1097/00001756-200308060-00006 [DOI] [PubMed] [Google Scholar]
- Zhang M., Pickart C.M., Coffino P. 2003. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 22:1488–1496 10.1093/emboj/cdg158 [DOI] [PMC free article] [PubMed] [Google Scholar]