Aurora B phosphorylation antagonizes the interaction between the Ska complex and the KMN network, thereby controlling Ska recruitment to kinetochores and stabilization of kinetochore–microtubule attachments.
Abstract
The KMN network (named according to the acronym for KNL1, Mis12, and Ndc80) and the more recently identified Ska complex (Ska1–3) have been shown to mediate kinetochore (KT)–microtubule (MT) attachments. How these two complexes cooperate to achieve stable end-on attachments remains unknown. In this paper, we show that Aurora B negatively regulates the localization of the Ska complex to KTs and that recruitment of the Ska complex to KTs depends on the KMN network. We identified interactions between members of the KMN and Ska complexes and demonstrated that these interactions are regulated by Aurora B. Aurora B directly phosphorylated Ska1 and Ska3 in vitro, and expression of phosphomimetic mutants of Ska1 and Ska3 impaired Ska KT recruitment and formation of stable KT–MT fibers (K-fibers), disrupting mitotic progression. We propose that Aurora B phosphorylation antagonizes the interaction between the Ska complex and the KMN network, thereby controlling Ska recruitment to KTs and stabilization of KT–MT attachments.
Introduction
Chromosome alignment and segregation require that all kinetochores (KTs) establish stable bioriented attachments to spindle microtubules (MTs). Central to this process and providing the core KT–MT attachment interface at outer KTs is the conserved KMN network, composed of the KNL1 protein and the four-subunit Mis12 (Mis12, Mis13, Mis14, and Nnf1) and Ndc80 (Hec1, Nuf2, Spc24, and Spc25) complexes (Santaguida and Musacchio, 2009). Both the Ndc80 complex and KNL1 bind directly to MTs in vitro, whereas the Mis12 complex serves as a scaffold to form the KMN network and enhances the MT binding activities of the other components (Cheeseman et al., 2006). KTs are also important for discriminating bipolar from inappropriate MT attachments and for generating a signal that controls the timing of anaphase onset through the spindle assembly checkpoint (Musacchio and Salmon, 2007). The mitotic kinase Aurora B is critical for both error correction through destabilization of incorrect attachments (Lampson et al., 2004; Nezi and Musacchio, 2009) and for checkpoint signaling (Maresca and Salmon, 2010; Santaguida et al., 2011). Other proteins localized to the KT–MT interface during mitosis also contribute to the formation of stable and functional connections. Prominent among these is the Ska complex, composed of Ska1, 2, and 3, which has been proposed to be required for stable KT–MT attachments (Hanisch et al., 2006; Gaitanos et al., 2009; Raaijmakers et al., 2009; Theis et al., 2009; Welburn et al., 2009). The Ska complex has also been implicated in silencing of the spindle checkpoint (Hanisch et al., 2006; Daum et al., 2009) and in maintenance of sister chromatid cohesion (Daum et al., 2009; Theis et al., 2009). It comprises two copies of each subunit and directly binds to MTs in vitro (Welburn et al., 2009). Efficient depletion of the Ska complex leads to severe attachment defects and unstable K-fibers, reminiscent of the KT-null phenotype observed upon Ndc80 depletion (Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009; however, see Daum et al. [2009]). As recruitment of the Ska complex to KTs has been shown to depend on the Ndc80 complex (Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009), the phenotype observed in Ndc80-depleted cells might reflect the loss of both complexes.
The available evidence indicates that the Ska complex acts in concert with the KMN network for generating stable end-on attachments. However, no interaction between the KMN network and the Ska complex has been described, and how the KT recruitment of the Ska complex is regulated remains unknown. To understand how functional KT–MT attachments are stabilized in mitosis, it is therefore critical to determine whether and how the Ska complex interacts with members of the KMN network and how this interaction is regulated in time and space.
Results and discussion
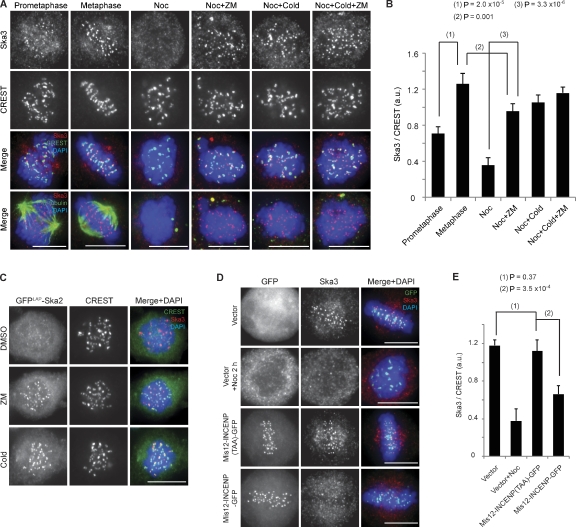
To obtain insight into the regulation of KT–MT attachment by the Ska complex, we first investigated how its KT localization is controlled. In line with a dependency on MT occupancy at KTs (Hanisch et al., 2006; Gaitanos et al., 2009), Ska3 staining intensity was about twofold higher at metaphase KTs than prometaphase KTs (Fig. 1, A and B). Moreover, Ska levels were clearly reduced at misaligned KTs when compared with aligned KTs (Fig. S1 A), indicating that the Ska complex preferentially accumulates at fully attached and bioriented KTs. As phosphorylation of Aurora B substrates at outer KTs decreases as KTs become bioriented and tension is established (Liu et al., 2009; Welburn et al., 2010), we asked whether Aurora B might be responsible for removing the Ska complex from unattached KTs. We monitored Ska3 localization in nocodazole-treated cells incubated with the Aurora B inhibitor ZM447439 (Ditchfield et al., 2003). In parallel, we also tested the effect of inhibiting the mitotic kinases Mps1 (by Mps1-IN-1 [Kwiatkowski et al., 2010] or reversine [Santaguida et al., 2010]) or Plk1 (by ZK-thiazolidinone [TAL]; Santamaria et al., 2007). Cold-treated cells were included as a positive control, as exposure to low temperature restores the KT localization of Ska proteins in nocodazole-treated cells (Hanisch et al., 2006; Gaitanos et al., 2009). Indeed, inhibition of Aurora B, but not Mps1 or Plk1, restored 70% of the Ska3 KT signal detected in metaphase cells (Figs. 1 [A and B] and S1 [B and C]). Similar results were obtained when GFPLAP-Ska2 and endogenous Ska1 were examined (Figs. 1 C and S1 D). Together, these results strongly suggest that Aurora B negatively regulates the association of the Ska complex with KTs, reminiscent of recent data on the Astrin–small kinetochore-associated protein complex (Schmidt et al., 2010). They further indicate that Aurora B plays a major role in regulating Ska recruitment to KTs, whereas KT–MT attachment seems to contribute to Ska recruitment independently of Aurora B’s effect. To further strengthen this notion, we expressed a Mis12-INCENP (inner centromere protein) fusion protein, shown to increase phosphorylation of Aurora B substrates by recruiting additional Aurora B to outer KTs (Liu et al., 2009). Expression of Mis12-INCENP, but not a Mis12-INCENP (TAA) mutant that is unable to activate Aurora B (Sessa et al., 2005), resulted in a twofold reduction of Ska3 staining at KTs (Fig. 1, D and E).
Figure 1.
Aurora B antagonizes Ska complex localization to KTs. (A) HeLa S3 cells were either treated with 3.3 µM nocodazole (Noc) for 1 h before a 2-h incubation at 4°C (Cold) with ZM447439 (ZM) or left untreated. (B) A bar graph showing the quantification of Ska3 staining intensity at KTs (normalized against CREST) of cells treated as in A (>100 KTs from five cells; error bars indicate the SD of five cells). a.u., arbitrary unit. (C) GFPLAP-Ska2–expressing HeLa cells were treated with 3.3 µM nocodazole for 1 h followed by 2 h with DMSO, ZM, or cold. (D) HeLa S3 cells were transfected with plasmids encoding GFP alone, Mis12-INCENP-GFP WT, or TAA. (E) A bar graph showing the quantification of Ska3 staining intensity at KTs (normalized against CREST) of cells treated as in D (>100 KTs from five cells; SD of five cells). Bars, 10 µm.
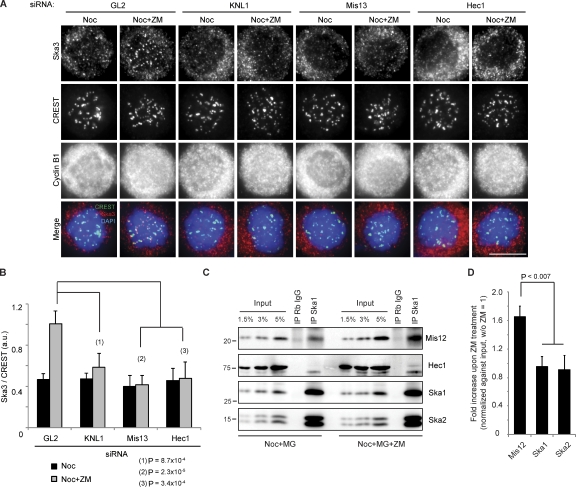
Next, we investigated how Aurora B influences the KT recruitment of the Ska complex. As it has been shown that KT localization of Ska proteins depends on the Ndc80 complex (Hanisch et al., 2006; Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009), we asked whether other KMN components, notably KNL1 and the Mis12 complex, are also required. Indeed, depletion of not only Hec1 but also KNL1 or Mis13 abolished the KT localization of Ska3 without affecting Ska protein levels (Fig. S1, E–G). To exclude that this phenotype was a result of an inability of cells to form proper KT–MT attachments, cells depleted of KNL1, Mis13, Hec1, or GL2 (Photinus pyralis luciferase gene; as a control) were treated with nocodazole followed by ZM447439. Importantly, Aurora B inhibition did not restore Ska3 to the KT in KNL1, Mis13, or Hec1-depleted cells, although it did so in control (GL2 treated) cells (Fig. 2, A and B). This strongly argues that the KMN network is directly involved in recruiting the Ska complex to KTs and that Aurora B regulates the interaction between the KMN and Ska complexes. To corroborate this conclusion, we investigated whether KMN components could be coimmunoprecipitated with Ska proteins and whether Aurora B inhibition might strengthen this interaction. Indeed, Mis12 could readily be coimmunoprecipitated with the Ska complex, and the interaction was clearly enhanced when cells were treated with ZM447439 (Fig. 2, C and D). Reciprocally, the Ska complex could also be coimmunoprecipitated with Mis12 (Fig. S2 A). Aurora B inhibition also revealed a faint interaction between the Ska complex and Hec1 (Fig. 2 C), but no KNL1 could be detected in our Ska1 immunoprecipitates (not depicted). To see whether any of the subunits of the Mis12 and Ndc80 complexes can interact with the Ska proteins, directed yeast two-hybrid experiments were performed. Importantly, we observed multiple interactions between members of the Ndc80 and Mis12 complexes and the Ska complex, namely Hec1-Ska1, Mis13-Ska2, and Spc24-Ska3 (Fig. S2, B–E). Furthermore, we could confirm binding between Ska2 and Mis13 by GST pull-down experiments (Fig. S2 F).
Figure 2.
Aurora B regulates the interaction between Ska and KMN. (A) HeLa S3 cells were treated for 48 h with GL2, KNL1, or Hec1 siRNAs or for 72 h with Mis13 siRNA before treatment with 3.3 µM nocodazole (Noc) for 1 h and a 2-h incubation with DMSO or ZM. Bar, 10 µm. (B) A bar graph showing the quantification of Ska3 staining intensity, as in Fig. 1 E, of cells treated as in A (>100 KTs from five cells; SD of five cells). (C) 0.33 µM nocodazole-arrested mitotic cells was treated with MG132 or MG132 plus ZM. After 2 h, cells were collected, and immunoprecipitations (IP) were performed with either rabbit IgGs (Rb IgG; as a control) or anti-Ska1 antibodies. Molecular mass is indicated in kilodaltons. (D) A bar graph showing the quantification of the intensities of coprecipitated proteins upon ZM treatment (relative to treatment without ZM; SD of three independent experiments).
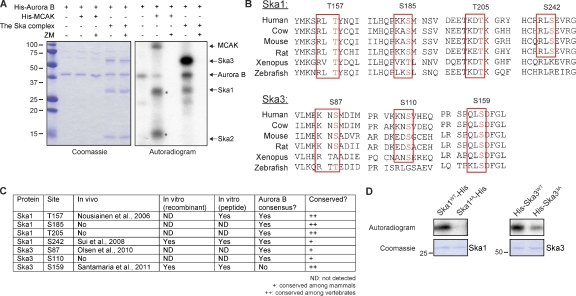
Following up on earlier evidence that mitotic phosphorylation of Ska3 is partly Aurora B dependent (Theis et al., 2009), we assayed the ability of Aurora B to phosphorylate a reconstituted Ska complex. Under these in vitro conditions, Ska1 and Ska3, but not Ska2, were readily phosphorylated by Aurora B (Fig. 3 A). This phosphorylation was completely abolished by ZM447439 treatment, attesting to its specificity (Fig. 3 A). Examination of the Ska1 and Ska3 protein sequences revealed four conserved Aurora B consensus sites (K/R-X-T/S) in Ska1 (T157, S185, T205, and S242) and two in Ska3 (S87 and S110; Fig. 3, B and C). Furthermore, mass spectrometric analysis of the in vitro phosphorylated Ska complex identified a highly conserved phosphorylation site in Ska3 (S159). Although this site does not match the canonical Aurora B consensus (Fig. 3, B and C), a peptide spanning S159 of Ska3 was clearly phosphorylated by Aurora B (Fig. S3 A). In addition, several of the putative Aurora B sites in Ska proteins have been shown to be phosphorylated in vivo in phosphoproteomics studies (Fig. 3 C; Nousiainen et al., 2006; Sui et al., 2008; Olsen et al., 2010; Santamaria et al., 2011). Thus, we generated mutants covering the potential Aurora B sites in Ska proteins. Kinase assays performed on nonphosphorylatable mutants (denoted as Ska14A and Ska33A) confirmed that these sites represent major targets of Aurora B in Ska1 and Ska3 (Fig. 3 D).
Figure 3.
Aurora B phosphorylates Ska1 and Ska3 in vitro. (A) In vitro kinase assay with Aurora B alone or Aurora B plus ZM on the Ska complex or mitotic centromere-associated kinesin (MCAK; as a positive control; Andrews et al., 2004; Lan et al., 2004). A Coomassie-stained gel and autoradiogram (left and right, respectively) are shown. Asterisks indicate unspecific bands in the mitotic centromere-associated kinesin preparation. Molecular mass is indicated in kilodaltons. (B) Sequence alignment of the potential Aurora B sites in Ska1 and Ska3 from the indicated species; alignments were performed with CLUSTALW (network protein sequence analysis). Potential Aurora B sites conserved across species are marked with a rectangle. (C) A table summarizing collected information for the seven potential Aurora B sites mutated in Ska1 and Ska3. (D) In vitro Aurora B kinase assays of Ska1WT-His and His-Ska3WT and the corresponding nonphosphorylatable mutants. An autoradiogram and Coomassie-stained gel (top and bottom, respectively) are shown.
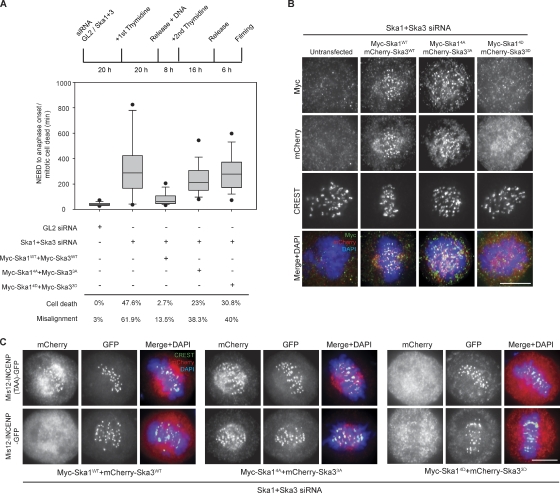
To assess the role of Aurora B phosphorylation on the Ska complex in vivo, we investigated the functionality of the phosphomimetic (to aspartate) and nonphosphorylatable (to alanine) mutants of Ska1 and Ska3 in HeLa S3 cells stably expressing H2B-GFP by combining an siRNA-based complementation approach with live-cell imaging (Figs. 4 A and S3, B–D). Consistent with previous work (Gaitanos et al., 2009), codepletion of Ska1 and Ska3 yielded a significant increase in mitotic timing (mean duration of 338 min from nuclear envelope breakdown [NEBD] to anaphase onset or mitotic cell death), and a significant proportion of cells (47.6%) died after prolonged arrest in mitosis but before anaphase. Of all Ska-depleted cells, 62% showed prolonged prometaphase with obvious chromosome congression defects (Figs. 4 A and S3, B–D), whereas the remaining cells experienced a delay without obviously misaligned chromosomes (Fig. S3 D). For comparison, GL2-treated control cells proceeded rapidly from NEBD to anaphase (mean of 42 min), only 3% displayed misalignment, and virtually no mitotic cell death was observed (Figs. 4 A and S3, B and D; and Videos 1 and 2). Although prolonged mitosis can lead to chromosomes scattering as a result of cohesion fatigue (Daum et al., 2009, 2011; Gassmann et al., 2010), we emphasize that the misalignment defects in Ska-depleted cells could be observed within 2 h after NEBD and before formation of metaphase plates (see example in Fig. S3 B). Furthermore, in Ska1 + Ska3–depleted cells, positive Mad1 signals could be seen not only on unaligned KTs but also on some apparently aligned KTs (unpublished data), which is in line with the notion that KT–MT attachments failed to fully stabilize. These defects were largely rescued by coexpressing wild-type (WT) Ska1 (Ska1WT) and Ska3WT (mean of 85 min for mitotic timing, with only 3% cell death and 14% of cells with misaligned chromosomes; Figs. 4 A and S3 B and Video 3). In contrast, expression of neither the nonphosphorylatable mutants (Ska14A + Ska33A) nor the phosphomimetic mutants (Ska14D + Ska33D) rescued the observed depletion phenotypes (mean of 246 min for mitotic timing, 23 and 38% of cell death and misalignment, respectively, for Ska14A + Ska33A; mean of 297 min for mitotic timing, 31 and 40% of cell death and misalignment, respectively, for Ska14D + Ska33D; Figs. 4 A and S3, B and D; and Videos 4 and 5). These results suggest that precise temporal control of Ska1/Ska3 phosphorylation/dephosphorylation is essential for mitotic progression.
Figure 4.
Aurora B phosphorylation regulates the function and localization of the Ska complex. (A) A schematic representation of the Ska rescue protocol to assess mitotic progression. A box-and-whisker plot showing the elapsed time (minutes) that cells spent in mitosis from NEBD to anaphase onset or mitotic cell death (≥60 cells from two independent experiments) is shown. Percentages of mitotic cell death and cells with misaligned chromosomes are displayed below the plot. (B) HeLa S3 cells were treated as in A but fixed after a 10-h release from the second thymidine block. (C) Cells were treated as in B, but Mis12-INCENP-GFP WT or TAA plasmids were cotransfected with the Ska plasmids. Bars, 10 µm.
As Aurora B activity controls Ska localization at KTs (Fig. 1), we next examined the localization of the aforementioned mutant proteins. The phosphomimetic mutants (Ska14D + Ska33D) failed to localize to KTs (Fig. 4 B), confirming that phosphorylation by Aurora B significantly reduces binding of the Ska complex to KTs. However, they still interacted with Hec1 and Spc24 in yeast two-hybrid assays, indicating they do not effectively mimic phosphorylation in the latter context (unpublished data). In contrast, the nonphosphorylatable Ska mutants (Ska14A + Ska33A) localized to KTs identically to their WT counterparts. Considering that the association of the Ska complex with KTs is both highly dynamic (Raaijmakers et al., 2009) and maximal during metaphase (Fig. 1 A), we reasoned that the absence of Aurora B phosphorylation on the Ska14A and Ska33A mutants might induce premature KT recruitment (or prevent normal turnover) of the Ska complex at KTs. To test whether the nonphosphorylatable Ska mutants would resist Aurora B–dependent removal from KTs, Ska1 and Ska3 constructs were cotransfected with Mis12-INCENP WT or TAA plasmids (Fig. 4 C). In contrast to the SkaWT proteins, which were displaced from KTs in cells expressing Mis12-INCENP, the nonphosphorylatable mutants persisted at KTs even in the presence of increased Aurora B activity at outer KTs (Fig. 4 C), clearly indicating that mutations preventing Aurora B phosphorylation on the Ska complex severely compromise its Aurora B–dependent removal from KTs.
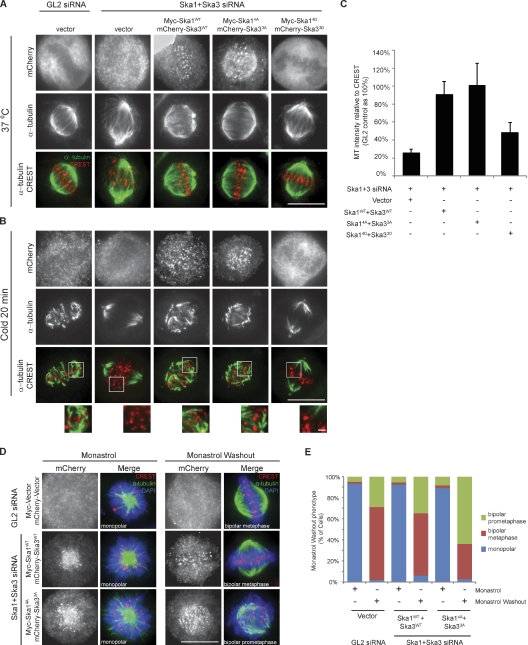
As shown previously (Gaitanos et al., 2009), K-fiber stability is lost upon Ska depletion. Thus, we assayed the stability of K-fibers after exposure of cells expressing either Ska WT or phosphorylation site mutants to cold. As expected, expression of SkaWT proteins in Ska-depleted cells rescued K-fiber stability (Fig. 5, A–C). In contrast, the phosphomimetic mutants failed to restore K-fiber stability, which is in line with their reduced KT recruitment and inability to rescue the Ska depletion phenotype. However, the nonphosphorylatable mutants not only supported stable K-fiber formation (Fig. 5, A–C) but also generated inter-KT tension comparable with that observed in cells expressing SkaWT (inter-KT distance of 1.25 and 1.21 µm for Ska14A + Ska33A and SkaWT, respectively; Fig. S3, E and F). This then raised the question of why the expression of these mutants in cells caused severe mitotic defects. Given that the nonphosphorylatable mutants persisted at KTs with high Aurora B activity (Fig. 4 C), we considered it plausible that these proteins might produce hyper-stable KT–MT attachments. Thus, we conducted monastrol washout assays to test the ability of these cells to correct syntelic attachments (Kapoor et al., 2000). 1 h after monastrol washout, only 30% of cells expressing the nonphosphorylatable Ska mutants had achieved metaphase, whereas >60% of control (GL2 treated) and SkaWT-expressing cells had done so (Fig. 5, D and E). These results support the view that nonphosphorylatable Ska mutants cause premature and/or excessive stabilization of KT–MT attachments, which in turn interferes with error correction and gives rise to mitotic defects. Finally, we tested whether Aurora B might also modulate the MT binding affinity of the Ska complex. Aurora B did not detectably influence the amount of Ska protein pelleted with MTs (Fig. S3 G), although it strongly reduced the amount of Ndc80Bonsai examined for control (Ciferri et al., 2008). Together, these results demonstrate that Aurora B–mediated regulation of Ska complex recruitment to KTs is crucial to stabilize KT–MT interactions.
Figure 5.
Aurora B phosphorylation of Ska proteins regulates K-fiber stability. (A and B) HeLa S3 cells were treated as in Fig. 4 B. Cells were left at 37°C (A) or placed at 4°C (B) for 20 min before being stained. (insets) Magnifications of a few KTs for each condition. (C) A bar graph showing the quantification of α-tubulin staining intensity around KTs (normalized against CREST) of cells treated as in B (>12 cells, >25 KTs per cell; SD of three independent experiments). (D) Cells were treated as in Fig. 4 B except that monastrol was added after the second thymidine release. After 10 h, cells were either fixed or washed and released into fresh medium containing MG132 for 1 h before fixation. (E) A bar graph showing the quantification of cells with monopolar/bipolar spindles (>60 cells per condition, ≥20 cells in three independent experiments). Bars: (A, B, and D) 10 µm; (B, insets) 1 µm.
Phosphorylation of the KMN subunits Hec1, Mis13, and KNL1 by Aurora B has previously been shown to reduce the KMN’s affinity for MT binding (Cheeseman et al., 2006; DeLuca et al., 2006; Ciferri et al., 2008; Guimaraes et al., 2008; Welburn et al., 2010). Here, we describe an additional mechanism for how Aurora B can negatively regulate KT–MT attachments. We propose that the Ska complex is recruited to KTs to stabilize end-on attachments mediated by the KMN network and that Aurora B regulates this recruitment. At poorly attached and/or tensionless KTs, the Ska complex is spatially close to Aurora B and therefore readily phosphorylated (Liu et al., 2009; Wang et al., 2011). This then interferes with KMN–Ska interactions and reduces the recruitment of the Ska complex to KTs. Once sister chromatids are bioriented and tension is established, members of the KMN and Ska complexes are dephosphorylated, leading to full stabilization of the KT–MT attachments. We do not exclude that Aurora B phosphorylation on Ska proteins could produce additional effects, such as influencing Ndc80 activity or preventing Ska proteins from interacting with proteins other than KMN. Likewise, Aurora B may phosphorylate additional targets that contribute to the interaction between KMN and Ska.
The cooperation of the KMN network and the Ska complex in the formation of stable end-on attachments is reminiscent of the relationship between the Ndc80 and Dam1 complexes in budding yeast. Thus, the Ska complex has been proposed to be the functional counterpart of the fungal Dam1 complex in metazoan organisms (Hanisch et al., 2006; Gaitanos et al., 2009; Welburn et al., 2009). Although this view is far from being universally accepted, our present results strengthen the view that different organisms use an evolutionarily conserved mechanism to form and maintain KT–MT attachments. Similar to the results reported here, the interaction between Dam1 and Ndc80 is also regulated by Aurora B in vitro (Lampert et al., 2010; Tien et al., 2010), and Ndc80 mutants that cannot bind Dam1 show defects in forming end-on attachments in Saccharomyces cerevisiae (Maure et al., 2011). Surprisingly, Dam1 in Schizosaccharomyces pombe and Ska3 in chicken DT40 cells are nonessential for viability (Sanchez-Perez et al., 2005; Ohta et al., 2010), arguing that in these cells, the KMN network may be sufficient, albeit perhaps less efficient, to support functional KT–MT attachments. In the future, it will be interesting to determine whether the Ska complex is essential in mammalian cells. Also, structural information on KMN network–Ska complex interactions will hopefully contribute to our molecular understanding of how these proteins cooperate to perform their functions at metazoan KTs.
Materials and methods
Cloning, plasmid, and siRNA transfection
The cDNAs of Ska1 and Ska3 were cloned into pcDNA5/FRT/TO or pcDNA3.1 vectors (Invitrogen) encoding an N-terminal 3xmyc tag or an N-terminal mCherry tag. An siRNA-resistant version of Ska1 containing six silent mutations and the phosphorylation site mutants of Ska1 and Ska3 were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies). Mis12-INCENP-GFP and Mis12-INCENP (TAA)-GFP plasmids (Liu et al., 2009) were provided by S.M. Lens (University Medical Center Utrecht, Utrecht, Netherlands). Plasmid transfections were performed using TransIT-LT1 reagent (Mirus Bio Corporation) according to the manufacturer’s instructions. siRNA duplexes were transfected using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. siRNA duplexes for GL2 (5′-CGTACGCGGAATACTTCGA-3′), Ska1 (5′-CCCGCTTAACCTATAATCAAA-3′; Hanisch et al., 2006), Ska3 (5′-AGACAAACATGAACATTAA-3′; Gaitanos et al., 2009), hSpindly (5′-GGAGAAATTTAAGAATTTA-3′), Hec1 (5′-GTTCAAAAGCTGGATGATC-3′; Chan et al., 2009), KNL1 (5′-GGAATCCAATGCTTTGAGA-3′; Liu et al., 2010), and Mis13 (5′-GGCGTTTCAGAGGAAAGAA-3′; Obuse et al., 2004) were previously described.
Protein purification, GST pull-down, and kinase assays
All His6-tagged proteins or GST-Ska2 were expressed in Escherichia coli and purified by Ni–nitrilotriacetic acid agarose beads (QIAGEN) or glutathione Sepharose (GE Healthcare), respectively. The plasmid encoding His6-tagged Aurora B (Yang et al., 2008) was a gift from X. Yao (University of Science and Technology of China). The full-length Ska complex comprising Ska1-His6, untagged Ska2 (a gift from A.A. Jeyaprakash and E. Conti, Max Planck Institute of Biochemistry, Martinsried, Germany), and His6-Ska3 was purified as previously described (Gaitanos et al., 2009). In brief, cells transformed with Ska1-His6 and Ska2 were mixed with cells transformed with His6-Ska3. Cells were then lysed in buffer containing 10 mM Tris-HCl, pH 8, 500 mM NaCl, 1 mM DTT, 0.05% NP-40, and 1 mM EDTA. The protein complexes were isolated by Ni–nitrilotriacetic acid beads and further purified by RESOURCE Q and Superdex 200 columns using fast protein liquid chromatography (ÄKTA Explorer; GE Healthcare). Myc-Mis13 was produced by using the TNT T7 coupled transcription/translation system (Promega) and incubated with glutathione Sepharose coupled with GST-Ska2 or GST alone for 1 h. The Sepharose beads were then washed five times with PBS, and bound species were resolved by SDS-PAGE and analyzed by Western blotting. In vitro kinase assays on recombinant proteins were performed at 30°C in buffer (25 mM Hepes, 50 mM NaCl, 1 mM DTT, 2 mM EGTA, 5 mM MgSO4, 10 µM ATP, and 5 µCi γ-[32P]ATP) for 30 min. Samples were then resolved by SDS-PAGE and visualized by autoradiography. For kinase assays on membranes, 12-mer peptides containing putative Aurora B sites (S/T) of Ska1 and Ska3 at position 7 were generated using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on a MultiPep robotic spotter (Intavis) according to the manufacturer’s directions and immobilized on cellulose membranes. The membranes were then incubated with Aurora B in the aforementioned kinase buffer, and kinase assays were performed as previously described (Santamaria et al., 2011).
Cell culture and synchronization
HeLa cells were cultured in a 5% CO2 atmosphere in DME (Invitrogen) supplemented with 10% heat-inactivated FCS and penicillin-streptomycin (100 IU/ml and 100 µg/ml, respectively). Nocodazole (0.33 or 3.3 µM), thymidine (2 mM), and monastrol (150 µM) were obtained from Sigma-Aldrich, and ZM447439 (10 µM) was obtained from Tocris Bioscience. MG132 (10 µM) was obtained from EMD. Reversine (0.5 µM) was obtained from Cayman Chemical. Mps1-IN-1 (2 µM; Kwiatkowski et al., 2010) was a gift from N.S. Gray (Dana-Farber Cancer Institute, Boston, MA).
Cell lysates, immunoprecipitation, and Western blot analysis
Cell lysates were prepared with Hepes lysis buffer (50 mM Hepes, 150 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 30 µg/ml DNase, 30 µg/ml RNase, and protease and phosphatase inhibitors), and Western blotting was performed as previously described (Chan et al., 2009). For coimmunoprecipitation, lysates were prepared using Hepes lysis buffer (buffers containing 0.1% or no Triton X-100 were used for Ska1 or Mis12 coimmunoprecipitation, respectively). Immunoprecipitations on cell lysates were performed using 5 µl of solid beads (Affi-Prep Protein A Matrix; Bio-Rad Laboratories) chemically cross-linked to 2–3 µg/µl of antibody against 2 mg of clarified lysates for 2 h at 4°C. The beads were washed four times with Hepes lysis buffer, and bound species were resolved by SDS-PAGE and analyzed by Western blotting. ImageJ (National Institutes of Health) was used to quantify the relative intensities of the coprecipitated proteins in Ska1 immunoprecipitations. First, we subtracted the intensity of the measured protein precipitated with control IgGs from that precipitated with Ska1 antibodies. Intensities were then normalized by dividing them by the mean intensity of the inputs. The value from the Noc + MG132 samples was set as 1.
Immunofluorescence microscopy
HeLa S3 cells or HeLa cells stably expressing GFPLAP-Ska2 (a gift from I.M. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, MA; Welburn et al., 2009) were grown on coverslips and simultaneously fixed and permeabilized for 10 min at room temperature in PTEMF buffer (20 mM Pipes, pH 6.8, 3.7% formaldehyde, 0.2% Triton X-100, 10 mM EGTA, and 1 mM MgCl2), as previously described (Silljé et al., 2006). DNA was visualized with 2 µg/ml DAPI. Images were acquired with a microscope (DeltaVision; Applied Precision) equipped with Plan Apochromat 60×/1.42 oil immersion objective (Olympus) and a camera (CoolSNAP HQ2; Photometrics). Images were obtained with 0.2-µm–distanced optical sections in z axis. Deconvolution of each section and projection of all sections into one picture were performed with softWoRx software (Applied Precision). For quantification of Ska KT intensity and K-fiber stability, circles with a diameter of 5 and 10 pixels, respectively, centered on each KT (the diameter of a KT was around 5 pixels) were drawn, and the intensity of Ska3 or α-tubulin within the circles was measured and normalized against the corresponding CREST (calcinosis, Raynaud’s phenomenon, esophageal dysfunction, sclerodactyly, and telangiectasia) using ImageJ. Statistical significances were verified by a two-tailed Student’s t test.
Antibodies
The following antibodies were used: mouse anti–α-tubulin (1:5,000 for Western blotting and 1:1,000 for immunofluorescence; Sigma-Aldrich), mouse anti-Hec1 (1:1,000; GeneTex Inc.), rabbit anti-Mis13 (1:500; Yang et al., 2008), rabbit anti-KNL1 (1:500; Abcam), rabbit anti-Ska1 (1:1,000; Hanisch et al., 2006), rabbit anti-Ska2 (1:1,000; Hanisch et al., 2006), rabbit anti-Ska3 (1:1,000; Gaitanos et al., 2009), rabbit anti-mCherry (1:5,000; generated by immunization of rabbits [Charles River] with His6-mCherry expressed in E. coli; Hübner et al., 2010), human CREST autoimmune serum (1:2,000; ImmunoVision, Inc.), mouse anti–Cyclin B1 (1:1,000; Millipore), and mouse anti-Myc (1:5; 9E10 tissue culture supernatant). For immunofluorescence analysis, primary antibodies were detected with Cy2-, Cy3-, and Cy5-conjugated donkey anti–mouse, –rabbit, or –human IgGs (1:1,000; Dianova).
Directed yeast two-hybrid analysis
cDNAs encoding the respective prey or bait proteins were cloned in-frame with the GAL activation domain of pACT2 (Spc24) or pGAD-C1 (in the case of Ska1, Ska2, and Ska3) vectors or the GAL-binding domain of pFBT9 vector (in the case of Hec1, Nuf2, Spc24, and Spc25). All vectors were obtained from Takara Bio Inc. Plasmids encoding members of the Mis12 complex (Mis12, Mis13, Mis14, and Nnf1; Kiyomitsu et al., 2010) were a gift from M. Yanagida (Kyoto University, Kyoto, Japan). Directed yeast two-hybrid experiments were performed as previously described (Hanisch et al., 2006).
Time-lapse microscopy
After siRNA and plasmid transfection, HeLa S3 cells stably expressing histone H2B-GFP (Silljé et al., 2006) were imaged using a microscope (Eclipse Ti; Nikon) equipped with a CoolLED pE-1 excitation system and a 20×/0.75 air Plan Apochromat objective (Nikon) at 37°C. Images were acquired at multiple positions every 3 min for 18 h. GFP signal was acquired at each time point with a 10-ms exposure time. mCherry signal was acquired every 10 time points with a 30-ms exposure time. MetaMorph software (7.7; Molecular Devices) was used to collect and process the data. The results are displayed as box-and-whisker plots; boxes represent 25–75% of the cells, the line within the box indicates the median, top and bottom whiskers represent the 10th and 90th percentiles, and dots represent the 5th and 95th percentiles, respectively.
MT pelleting assay
MT cosedimentation assays were performed as reported elsewhere (Ciferri et al., 2008). In a typical reaction, 3 µM Ska complex or Ndc80Bonsai complex (provided by A. Musacchio, Max Planck Institute of Molecular Physiology, Dortmund, Germany; purified as reported in Ciferri et al. [2008]) was incubated with increasing concentration of Aurora B and 10 mM ATP and 20 mM MgSO4 for 30 min at 30°C. 6 µM taxol-stabilized MTs was added, and incubation was continued for a further 10 min at room temperature. Reactions were transferred onto 100 µl of cushion buffer (BRB80 buffer containing 50% glycerol and 50 µM taxol) and ultracentrifuged for 10 min at 80,000 rpm using a TLA-100 rotor (Beckman Coulter). Equal volumes of pellets and supernatants were analyzed by SDS-PAGE.
Online supplemental material
Fig. S1 shows the localization of Ska1 in hSpindly-depleted or Aurora B–inhibited cells and Ska3 staining in mitotic kinase–inhibited and KMN-depleted cells. Fig. S2 shows the results from the Mis12 coimmunoprecipitation, directed yeast two-hybrid experiments, and GST pull down. Fig. S3 shows the information from the live-cell imaging experiments, the measurement of inter-KT distances, and the MT pelleting assay. Videos 1–5 show the mitotic progression of cells treated with Ska1 + Ska3 siRNAs followed by transfection with vector control, WT, or mutant Ska plasmids. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109001/DC1.
Acknowledgments
We thank N.S. Gray, X. Yao, M. Yanagida, S.M. Lens, A. Musacchio, and I.M. Cheeseman for reagents, H.H. Silljé, A. Hanisch, and A. Wehner for initial yeast two-hybrid screens, and T. Glatter for help with the mass spectrometry analysis. We also thank T.N. Gaitanos and L.L. Fava for critical reading of the manuscript and all members of our laboratory for insightful discussion. A. Arockia Jeyaprakash thanks E. Conti for support and the Max Planck Gesellschaft for funding.
This work was supported by the University of Basel and the Swiss Cancer League (KFS 02657-08-2010 to A. Santamaria and E.A. Nigg). All authors declare no conflict of interest.
Footnotes
Abbreviations used in this paper:
- KT
- kinetochore
- MT
- microtubule
- NEBD
- nuclear envelope breakdown
- WT
- wild type
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., Santamaria A. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J.R., Wren J.D., Daniel J.J., Sivakumar S., McAvoy J.N., Potapova T.A., Gorbsky G.J. 2009. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19:1467–1472 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J.R., Potapova T.A., Sivakumar S., Daniel J.J., Flynn J.N., Rankin S., Gorbsky G.J. 2011. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21:1018–1024 10.1016/j.cub.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., Nigg E.A. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., Desai A. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A., Silljé H.H., Nigg E.A. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner N.C., Wang L.H., Kaulich M., Descombes P., Poser I., Nigg E.A. 2010. Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma. 119:149–165 10.1007/s00412-009-0244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Iwasaki O., Obuse C., Yanagida M. 2010. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J. Cell Biol. 188:791–807 10.1083/jcb.200908096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N., Jelluma N., Filippakopoulos P., Soundararajan M., Manak M.S., Kwon M., Choi H.G., Sim T., Deveraux Q.L., Rottmann S., et al. 2010. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 6:359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2010. Welcome to a new kind of tension: Translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123:825–835 10.1242/jcs.064790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure J.F., Komoto S., Oku Y., Mino A., Pasqualato S., Natsume K., Clayton L., Musacchio A., Tanaka T.U. 2011. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr. Biol. 21:207–213 10.1016/j.cub.2010.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nezi L., Musacchio A. 2009. Sister chromatid tension and the spindle assembly checkpoint. Curr. Opin. Cell Biol. 21:785–795 10.1016/j.ceb.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Nousiainen M., Silljé H.H., Sauer G., Nigg E.A., Körner R. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA. 103:5391–5396 10.1073/pnas.0507066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141 10.1038/ncb1187 [DOI] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves Fde.L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 142:810–821 10.1016/j.cell.2010.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J.A., Tanenbaum M.E., Maia A.F., Medema R.H. 2009. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J. Cell Sci. 122:2436–2445 10.1242/jcs.051912 [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S.J., Crawley K., Karig I., Buck V., Meadows J.C., Franco-Sanchez A., Fleig U., Toda T., Millar J.B. 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24:2931–2943 10.1038/sj.emboj.7600761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., et al. 2007. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell. 18:4024–4036 10.1091/mbc.E07-05-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M., Schmidt A., Silljé H.H., Körner R., Nigg E.A. 2011. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics. 10:M110:004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Kiyomitsu T., Hori T., Backer C.B., Fukagawa T., Cheeseman I.M. 2010. Aurora B kinase controls the targeting of the Astrin–SKAP complex to bioriented kinetochores. J. Cell Biol. 191:269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., Musacchio A. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391 [DOI] [PubMed] [Google Scholar]
- Silljé H.H., Nagel S., Körner R., Nigg E.A. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742 [DOI] [PubMed] [Google Scholar]
- Sui S., Wang J., Yang B., Song L., Zhang J., Chen M., Liu J., Lu Z., Cai Y., Chen S., et al. 2008. Phosphoproteome analysis of the human Chang liver cells using SCX and a complementary mass spectrometric strategy. Proteomics. 8:2024–2034 [DOI] [PubMed] [Google Scholar]
- Theis M., Slabicki M., Junqueira M., Paszkowski-Rogacz M., Sontheimer J., Kittler R., Heninger A.K., Glatter T., Kruusmaa K., Poser I., et al. 2009. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 28:1453–1465 10.1038/emboj.2009.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., Lampson M.A. 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R., III, Cheeseman I.M. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., et al. 2008. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283:26726–26736 10.1074/jbc.M804207200 [DOI] [PMC free article] [PubMed] [Google Scholar]