JunB helps set in motion the transcriptional program necessary for the epithelial–mesenchymal transition and tissue fibrosis in response to TGF-β.
Abstract
The process of epithelial–mesenchymal transition (EMT) in response to transforming growth factor–β (TGF-β) contributes to tissue fibrosis, wound healing, and cancer via a mechanism that is not fully understood. This study identifies a critical role of JunB in the EMT and profibrotic responses to TGF-β. Depletion of JunB by small interfering ribonucleic acid abrogates TGF-β–induced disruption of cell–cell junctions, formation of actin fibers, focal adhesions, and expression of fibrotic proteins. JunB contributes to Smad-mediated repression of inhibitor of differentiation 2 through interaction with transcription repressor activating transcription factor 3. Importantly, JunB mediates the TGF-β induction of profibrotic response factors, fibronectin, fibulin-2, tropomyosin (Tpm1), and integrin-β3, which play critical roles in matrix deposition, cell–matrix adhesion, and actin stress fibers. In summary, JunB provides important input in setting the transcriptional program of the EMT and profibrotic responses to TGF-β. Thus, JunB represents an important target in diseases associated with EMT, including cancer and fibrosis.
Introduction
The process of epithelial–mesenchymal transition (EMT) is implicated in cancer progression, wound healing, and tissue fibrosis as well as normal embryonic development (Kalluri and Neilson, 2003; Thiery, 2003; Lee et al., 2006). In tissue fibrosis and wound healing, EMT is thought to contribute to generation of myofibroblasts and myofibroblast-like cells that mediate deposition of ECM proteins, such as collagens and fibronectin. In cancer, EMT leads to generation of more aggressive and invasive carcinoma cells as well as cancer stem cells. EMT involves disassembly of the polarized epithelial architecture and remodeling of the cell cytoskeleton, including intermediate and actin filaments. TGF-β cytokines have emerged as major regulators of EMT in human diseases and embryonic development (Zavadil and Böttinger, 2005). TGF-β can induce EMT in normal and carcinoma cells, disrupting cell junctions and inducing actin fibers linked to focal adhesions (Miettinen et al., 1994; Fialka et al., 1996; Oft et al., 1996; Piek et al., 1999; Bakin et al., 2004; Brown et al., 2004). Under physiological conditions, TGF-β functions as a potent tumor suppressor, regulating normal tissue homeostasis, cell proliferation, and matrix deposition (Stover et al., 2007). Malignant cancers are unresponsive to antimitogenic effects of TGF-β and produce elevated levels of TGF-β (Walker and Dearing, 1992; Wikström et al., 1998; Maehara et al., 1999). This has been linked to the induction of EMT in carcinoma cells, promoting tumor invasion, resistance to therapy, and metastatic spread (Maehara et al., 1999; Huber et al., 2005; Lee et al., 2006; Stover et al., 2007). The mechanisms underlying TGF-β–induced EMT and fibrotic responses are not fully understood.
TGF-β cytokines are deposited in the matrix in a latent/inactive form and are released in active form by various environmental signals (Annes et al., 2003). Active TGF-β binds to the receptor complex and stimulates a set of signaling events, leading to changes in gene expression and cell behavior (Pardali and Moustakas, 2007). The EMT response to TGF-β requires transcription and de novo protein synthesis (Bakin et al., 2004). Smad transcription factors, PI3 kinase, and MAPKs p38 and ERK have been implicated in EMT (Zavadil and Böttinger, 2005). Receptor-associated Smad3 and Smad4 play a major role in the EMT response (Bakin et al., 2004; Levy and Hill, 2005; Valcourt et al., 2005). Small GTPases Rac1 and RhoA contribute to EMT by activating p38 MAPK, PI3K-Akt, and Rho kinase signaling (Bakin et al., 2000, 2002; Bhowmick et al., 2001; Zavadil and Böttinger, 2005). Recent studies suggest that TGF-β–induced EMT involves Smad-dependent down-regulation of inhibitor of differentiation 2/3 (Id2/3) helix-loop-helix transcription factors (Kondo et al., 2004; Kowanetz et al., 2004). In some cell systems, TGF-β up-regulates Twist, Snail, Slug, and Hmga2 (Moustakas and Heldin, 2007). Forced expression of Hmga2, Snail, or Twist alone can induce EMT, down-regulating E-cadherin and increasing cell migration (Moustakas and Heldin, 2007). Formation of actin stress fibers is a main characteristic of TGF-β–induced EMT. Smads regulate expression of proteins, mediating the formation of actin fibers (tropomyosin Tpm1, α-actinin Actn1, and calponin Cnn2) and focal adhesions, including palladin and integrins (Bakin et al., 2004; Valcourt et al., 2005; Zheng et al., 2008; Safina et al., 2009; Bianchi et al., 2010). Tropomyosin-mediated actin fibers control tumor cell invasion and anchorage-independent growth (Pawlak and Helfman, 2001; Zheng et al., 2008; Safina et al., 2009). Actin fibers and focal adhesions are also actively involved in the deposition and remodeling of the ECM and may facilitate tissue fibrosis.
Activating protein 1 (AP1) transcription factors contribute to various TGF-β biological responses (Moustakas and Heldin, 2007). The AP1 factors are dimeric complexes of the basic leucine zipper proteins representing the FOS, JUN, activating transcription factor (ATF)/cAMP response element-binding, or musculoaponeurotic fibrosarcoma families (Eferl and Wagner, 2003). The leucine zipper domain mediates hetero- and homodimerization of these proteins, whereas the basic regions are responsible for DNA binding. Fos and Jun can induce EMT and promote invasion in epithelial cell lines (Ozanne et al., 2007), disrupting epithelial cell polarity without down-regulation of E-cadherin (Fialka et al., 1996). Likewise, constitutive MEK1-JUN signaling dissolves cell–cell junctions but does not suppress E-cadherin (Pinkas and Leder, 2002). Jun and JunB share extensive homology within the leucine zipper and basic domains, and JunB can rescue a lethal phenotype in Jun-null mice (Passegué et al., 2002). Despite their homology, these proteins display different transcriptional activity. Jun is a strong transcriptional activator, whereas JunB is a modest trans-activator and may even repress transcription (Finch et al., 2002; Eferl and Wagner, 2003). Both Jun and JunB can be induced by TGF-β in epithelial cells, whereas mesenchymal cells respond with up-regulation of JunB but not Jun (Chung et al., 1996). Jun and JunB can interact with Smad factors (Zhang et al., 1998) and positively or negatively regulate Smad target genes (Verrecchia et al., 2001a; Selvamurugan et al., 2004; Wang et al., 2008). Although Jun family proteins have been implicated in TGF-β responses and/or the EMT process, their role in TGF-β–induced EMT has not been investigated.
The present work addresses the function of JunB in TGF-β–induced EMT and fibrotic responses in mammary and kidney epithelial cells. The study shows that JunB is an immediate early response gene of TGF-β–Smad signaling, and JunB is required for the repression of Id2, a negative regulator of EMT, as well as for the up-regulation of factors mediating EMT and profibrotic responses. Thus, JunB may represent an important target in human diseases associated with EMT such as cancer and fibrosis.
Results
TGF-β–induced EMT is associated with Smad-dependent up-regulation of JunB
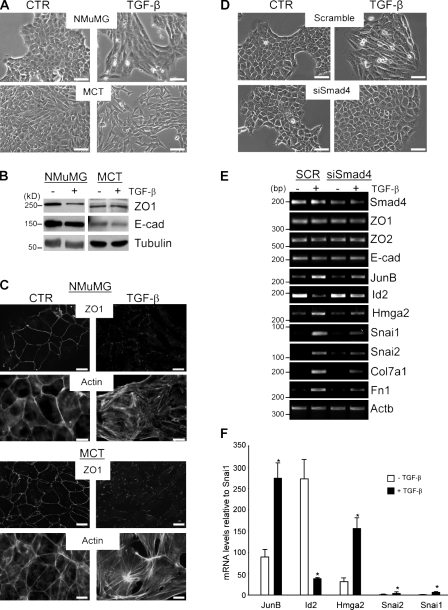
Two well-established cell models of EMT and fibrotic responses have been used in this study: the mouse mammary epithelial cell line NMuMG (Miettinen et al., 1994; Piek et al., 1999; Bakin et al., 2000; Bhowmick et al., 2001) and the murine kidney proximal tubular cell line murine cortical tubule (MCT; Wolf et al., 1999; Kalluri and Neilson, 2003). Treatment with TGF-β induced morphological EMT in both NMuMG and MCT cells within 24 h (Fig. 1 A). Immunofluorescence microscopy confirmed the disruption of cell–cell junctions and induction of actin stress fibers, characteristics of the TGF-β–induced EMT (Fig. 1 C). Immunoblotting showed down-regulation of a tight junction protein ZO1 in NMuMG cells but not in MCT, whereas adherence junction protein E-cadherin was not reduced by TGF-β in either cell line (Fig. 1 B). RT-PCR demonstrated that mRNA levels of ZO1 and ZO2 and E-cadherin were not affected by TGF-β in NMuMG (Fig. 1 E) and MCT cells (Fig. S1). These findings are in agreement with previous studies showing that morphological transition, dissolution of cell junctions, and formation of actin fibers occur within 24–48 h of TGF-β treatment without down-regulation of E-cadherin (Tian and Phillips, 2003; Brown et al., 2004; Maeda et al., 2005; Safina et al., 2009). Suppression of Smad4 blocked TGF-β–induced EMT in both cell lines (shown for NMuMG cells; Fig. 1 D), validating Smad dependency of this TGF-β response, as expected from previous studies (Bakin et al., 2004; Valcourt et al., 2005; Deckers et al., 2006).
Figure 1.
Smad4 is required for TGF-β–induced EMT. (A–C) NMuMG and MCT cells were treated with 2 ng/ml TGF-β1 for 24 h. (A) Phase-contrast images. CTR, control. Bars, 200 µm. (B) Immunoblotting of ZO1 and E-cadherin (E-cad); tubulin is a loading control. (C) Immunofluorescence images of ZO1, E-cadherin, and actin. Bars, 20 µm. (D and E) NMuMG cells were transfected with siRNA to Smad4 (siSmad4) or scramble (SCR) control. Cells were treated with 2 ng/ml TGF-β1 for 24 h. (D) Phase-contrast images. Bars, 200 µm. (E) RT-PCR of Smad4, ZO1, ZO2, E-cadherin, JunB, Id2, Hmga2, Snai1, Snai2, Col7a1, Fn1, and Actb, a loading control. Approximate size markers are shown based on the migration of markers relative to analogous RT-PCR products run on a different gel. (F) Quantitative RT-PCR for JunB, Id2, Hmga2, Snai1, and Snai2 was performed in triplicate using total RNA from NMuMG cells treated with 2 ng/ml TGF-β1 for 24 h. The CT values were normalized to 18S rRNA, and data are expressed as fold difference relative to Snai2. *, P < 0.05 using a Student’s t test. Error bars represent the mean value ± SD.
To dissect the transcriptional program of TGF-β–induced EMT, mRNA levels of several transcription factors were assessed by semiquantitative RT-PCR. The analysis revealed that JunB, as well as Id2, Hmga2, Snai1/Snail, and Snai2/Slug, is regulated via Smad4 (Fig. 1 E), although Snai1 and Snai2 are expressed at several-fold lower levels compared with JunB. Snai1 and Snai2 were up-regulated at 24 h but not at early time points of TGF-β treatment (Fig. S2 A). Quantitative PCR confirmed low levels of Snai1 and Snai2 (close to the background), whereas JunB, Id2, and Hmga2 transcripts are expressed at least 50-fold higher (Fig. 1 F). These findings are consistent with lack of E-cadherin repression. In addition, TGF-β–Smad signaling strongly induced fibrotic factors collagen 7a1 (Col7a1) and fibronectin (Fig. 1 E). JunB belongs to the Jun family of AP1 transcription factors, which together with Smads can regulate Col7a1 in keratinocytes (Naso et al., 2003). Thus, JunB is up-regulated by TGF-β–Smad signaling and may contribute to the TGF-β–induced EMT and fibrotic responses.
JunB is a direct target and a partner of Smad-dependent transcription
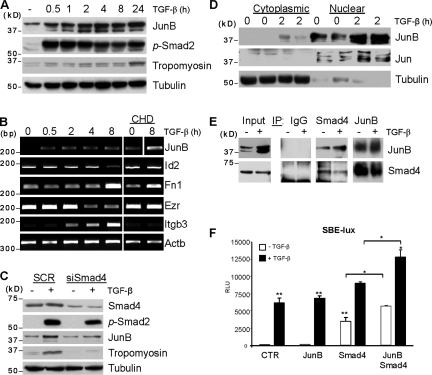
Early induction of JunB suggests its active role in the onset of the EMT program, although the dynamics of JunB regulation by TGF-β could be cell type specific (Mauviel et al., 1996). Here, we assessed the JunB regulation and the link of JunB to Smad transcription factors. Immunoblotting showed up-regulation of JunB protein levels within 30 min of the TGF-β treatment, which is within the time frame of Smad2 phosphorylation (Fig. 2 A). In comparison, the induction of tropomyosin (Tpm1), a late-response target of TGF-β (Bakin et al., 2004), was delayed by 4 h. To determine whether JunB is a direct target, we examined JunB regulation by TGF-β in the presence of cycloheximide (CHD), an inhibitor of de novo protein synthesis. RT-PCR showed that CHD did not inhibit up-regulation of JunB mRNA, whereas suppression of Id2 and Ezr/Vil2 (ezrin) and up-regulation of Itgb3 and partly Fn1 were blocked (Fig. 2 B). An increase in JunB basal levels is likely to be associated with CHD-induced stress response. Next, we examined the contribution of Smads to JunB regulation. Depletion of Smad4 blocked induction of both JunB and Tpm1 (Fig. 2 C), indicating that JunB is a direct target of TGF-β–Smad signaling. To assess localization of JunB, we performed cellular fractionation experiments. Nuclear cytoplasmic fractionations showed that JunB and Jun are predominantly located in nuclear fractions, and TGF-β increased JunB levels in the nucleus (Fig. 2 D).
Figure 2.
JunB is a direct target of TGF-β–Smad signaling and forms a complex with Smad4. (A and B) NMuMG cells were treated with 2 ng/ml TGF-β1 for the indicated times in the absence or presence of 10 µM CHD. (A) Immunoblotting of JunB, phospho-Smad2 (p-Smad2), and tropomyosin; tubulin is a loading control. (B) RT-PCR of JunB and Actb, a loading control. (C) Immunoblotting of Smad4, phospho-Smad2/3, JunB, tropomyosin, and tubulin. NMuMG cells were transfected with siRNA to Smad4 (siSmad4) and treated with 2 ng/ml TGF-β1 for 24 h. SCR, scramble. (D) Immunoblotting of Smad4 and JunB in cytoplasmic and nuclear fractions. Tubulin and Jun are controls for cytoplasmic and nuclear fractions, respectively. (E) NMuMG cells were treated with 2 ng/ml of TGF-β1 for 2 h; cell extracts were immunoprecipitated (IP) with anti-Smad4, anti-JunB, or control IgG antibodies. Immune complexes were resolved on a denaturing gel and immunoblotted for Smad4 and JunB. (F) NMuMG cells were cotransfected with the Smad-dependent luciferase reporter SBE-Lux, a CMV-Rl, and combinations of Smad4 and JunB. Luciferase activities were measured after treatment of cells with 2 ng/ml TGF-β1 for 24 h. CTR, control; RLU, relative luciferase unit. Experiments were performed in triplicate and repeated at least twice. *, P < 0.05; **, P < 0.005 using a Student’s t test. Error bars represent the mean value ± SD.
Binding of Smads to DNA can be facilitated by direct interaction with AP1 transcription factor (Zhang et al., 1998). To assess JunB–Smad interactions, we performed immunoprecipitation studies with NMuMG cells. Cell lysates were precipitated with Smad4 antibody followed by immunoblotting for JunB and vice versa. The study found JunB in Smad4 immune complexes and Smad4 in JunB precipitates (Fig. 2 E), indicating that JunB and Smad4 are present in the same complexes. This interaction is independent of TGF-β treatment and is likely to occur in the nucleus, given nuclear localization of JunB (Fig. 2 D). Immunoblotting revealed two bands of JunB in total and nuclear fractions, whereas only the upper band was seen in Smad4 immunoprecipitates (Fig. 2, D and E). The upper band may represent JunB with posttranslational modifications (this question is under investigation). To determine whether JunB contributes to Smad-mediated transcription, cells were transfected with Smad-dependent luciferase reporter (Smad-binding element [SBE]–Lux) along with JunB, Smad4, or both. JunB alone had no effect on a basal activity of the reporter, whereas Smad4 significantly increased basal reporter activity (Fig. 2 F). Cotransfection of both JunB and Smad4 greatly enhanced the basal and TGF-β–induced reporter activities. Together, these findings demonstrate that transcription factor JunB is an immediate early target of TGF-β–Smad signaling. TGF-β increases JunB levels in the nucleus, where JunB forms a complex with Smad4 and enhances Smad-dependent transcription. Thus, JunB–Smad interaction may contribute to the TGF-β–induced EMT program.
JunB is required for the disruption of cell–cell junctions in response to TGF-β
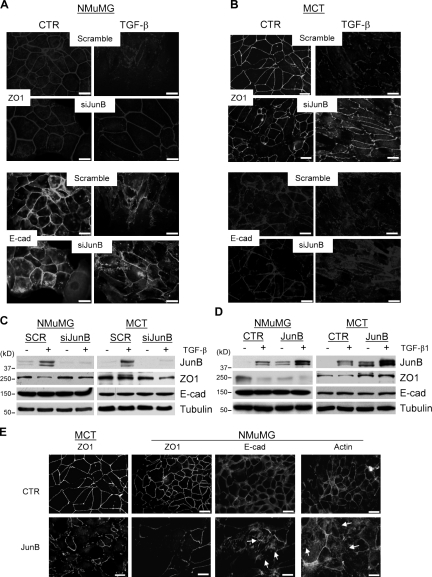
To assess the role of JunB in TGF-β–induced EMT, NMuMG and MCT cells were depleted for JunB by siRNA to JunB (Fig. 3 C). In both cell lines, suppression of JunB prevented TGF-β–induced disruption of cell junctions (Fig. 3, A and B) and changes in cell morphology (Fig. S1), whereas these TGF-β responses were clearly observed in cells transfected with scramble control siRNA. Down-regulation of ZO1 protein in NMuMG cells was blocked by depletion of JunB, whereas ZO1 mRNA levels were unchanged (Fig. S1). E-cadherin protein and mRNA levels were not affected (Figs. 3 C and S1). Thus, JunB is required for the TGF-β–induced morphological transition and disruption of cell junctions.
Figure 3.
TGF-β–mediated dissolution of cell junctions requires JunB. (A–D) NMuMG and MCT cells were transfected with siRNA to JunB (siJunB) or infected with retrovirus encoding JunB or empty vector (control [CTR]) followed by treatment with 2 ng/ml TGF-β1 for 24 h. (A and B) Immunofluorescence images of ZO1 and E-cadherin (E-cad) in NMuMG and MCT cells. Bars, 20 µm. (C and D) Immunoblotting of JunB, E-cadherin, ZO1, and tubulin, a loading control. SCR, scramble. (E) Immunofluorescence images of ZO1, E-cadherin, and actin in NMuMG or MCT cells expressing JunB or empty vector. Arrows indicate the disruption of continuous E-cadherin staining or formation of actin stress fibers in cells overexpressing JunB. Bars, 20 µm.
To determine whether JunB is sufficient to induce the transition, NMuMG and MCT cells were transduced with EGFP and JunB using a bicistronic retroviral vector. Immunoblotting revealed comparable levels of JunB in TGF-β–treated cells and in JunB-transduced cells (Fig. 3 D). Protein levels of E-cadherin were not altered by forced expression of JunB in either cell line. Overexpression of JunB reduced ZO1 protein in NMuMG cells but not in MCT cells (Fig. 3 D), which is in agreement with TGF-β response in parental cells (Fig. 1 B). In both cell lines, JunB severely disrupted tight junctions (ZO1), adherens junctions (E-cadherin), and continuous adhesion beltlike actin fibers, increasing linear actin fibers (Fig. 3 E). Thus, forced expression of JunB in polarized epithelial cells impairs organization of tight and adherens junctions and disrupts the actin cytoskeleton architecture.
JunB contributes to the formation of actin stress fibers and focal adhesions in response to TGF-β
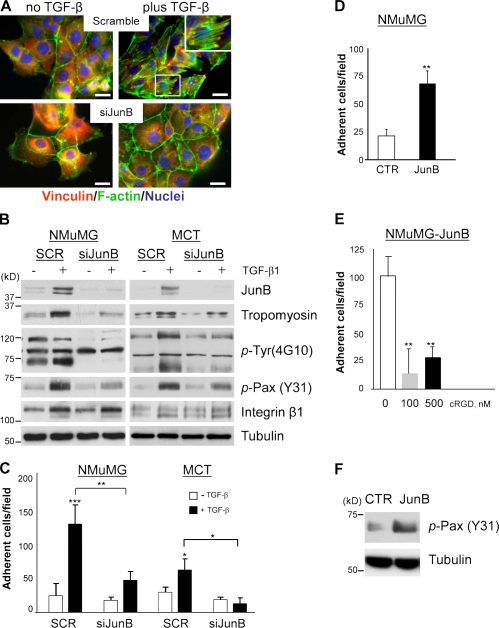
The formation of an extensive network of actin stress fibers and focal adhesions is a distinct characteristic of TGF-β–induced EMT (Miettinen et al., 1994; Piek et al., 1999; Bakin et al., 2000). It has been also suggested that the actin fibers and focal adhesions are critical mediators of EMT (Bakin et al., 2004; Bianchi et al., 2010). Given that forced expression of JunB altered the actin cytoskeleton (Fig. 3 E), we asked whether suppression of JunB would block the induction of actin fibers and focal adhesions by TGF-β. NMuMG cells were depleted for JunB by siRNA, and actin filaments were assessed before and after TGF-β treatment. Formation of actin stress fibers and localization of vinculin to focal adhesions were clearly observed in control NMuMG cells treated with TGF-β, whereas this response was blocked in cells transfected with siRNA to JunB (Fig. 4 A). The induction of actin stress fibers by TGF-β–Smad signaling requires tropomyosins (Tpm1; Bakin et al., 2004). Given the link between JunB and Smad signaling (Fig. 2), we examined whether JunB contributes to Tpm1 expression. Immunoblotting showed that Tpm1 protein levels were induced by TGF-β in NMuMG and MCT control cells but not in JunB-depleted cells (Fig. 4 B). Tropomyosins stabilize actin fibers and enhance cell adhesion and integrin-mediated phosphorylation of tyrosine residues (phospho-Tyr) in paxillin (Pax; Zheng et al., 2008; Safina et al., 2009). Probing with the 4G10 monoclonal antibody to phospho-Tyr showed that TGF-β increases phospho-Tyr levels in proteins with a molecular mass of 120–130 and 60–70 kD, whereas a prominent band at 90 kD was not sensitive to TGF-β (Fig. 4 B). Depletion of JunB reduced phospho-Tyr levels in 120–130- and 60–70-kD proteins (Fig. 4 B). Pax is a 68-kD protein that plays a critical role in focal adhesions and is phosphorylated by integrin signaling (Brown and Turner, 2004). Immunoblotting revealed a marked reduction of phospho-Tyr31–Pax levels by JunB depletion, although integrin-β1 levels were not altered (Fig. 4 B). Together, these findings show that JunB is required for TGF-β induction of actin stress fibers, formation of focal adhesions, and integrin signaling. JunB contributes to the regulation of Tpm1, a critical component of this response.
Figure 4.
JunB is required for TGF-β induction of actin fibers and cell adhesion. (A and B) NMuMG and MCT cells were transfected with siRNA to JunB (siJunB) or scramble (SCR) control and treated with 2 ng/ml TGF-β1 for 24 h. (A) Immunofluorescence images of vinculin, actin filaments, and nuclei in NMuMG cells. Bars, 20 µm. (inset) An enlargement of the selected area showing actin fibers linked to focal adhesions (vinculin). (B) Immunoblotting of JunB, tropomyosin (TM311 antibody), phospho-Tyr (p-Tyr; 4G10 antibody), phospho-Pax (p-Pax; Y31), and integrin-β1; tubulin is a loading control. (C–E) Adhesion of cells to fibronectin-coated wells. Cells were pretreated with 2 ng/ml TGF-β1 for 24 h where indicated. Adherent cells were fixed and counted from 6 replicate wells. Experiments were repeated at least twice. *, P < 0.05; **, P < 0.005; ***, P < 0.001 using a Student’s t test. Error bars represent the mean value ± SD. (C) Adhesion of NMuMG and MCT cells transfected with siRNA to JunB. (D) Adhesion of empty vector (control [CTR]) and JunB-expressing NMuMG cells. (E) Adhesion of JunB-expressing NMuMG cells that were untreated (0) or treated with 100 and 500 nM of cyclic RGD (cRGD) peptide. (F) Immunoblotting of phospho-Pax (Y31) in NMuMG cells expressing JunB or empty vector; tubulin is a loading control.
Our findings suggest that JunB may contribute to the TGF-β induction of cell adhesion to matrix. To test this idea, the adhesion of NMuMG and MCT cells onto fibronectin-coated plates was examined after pretreatment with TGF-β1 for 24 h. A fourfold increase in adhesion by TGF-β treatment was found in control NMuMG and MCT cells, whereas siRNA to JunB abrogated this response (Fig. 4 C). Accordingly, NMuMG cells overexpressing JunB exhibited threefold greater adhesion onto fibronectin than control cells (Fig. 4 D). The addition of a cyclic RGD peptide, mimicking the integrin-recognized epitope in fibronectin, abrogated JunB-enhanced adhesion (Fig. 4 E). JunB also increased basal levels of phospho-Tyr31–Pax (Fig. 4 F). Together, these results demonstrate that JunB contributes to the formation of actin fibers, cell–matrix adhesions, and integrin signaling in response to TGF-β.
JunB in the regulation of the EMT transcription program in response to TGF-β
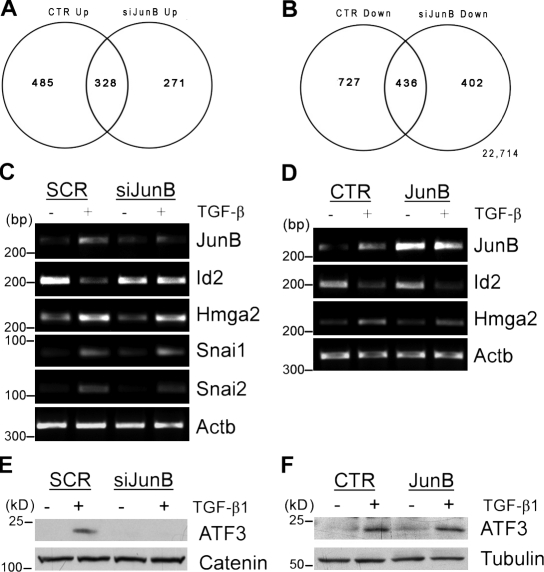
Thus far, our findings revealed that JunB contributes to major events of EMT induced by TGF-β–Smad signaling: the dissolution of cell junctions and formation of cell–matrix contacts. The EMT response to TGF-β involves a complex transcriptional program mediated by Smad3 and Smad4, affecting expression of regulatory and structural genes (Zavadil and Böttinger, 2005; Moustakas and Heldin, 2007; Safina et al., 2009). To assess the role of JunB in the TGF-β transcription program of EMT, an unbiased gene expression profiling study was performed using Affymetrix microarrays. Total RNA samples were prepared from scramble control and siRNA JunB–transfected cells that were treated with TGF-β1 for 24 h, the time needed for the disruption of cell junctions and formation of actin fibers. Depletion of JunB altered expression of 891 genes (3.67%; 531 up-regulated and 360 down-regulated genes compared with control NMuMG cells). TGF-β1 regulated nearly 2,000 genes (8.1%; 1,976 gene probes) by at least twofold in control cells (Fig. 5, A and B). Treatment with TGF-β1 up-regulated 813 genes in control and 599 genes in siRNA JunB cells. Nearly 40% of TGF-β1 target genes were commonly up-regulated (328 gene probes) and commonly repressed (436 gene probes) in siRNA JunB and control cells. Thus, the regulation of almost 60% of the TGF-β target genes may depend on JunB. Pathway analysis revealed the JunB-dependent regulation of a large number of genes involved in cell proliferation as well as cell adhesion and motility (Table 1).
Figure 5.
JunB contributes to the regulation of TGF-β target genes implicated in EMT. (A and B) Venn diagrams show the comparison of gene regulation by TGF-β in control and siRNA JunB–transfected (siJunB) cells. NMuMG cells were transfected with siRNA to JunB or scramble control and treated with 2 ng/ml TGF-β1 for 24 h. (C) RT-PCR analysis of total RNA from control and siRNA JunB cells for JunB, Id2, Hmga2, Snai1, Snai2, and Actb, a loading control. Approximate size markers are shown based on the migration of markers relative to analogous RT-PCR products run on a different gel. SCR, scramble. (E) Immunoblotting of ATF3 and Jun in whole-cell lysates from control and siRNA JunB cells; α-catenin is a loading control. (D and F) Control (CTR) or JunB-overexpressing NMuMG cells were treated with 2 ng/ml TGF-β1 for 24 h. (D) RT-PCR for mRNA levels of JunB, Id2, Hmga2, and Actb, a loading control. Approximate size markers are shown based on the migration of markers relative to analogous RT-PCR products run on a different gel. (F) Immunoblots of ATF3 and tubulin, a loading control.
Table 1.
Function categories of JunB-dependent genes
| Category | No. of genes | Raw p-value | Adjusted p-valuea |
| DNA metabolism | 33 | 8.14E−07 | 1.55E−04 |
| Cell cycle | 70 | 8.16E−07 | 7.75E−05 |
| DNA replication | 15 | 3.00E−04 | 0.01884957 |
| Cell structure and motility | 67 | 5.29E−04 | 0.024801105 |
| Intracellular signaling cascade | 55 | 0.00188 | 0.069146938 |
| Protein glycosylation | 17 | 0.00428 | 0.126966795 |
| Cell cycle control | 29 | 0.00603 | 0.1513707 |
| Cell adhesion | 39 | 0.00609 | 0.134952822 |
| Amino acid metabolism | 19 | 0.00686 | 0.135164018 |
Adjusted for false discovery rate using the Benjamin multiple testing.
We focused on helix-loop-helix protein Id2, which is involved in cell proliferation and differentiation as well as it is a key negative regulator of TGF-β–induced EMT in epithelial cells (Kondo et al., 2004; Kowanetz et al., 2004). To validate the profiling data, RT-PCR analysis was performed for Id2 and several genes implicated in EMT, including chromatin-associated factor Hmga2 and transcription repressors Snai1/Snail and Snai2/Slug (Thuault et al., 2006). This study confirmed that depletion of JunB prevents repression of Id2 and showed that levels of Hmga2, Snai1, and Snai2 were not affected (Fig. 5 C). To complement siRNA studies, we examined mRNA levels of these genes in JunB-overexpressing cells. Surprisingly, expression of Id2 was not affected, although JunB protein levels were comparable with TGF-β–treated cells (Figs. 3 D and 5 D). Down-regulation of Id factors requires transcription repressor ATF3 (Kang et al., 2003; Bakin et al., 2005), which is regulated by TGF-β in NMuMG cells at the protein level (Bakin et al., 2005). We found that depletion of JunB blocks up-regulation of ATF3, whereas forced expression of JunB had little effect on ATF3 (Fig. 5, E and F). This may account for the lack of repression of Id2 in JunB-overexpressing cells. Together, these findings show that JunB contributes to the regulation of TGF-β target genes involved in key steps of EMT such as repression of Id2, an antagonist of EMT, and formation of matrix adhesions (fibronectin and tropomyosin). JunB does not affect Hmga2 and Snails, and this is consistent with the absence of E-cadherin repression (Fig. 1).
Transcriptional repression of Id2 by TGF-β is mediated through AP1 sites
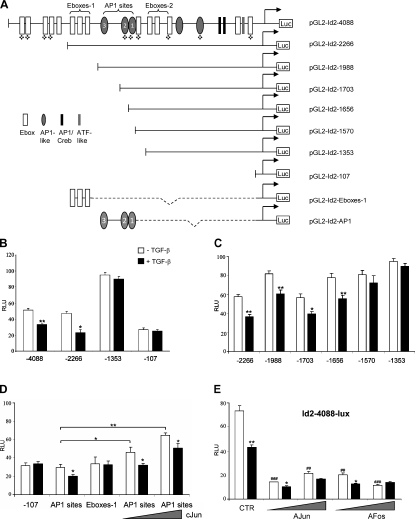
To investigate the transcriptional regulation of Id2 by TGF-β, we explored the potential regulatory elements in the mammalian Id2 promoter. The analysis of the published data and a 4-kb region of the Id2 promoter are summarized in Fig. 6 A. Previous studies have reported that Myc family helix-loop-helix proteins are capable of inducing Id2 expression through E-box clusters 1 and 2 (E-boxes-1 and -2) located upstream of the −2.2-kb region (Neuman et al., 1995; Lasorella et al., 2000). To examine the role of these and other potential regulatory elements in TGF-β–mediated repression of Id2, a 4-kb fragment of mouse Id2 promoter was cloned into pGL2 luciferase reporter vector, and several deletion constructs were generated. The luciferase reporter activity of the full-length 4-kb construct was reduced by 40% by treatment with TGF-β1, whereas the Id2–2266 construct showed comparable basal activity and was repressed by 50% (Fig. 6 B). However, the Id2–1353 construct was not repressed by TGF-β (Fig. 6 B). The Id2–1353 construct lacks clusters of E-boxes-1 and AP1-like sites but includes the E-boxes-2 cluster that is thought to be important for repression of Id2 by TGF-β via Mad proteins (Siegel et al., 2003). In addition, mRNA levels of Mads are not regulated by TGF-β in studied systems (unpublished data). Thus, our results suggest that the E-boxes-1 and AP1 sites within the −1353–2266 region could be responsible for TGF-β–mediated repression of Id2, whereas E-boxes-2 sites are unlikely to be involved in this response. Two of the three AP1 sites are conserved among human and mouse Id2 promoters, whereas E-boxes-1 elements are present only in the human Id2 promoter (Figs. 6 A and 7 D). To test the contribution of these elements to Id2 repression by TGF-β, we generated constructs with consecutive deletions of E-boxes-1 and AP1 sites (Fig. 6 C). Deletion of E-boxes-1 (pGL2-Id2–1988) had no effect on the ability of TGF-β to repress the reporter activity. The constructs with removed AP1 site 1 (pGL2-Id2–1703) or AP1 sites 1 and 2 (pGL2-Id2–1656) were still repressed by TGF-β (Fig. 6 C). However, when all three sites were deleted, the repression by TGF-β was lost. To validate these findings, DNA fragments containing the E-boxes-1 or three AP1 sites were inserted upstream of the pGL2-Id2–107 construct carrying the 2107-bp region of the mouse Id2 promoter (Fig. 6 A). The pGL2-Id2–107 construct is not repressed by TGF-β (Fig. 6 B). Addition of AP1 sites conferred the repression by TGF-β, whereas the E-boxes were not effective (Fig. 6 D). To confirm that the AP1-like sites are indeed regulated by AP1 factors, the pGL2-Id2-AP1 construct was cotransfected with an expression vector for c-Jun. The activity of the pGL2-Id2-AP1 reporter was increased by c-Jun in a dose-dependent manner, although TGF-β still exerted a negative effect on the reporter (Fig. 6 D). In agreement with these findings, cotransfection of dominant-negative c-Jun (A-Jun) or c-Fos (A-Fos) repressed the full-length Id2 reporter (Fig. 6 E). Together, these findings demonstrate that negative regulation of Id2 by TGF-β is mediated via AP1 binding sites within the −1570–1703 region of the Id2 promoter.
Figure 6.
Repression of Id2 by TGF-β is mediated via AP1 sites. (A) A schematic representation of deletion constructs spanning the −4088–224 region of the mouse Id2 promoter. Quatrefoils indicate sites conserved among mouse, rat, and human. (B–E) NMuMG cells were transfected with Id2 promoter luciferase reporters and a Renilla luciferase reporter (CMV-Rl). Cells were treated with 2 ng/ml TGF-β1 for 16 h (shaded bars). Luc values were normalized to Renilla values. RLU, relative luciferase unit. Experiments were performed in triplicate and repeated at least twice. *, P < 0.05; **/##, P < 0.005; ###, P < 0.0005 using a Student’s t test. ## and ### indicate t test values for control and experimental points in the absence of TGF-β1. Error bars represent the mean value ± SD. (D and E) NMuMG cells were transfected with indicated Id2 promoter luciferase reporters along with empty vector control, 0.5 or 2 µg of cJun, A-Jun, or A-Fos, and thymidine kinase–Rl for normalization. Cells were treated with 2 ng/ml TGF-β1 for 16 h before lysis and analyzed for luciferase activity.
Figure 7.
JunB and ATF3 cooperate to mediate transcriptional repression of Id2 by TGF-β. (A) NMuMG cells were cotransfected with AP1-Lux luciferase reporter, a Renilla luciferase reporter under the control of the herpes simplex virus thymidine kinase promoter, and increasing amounts of JunB and ATF3 (1, 2, and 4 µg) or both factors together (2 µg each). Cells were treated with 2 ng/ml TGF-β1 for 16 h where indicated. CTR, control; RLU, relative luciferase unit. Experiments were performed in triplicate and repeated at least twice. Error bars represent the mean value ± SD. (B and C) NMuMG–Tet-Off–ATF3 control or JunB-overexpressing cells were grown in the absence or presence of 1 µg/ml tetracycline for 24 h followed by treatment with 2 ng/ml TGF-β1 for 24 h. Total RNA and whole-cell extracts were used for quantitative RT-PCR (B) and immunoblotting (C). (B) Quantitative PCR for Id2 was performed in triplicate. The CT values were normalized to 18S rRNA and presented as fold difference relative to untreated control. Error bars represent the mean value ± SD. (C) Immunoblotting of Id2, JunB, ATF3, and tubulin, a loading control. (D) Location of conserved AP1 sites within the mouse Id2 promoter relative to primers used for ChIP studies. Boxed regions indicate the putative binding sites for AP1 transcription factor. (A, B, and D) *, P < 0.05 using a Student’s t test. (E) ChIP experiments were performed with NMuMG cells after treatment with 2 ng/ml TGF-β1 for 8 h. Amplifications were performed using DNA samples before precipitation (Input) and after precipitation with control IgG or antibodies to JunB or ATF3.
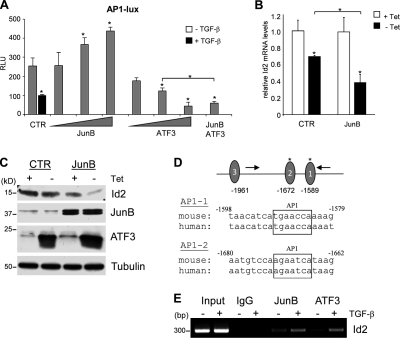
JunB and ATF3 cooperate to mediate transcriptional repression of Id2 by TGF-β
Thus far, our results show that TGF-β increases levels of JunB, a component of the AP1 transcription factor, and suppresses AP1-dependent transcription. JunB is also required for up-regulation of ATF3, a basic leucine zipper protein (Fig. 5 E). ATF3/LRF-1 lacks trans-activating activity and represses AP1-dependent transcription by forming heteromeric complexes with Jun family proteins, including JunB (Hsu et al., 1993; Chen et al., 1994). To determine whether JunB and ATF3 repress AP1-dependent transcription, cells were transfected with the AP1 luciferase reporter and JunB, ATF3, or both. Increasing amounts of JunB moderately stimulated the AP1 reporter, whereas ATF3 repressed luciferase activity (Fig. 7 A). Cotransfection of JunB and ATF3 resulted in a cooperative repression of the AP1 reporter activity (Fig. 7 A). To assess the ability of JunB and ATF3 to repress endogenous Id2, we generated NMuMG cells with constitutive expression of JunB and Tet-Off–regulated expression of ATF3. Forced expression of ATF3 alone reduced Id2 mRNA and protein levels, and this was further enhanced in cells expressing JunB (Fig. 7, B and C). Finally, to determine whether JunB and ATF3 bind to endogenous Id2 promoter, we analyzed the occupancy of the AP1-containing region using chromatin immunoprecipitation (ChIP) experiments (Fig. 7 D). The ChIP assays showed that TGF-β stimulated the AP1 site occupancy by JunB and ATF3 (Fig. 7 E). Together, these finding argue that JunB and ATF3 cooperate in repression of the Id2 promoter activity by TGF-β via AP1 regulatory elements.
JunB mediates transcriptional regulation of proteins involved in the TGF-β fibrotic response
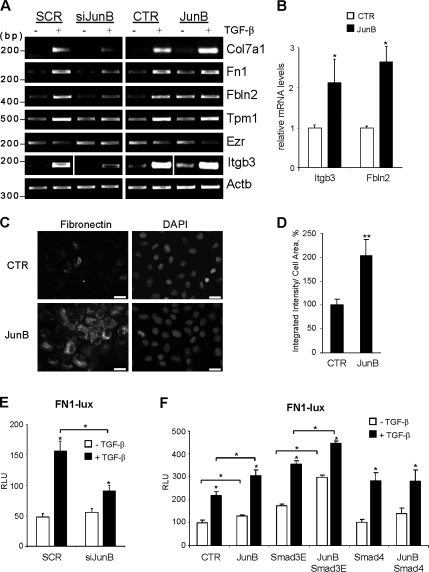
Our findings indicate that JunB contributes to the regulation of fibronectin, Col7a1, and tropomyosin (Figs. 1 and 4), which are ultimately involved in EMT and fibrosis. To validate our data, we performed RT-PCR analysis and explored the contribution of JunB to the regulation of fibronectin, an important component of TGF-β–mediated EMT (Yi et al., 2002; Câmara and Jarai, 2010). Semiquantitative RT-PCR of total RNA from JunB-depleted cells confirmed that JunB is critical for up-regulation of Col7a1, fibronectin-1 (Fn1), and fibulin-2 (Fbln2) as well as genes involved in the actin cytoskeleton (tropomyosin [Tpm1] and ezrin [Ezr/Vil2]) and cell–matrix adhesion (integrin-β3 [Itgb3]; Fig. 8 A). Tropomyosin is required for the TGF-β induction of actin stress fibers and enhances focal adhesions and integrin signaling (Zheng et al., 2008). Villin-2/ezrin links the actin cytoskeleton to the peripheral plasma membrane, regulating cell surface structure, adhesion, and migration (Louvet-Vallée, 2000). Complementary analysis of JunB-expressing cells showed that JunB increases basal levels of Col7a1, Fn1, Fbln2, and Itgb3 transcripts, whereas Tpm1 and Ezr are not modulated (Fig. 8 A). The regulation of two novel JunB targets, Itgb3 and Fbln2, in JunB-expressing cells was confirmed by quantitative RT-PCR (Fig. 8 B). These findings identify novel JunB targets that represent ECM components, their receptors, and proteins stabilizing cell adhesion structures.
Figure 8.
JunB contributes to the induction of profibrotic proteins by TGF-β. (A) RT-PCR analysis of total RNA from NMuMG cells transfected with scramble (SCR) control or siRNA JunB (siJunB) and NMuMG cells expressing JunB or EGFP-control (CTR). Cells were treated with 2 ng/ml TGF-β1 for 24 h and analyzed for Col7a1, Fn1, Fbln2, tropomyosin-1 (Tpm1), ezrin (Ezr/Vil2), and Itgb3; Actb is a loading control. (B) Quantitative PCR was performed on EGFP-control or JunB-expressing NMuMG cells in triplicates. The CT values were normalized to 18S rRNA and presented as fold difference relative to EGFP-control. Error bars represent the mean value ± SD. (C) Immunofluorescence of fibronectin in control or JunB-expressing cells. Bars, 20 µm. (D) The integrated intensities of fibronectin fluorescence were determined for at least 30 cells per field in three fields of control and JunB-expressing cells. Error bars represent the mean value ± SD. (E) The relative luciferase activity of the FN1-Lux reporter in NMuMG cells transfected with siRNA scramble control or JunB after treatment of cells with 2 ng/ml TGF-β1 for 16 h. Luc values were normalized with Renilla luciferase values (CMV-Rl). RLU, relative luciferase unit. Experiments were performed in triplicate and repeated at least twice. Error bars represent the mean value ± SD. (F) The FN1-Lux reporter activity was measured in cells transfected with constitutively active Smad3-3E mutant (Smad3E), Smad4, and JunB after treatment with 2 ng/ml TGF-β1 for 16 h. (B and D–F) *, P < 0.05; **, P < 0.005 using a Student’s t test.
Given that fibronectin plays a central role in tissue fibrosis (Hsu et al., 2008; Kadler et al., 2008) and enhances EMT (Yi et al., 2002; Câmara and Jarai, 2010), we further investigated the link of JunB to fibronectin expression in response to TGF-β. To confirm that JunB regulates fibronectin production, we examined fibronectin levels and localization in JunB-overexpressing NMuMG cells. Immunofluorescence showed that JunB-expressing cells have significant levels of fibronectin in both the cytoplasm and extracellular space compared with control cells (Fig. 8 C), with a twofold increase in fibronectin protein within the cell area (Fig. 8 D). To examine whether JunB regulates fibronectin transcription, luciferase reporter assays were performed with a fragment of the human FN1 promoter. Depletion of JunB by siRNA markedly decreased the reporter activity in response to TGF-β but did not affect the basal activity (Fig. 8 E). Transfection of JunB increased the basal and TGF-β–induced activity of the reporter as compared with the control (Fig. 8 F). Next, we tested whether JunB cooperates with Smads in Fn1 regulation using a cotransfection approach. Immunoblotting confirmed comparable amounts of Smads and JunB in cotransfection experiments (unpublished data). Transfection of Smad3E increased basal promoter activity, whereas cotransfection of JunB and Smad3E increased the basal and TGF-β–induced activity of the promoter (Fig. 8 F). Transfection of Smad4 alone was not effective, whereas cotransfection with JunB increased the TGF-β–induced activity of the promoter (Fig. 8 F). Thus, it appears that JunB cooperates with Smad3 and Smad4 in the regulation of fibronectin.
Discussion
This study provides evidence that AP1 transcription factor JunB plays an important role in TGF-β–induced EMT and profibrotic responses. Knockdown of JunB by siRNA blocks the dissolution of cell–cell junctions, production of profibrotic proteins, and formation of cell–matrix adhesions. Consistent with these findings, JunB-depleted cells show a significant reduction in cell–matrix adhesion. JunB is an immediate early target gene in TGF-β–Smad signaling. JunB in cooperation with ATF3 mediates the repression of Id2, an antagonist of the TGF-β–induced EMT. Basic leucine zipper proteins JunB and ATF3 repress the Id2 promoter activity in response to TGF-β via AP1 regulatory elements. In addition, JunB contributes to up-regulation of the key components of actin stress fibers and cell–matrix adhesions. Thus, JunB provides a critical input in the onset of the TGF-β transcriptional program of EMT and profibrotic responses.
Components of the AP1 transcription factor are implicated in tumor invasion and metastasis (Ozanne et al., 2007). Among AP1 proteins, JunB and JunD have no transforming activity, whereas Jun, Fos, and FosB can transform cells in culture (Eferl and Wagner, 2003). Here, forced expression of JunB induced partial EMT in two epithelial cell lines, disrupting tight junctions and adherens junctions. In comparison, overexpression of Jun induces loss of epithelial polarity, disrupting cell junctions in the mammary epithelial cell line EpH4 (Fialka et al., 1996). Both JunB and Jun do not affect expression of E-cadherin (Fig. 1 E; Fialka et al., 1996). Consistent with these results, TGF-β and JunB did not induce appreciable levels of Snail and Slug, transcriptional repressors of E-cadherin (Figs. 1 and 5). Interestingly, the EMT conversion by Jun in EpH4 cells results in down-regulation of JunB and PAI-1 (Fialka et al., 1996), whereas these two proteins are induced by TGF-β during EMT in mammary NMuMG and kidney MCT epithelial cells. In addition, TGF-β–JunB signaling down-regulates ezrin/Vil2 (Fig. 7 A), whereas Jun increases ezrin expression (Gao et al., 2009), and ezrin is induced upon cell transformation by the Fos–Jun complex (Miao and Curran, 1994; Jooss and Müller, 1995; Lamb et al., 1997). Thus, despite the phenotypic similarities in morphological transitions by Jun and TGF-β–JunB signaling, the underlying molecular mechanisms are likely to be different.
Expression of JunB is regulated by various stimuli, including TGF-β, activin, IL6, IL11, and bone morphogenetic proteins 2/4 (BMP2/4; Pertovaara et al., 1989; Baumann et al., 1991; Laiho et al., 1991; Nakajima and Wall, 1991; Hashimoto et al., 1993; Yin et al., 1993; Hollnagel et al., 1999). However, it appears that only TGF-β, but not BMP and activin, induces the EMT-like response (Piek et al., 1999). TGF-β induces a prolonged up-regulation of JunB for at least 24 h (Fig. 2). In comparison, BMP causes only a transient up-regulation of JunB and a sustained increase in c-Jun (Hollnagel et al., 1999). The differential regulation of Jun and JunB or their partners may govern the ability of these growth factors to induce EMT. For example, JunB and Jun can form distinct dimeric complexes with other basic leucine zipper proteins of the Fos, ATF, and musculoaponeurotic fibrosarcoma families (Eferl and Wagner, 2003).
Our findings, for the first time, demonstrate a key role of JunB in repression of Id2 in response to TGF-β. Id2 controls cell proliferation and inhibits TGF-β–induced EMT (Kondo et al., 2004; Kowanetz et al., 2004). Previous studies have shown that BMPs up-regulate expression of helix-loop-helix proteins Id1, Id2, and Id3 (Hollnagel et al., 1999), whereas TGF-β represses these genes (Kang et al., 2003; Kondo et al., 2004; Kowanetz et al., 2004). It has been shown that Id1 repression is mediated through a Smad3–ATF3 complex (Kang et al., 2003), whereas the Id2 repression is thought to be mediated by Mad binding at E-boxes within the Id2 promoter (Siegel et al., 2003). Here, we found no evidence for this latter mechanism in NMuMG cells. Instead, our findings argue that AP1 sites within the −1570–1703 region of the Id2 promoter are important for repression of Id2 by JunB and ATF3 (Fig. 6). TGF-β–Smad signaling up-regulates JunB, which is required for sustained induction of ATF3 (Fig. 5). The JunB–ATF3 complex has been shown to repress AP1-driven transcription (Hsu et al., 1992, 1993). Our work reveals that JunB cooperates with ATF3 in repressing endogenous Id2 expression and the Id2 promoter reporter (Fig. 7). ChIP assays show that TGF-β stimulates binding of both ATF3 and JunB at the genomic region containing the AP1 sites (Fig. 7 E). This mechanism is consistent with a previous report that ATF3 is a negative regulator of Id1 (Kang et al., 2003), whereas our data indicate that JunB is likely to be a partner of ATF3 in the AP1 repressor complex.
TGF-β regulates cell adhesion and matrix receptors (integrins) in various cell types (Heino et al., 1989; Ignotz et al., 1989; Spurzem et al., 1993). Our results show that JunB mediates TGF-β–stimulated cell–matrix adhesion in epithelial cell systems, contributing to the induction of Itgb3 and actin-stabilizing protein tropomyosin (Figs. 3 and 8). In addition, JunB facilitates the expression of matrix proteins such as fibronectin, Fbln2, and Col7a1 (Fig. 8). Accordingly, basal and TGF-β–induced integrin signaling is markedly affected by modulation of JunB levels (Fig. 3). It appears that JunB cooperates with Smad3 and Smad4 in the activation of the target genes such as fibronectin. In some cell systems, however, JunB may reduce Smad-dependent transcription (Verrecchia et al., 2001b). Smad proteins form distinct oligomeric complexes in response to TGF-β (Jayaraman and Massague, 2000), and it is conceivable that JunB interacts with a subset of Smad complexes, affecting DNA binding affinity and the pattern of gene regulation (Zhang et al., 1998). Thus, JunB provides an additional level in the regulation of specific TGF-β target genes contributing to actin fibers, focal adhesions, and matrix deposition.
JunB mediates the TGF-β induction of actin stress fibers and cell–matrix adhesions that are implicated in the tumor suppressor function of TGF-β (Safina et al., 2009; Bianchi et al., 2010). On the other hand, E-cadherin expression is not affected by either TGF-β or JunB in the studied model cell systems. This is consistent with previous studies demonstrating that repression of E-cadherin is not required for the EMT response to TGF-β (Tian and Phillips, 2003; Maeda et al., 2005; Safina et al., 2009). The role of JunB in the tumor suppressor function of TGF-β apparently contradicts the current idea of EMT as a process that underlies tumor invasion and metastasis. This novel JunB activity is consistent with the antitumor function of JunB in leukemia (Eferl and Wagner, 2003; Steidl et al., 2006). Indeed, JunB-deficient mice develop B-lymphoid leukemia, and loss of JunB is found in human leukemia (Ott et al., 2007). In addition, JunB inhibits Ras transformation and cell proliferation, reducing expression of cyclin D1 and increasing p16/INK4A (Eferl and Wagner, 2003). Consistent with this finding, tropomyosin, a JunB target, and oncogenic Ras have opposite effects on cell invasion and anchorage-independent growth, affecting actin stress fibers and cell–matrix adhesion (Safina et al., 2009). JunB may interfere with function of Fra1/Fosl1 and Jun, mediators of Ras transformation, in the regulation of Tpm1 and actin fibers. Indeed, Fos-induced transformation suppresses Tpm1 expression but increases expression of ezrin/Vil2 (Jooss and Müller, 1995; Gao et al., 2009), which is repressed by the TGF-β–Smad-JunB axis (Fig. 7). Fra1 and Jun are frequently up-regulated in advanced-stage cancers (Eferl and Wagner, 2003) and may modify the EMT response to TGF-β in cancer progression.
In summary, our study reveals an important role for JunB in the EMT program and profibrotic responses to TGF-β (Fig. 9). JunB functions as an immediate early factor that enables the transcriptional control of the EMT antagonist Id2 and key factors contributing to actin cytoskeleton remodeling, formation of adhesion structures, and matrix composition. The TGF-β profibrotic response is an integral part of the EMT process and may underlie the disruption of cell–cell junctions (Bianchi et al., 2010). Thus, JunB may represent an important biomarker and a therapeutic target in treatment of cancer and human diseases linked to EMT and tissue fibrosis.
Figure 9.
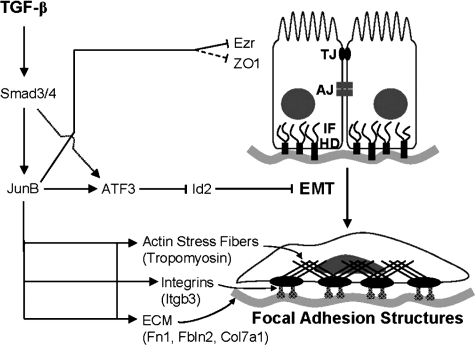
The function of JunB in the EMT response to TGF-β. TGF-β–Smad signaling up-regulates JunB, which, in turn, contributes to a negative and positive regulation of TGF-β target genes. JunB promotes a sustained induction of ATF3 and, in cooperation with ATF3, mediates repression of Id2, a negative regulator of TGF-β–induced EMT. In addition, JunB contributes to the regulation of proteins that are essential for the formation of actin stress fibers (tropomyosin), focal adhesions (Itgb3), and components of the ECM (Fn1, Fbln2, and Col7a1). JunB also contributes to suppression of ezrin (Ezr/Vil2) and ZO1. TJ, tight junctions; AJ, adherens junctions; IF, intermediate filaments; HD, hemidesmosomes.
Materials and methods
Cell culture
The mouse mammary epithelial NMuMG cell line was obtained from American Type Culture Collection, and the mouse kidney epithelial MCT cell line was a gift from H. Moses (Vanderbilt University, Nashville, TN). Cells were maintained in DME (Invitrogen) at 37°C under an atmosphere of 5% carbon dioxide. The medium was supplemented with 10% FBS (Cellgro; Mediatech, Inc.), 100 U/ml penicillin, and 0.1mg/ml streptomycin (Invitrogen). NMuMG cells were also supplemented with 10 µg/ml insulin (Sigma-Aldrich).
Antibodies and other reagents
TGF-β1 was purchased from R&D Systems. The following antibodies were used: mouse monoclonal to integrin-β1 and E-cadherin (BD); mouse monoclonal to vinculin, tropomyosin (TM311), and tubulin (Sigma-Aldrich); rabbit polyclonal to phosphorylated Smad2, Pax, and p44/42 (Erk1/2; Cell Signaling Technology); rabbit polyclonal to JunB, c-Jun, ATF3, and Id2 and mouse monoclonal to Smad4 (Santa Cruz Biotechnology, Inc.); and rabbit polyclonal to ZO1 and α-catenin (Invitrogen). Antibody 4G10 to phospho-Tyr was a gift from R. Mernaugh (Vanderbilt University, Nashville, TN). Alexa Fluor green and Texas red–conjugated phalloidins and Hoechst 3342 were purchased from Invitrogen. Anti–rabbit or –mouse IgG antibodies conjugated to HRP were obtained from GE Healthcare. CHD was purchased from EMD.
Plasmids and retroviral constructs
The A-Jun and A-Fos expression constructs encoding dominant-negative forms of c-Jun and c-Fos (Olive et al., 1997) were provided by C. Vinson (National Cancer Institute, Bethesda, MD). The ATF3 expression construct pCG-ATF3 was a gift from T. Hai (Ohio State University, Columbus, OH). Mouse JunB cDNA in a pCMV-Sport6 vector was purchased from Thermo Fisher Scientific. The retroviral vector pBMN-IRES-EGFP was provided by G. Nolan (Stanford University, Stanford, CA). pBMN-mJunB-IRES-EGFP was generated by subcloning mouse JunB cDNA into pBMN-IRES-EGFP at EcoRI–XhoI sites. The SBE-Lux reporter, containing 12 repeats of the Smad-binding sequence (Dennler et al., 1998), was provided by J.-M. Gauthier (Laboratoire Glaxo Wellcome, Marly Le Roi, France). AP1-Lux, containing six AP1 sites, was obtained from Takara Bio Inc. FN1-Lux, containing the −510–69-bp region of the human FN1 promoter in a pGL3 vector, was a gift from H. Moses. pGL2-Id2–2266 and pGL2-Id2–107 constructs (Karaya et al., 2005) were provided by Y. Yokota (University of Fukui, Fukui City, Japan). Constructs pGL2-Id2–4088, –1988, –1703, –1656, and –1570 were generated by PCR using mouse genomic bacterial artificial chromosome clone RP23-356E7 (Roswell Park Cancer Institute) containing the mouse Id2 promoter. These constructs included the 224-bp coding region and were cloned at KpnI–XhoI sites. Constructs pGL2-Id2-AP1 and pGL2-Id2–Eboxes-1 were generated by the cloning of PCR fragments from the −4088–224 region into pGL2-Id2–107 at KpnI–XhoI sites. pGL2-Id2–1353 was produced by excision of the −2256–1354 region from pGL2-Id2–2266 using PstI. The identity of constructs was verified by sequencing.
Retroviral infection of cells
Retroviruses were prepared by transfection of Phoenix cells with 10 µg DNA per 100-mm dish of pBMN-IRES-EGFP or pBMN-mJunB-IRES-EGFP using FuGENE 6 reagent (Roche). Supernatants were collected and filtered through 0.4-µm filters and then used to infect NMuMG and MCT cells in the presence of 8 µg/ml Polybrene (Sigma-Aldrich). 3 d later, GFP-positive cells were selected by flow cytometry.
Generation of NMuMG–Tet-Off–ATF3-EGFP/JunB cells
NMuMG–Tet-Off cells were generated by cotransfection with pBabe-Puro and ptTA-IRES-Neo plasmids (Yu et al., 1999) with FuGENE 6 according to manufacturer’s protocol. Puromycin-resistant clones exhibiting TGF-β responses equal to parental cells were selected in the presence of 1 µg/ml puromycin. Individual NMuMG-tTA clones with a tight regulation of the Tet-responsive pTRE-Lux reporter (BD) and intact TGF-β responses were chosen to generate inducible cell lines. Mouse ATF3 cDNA was excised from pCG-ATF3 and subcloned into pBluescript II (KS+) (Agilent Technologies) at BamHI–XbaI sites and then shuttled into pTRE2hyg (BD) at NotI–SalI sites to generate pTRE2hyg-ATF3. The NMuMG–tTA-22 was transfected with pTRE2hyg-ATF3 using FuGENE 6. Cells were selected with 200 µg/ml hygromycin in the presence of 2 µg/ml tetracycline. Clone 21 of NMuMG–Tet-Off–ATF3 cells was transduced with pBMN-IRES-EGFP or pBMN-mJunB-IRES-EGFP retroviruses in the presence of tetracycline, and GFP-positive cells were selected by flow cytometry. To induce expression of ATF3, NMuMG cells were washed twice with 1× PBS while in suspension and then seeded in tissue culture dishes, and the media was replenished the next day. Induction of ATF3 was confirmed by immunoblotting.
Luciferase reporter assay
The following firefly luciferase (Luc) reporters were used to measure transcriptional responses: SBE-Lux, AP1-Lux, FN1-Lux, pGL2-Id2–4088, pGL2-Id2–2266, pGL2-Id2–1988, pGL2-Id2–1703, pGL2-Id2–1656, pGL2-Id2–1570, pGL2-Id2–1353, pGL2-Id2–107, pGL2-Id2-AP1, and pGL2-Id2–E-boxes-1. NMuMG cells were transfected with 1 µg Luc reporter and 0.025 µg pCMV–Renilla reniformis luciferase (Rl; Promega) or 0.2 µg pTK-Rl (provided by J. Black, Roswell Park Cancer Institute, Buffalo, NY) in 6-well plates (106 cells/well) using FuGENE 6 reagent and according to the manufacturer’s protocol. The next day, cells were transferred into a 48-well plate (2 × 104 cells/well). The cells were treated with 2 ng/ml TGF-β1 for 24 h. All TGF-β1 treatments were performed in the presence of 5% FBS. Luc and Rl activities were determined in cell lysates using the Dual-Luciferase reporter assay system (Promega), according to the manufacturer’s protocol, in a microplate luminometer (Veritas; Promega). Firefly activity was normalized to Renilla activity and presented as relative luciferase units. All assays were performed in triplicate wells, and each experiment was repeated at least twice.
Fluorescence microscopy
Cells were grown on glass coverslips (22 × 22 mm) and treated with 2 ng/ml TGF-β1 for 24 h. The cells were fixed with 4% PFA and permeabilized with 0.05% Triton X-100 (for E-cadherin, vinculin, and fibronectin staining) or with 100% methanol for ZO1 staining and then blocked with 3% milk in PBS for 30 min at room temperature. The cells were incubated for 1 h with antibodies to vinculin, ZO1, E-cadherin, and/or fibronectin (1:400 dilution) in 1% milk/PBS followed by incubation for 30 min with Texas red–conjugated secondary antibody (1:500) at room temperature. Actin filaments were stained with phalloidin conjugated to Alexa Fluor 488 or 568 (1:200). Fluorescence images were taken with a Plan Apochromat 60×/1.40 NA oil objective lens at ambient temperature using an inverted microscope (TE2000-E; Nikon) equipped with a charge-coupled device camera (CoolSNAP HQ; Photometrics). The images were acquired using MetaVue imaging software (v6.2; Molecular Devices).
Immunoprecipitation and immunoblotting
NMuMG cells were grown in 5% serum DME and treated with 2 ng/ml TGF-β1 for 2 h. To prepare whole-cell extracts, cells were washed with PBS and then lysed in NP-40 lysis buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 10% glycerol, 20 mM NaF, and 1 mM Na orthovanadate supplemented with 1 mM PMSF and a protease inhibitor cocktail (Roche). Protein concentrations were measured using a detergent-compatible protein assay (Bio-Rad Laboratories) according to the manufacturer’s protocol. Lysates were precleared with 30 µl protein A/G agarose beads (Santa Cruz Biotechnology, Inc.). Precleared lysates (400 µg/antibody) were incubated with 2 µg of antibodies to Smad4 or JunB or an appropriate control IgG in 500 µl NP-40 lysis buffer overnight with rotation at 4°C. Lysates were incubated with 40 µl protein A/G agarose beads for 1 h, rotating at 4°C. The immunocomplexes were washed twice in NP-40 lysis buffer and once with PBS. Finally, the pellets were resuspended in equal volume of 2× loading buffer.
Immunoblot analysis of whole-cell extracts and immunoprecipitates was performed as previously described (Bakin et al., 2002). Protein samples were separated on a denaturing polyacrylamide gel and transferred onto nitrocellulose membranes. Membranes were incubated overnight at 4°C with primary antibodies in 5% milk followed by a 1-h incubation with appropriate secondary antibodies. Immune complexes were visualized using the West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) or ECL (GE Healthcare).
Nuclear and cytoplasmic fractionation
NMuMG cells in 5% serum DME were treated with 2 ng/ml TGF-β1. Cells were washed with PBS, scraped, and collected into 1.5-ml tubes. Cell pellets were resuspended in a hypotonic solution (10 mM Hepes, pH 7.3, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.4% NP-40, 50 mM NaF, and 1 mM DTT supplemented with 0.5 mM PMSF and a protease inhibitor cocktail (Roche). Mixtures were placed on ice for 5 min before centrifugation at 15,000 rpm for 1 min at 4°C. Cytoplasmic fractions were transferred to fresh tubes. Nuclear pellets were washed with hypotonic solution and resuspended in a high-salt buffer (20 mM Hepes, pH 7.3, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, and 1 mM DTT) supplemented with 0.5 mM PMSF and a protease cocktail (Roche). Tubes were rotated at 4°C for 15 min and centrifuged at 16,100 g for 1 min at 4°C. Supernatants containing nuclear fractions were transferred to fresh tubes. Protein extracts (50 µg/lane) were further subjected to immunoblotting.
Affymetrix microarrays
Microarray analysis was performed using the Mouse MOE430_2AB chips (Affymetrix). Total RNA samples were isolated from NMuMG and NMuMG cells transfected with siRNA to JunB that were treated or left untreated with 2 ng/ml TGF-β1 for 24 h. The MAS5.0 algorithm in the Affy package of Bioconductor in the R statistical computing environment was used to generate expression summary values followed by trimmed mean global normalization to bring the mean expression values of all chips to the same scale. For data quality control, MAS5.0 present/absent calls were used to filter out probe sets whose expression intensities were close to background noise (i.e., absent) across all samples. Genes that were altered with a minimal of twofold change were considered significant for further analysis. The function classification and statistical overrepresentation of gene function categories (e.g., Gene Ontology terms) were analyzed using the National Center for Biotechnology Information DAVID package (Reich et al., 2006).
RT-PCR analysis
Total RNA was extracted using a PerfectPure RNA Cultured Cell kit (5 PRIME, Inc.). Primer sequences are listed in Tables S1 (semiquantitative) and S2 (quantitative PCR). Amplification of transcripts was performed using the one-step RT-PCR system (Invitrogen) with 100 ng total RNA according to the manufacturer’s protocol. The optimal number of PCR cycles was determined for each primer set to ensure a linear range of amplification. PCR products were resolved on 1% agarose gel and visualized by ethidium bromide staining. For quantitative RT-PCR, first-strand cDNA was synthesized from 3 µg total RNA using a random hexamer mixture and Superscript III (both from Invitrogen) in a 20-µl reaction according to the manufacturer’s protocol. Quantitative real-time PCR was performed using 1 µl of first-strand cDNA. Each primer set was designed to span an intron to avoid amplification of contaminating genomic DNA. PCR was performed with SYBR green real-time PCR master mix (Invitrogen) using the default parameters of the 7700 sequence detection system (ABI PRISM; Applied Biosystems). To compare gene expressions between samples, the threshold cycle (CT) value was normalized using the mean CT for the reference gene, 18S ribosomal RNA (rRNA). The normalized mRNA level was defined as ΔCT = CT (test gene) − CT (mean for the reference gene). The final data were expressed as the fold difference between the test sample and the control sample, which was defined as 2(ΔCT test − ΔCT control). All reactions were performed in triplicate, and the experiments were repeated at least twice. The results are presented as the mean of at least two experiments.
siRNA
RNA duplexes against JunB and Smad4 were prepared at Invitrogen. The siRNA sequences are sense 5′-ACACCAACCUCAGCAGUUAUU-3′ and antisense 5′-UAACUGCUGAGGUUGGUGUUU-3′ for JunB and sense 5′-AUGGGCAAAGGAGUGCAGUUGGAUU-3′ and antisense 5′-AAUCCAACUGCACUCCUUUGCCUAU-3′ for Smad4. A scramble control RNA duplex labeled with Rhodamine was obtained from QIAGEN. Cells were transfected with RNA duplexes (200 nM for Smad4 and 50 nM for JunB) using Lipofectamine 2000 from Invitrogen following the manufacturer’s protocol. The media was replenished after 8 h, and cells were grown for 1 d before proceeding with further experiments.
Cell adhesion
Fibronectin was purchased from Sigma-Aldrich. The wells of 96-well plates were coated by incubating with fibronectin (10 µg/ml in PBS) for 1 h at 37°C and then blocked with 10 mg/ml BSA. Cells were seeded at 2 × 104/well in 5% serum DME in six replicates. After 30 min for NMuMG cells and 15 min for MCT cells, nonadherent and loosely attached cells were removed from the wells by gently washing with PBS. Attached cells were fixed with 5% glutaraldehyde and washed with PBS. Cells were counted at 100× magnification. The results were expressed as the mean value ± SD from two independent experiments.
ChIP assay
NMuMG cells were treated with 2 ng/ml TGF-β1 for 8 h. ChIP was performed using the EZ ChIP kit (Millipore) according to manufacturer’s protocol. Protein–DNA complexes were cross-linked with 1% formaldehyde (37% stock; Sigma-Aldrich) for 10 min at 37°C. The reaction was terminated by addition of glycine (Sigma-Aldrich). Cells were collected in cold PBS containing protease inhibitor cocktail (Millipore). After centrifugation, cell pellets were resuspended in SDS lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, and 1% SDS) supplemented with protease inhibitor cocktail (Millipore) and sonicated five times for 10 s. Cellular debris was pelleted by centrifugation, and the supernatant was transferred to a new tube and diluted 1:10 in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, and 1.1% Triton X-100). Samples were precleared with protein G agarose beads for 1 h at 4°C with rotation. After brief centrifugation, the supernatants were transferred to fresh tubes. Rabbit polyclonal IgG, anti-JunB, or anti-ATF3 antibodies (Santa Cruz Biotechnology, Inc.) were added, and the samples were incubated at 4°C overnight with rotation followed by a 1-h incubation with protein G agarose beads at 4°C with rotation. Beads were washed with 1× ChIP dilution buffer, and the cross-linking was reversed by addition of sodium chloride to a final concentration of 200 mM and incubation at 65°C for 5 h. The samples were treated with RNase A and proteinase K (Millipore) for 1 h at 45°C followed by purification with a spin filter column (Millipore). DNA was then subjected to PCR analysis. Primer sequences used for PCR amplification are listed in Tables S1 and S2.
Online supplemental material
Fig. S1 shows cell morphology and mRNA levels of E-cadherin and ZO1/Tjp1 in NMuMg and MCT cells transfected with siRNA to JunB and treated with TGF-β. Fig. S2 shows mRNA levels of Snai1 and Snai2 at various time points of TGF-β treatment and that JunB depletion does not affect phosphorylation of Smad2 and ERK1/2 in NMuMG cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109045/DC1.
Acknowledgments
We thank Dr. Heinz Baumann for his helpful discussion of the manuscript and Drs. Hal Moses and Ray Mernaugh for providing reagents.
This work was supported by Public Health Service grant R01 CA95263 (to A.V. Bakin) and in part by the Human Studies Review Board–New York State Department of Health Peter T. Rowley Breast Cancer Project and Roswell Park Cancer Institute Cancer Center Support grant CA 16056.
Footnotes
Abbreviations used in this paper:
- ATF
- activating transcription factor
- BMP
- bone morphogenetic protein
- CHD
- cycloheximide
- ChIP
- chromatin immunoprecipitation
- EMT
- epithelial–mesenchymal transition
- MCT
- murine cortical tubule
- Rl
- Renilla reniformis luciferase
- rRNA
- ribosomal RNA
- SBE
- Smad-binding element
References
- Annes J.P., Munger J.S., Rifkin D.B. 2003. Making sense of latent TGFbeta activation. J. Cell Sci. 116:217–224 10.1242/jcs.00229 [DOI] [PubMed] [Google Scholar]
- Bakin A.V., Tomlinson A.K., Bhowmick N.A., Moses H.L., Arteaga C.L. 2000. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275:36803–36810 10.1074/jbc.M005912200 [DOI] [PubMed] [Google Scholar]
- Bakin A.V., Rinehart C., Tomlinson A.K., Arteaga C.L. 2002. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci. 115:3193–3206 [DOI] [PubMed] [Google Scholar]
- Bakin A.V., Safina A., Rinehart C., Daroqui C., Darbary H., Helfman D.M. 2004. A critical role of tropomyosins in TGF-beta regulation of the actin cytoskeleton and cell motility in epithelial cells. Mol. Biol. Cell. 15:4682–4694 10.1091/mbc.E04-04-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A.V., Stourman N.V., Sekhar K.R., Rinehart C., Yan X., Meredith M.J., Arteaga C.L., Freeman M.L. 2005. Smad3-ATF3 signaling mediates TGF-beta suppression of genes encoding Phase II detoxifying proteins. Free Radic. Biol. Med. 38:375–387 10.1016/j.freeradbiomed.2004.10.033 [DOI] [PubMed] [Google Scholar]
- Baumann H., Jahreis G.P., Morella K.K., Won K.A., Pruitt S.C., Jones V.E., Prowse K.R. 1991. Transcriptional regulation through cytokine and glucocorticoid response elements of rat acute phase plasma protein genes by C/EBP and JunB. J. Biol. Chem. 266:20390–20399 [PubMed] [Google Scholar]
- Bhowmick N.A., Ghiassi M., Bakin A.V., Aakre M., Lundquist C.A., Engel M.E., Arteaga C.L., Moses H.L. 2001. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell. 12:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Gervasi M.E., Bakin A. 2010. Role of β5-integrin in epithelial-mesenchymal transition in response to TGF-β. Cell Cycle. 9:1647–1659 10.4161/cc.9.8.11517 [DOI] [PubMed] [Google Scholar]
- Brown K.A., Aakre M.E., Gorska A.E., Price J.O., Eltom S.E., Pietenpol J.A., Moses H.L. 2004. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 6:R215–R231 10.1186/bcr778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.C., Turner C.E. 2004. Paxillin: Adapting to change. Physiol. Rev. 84:1315–1339 10.1152/physrev.00002.2004 [DOI] [PubMed] [Google Scholar]
- Câmara J., Jarai G. 2010. Epithelial-mesenchymal transition in primary human bronchial epithelial cells is Smad-dependent and enhanced by fibronectin and TNF-alpha. Fibrogenesis Tissue Repair. 3:2 10.1186/1755-1536-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.P., Liang G., Whelan J., Hai T. 1994. ATF3 and ATF3 delta Zip. Transcriptional repression versus activation by alternatively spliced isoforms. J. Biol. Chem. 269:15819–15826 [PubMed] [Google Scholar]
- Chung K.Y., Agarwal A., Uitto J., Mauviel A. 1996. An AP-1 binding sequence is essential for regulation of the human alpha2(I) collagen (COL1A2) promoter activity by transforming growth factor-beta. J. Biol. Chem. 271:3272–3278 10.1074/jbc.271.6.3272 [DOI] [PubMed] [Google Scholar]
- Deckers M., van Dinther M., Buijs J., Que I., Löwik C., van der Pluijm G., ten Dijke P. 2006. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 66:2202–2209 10.1158/0008-5472.CAN-05-3560 [DOI] [PubMed] [Google Scholar]
- Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J.M. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091–3100 10.1093/emboj/17.11.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., Wagner E.F. 2003. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer. 3:859–868 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Fialka I., Schwarz H., Reichmann E., Oft M., Busslinger M., Beug H. 1996. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J. Cell Biol. 132:1115–1132 10.1083/jcb.132.6.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch S., Joseloff E., Bowden T. 2002. JunB negatively regulates AP-1 activity and cell proliferation of malignant mouse keratinocytes. J. Cancer Res. Clin. Oncol. 128:3–10 10.1007/s00432-001-0298-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.-Y., Li E.-M., Cui L., Lu X.-F., Meng L.-Y., Yuan H.-M., Xie J.-J., Du Z.-P., Pang J.-X., Xu L.-Y. 2009. Sp1 and AP-1 regulate expression of the human gene VIL2 in esophageal carcinoma cells. J. Biol. Chem. 284:7995–8004 10.1074/jbc.M809734200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Gaddy-Kurten D., Vale W. 1993. Protooncogene junB as a target for activin actions. Endocrinology. 133:1934–1940 10.1210/en.133.5.1934 [DOI] [PubMed] [Google Scholar]
- Heino J., Ignotz R.A., Hemler M.E., Crouse C., Massagué J. 1989. Regulation of cell adhesion receptors by transforming growth factor-beta. Concomitant regulation of integrins that share a common beta 1 subunit. J. Biol. Chem. 264:380–388 [PubMed] [Google Scholar]
- Hollnagel A., Oehlmann V., Heymer J., Rüther U., Nordheim A. 1999. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274:19838–19845 10.1074/jbc.274.28.19838 [DOI] [PubMed] [Google Scholar]
- Hsu E., Yasuoka H., Feghali-Bostwick C.A. 2008. Gene expression in pulmonary fibrosis. Crit. Rev. Eukaryot. Gene Expr. 18:47–56 [DOI] [PubMed] [Google Scholar]
- Hsu J.C., Bravo R., Taub R. 1992. Interactions among LRF-1, JunB, c-Jun, and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol. Cell. Biol. 12:4654–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-C., Cressman D.E., Taub R. 1993. Promoter-specific trans-activation and inhibition mediated by JunB. Cancer Res. 53:3789–3794 [PubMed] [Google Scholar]
- Huber M.A., Kraut N., Beug H. 2005. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17:548–558 10.1016/j.ceb.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Ignotz R.A., Heino J., Massagué J. 1989. Regulation of cell adhesion receptors by transforming growth factor-beta. Regulation of vitronectin receptor and LFA-1. J. Biol. Chem. 264:389–392 [PubMed] [Google Scholar]
- Jayaraman L., Massague J. 2000. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J. Biol. Chem. 275:40710–40717 10.1074/jbc.M005799200 [DOI] [PubMed] [Google Scholar]
- Jooss K.U., Müller R. 1995. Deregulation of genes encoding microfilament-associated proteins during Fos-induced morphological transformation. Oncogene. 10:603–608 [PubMed] [Google Scholar]
- Kadler K.E., Hill A., Canty-Laird E.G. 2008. Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 20:495–501 10.1016/j.ceb.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Neilson E.G. 2003. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112:1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Chen C.R., Massagué J. 2003. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell. 11:915–926 10.1016/S1097-2765(03)00109-6 [DOI] [PubMed] [Google Scholar]
- Karaya K., Mori S., Kimoto H., Shima Y., Tsuji Y., Kurooka H., Akira S., Yokota Y. 2005. Regulation of Id2 expression by CCAAT/enhancer binding protein beta. Nucleic Acids Res. 33:1924–1934 10.1093/nar/gki339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Cubillo E., Tobiume K., Shirakihara T., Fukuda N., Suzuki H., Shimizu K., Takehara K., Cano A., Saitoh M., Miyazono K. 2004. A role for Id in the regulation of TGF-beta-induced epithelial-mesenchymal transdifferentiation. Cell Death Differ. 11:1092–1101 10.1038/sj.cdd.4401467 [DOI] [PubMed] [Google Scholar]
- Kowanetz M., Valcourt U., Bergström R., Heldin C.H., Moustakas A. 2004. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol. Cell. Biol. 24:4241–4254 10.1128/MCB.24.10.4241-4254.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiho M., Rönnstrand L., Heino J., Decaprio J.A., Ludlow J.W., Livingston D.M., Massagué J. 1991. Control of junB and extracellular matrix protein expression by transforming growth factor-beta 1 is independent of simian virus 40 T antigen-sensitive growth-sensitive growth-inhibitory events. Mol. Cell. Biol. 11:972–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.F., Ozanne B.W., Roy C., McGarry L., Stipp C., Mangeat P., Jay D.G. 1997. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr. Biol. 7:682–688 10.1016/S0960-9822(06)00295-8 [DOI] [PubMed] [Google Scholar]
- Lasorella A., Noseda M., Beyna M., Yokota Y., Iavarone A. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 407:592–598 10.1038/35036504 [DOI] [PubMed] [Google Scholar]
- Lee J.M., Dedhar S., Kalluri R., Thompson E.W. 2006. The epithelial–mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 172:973–981 10.1083/jcb.200601018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L., Hill C.S. 2005. Smad4 dependency defines two classes of transforming growth factor beta (TGF-beta) target genes and distinguishes TGF-beta-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 25:8108–8125 10.1128/MCB.25.18.8108-8125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet-Vallée S. 2000. ERM proteins: From cellular architecture to cell signaling. Biol. Cell. 92:305–316 10.1016/S0248-4900(00)01078-9 [DOI] [PubMed] [Google Scholar]
- Maeda M., Johnson K.R., Wheelock M.J. 2005. Cadherin switching: Essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J. Cell Sci. 118:873–887 10.1242/jcs.01634 [DOI] [PubMed] [Google Scholar]
- Maehara Y., Kakeji Y., Kabashima A., Emi Y., Watanabe A., Akazawa K., Baba H., Kohnoe S., Sugimachi K. 1999. Role of transforming growth factor-beta 1 in invasion and metastasis in gastric carcinoma. J. Clin. Oncol. 17:607–614 [DOI] [PubMed] [Google Scholar]
- Mauviel A., Chung K.-Y., Agarwal A., Tamai K., Uitto J. 1996. Cell-specific induction of distinct oncogenes of the Jun family is responsible for differential regulation of collagenase gene expression by transforming growth factor-beta in fibroblasts and keratinocytes. J. Biol. Chem. 271:10917–10923 10.1074/jbc.271.18.10917 [DOI] [PubMed] [Google Scholar]
- Miao G.G., Curran T. 1994. Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol. Cell. Biol. 14:4295–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen P.J., Ebner R., Lopez A.R., Derynck R. 1994. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: Involvement of type I receptors. J. Cell Biol. 127:2021–2036 10.1083/jcb.127.6.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A., Heldin C.-H. 2007. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 98:1512–1520 10.1111/j.1349-7006.2007.00550.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Wall R. 1991. Interleukin-6 signals activating junB and TIS11 gene transcription in a B-cell hybridoma. Mol. Cell. Biol. 11:1409–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naso M., Uitto J., Klement J.F. 2003. Transcriptional control of the mouse Col7a1 gene in keratinocytes: Basal and transforming growth factor-beta regulated expression. J. Invest. Dermatol. 121:1469–1478 10.1111/j.1523-1747.2003.12640.x [DOI] [PubMed] [Google Scholar]
- Neuman K., Nornes H.O., Neuman T. 1995. Helix-loop-helix transcription factors regulate Id2 gene promoter activity. FEBS Lett. 374:279–283 10.1016/0014-5793(95)01128-2 [DOI] [PubMed] [Google Scholar]
- Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. 1996. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10:2462–2477 10.1101/gad.10.19.2462 [DOI] [PubMed] [Google Scholar]
- Olive M., Krylov D., Echlin D.R., Gardner K., Taparowsky E., Vinson C. 1997. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272:18586–18594 10.1074/jbc.272.30.18586 [DOI] [PubMed] [Google Scholar]
- Ott R.G., Simma O., Kollmann K., Weisz E., Zebedin E.M., Schorpp-Kistner M., Heller G., Zöchbauer S., Wagner E.F., Freissmuth M., Sexl V. 2007. JunB is a gatekeeper for B-lymphoid leukemia. Oncogene. 26:4863–4871 10.1038/sj.onc.1210285 [DOI] [PubMed] [Google Scholar]
- Ozanne B.W., Spence H.J., McGarry L.C., Hennigan R.F. 2007. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 26:1–10 10.1038/sj.onc.1209759 [DOI] [PubMed] [Google Scholar]
- Pardali K., Moustakas A. 2007. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta. 1775:21–62 [DOI] [PubMed] [Google Scholar]
- Passegué E., Jochum W., Behrens A., Ricci R., Wagner E.F. 2002. JunB can substitute for Jun in mouse development and cell proliferation. Nat. Genet. 30:158–166 10.1038/ng790 [DOI] [PubMed] [Google Scholar]
- Pawlak G., Helfman D.M. 2001. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11:41–47 10.1016/S0959-437X(00)00154-4 [DOI] [PubMed] [Google Scholar]
- Pertovaara L., Sistonen L., Bos T.J., Vogt P.K., Keski-Oja J., Alitalo K. 1989. Enhanced jun gene expression is an early genomic response to transforming growth factor beta stimulation. Mol. Cell. Biol. 9:1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek E., Moustakas A., Kurisaki A., Heldin C.H., ten Dijke P. 1999. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112:4557–4568 [DOI] [PubMed] [Google Scholar]
- Pinkas J., Leder P. 2002. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 62:4781–4790 [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- Safina A.F., Varga A.E., Bianchi A., Zheng Q., Kunnev D., Liang P., Bakin A.V. 2009. Ras alters epithelial-mesenchymal transition in response to TGFbeta by reducing actin fibers and cell-matrix adhesion. Cell Cycle. 8:284–298 10.4161/cc.8.2.7590 [DOI] [PubMed] [Google Scholar]
- Selvamurugan N., Kwok S., Partridge N.C. 2004. Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells. J. Biol. Chem. 279:27764–27773 10.1074/jbc.M312870200 [DOI] [PubMed] [Google Scholar]
- Siegel P.M., Shu W., Massagué J. 2003. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J. Biol. Chem. 278:35444–35450 10.1074/jbc.M301413200 [DOI] [PubMed] [Google Scholar]
- Spurzem J.R., Sacco O., Rickard K.A., Rennard S.I. 1993. Transforming growth factor-beta increases adhesion but not migration of bovine bronchial epithelial cells to matrix proteins. J. Lab. Clin. Med. 122:92–102 [PubMed] [Google Scholar]
- Steidl U., Rosenbauer F., Verhaak R.G., Gu X., Ebralidze A., Otu H.H., Klippel S., Steidl C., Bruns I., Costa D.B., et al. 2006. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat. Genet. 38:1269–1277 10.1038/ng1898 [DOI] [PubMed] [Google Scholar]
- Stover D.G., Bierie B., Moses H.L. 2007. A delicate balance: TGF-beta and the tumor microenvironment. J. Cell. Biochem. 101:851–861 10.1002/jcb.21149 [DOI] [PubMed] [Google Scholar]
- Thiery J.P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740–746 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C.H., Moustakas A. 2006. Transforming growth factor-β employs HMGA2 to elicit epithelial–mesenchymal transition. J. Cell Biol. 174:175–183 10.1083/jcb.200512110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.C., Phillips A.O. 2003. TGF-beta1-mediated inhibition of HK-2 cell migration. J. Am. Soc. Nephrol. 14:631–640 10.1097/01.ASN.0000053418.56286.5E [DOI] [PubMed] [Google Scholar]
- Valcourt U., Kowanetz M., Niimi H., Heldin C.-H., Moustakas A. 2005. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 16:1987–2002 10.1091/mbc.E04-08-0658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrecchia F., Tacheau C., Schorpp-Kistner M., Angel P., Mauviel A. 2001a. Induction of the AP-1 members c-Jun and JunB by TGF-beta/Smad suppresses early Smad-driven gene activation. Oncogene. 20:2205–2211 10.1038/sj.onc.1204347 [DOI] [PubMed] [Google Scholar]
- Verrecchia F., Vindevoghel L., Lechleider R.J., Uitto J., Roberts A.B., Mauviel A. 2001b. Smad3/AP-1 interactions control transcriptional responses to TGF-beta in a promoter-specific manner. Oncogene. 20:3332–3340 10.1038/sj.onc.1204448 [DOI] [PubMed] [Google Scholar]
- Walker R.A., Dearing S.J. 1992. Transforming growth factor beta 1 in ductal carcinoma in situ and invasive carcinomas of the breast. Eur. J. Cancer. 28:641–644 10.1016/S0959-8049(05)80116-9 [DOI] [PubMed] [Google Scholar]
- Wang Y., Fortin J., Lamba P., Bonomi M., Persani L., Roberson M.S., Bernard D.J. 2008. Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity. Endocrinology. 149:5577–5591 10.1210/en.2008-0220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström P., Stattin P., Franck-Lissbrant I., Damber J.E., Bergh A. 1998. Transforming growth factor beta1 is associated with angiogenesis, metastasis, and poor clinical outcome in prostate cancer. Prostate. 37:19–29 [DOI] [PubMed] [Google Scholar]
- Wolf G., Kalluri R., Ziyadeh F.N., Neilson E.G., Stahl R.A.K. 1999. Angiotensin II induces alpha3(IV) collagen expression in cultured murine proximal tubular cells. Proc. Assoc. Am. Physicians. 111:357–364 10.1046/j.1525-1381.1999.99117.x [DOI] [PubMed] [Google Scholar]
- Yi J.Y., Hur K.C., Lee E., Jin Y.J., Arteaga C.L., Son Y.S. 2002. TGFbeta1 -mediated epithelial to mesenchymal transition is accompanied by invasion in the SiHa cell line. Eur. J. Cell Biol. 81:457–468 10.1078/0171-9335-00265 [DOI] [PubMed] [Google Scholar]
- Yin T., Taga T., Tsang M.L., Yasukawa K., Kishimoto T., Yang Y.C. 1993. Involvement of IL-6 signal transducer gp130 in IL-11-mediated signal transduction. J. Immunol. 151:2555–2561 [PubMed] [Google Scholar]
- Yu J., Zhang L., Hwang P.M., Rago C., Kinzler K.W., Vogelstein B. 1999. Identification and classification of p53-regulated genes. Proc. Natl. Acad. Sci. USA. 96:14517–14522 10.1073/pnas.96.25.14517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J., Böttinger E.P. 2005. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774 10.1038/sj.onc.1208927 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X.H., Derynck R. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 394:909–913 (published erratum appears in Nature. 1998. 396:491) 10.1038/29814 [DOI] [PubMed] [Google Scholar]
- Zheng Q., Safina A., Bakin A.V. 2008. Role of high-molecular weight tropomyosins in TGF-beta-mediated control of cell motility. Int. J. Cancer. 122:78–90 10.1002/ijc.23025 [DOI] [PubMed] [Google Scholar]