Abstract
Regeneration of complex structures after injury requires dramatic changes in cellular behavior. Regenerating tissues initiate a program that includes diverse processes such as wound healing, cell death, dedifferentiation, and stem (or progenitor) cell proliferation; furthermore, newly regenerated tissues must integrate polarity and positional identity cues with preexisting body structures. Gene knockdown approaches and transgenesis-based lineage and functional analyses have been instrumental in deciphering various aspects of regenerative processes in diverse animal models for studying regeneration.
Regeneration: Sometimes truth is stranger than (science) fiction
Fans of Doctor Who, the quintessential British science fiction television series, have been able to enjoy the adventures of its main character (the Doctor) for over three decades. As a Time Lord from the planet Gallifrey, the Doctor can regenerate his body completely as he nears death. The Doctor’s regenerative abilities, however, are not without flaw: after he regenerates, he takes on different features and characteristics. This imperfect process enables the series to continue with a new actor portraying the Doctor upon the departure of his predecessor. The Doctor’s slightly defective regenerative power is not only a boon for the show’s fans (and producers), but it is also a rare example of science fiction being outdone by the real world. On Earth, a wide range of actual organisms can regenerate missing parts after injury. Unlike the fictional Doctor, these animals can rebuild new structures that are indistinguishable from those they are replacing1. With the goal of understanding the factors that enable some organisms, but not others, to restore missing structures, biologists have been studying regeneration for well over two centuries (Morgan, 1901; Lenhoff and Lenhoff, 1986).
Although the Doctor’s regenerative abilities remain a mystery, recent research on various earthlings has started to reveal the mechanisms leading to regeneration of complex structures. During the past decade the application of molecular genetic techniques, including double-stranded RNA-mediated genetic interference (RNAi) and transgenesis, has revitalized studies of classical animal models of regeneration (Poss, 2010). Here we highlight a sample of recent work addressing some of the critical questions facing regeneration researchers: What signals initiate regeneration? What is the nature of the cells that produce the regenerate? What role does the nervous system play in this process? The answers to these and related questions should help developmental biologists, tissue engineers, and clinicians to understand and one day overcome the limits on human regeneration.
Back from the dead: Apoptosis and regeneration
After amputation, local responses at the site of the wound play important roles in the initiation of regenerative processes. Wound healing (Gurtner et al., 2008), ion flux (Levin, 2009), and interactions between the wound epidermis and the underlying tissue appear critical for regenerative outgrowth. Recent work suggests that programmed cell death may play a role in triggering regenerative responses in many different organisms, including Hydra (Bergmann and Steller, 2010).
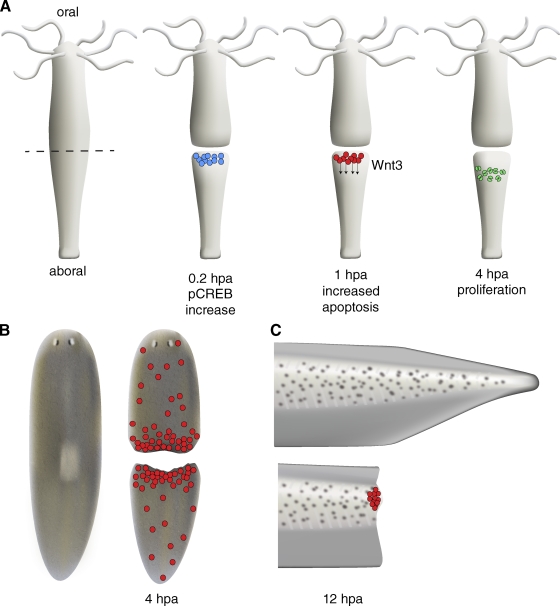
Hydra, a freshwater polyp belonging to the Phylum Cnidaria (which includes jellyfish, sea anemones, and corals), was the subject of the first scientific investigations of regeneration in animals (Lenhoff and Lenhoff, 1986). The Hydra body consists of ectodermal and endodermal cell layers separated by an extracellular matrix; neurons and interstitial cells (stem cells that produce neurons, gland cells, and germ cells, among other cell types) reside between these two layers. The animal is radially symmetric, and is polarized along the oral (head and tentacles)/aboral (foot) axis (Fig. 1 A). Small fragments of Hydra tissue can regenerate a complete organism; even dissociated single cells can reaggregate, reestablish polarity, and form a new animal (Noda, 1971; Gierer et al., 1972).
Figure 1.
Increased apoptosis is associated with early phases of regeneration. (A) After mid-gastric bisection in Hydra, MAPK signaling leads to rapid activation of the transcription factor CREB in fragments regenerating a head (blue cells). MAPK/CREB activity is required for stimulating a wave of apoptosis in interstitial cells near the site of injury (red cells). These apoptotic cells secrete Wnt3, inducing a zone of proliferation (green cells) below the region of apoptosis. (B) Planarians and (C) Xenopus larval tails also show a rapid, localized increase in apoptosis after amputation. In Xenopus, like Hydra, apoptosis may provide important signals during early phases of regeneration; inhibiting apoptosis during the first 24 h post amputation (hpa) blocks regeneration.
An apoptotic program is initiated asymmetrically after transection midway through the body column. The fragment that will regenerate a new head displays a robust apoptotic response near the wound site, whereas the fragment regenerating a new foot does not (Fig. 1 A; Chera et al., 2009). Inhibition of apoptosis blocks head regeneration, and activation of apoptosis at a foot-regenerating wound leads to ectopic head formation (Chera et al., 2009). Thus, induction of apoptosis is both necessary and sufficient for head regeneration. The apoptotic response stimulates a synchronous burst of proliferative activity in neighboring cells and leads to the establishment of a head organizer via secretion of Wnt3 ligand by apoptotic interstitial cells (Chera et al., 2009). RNAi knockdown of Wnt3 prevents the proliferative burst induced by apoptosis, whereas treatment with exogenous Wnt3 rescues proliferation when apoptosis is inhibited (Chera et al., 2009). The induction of apoptosis after injury appears to require the mitogen-activated protein kinase (MAPK) pathway, including ribosomal S6 kinase (RSK), and cAMP response element–binding protein (CREB). RNAi knockdown of RSK and CREB blocks apoptosis at the head-regenerating tip, as does treatment with U0126, a pharmacological inhibitor of MEK, the kinase that phosphorylates MAPK (Chera et al., 2011). In fragments that will regenerate a new head, the transcription factor CREB is phosphorylated by RSK within minutes of amputation (Fig. 1 A; Chera et al., 2011). Intriguingly, MAPK/CREB-induced apoptosis is not required in fragments regenerating a foot, and cell proliferation is not up-regulated in foot regenerates. Rather, foot regeneration proceeds through cellular rearrangement and transdifferentiation (morphallactic regeneration). The exact nature of the signal that leads to asymmetric MAPK activation (and, thus, apoptosis) at the site of injury in head-regenerating fragments remains to be determined.
The apoptosis-induced compensatory proliferation (Fan and Bergmann, 2008) observed in Hydra may be a conserved mechanism for stimulating proper wound healing and regeneration. Apoptotic cells have been observed during early phases of regeneration in several animals that can regenerate missing tissues: planarians (Fig. 1 B), Xenopus (Fig. 1 C), and newts (Hwang et al., 2004; Vlaskalin et al., 2004; Tseng et al., 2007; Chera et al., 2009; Pellettieri et al., 2010). Although the requirement for apoptosis during regeneration has not been addressed in planarians and newts, treatment of Xenopus larvae with caspase inhibitors during the first 24 h after amputation blocks tail regeneration (Tseng et al., 2007). As observed in Hydra, inhibition of apoptosis impedes proliferation; in addition, nerves fail to extend appropriately toward the site of amputation. Because nerves release important regenerative signals (see penultimate section), it is unclear whether reduced proliferation results directly from lack of signals derived from apoptotic cells, or indirectly from defective innervation. Furthermore, because these assays rely on caspase inhibition it is possible that nonapoptotic roles for caspases (Kuranaga and Miura, 2007) may be involved rather than apoptosis by itself.
Apoptotic cells have been shown to provide a number of signals that can regulate wound healing and regeneration. In Drosophila, the larval imaginal discs are capable of regenerating: wing discs can produce appropriately sized adult wings after radiation-induced killing of over 50% of the cells (Haynie and Bryant, 1977). MAPK signaling through Jun kinase is important both for initiating apoptosis and production of Wnt (Wg) and BMP (Dpp) mitogens (Pérez-Garijo et al., 2009; Bergantiños et al., 2010; Morata et al., 2011). The extent to which these factors are required for stimulating compensatory proliferation remains unclear. After radiation-induced apoptosis, neither dpp nor wg were required (Pérez-Garijo et al., 2009). However, after ectopic expression of a pro-apoptotic gene, wg was required for the proliferative response (Smith-Bolton et al., 2009). Furthermore, wg is also required for disc regeneration after surgical transection (Schubiger et al., 2010). Additional roles for apoptosis have been reported in epidermal wound healing and liver regeneration in mouse. Caspase 3 and caspase 7 mutant mice have defects in both processes, and these mutants show reduced cell proliferation in these contexts (Li et al., 2010). Caspases 3 and 7 can activate Ca2+-independent phospholipase A2, leading to production of arachidonic acid and prostaglandin in apoptotic cells, the latter of which can stimulate proliferation (Li et al., 2010).
Injury-induced apoptotic signals are also required to maintain tissue homeostasis. When cells of the adult Drosophila midgut are injured by toxins or induced to undergo apoptosis, intestinal enterocytes secrete the cytokine Unpaired, which stimulates proliferation of intestinal stem cells through activation of the Jak/Stat pathway (Jiang et al., 2009). Similarly, in the mouse intestine massive induction of apoptosis (via intestine-specific knockout of the p53 inhibitor Mdm2) is eventually compensated in adults by increased proliferation and expansion of the stem cell pool (Valentin-Vega et al., 2008). Apoptotic cells also contribute to homeostasis in epithelia by lipid-based signaling (sphingosine-1-phosphate) that triggers actomyosin contraction in the surrounding cells, leading to the extrusion of the dying cells (Gu et al., 2011). These observations suggest many potential roles for dead and dying cells to alter cell behavior at sites of injury.
Conjuring up spare parts: Cellular sources of regeneration
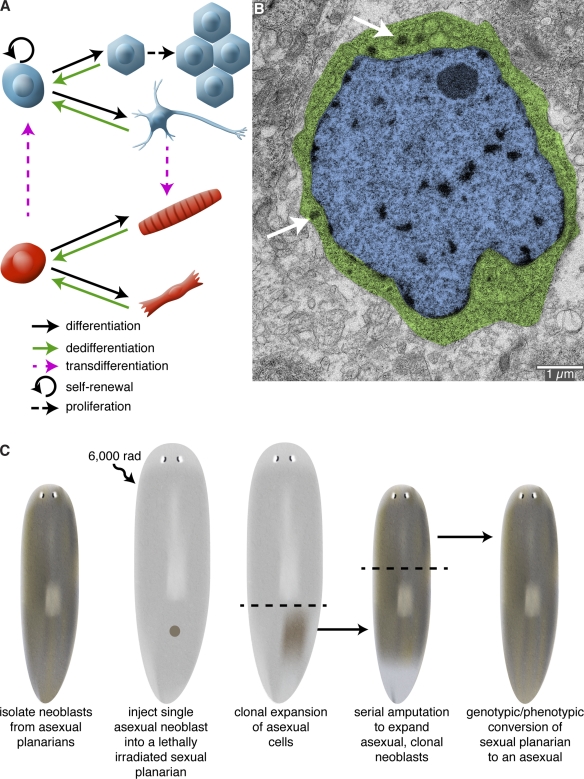
In most regenerating organisms, replacing an amputated structure requires the production of new cells. Therefore, one of the main functions of early signaling events after injury is to stimulate the production of additional cells that are capable of rebuilding lost structures. New cells coalesce near the site of injury, giving rise to a mass of undifferentiated cells called the regeneration blastema. Subsequent signals then regulate outgrowth and patterning of the newly formed tissue. To understand how early signaling events initiate regeneration and stimulate blastema formation, it is crucial to identify the cells upon which these signals act. New cells can be generated in a variety of ways, including proliferation of a resident stem cell population, division of terminally differentiated cells, or dedifferentiation/transdifferentiation of mature cells to a stem cell–like precursor or another cell type (Fig. 2 A). The extent to which each mode is used varies between species and even across tissues within the same species.
Figure 2.
Cellular sources of regeneration. (A) The ability to regenerate amputated structures often requires the production of new cells. These new cells can be derived from amplification and differentiation of resident stem cells, proliferation of differentiated cells, dedifferentiation of cells to a more primitive state, or transdifferentiation of one cell type to another cell type. (B) Transmission electron micrograph of a neoblast from the planarian Schmidtea mediterranea. Planarians owe their impressive regenerative abilities to these adult stem cells. Neoblasts are the only mitotic somatic cells and are defined by their high nuclear (blue) to cytoplasm (green) ratio and the presence of cytoplasmic ribonucleoprotein complexes called chromatoid bodies (arrows). Image courtesy of Ana Vieira (University of Illinois at Urbana-Champaign, Urbana, IL). (C) Rescue of a lethally irradiated planarian by introduction of a single neoblast. S. mediterranea exists as two genetically distinct strains: an asexual strain (brown) that reproduces by transverse fission; and a sexual, hermaphroditic strain (gray) that reproduces by cross-fertilization. Wagner et al. (2011) have shown that lethally irradiated sexual animals can be rescued by injection of a single asexual neoblast. Because the asexual donor neoblast is the only source of new cells, the sexual host is eventually converted into an asexual animal after repeated rounds of amputation, regeneration, and tissue turnover.
Stem cells and transdifferentiation both contribute to the regenerative abilities of Hydra. These animals possess three distinct stem cell populations: ectodermal, interstitial, and endodermal stem cells (Galliot et al., 2006). Development of transgenesis in Hydra (Wittlieb et al., 2006) has enabled in vivo tracking of the stem cell lineages: for example, investigating the differentiation of interstitial stem cells and migration of their progeny in intact animals (Khalturin et al., 2007). Transgenesis has also aided the analysis of transdifferentiation. By expressing eGFP in the zymogen gland cells (a derivative of the interstitial stem cells), Siebert et al. (2008) showed that these cells move up the body column and down-regulate the expression of a zymogen gland cell–specific marker. By using histology, electron microscopy, and in situ hybridization with cell type–specific markers, they identified cells with characteristics of both zymogen gland cells and another cell type (granular mucous cells), suggesting that zymogen gland cells could convert to granular mucous cells as they were displaced up the body column and entered the head region (Siebert et al., 2008). The ability of Hydra cells to transdifferentiate permits these animals to regenerate even in the absence of cell proliferation (Cummings and Bode, 1984).
Freshwater planarians (another classical model for studying regeneration) owe their amazing regenerative abilities to stem cells. In these animals, a population of mesenchymal stem cells, called neoblasts, is the source of cells for regeneration (Baguñà et al., 1989). Neoblasts are the only dividing somatic cells and have been defined by their high nuclear/cytoplasmic ratio and sensitivity to γ-radiation, as well as the expression of several post-transcriptional regulators and markers of proliferation (Fig. 2 B; Baguñà et al., 1989; Newmark and Sánchez Alvarado, 2000; Orii et al., 2005; Reddien et al., 2005; Salvetti et al., 2005; Guo et al., 2006; Yoshida-Kashikawa et al., 2007; Rouhana et al., 2010). Classical experiments showed that injection of neoblasts could restore regenerative abilities and long-term viability to lethally irradiated planarians; furthermore, injection of neoblasts derived from a sexual planarian strain could “transform” lethally irradiated asexual individuals into sexuals (Baguñà et al., 1989). Because these injections used thousands of cells, neoblasts as a population have long been considered pluripotent. However, whether individual neoblasts are pluripotent or if there are subsets of lineage-committed neoblasts has remained an open question. Recent work by Wagner et al. (2011) provides convincing evidence that a subset of neoblasts is pluripotent. They injected single asexual donor neoblasts into lethally irradiated sexual hosts (the irradiated sexual animals survive longer after irradiation than the asexuals do, permitting sufficient time for clonal expansion of the injected cells). Single neoblast injections were able to restore viability to the sexual animals and convert them to an asexual mode of reproduction. Restriction fragment length polymorphism and haplotype sequencing confirmed the genotypic conversion of the host to that of the donor strain (Fig. 2 C; Wagner et al., 2011). Characterizing this pluripotent subset of the neoblast population will be an important avenue for future research.
The source of regenerative cells in vertebrates varies between tissues and organisms, and in some cases remains a matter of continued debate. Many vertebrate tissues contain adult stem cells that play important roles in tissue turnover and homeostasis. Nonetheless, division, dedifferentiation, and transdifferentiation of differentiated cells contribute to regeneration in several different contexts. For example, whereas liver progenitor cells appear to be major sources of new hepatocytes under conditions of extreme damage or chronic disease, restoration of liver mass after partial hepatectomy or mild liver injury is largely accomplished through proliferation of remaining hepatocytes (Riehle et al., 2011). Similarly, recent lineage-tracing experiments indicate that after damage to the zebrafish heart, existing cardiomyocytes undergo dedifferentiation and proliferate to generate new cardiomyocytes for replacing lost heart mass (Jopling et al., 2010; Kikuchi et al., 2010). Pigmented epithelial cells in the newt dorsal iris can regenerate a new lens via transdifferentiation: these cells from the dorsal iris can dedifferentiate, reenter the cell cycle, and differentiate to produce new lens cells (Henry and Tsonis, 2010).
Dedifferentiation also contributes new cells during appendage regeneration in Urodele amphibians (newts and axolotls). Near the site of amputation, syncytial skeletal myotubes fragment and produce mononucleate cells that reenter the cell cycle (Lo et al., 1993; Echeverri et al., 2001; Kumar et al., 2004). Skeletal muscle from adult newts also contains Pax7-positive muscle stem cells (satellite cells) that become activated and contribute to new tissues during regeneration (Morrison et al., 2006, 2010). The respective contributions of muscle dedifferentiation and satellite cell activation during limb regeneration in the axolotl and newt remain open questions. In contrast, larval tail regeneration in Xenopus laevis appears to be driven largely by progenitor cells; in lineage-tracing experiments the extent of labeling of new muscle correlates with the amount of satellite cell labeling before amputation (Slack et al., 2004). Lineage tracing experiments in both axolotl and Xenopus indicate that multiple cell types contribute to formation of the blastema (Slack et al., 2004; Kragl et al., 2009). However, it is still unclear whether these other tissues contribute cells through proliferation of progenitor cells or dedifferentiation of mature cells. Such an understanding will be crucial for deciphering the mechanisms by which early regenerative signals trigger blastema formation.
To thine own self be true: Cellular memory during regeneration
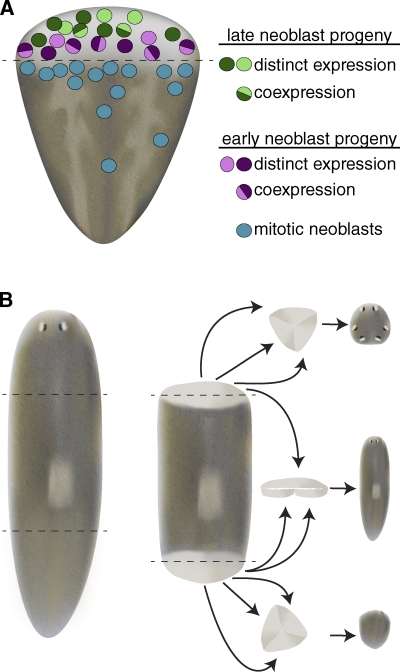
In addition to identifying the cellular sources of regeneration in different systems, researchers have been examining whether blastemal cells are pluripotent, multipotent, or have more limited potential. As discussed already, pluripotent neoblasts are the source of new cellular material that drives regeneration in planarians. However, cell proliferation is restricted largely to regions outside the blastema (Reddien and Sánchez Alvarado, 2004; Wenemoser and Reddien, 2010); thus, post-mitotic neoblast progeny are the predominant cells within the blastema that rebuild lost tissues. Eisenhoffer et al. (2008) have exploited the sensitivity of neoblasts to γ-radiation to identify numerous neoblast as well as early and late neoblast progeny markers. Interestingly, not all early neoblast progeny express the same panel of markers (Fig. 3 A). Although this difference could reflect transient temporal alterations in gene expression in early neoblast progeny, it more likely indicates that these cells are in various early stages of lineage commitment and/or en route to different terminal cell types. This heterogeneity has also been suggested by gene expression profiling on individual neoblasts (and their progeny) isolated using fluorescence-activated cell sorting (Hayashi et al., 2010). A subset of radiation-sensitive cells with G2 DNA content and expressing various neoblast markers also expressed a marker of muscle differentiation, suggesting that not all lineage-committed neoblasts are post-mitotic (Hayashi et al., 2010). Consistent with this result, S phase or G2 neoblasts have been observed to express markers of differentiated cell types, including excretory and neuronal markers (Nishimura et al., 2011; Scimone et al., 2011).
Figure 3.
Neoblast progeny are specified during early phases of regeneration yet retain developmental plasticity. (A) The planarian blastema is composed of a heterogeneous mixture of differentiating cells. The blastema is largely devoid of proliferating neoblasts, and instead is composed of cells expressing early and late neoblast progeny markers with distinct spatial distributions. Although some cells coexpress early (or late) neoblast progeny markers, other cells show distinct expression profiles for various early neoblast progeny markers, suggesting early neoblast progeny may be en route to committing to terminal cell types (adapted from Eisenhoffer et al., 2008). (B) 3-d anterior blastemas (top) are capable of forming anterior structures, including photoreceptors and cephalic ganglia, after being surgically isolated from intact tissue and cultured in vitro. Similarly, 3-d posterior blastemas (bottom) repigment and form muscle, but do not generate anterior structures, suggesting that by 3 d of regeneration head or tail specification has occurred. When 3-d anterior and posterior blastemas are juxtaposed (middle) they can generate a planarian containing mid-body structures (including a pharynx), in addition to head and tail tissue. These results indicate that at least some of the post-mitotic neoblast progeny that compose the blastema are capable of altering their fates (adapted from Sengel, 1960).
In planarians, specification of blastema positional identity (i.e., as anterior or posterior) likely occurs early during regeneration. Sengel (1960) removed anterior blastemas after 3 d of regeneration and cultured them as explants in vitro; these blastema explants produced only anterior tissues, including photoreceptors and cephalic ganglia (Fig. 3 B; Sengel, 1960). By contrast, explanted posterior blastemas did not produce anterior tissues. Therefore, these early experiments suggested that specification of blastemas to produce heads or tails occurs during early stages of regeneration. This idea has been confirmed by recent molecular studies showing rapid up-regulation of polarity signals (Petersen and Reddien, 2009, 2011; Gurley et al., 2010). Surprisingly, when anterior blastemas were co-cultured together with posterior blastemas, they were now able to produce small planarians containing head, central body, and tail structures (Fig. 3 B). These results suggest that, although neoblast progeny are specified early during regeneration, they may retain some developmental plasticity; in response to altered position cues (e.g., juxtaposition of anterior and posterior tissues) they may be able to change their fates and generate different cell types and tissues than they would have formed otherwise. Alternatively, the blastema explants may have contained some neoblasts that responded to the juxtaposition of anterior and posterior tissues to generate distinct body structures.
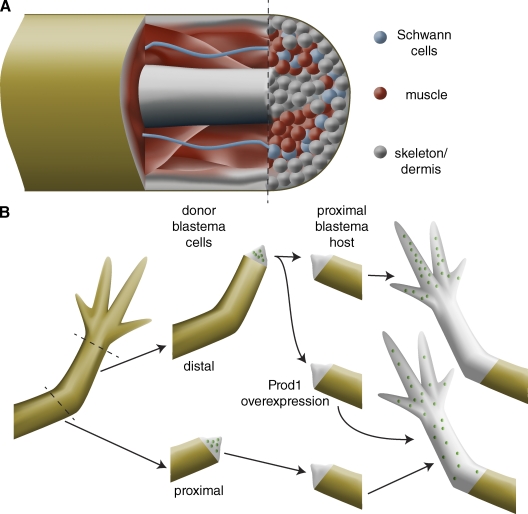
Recent experiments using transgene-based lineage tracing suggest that vertebrate blastema cells can “remember” the tissue from which they are derived, and that their fates are largely restricted to forming similar tissues in the regenerate. For example, transgenesis in axolotl and Xenopus has been combined with embryonic grafting to specifically label various tissues, including muscle, Schwann cells, spinal cord, and dermis; this tissue-specific labeling enables the contribution of various tissues to be analyzed during regeneration (Fig. 4 A; Gargioli and Slack, 2004; Kragl et al., 2009). These experiments revealed that regenerated muscle tissue is derived solely from muscles present in the limb before amputation. However, in the axolotl, interconversion of dermis and cartilage was observed. Both of these tissues are derivatives of lateral plate mesoderm, suggesting that these tissues may dedifferentiate to produce progenitors restricted to lateral plate fates or that the dermis may contain an uncommitted stem cell population (Kragl et al., 2009).
Figure 4.
Cellular memory during vertebrate limb regeneration. (A) Although the regeneration blastema appears to be a homogeneous mass of undifferentiated cells, lineage-tracing experiments in the axolotl limb, the Xenopus tail, and the zebrafish caudal fin indicate that blastema cells only contribute to tissues of similar developmental origin as that from which they are derived. For example, muscle (red) only gives rise to new muscle. Although dermis (gray) can give rise to new skeletal elements, these tissues are both lateral plate mesoderm derivatives, suggesting only limited dedifferentiation. Therefore, the blastema is composed of a heterogeneous mixture of lineage-restricted cells and is not a homogeneous population of multipotent cells. (B) Blastema cells retain their proximo-distal identity: distal amputations produce blastemas that only regenerate distal structures, whereas more proximal amputations produce blastemas that regenerate medial and distal structures. When cells from a distal blastema (green dots) are transplanted into a proximal blastema, they contribute only to distal structures; by contrast, cells from a proximal blastema (green dots) contribute to structures along the length of the proximo-distal axis. Overexpression of the cell surface protein Prod1 transforms distal blastema cells to more proximal fates.
Lineage restriction has also been observed during fin regeneration in zebrafish. Tu and Johnson (2011) have used transposon-based clonal analysis to examine the potency of various cell lineages during development, growth, and regeneration (Tu and Johnson, 2010, 2011). This method stably labels one to a few cells in the developing fin bud, enabling the progeny of single fin bud cells to be monitored. Examination of hundreds of animals indicated that fin bud cells are greatly restricted in developmental potential, only generating one to a few cell types in the adult fin (Tu and Johnson, 2011). As in axolotl and Xenopus, zebrafish caudal fin cells remain lineage committed during regeneration, and do not contribute to lineages other than that from which they are derived (Tu and Johnson, 2011). Similar results were obtained in studies of bone regeneration in the zebrafish fin. Osteoblasts near the amputation site down-regulate osteoblast differentiation markers, lose their differentiated morphology, proliferate, and give rise to new bone in the regenerate (Knopf et al., 2011). These results reveal that regeneration in these contexts does not require dedifferentiation to a pluripotent state, and that the cells that make up the blastema are lineage restricted. These lineage-restricted progenitors even occupy distinct spatial domains in the blastema, indicating that the initial sorting and positioning of these cells may be crucial for producing a properly patterned appendage (Kragl et al., 2009; Tu and Johnson, 2011).
Although the lineage restrictions observed during limb regeneration (Kragl, et al., 2009) are similar to those observed during limb development (Pearse et al., 2007), there are dramatic differences between development and regeneration. In addition to nerve dependence (see next section) and differences in scale of the structures being formed, limb regeneration differs significantly from limb development in that some components of the proximo-distal axis of the limb are retained after amputation. To prevent duplication or deletion of limb structures, cells at the site of amputation must be able to determine their position along the proximo-distal axis and use this information to regenerate only the structures that have been lost. Interestingly, when distal (wrist) blastemas are transplanted onto proximal (shoulder) blastemas, the distal blastema cells contribute only to distal structures, suggesting that they retain memory of their proximo-distal origin (Fig. 4 B; Echeverri and Tanaka, 2005). Additionally, when proximal and distal blastemas are co-cultured, the proximal blastema encapsulates the distal blastema, suggesting that the adhesive properties of cells differ along the proximo-distal axis (Nardi and Stocum, 1984; da Silva et al., 2002). Lineage tracing and examination of proximal (e.g., nuclear localized MEIS) and distal limb markers (e.g., hoxA13 expression) indicate that although some cells (such as cartilage) retain positional identity, other cells (such as Schwann cells) do not (Kragl et al., 2009). Because of the important role positional identity plays in limb regeneration, it is critical to understand the sources that provide positional values along the limb axes.
Based on its differential expression along the proximo-distal axis, da Silva et al. (2002) identified a cell surface protein, referred to as Prod1, that is expressed at high levels proximally and low levels distally. Treatment with anti-Prod1 antibodies blocked the encapsulation of distal blastema by proximal blastema tissues, suggesting that Prod1 plays a role in cell–cell interactions that mediate identity along the proximo-distal axis. Furthermore, when distal blastema cells overexpressing Prod1 were transplanted into proximal blastemas, they contributed to proximal rather than distal structures (Echeverri and Tanaka, 2005). The mechanisms by which differences in Prod1 expression along the proximo-distal axis translate into faithful regeneration of the limb are not completely clear; however, Prod1 has been found to interact with a secreted protein called newt Anterior Gradient (nAG; see next section), providing a link between proximo-distal patterning and the role of innervation in regeneration (Kumar et al., 2007a).
“Anything’s possible if you’ve got enough nerve”: Nerve dependence in regeneration
The above, out-of-context quote (Rowling, 2003) introduces the important role that innervation plays during regeneration in many organisms. For example, earthworms in which a portion of the ventral nerve cord was removed near the amputation plane failed to regenerate new heads, whereas manipulations resulting in two anteriorly exposed nerve cords produced two heads (Morgan, 1901). In another annelid, the polychaete Spirographis spallanzani, deviation of the nerve cord toward the lateral body wall induced the formation of supernumerary structures: the anterior-directed cut nerve cord led to the formation of an ectopic head, whereas the posterior-directed cut nerve cord led to the production of an ectopic tail (Kiortsis and Moraitou, 1965). The nervous system also appears to be required for arm regeneration in Echinoderms because removal of the radial nerve in the starfish prevents arm regeneration (Huet, 1975). In planarians, the nervous system is required for proper regeneration of the gonads and accessory reproductive organs. Collins et al. (2010) identified a single neuropeptide that mediates the effects of the nervous system on the postembryonic development and maintenance of the reproductive organs. Furthermore, the formation of ectopic cephalic outgrowths in planarians with improperly connected cephalic ganglia and ventral nerve cords suggests that discontinuities between the brain and nerve cords result in the production of signals that trigger proliferative outgrowth (Cebrià and Newmark, 2007).
The role of innervation is best understood in the context of amphibian limb regeneration. Classical experiments by Singer showed that denervated limbs failed to regenerate, and that the amount of innervation was critical for proper regeneration (Carlson, 2007). Kumar et al. (2007b) identified the nAG protein as a ligand for Prod1, the cell surface protein involved in proximo-distal patterning. During regeneration nAG is expressed in the Schwann cells of the nerve sheath and, later, in gland cells of the wound epidermis. Expression in both cell types is dependent on proper innervation. Remarkably, introduction of a vector encoding nAG into denervated limbs was able to rescue regeneration to the digit stage, thus restoring proximo-distal patterning. Experiments in which nAG was added to blastemal cells in culture revealed that nAG exerts its effects upon regeneration by stimulating blastemal cell proliferation. The mechanism by which nAG stimulates blastema cell proliferation remains an open question. However, when Prod1 is expressed in cultured newt blastema cells, it leads to the activation of MMP9 expression and ERK signaling; it also interacts with the epidermal growth factor receptor (Blassberg et al., 2011). It will be important to determine whether nAG plays a role in mediating these activities of Prod1.
Given the wide distribution of regenerative abilities throughout the animal kingdom, it has been postulated that some regenerative mechanisms have been conserved throughout evolution (Sánchez Alvarado and Tsonis, 2006; Brockes and Kumar, 2008; Bely, 2010; Poss, 2010). According to this view, lack of regenerative abilities reflects evolutionary loss. For good reasons, most investigators have focused their studies on broadly conserved developmental regulators. However, Prod1 provides an intriguing example of a molecule that has apparently evolved within a specific evolutionary lineage, the Urodele amphibians (Garza-Garcia et al., 2010). Thus, it will also be important for investigators to consider the roles of such taxon-specific genes in regeneration in diverse organisms. Ultimately, we need to know a great deal more about the cellular and molecular mechanisms operating during various regenerative processes to better understand the distribution of regenerative abilities throughout the metazoa.
Concluding thoughts
As discussed in this Review, investigations of model organisms have begun to characterize the cellular sources of regeneration, define their potency, and identify molecules required for restorative events. Thus, there is now a great opportunity for cell biological studies to link gene function to cellular behavior. For example, the cell biology of dedifferentiation is poorly understood: How is the complex cytoskeleton of cardiomyocytes or skeletal muscles reconfigured during the course of dedifferentiation to enable assembly of a mitotic spindle? How are mononucleate cells produced by syncytial myotubes? In addition, we need to understand how individual cell behaviors are integrated across tissues to permit a coordinated response to tissue loss. For example, the observation that tissues from proximal limb blastema encapsulate those from distal limb blastema suggests that cell–cell interactions are critical for proper sorting and organization within the blastema (Nardi and Stocum, 1984; da Silva et al., 2002). The nature of such differential adhesion remains to be determined in regenerating tissues. Similarly, it will be important to characterize how the extracellular environment is altered during regeneration, and how different extracellular matrices shape cell behaviors in the blastema (Calve et al., 2010). One of the long-term goals of regeneration research is to understand why humans have such limited regenerative potential and what, if anything, can be done to improve it. The knowledge gained from studying cell biological questions in model organisms should help drive such future efforts of regenerative medicine.
Acknowledgments
We thank Jim Collins, David Forsthoefel, Rachel Roberts-Galbraith, and the anonymous reviewers for helpful comments on the manuscript, as well as Ana Vieira for providing the electron micrograph for Figure 2. We extend our sincere apologies to the many colleagues whose work we were unable to cite due to space limitations.
Research in P.A. Newmark’s laboratory is supported by the National Institutes of Health (R01-HD043403) and the National Science Foundation (IOS-0744689). P.A. Newmark is an investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- CREB
- cAMP response element–binding protein
- nAG
- newt anterior gradient
- RSK
- ribosomal S6 kinase
References
- Baguñà J., Saló E., Auladell C. 1989. Regeneration and pattern formation in planarians III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 107:77–86 [Google Scholar]
- Bely A.E. 2010. Evolutionary loss of animal regeneration: pattern and process. Integr. Comp. Biol. 50:515–527 10.1093/icb/icq118 [DOI] [PubMed] [Google Scholar]
- Bergantiños C., Corominas M., Serras F. 2010. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development. 137:1169–1179 10.1242/dev.045559 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Steller H. 2010. Apoptosis, stem cells, and tissue regeneration. Sci. Signal. 3:re8 10.1126/scisignal.3145re8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg R.A., Garza-Garcia A., Janmohamed A., Gates P.B., Brockes J.P. 2011. Functional convergence of signalling by GPI-anchored and anchorless forms of a salamander protein implicated in limb regeneration. J. Cell Sci. 124:47–56 10.1242/jcs.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes J.P., Kumar A. 2008. Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 24:525–549 10.1146/annurev.cellbio.24.110707.175336 [DOI] [PubMed] [Google Scholar]
- Calve S., Odelberg S.J., Simon H.G. 2010. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 344:259–271 10.1016/j.ydbio.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B.M. 2007. Principles of Regenerative Biology. Academic Press, Burlington, MA: 400 pp [Google Scholar]
- Cebrià F., Newmark P.A. 2007. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development. 134:833–837 10.1242/dev.02794 [DOI] [PubMed] [Google Scholar]
- Chera S., Ghila L., Dobretz K., Wenger Y., Bauer C., Buzgariu W., Martinou J.C., Galliot B. 2009. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell. 17:279–289 10.1016/j.devcel.2009.07.014 [DOI] [PubMed] [Google Scholar]
- Chera S., Ghila L., Wenger Y., Galliot B. 2011. Injury-induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration. Dev. Growth Differ. 53:186–201 10.1111/j.1440-169X.2011.01250.x [DOI] [PubMed] [Google Scholar]
- Collins J.J., III, Hou X., Romanova E.V., Lambrus B.G., Miller C.M., Saberi A., Sweedler J.V., Newmark P.A. 2010. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 8:e1000509 10.1371/journal.pbio.1000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings S.G., Bode H.R. 1984. Head regeneration and polarity reversal in Hydra attenuata can occur in the absence of DNA synthesis. Rouxs Arch. Dev. Biol. 194:79–86 10.1007/BF00848347 [DOI] [PubMed] [Google Scholar]
- da Silva S.M., Gates P.B., Brockes J.P. 2002. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev. Cell. 3:547–555 10.1016/S1534-5807(02)00288-5 [DOI] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E.M. 2005. Proximodistal patterning during limb regeneration. Dev. Biol. 279:391–401 10.1016/j.ydbio.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Echeverri K., Clarke J.D., Tanaka E.M. 2001. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol. 236:151–164 10.1006/dbio.2001.0312 [DOI] [PubMed] [Google Scholar]
- Eisenhoffer G.T., Kang H., Sánchez Alvarado A. 2008. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 3:327–339 10.1016/j.stem.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A. 2008. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol. 18:467–473 10.1016/j.tcb.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B., Miljkovic-Licina M., de Rosa R., Chera S. 2006. Hydra, a niche for cell and developmental plasticity. Semin. Cell Dev. Biol. 17:492–502 10.1016/j.semcdb.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Gargioli C., Slack J.M. 2004. Cell lineage tracing during Xenopus tail regeneration. Development. 131:2669–2679 10.1242/dev.01155 [DOI] [PubMed] [Google Scholar]
- Garza-Garcia A.A., Driscoll P.C., Brockes J.P. 2010. Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integr. Comp. Biol. 50:528–535 10.1093/icb/icq022 [DOI] [PubMed] [Google Scholar]
- Gierer A., Berking S., Bode H., David C.N., Flick K., Hansmann G., Schaller H., Trenkner E. 1972. Regeneration of hydra from reaggregated cells. Nat. New Biol. 239:98–101 10.1038/239098a0 [DOI] [PubMed] [Google Scholar]
- Gu Y., Forostyan T., Sabbadini R., Rosenblatt J. 2011. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J. Cell Biol. 193:667–676 10.1083/jcb.201010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Peters A.H., Newmark P.A. 2006. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 11:159–169 10.1016/j.devcel.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Gurley K.A., Elliott S.A., Simakov O., Schmidt H.A., Holstein T.W., Sánchez Alvarado A. 2010. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347:24–39 10.1016/j.ydbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. 2008. Wound repair and regeneration. Nature. 453:314–321 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Shibata N., Okumura R., Kudome T., Nishimura O., Tarui H., Agata K. 2010. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev. Growth Differ. 52:131–144 10.1111/j.1440-169X.2009.01157.x [DOI] [PubMed] [Google Scholar]
- Haynie J.L., Bryant P.J. 1977. The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 183:85–100 10.1007/BF00848779 [DOI] [PubMed] [Google Scholar]
- Henry J.J., Tsonis P.A. 2010. Molecular and cellular aspects of amphibian lens regeneration. Prog. Retin. Eye Res. 29:543–555 10.1016/j.preteyeres.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet M. 1975. [Role of the nervous system during the regeneration of an arm in a starfish: Asterina gibbosa Penn. (Echinodermata, Asteriidae)]. J. Embryol. Exp. Morphol. 33:535–552 [PubMed] [Google Scholar]
- Hwang J.S., Kobayashi C., Agata K., Ikeo K., Gojobori T. 2004. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene. 333:15–25 10.1016/j.gene.2004.02.034 [DOI] [PubMed] [Google Scholar]
- Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. 2009. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 137:1343–1355 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J.C. 2010. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 464:606–609 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K., Anton-Erxleben F., Milde S., Plötz C., Wittlieb J., Hemmrich G., Bosch T.C. 2007. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev. Biol. 309:32–44 10.1016/j.ydbio.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Werdich A.A., Anderson R.M., Fang Y., Egnaczyk G.F., Evans T., Macrae C.A., Stainier D.Y., Poss K.D. 2010. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 464:601–605 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiortsis V., Moraitou M. 1965. Factors of regeneration in Spirographis spallanzanii. Regeneration in Animals and Related Problems. Kiortsis V., Trampusch H.A.L., North Holland, Amsterdam: 250–261 [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C.W., Mahatma G., Fisher S., Brand M., Schulte-Merker S., Weidinger G. 2011. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell. 20:713–724 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H.H., Tanaka E.M. 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 460:60–65 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kumar A., Velloso C.P., Imokawa Y., Brockes J.P. 2004. The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol. 2:E218 10.1371/journal.pbio.0020218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Gates P.B., Brockes J.P. 2007a. Positional identity of adult stem cells in salamander limb regeneration. C. R. Biol. 330:485–490 10.1016/j.crvi.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J.W., Gates P.B., Garza-Garcia A.A., Brockes J.P. 2007b. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 318:772–777 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E., Miura M. 2007. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 17:135–144 10.1016/j.tcb.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Lenhoff S.G., Lenhoff H.M. 1986. Hydra and the Birth of Experimental Biology: Abraham Trembley’s Memoirs Concerning the Natural History of a Type of Freshwater Polyp with Arms Shaped like Horns. The Boxwood Press, Pacific Grove, CA: 192 pp [Google Scholar]
- Levin M. 2009. Bioelectric mechanisms in regeneration: Unique aspects and future perspectives. Semin. Cell Dev. Biol. 20:543–556 10.1016/j.semcdb.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Huang Q., Chen J., Peng Y., Roop D.R., Bedford J.S., Li C.Y. 2010. Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci. Signal. 3:ra13 10.1126/scisignal.2000634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo D.C., Allen F., Brockes J.P. 1993. Reversal of muscle differentiation during urodele limb regeneration. Proc. Natl. Acad. Sci. USA. 90:7230–7234 10.1073/pnas.90.15.7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G., Shlevkov E., Pérez-Garijo A. 2011. Mitogenic signaling from apoptotic cells in Drosophila. Dev. Growth Differ. 53:168–176 10.1111/j.1440-169X.2010.01225.x [DOI] [PubMed] [Google Scholar]
- Morgan T.H. 1901. Regeneration and Liability to Injury. Science. 14:235–248 10.1126/science.14.346.235 [DOI] [PubMed] [Google Scholar]
- Morrison J.I., Lööf S., He P., Simon A. 2006. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J. Cell Biol. 172:433–440 10.1083/jcb.200509011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J.I., Borg P., Simon A. 2010. Plasticity and recovery of skeletal muscle satellite cells during limb regeneration. FASEB J. 24:750–756 10.1096/fj.09-134825 [DOI] [PubMed] [Google Scholar]
- Nardi J.B., Stocum D.L. 1984. Surface properties of regenerating limb cells: Evidence for gradation along the proximodistal axis. Differentiation. 25:27–31 10.1111/j.1432-0436.1984.tb01334.x [DOI] [Google Scholar]
- Newmark P.A., Sánchez Alvarado A. 2000. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220:142–153 10.1006/dbio.2000.9645 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Inoue T., Yoshimoto K., Taniguchi T., Kitamura Y., Agata K. 2011. Regeneration of dopaminergic neurons after 6-hydroxydopamine-induced lesion in planarian brain. J. Neurochem. 119:1217–1231 10.1111/j.1471-4159.2011.07518.x [DOI] [PubMed] [Google Scholar]
- Noda K. 1971. Reconstitution of dissociated cells of Hydra. Zool Mag. 80:27–31 [Google Scholar]
- Orii H., Sakurai T., Watanabe K. 2005. Distribution of the stem cells (neoblasts) in the planarian Dugesia japonica. Dev. Genes Evol. 215:143–157 10.1007/s00427-004-0460-y [DOI] [PubMed] [Google Scholar]
- Pearse R.V., II, Scherz P.J., Campbell J.K., Tabin C.J. 2007. A cellular lineage analysis of the chick limb bud. Dev. Biol. 310:388–400 10.1016/j.ydbio.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Fitzgerald P., Watanabe S., Mancuso J., Green D.R., Sánchez Alvarado A. 2010. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338:76–85 10.1016/j.ydbio.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A., Shlevkov E., Morata G. 2009. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development. 136:1169–1177 10.1242/dev.034017 [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Reddien P.W. 2009. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. USA. 106:17061–17066 10.1073/pnas.0906823106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C.P., Reddien P.W. 2011. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 332:852–855 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D. 2010. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 11:710–722 10.1038/nrg2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P.W., Sánchez Alvarado A. 2004. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20:725–757 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sánchez Alvarado A. 2005. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 310:1327–1330 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Riehle K.J., Dan Y.Y., Campbell J.S., Fausto N. 2011. New concepts in liver regeneration. J. Gastroenterol. Hepatol. 26(Suppl 1):203–212 10.1111/j.1440-1746.2010.06539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O., Agata K. 2010. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341:429–443 10.1016/j.ydbio.2010.02.037 [DOI] [PubMed] [Google Scholar]
- Rowling J.K. 2003. Harry Potter and the Order of the Phoenix. Scholastic Press, NY: 896 pp [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M.T., Evangelista M., Gremigni V. 2005. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 132:1863–1874 10.1242/dev.01785 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Tsonis P.A. 2006. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 7:873–884 10.1038/nrg1923 [DOI] [PubMed] [Google Scholar]
- Schubiger M., Sustar A., Schubiger G. 2010. Regeneration and transdetermination: the role of wingless and its regulation. Dev. Biol. 347:315–324 10.1016/j.ydbio.2010.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M.L., Srivastava M., Bell G.W., Reddien P.W. 2011. A regulatory program for excretory system regeneration in planarians. Development. 138:4387–4398 10.1242/dev.068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengel C. 1960. Culture in vitro de blastèmes de régénération de Planaires. J. Embryol. Exp. Morphol. 8:468–476 [PubMed] [Google Scholar]
- Siebert S., Anton-Erxleben F., Bosch T.C. 2008. Cell type complexity in the basal metazoan Hydra is maintained by both stem cell based mechanisms and transdifferentiation. Dev. Biol. 313:13–24 10.1016/j.ydbio.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Slack J.M., Beck C.W., Gargioli C., Christen B. 2004. Cellular and molecular mechanisms of regeneration in Xenopus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:745–751 10.1098/rstb.2004.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bolton R.K., Worley M.I., Kanda H., Hariharan I.K. 2009. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell. 16:797–809 10.1016/j.devcel.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng A.S., Adams D.S., Qiu D., Koustubhan P., Levin M. 2007. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev. Biol. 301:62–69 10.1016/j.ydbio.2006.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S., Johnson S.L. 2010. Clonal analyses reveal roles of organ founding stem cells, melanocyte stem cells and melanoblasts in establishment, growth and regeneration of the adult zebrafish fin. Development. 137:3931–3939 10.1242/dev.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S., Johnson S.L. 2011. Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell. 20:725–732 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega Y.A., Okano H., Lozano G. 2008. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 15:1772–1781 10.1038/cdd.2008.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaskalin T., Wong C.J., Tsilfidis C. 2004. Growth and apoptosis during larval forelimb development and adult forelimb regeneration in the newt (Notophthalmus viridescens). Dev. Genes Evol. 214:423–431 10.1007/s00427-004-0417-1 [DOI] [PubMed] [Google Scholar]
- Wagner D.E., Wang I.E., Reddien P.W. 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 332:811–816 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Reddien P.W. 2010. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344:979–991 10.1016/j.ydbio.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittlieb J., Khalturin K., Lohmann J.U., Anton-Erxleben F., Bosch T.C. 2006. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA. 103:6208–6211 10.1073/pnas.0510163103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Kashikawa M., Shibata N., Takechi K., Agata K. 2007. DjCBC-1, a conserved DEAD box RNA helicase of the RCK/p54/Me31B family, is a component of RNA-protein complexes in planarian stem cells and neurons. Dev. Dyn. 236:3436–3450 10.1002/dvdy.21375 [DOI] [PubMed] [Google Scholar]