Abstract
Here we demonstrate that aptamers tethered to gold nanoparticles enable direct visualization of protein-oligonucleotide interactions during gel electrophoresis. This technique is used to confirm that an aptamer previously identified as binding to C-reactive protein (CRP) only binds to the monomeric form of CRP. While native, pentameric CRP (pCRP) is used in clinical assays to predict cardiovascular disease (CVD) risk, it is the monomeric isoform that is more strongly associated with pro-inflammatory and pro-atherogenic effects. To visualize this selectivity, the CRP-aptamer was conjugated to streptavidin-coated gold nanoparticles and the mobility of the free oligonucleotide-nanoparticle conjugate (ON-NP) and the protein/ON-NP complex bands were visualized and recorded during electrophoresis using a simple digital camera. At a concentration of 6 mg/mL, monomeric CRP showed a significant decrease in the observed ON-NP mobility, whereas no change in mobility was observed with pCRP up to 18 mg/mL. Advantages of this nanoparticle-based electrophoretic mobility shift assay (NP-EMSA) over traditional EMSA include real-time detection of protein-oligonucleotide interactions, the avoidance of harmful radioisotopes, and elimination of the need for expensive gel imagers. The availability of both the NP-EMSA technique and an mCRP specific probe will allow for improved clinical diagnostic to more accurately predict future CVD risk.
Keywords: Aptamer, C-reactive protein, EMSA, Nanoparticle, Real-time detection
C-reactive protein (CRP) is an inflammatory marker that is currently used in clinical assays to predict the risk for cardiovascular disease (CVD) [1, 2]. Native CRP circulates as a pentamer (pCRP) but undergoes an in situ conformational change upon binding to damaged cell membranes including low-density lipoproteins that are oxidized or at acidic pH [3, 4], apoptotic blebs [5], and platelet-derived microparticles [6]. The resulting isoform of CRP, termed modified CRP, expresses neoepitopes [2, 7] and has been shown to elicit pro-inflammatory and atherogenic effects [8–10]. Because commercially available antibodies recognize both isoforms, while the modified CRP isoform may be more predictive of CVD, there is a critical need for assays that can distinguish between the isoforms.
Aptamers are single-stranded oligonucleotides (ONs) selected against molecular targets from a random pool of ssDNA or RNA through an in vitro screening process called systemic evolution of ligands by exponential enrichment (SELEX) [11, 12]. ONs have several advantages over antibodies including high thermal stability, ease of synthesis, and opportunities for site-specific modification and labeling [13, 14], which makes them a useful tool for sensor design. Aptamers against several cardiovascular biomarkers, including thrombin [15–17] and CRP [18–20], have been selected and used as sensors for these proteins. Here, we describe an aptamer-based biosensor and show the aptamer’s specificity against the monomeric CRP isoform in contrast to previous reports that described it recognizing pCRP [19, 20].
The affinity of an ON to its target protein is commonly determined using electrophoretic mobility shift assays (EMSAs) [21, 22]. In EMSA, the ON is pre-mixed with increasing amounts of the target analyte and then separated by gel electrophoresis. The migration of the ON-target complex is generally slower than that of the unbound ON [21, 22], and the difference in mobility can be detected using radioisotope-labeled probes [22] or by visualization of fluorophore-labeled ONs on a gel imager [23]. EMSA is typically is a multi-step process where a gel is run and then the fluorescent or radioactive label is used to assess mobility of the complex. Additional steps of membrane transfer, probe hybridization, and film development often results in a process that takes hours. In all cases, imagers are required to observe binding.
Here, we describe a novel and direct visualization of ON–protein interactions using a nanoparticle-based EMSA (NP-EMSA) (Scheme 1). In this approach, the affinity of the ON to its target is visualized and monitored in real time during electrophoresis. A video in Supporting Information shows the NP movement during electrophoresis. Furthermore, we used this technique to compare the affinity of the previously isolated CRP-aptamer [18, 20] for the two common isoforms of CRP
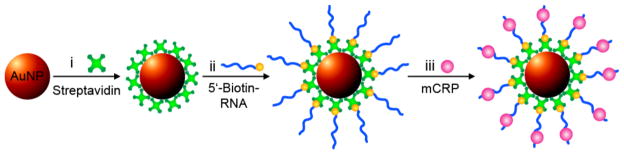
Scheme 1.
Preparation of ON-NP. Citrate-capped gold nanoparticles were incubated with (i) streptavidin followed by (ii) biotinylated aptamer to generate ON-NP. The nanoparticles were centrifuged between each step to remove unbound molecules in the supernatant. Binding of (iii) mCRP to the ON-NP was visually detected by EMSA.
Gold nanoparticles (AuNPs) (~18 nm) were prepared and characterized by dynamic light scattering (DLS), atomic force microscopy (AFM), and UV/Vis spectroscopy as described in Supporting Information. The nanoparticles were coated with streptavidin and biotinylated aptamer and purified (Scheme 1). The optimal amount of streptavidin and aptamer needed for full coating was calculated and confirmed using a flocculation assay described in Supporting Information.
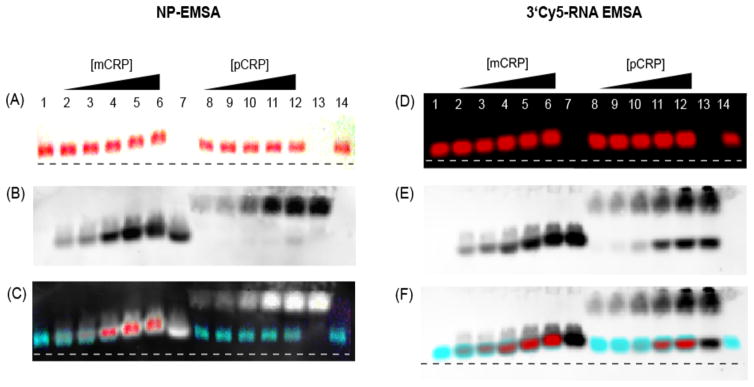
An increasing concentration (1.2–18 μg/mL) of either monomeric CRP (mCRP) or pCRP was added to the ON-conjugated AuNP (ON-NP) and incubated at room temperature for 30 min before electrophoresis. mCRP preparation and characterization is described in Supporting Information. The protein and ON-NP mixtures were loaded on a 0.8%, 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM ethylenediaminetetraacetic acid) gel and electrophoresis was carried out at 45 V for 30 min. Photographs of the gel were taken at 1-min interval using a digital camera (e.g. Fig. 1A), and the CRP was transblotted at 25 V for 90 min onto a nitrocellulose membrane after electrophoresis. CRP was detected on the nitrocellulose using a polyclonal anti-CRP antibody (Fig. 1B). As evident from the photograph (Fig. 1A) and the NP-EMSA movie (Supporting Information), the mobility of the ON-NP decreased with increasing concentrations of mCRP (lanes 2–6), indicating a complex formation between the ON-NP and mCRP. A conventional EMSA using a 3′Cy5-labeled aptamer (3′Cy5-RNA) was carried out in parallel and the 3′Cy5-RNA was detected using near-infrared (NIR) imaging on an Odyssey imager (Fig. 1D). The mobility of the 3′Cy5-RNA showed a similar trend as the NP-EMSA, with the 3′Cy5-RNA bands shifted upwards with increasing concentrations of mCRP (lanes 2–6). To confirm that the shift in ON-NP and 3′Cy5-RNA mobility was due to the formation of an ON-NP/mCRP complex, Western blots were performed (Fig. 1B and 1E) and CRP was detected using a polyclonal anti-CRP antibody. The mobility of mCRP by Western blot showed a similar upward shift with increasing amounts of mCRP in both gels (lanes 2–6), and the superimposed image of the gel and Western blot showed that the aptamers and mCRP bands overlapped in both EMSA formats (Fig. 1C and 1F).
Figure 1.
NP-EMSA and 3′Cy5-EMSA. Photographs of (A) NP-EMSA and (D) 3′Cy5-EMSA gel after electrophoresis. Western blots of the (B) NP-EMSA gel and (E) 3′Cy5-EMSA gel. Superimposed images of (C) NP-EMSA and (F) 3′Cy5-EMSA gel with their respective Western blots showing the overlapping regions of aptamer and mCRP. Lanes: (1) ON-NP or 3′Cy5-RNA, (2–6) ON-NP or 3′Cy5-RNA incubated with 0.12, 0.24, 0.60, 1.2 and 1.8 mg/mL mCRP, (7) 1.8 mg/mL mCRP, (8–12) ON-NP or 3′Cy5-RNA incubated with 0.12, 0.24, 0.6, 1.2 and 1.8 mg/mL pCRP, (13) 1.8 mg/mL pCRP, and (14) ON-NP or 3′Cy5-RNA incubated with 1.8 mg/mL BSA.
Using both EMSA formats, it was clear that the CRP aptamer has higher affinity for mCRP than pCRP. To determine whether the CRP aptamer bound exclusively to mCRP, a second series of EMSA experiments were carried out by incubating an increasing amount of pCRP with the ON-NP. No shift in mobility was observed using the same concentrations of pCRP (lanes 8–12) or with the BSA control (lane 14) (Fig. 1A and 1D), suggesting there was no affinity of the aptamer for pCRP or BSA. Western blots also showed that pCRP (lanes 8–12) did not co-migrate with the aptamer (Fig. 1B and 1E) and no overlapping bands of aptamer and pCRP were seen on the superimposed images (Fig. 1C and 1F), indicating that no aptamer–pCRP complex was formed. It should be noted, however, that a significant amount of mCRP was present in the 3′Cy5-RNA EMSA (Fig. 1E, arrow) compared with the NP-EMSA (Fig. 1B) due to the use of an older CRP stock. The presence of mCRP in the sample was not surprising because dissociation of pCRP has been reported to occur during storage [3]. Nevertheless, this demonstrated that the aptamer can distinguish mCRP from pCRP in a mixed sample (Fig. 1F). We have confirmed the specificity of the aptamer for mCRP using fluorescence anisotropy, dot blot assay, PAGE EMSA, and fluorescence microscopy [24].
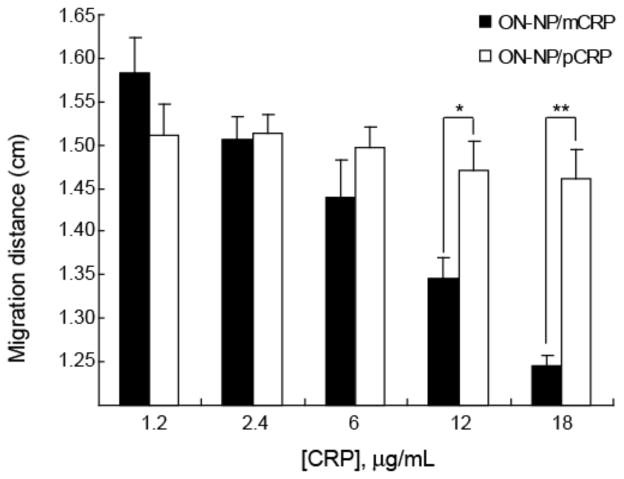
A quantitative assessment of the mobility of ON-NP/CRP complexes was performed to determine the difference and significance in mobility shifts and to quantify the limit of detection of the NP-EMSA technique. The migration distance (after 30 min of electrophoresis) was determined using Image J (http://imagej.nih.gov/) by measuring the distance from the center of the wells to the center of the bands for each well (Fig. 2). As indicated by the final migration distance (Fig. 2), the mobility of the ON-NP/mCRP at higher concentrations (12 and 18 mg/mL) was significantly slower compared to the mobility of ON-NP/pCRP. By comparing the final migration distance between the uncomplexed ON-NP (lane 1) and the ON-NP/mCRP at each concentration, a significant difference (p < 0.01) was observed at 18 μg/mL of mCRP (lane 4). The time-course migration profile of the ON-NP and ON-NP/CRP complex was then determined from each photograph of the gel taken at 1-min intervals as shown in Supporting Information. Unlike traditional EMSA, NP-EMSA allows for a real-time migration analysis to be performed from a single gel during electrophoresis.
Figure 2.
Migration of ON-NP/CRP complex. ON-NP/CRP complex were separated by electrophoresis at 45V for 30 min. The migration distances of ON-NP/mCRP (black) and ON-NP/pCRP (white) complex were measured from the center of the well to middle of the band at the end of the 30 min electrophoresis. Data was reported as the mean distance ± SD. n = 3; *, p<0.05; **, p<0.01. (B) Migration rates of ON-NP with increasing concentrations of mCRP and pCRP.
Because of the high extinction coefficient of AuNPs, they have been widely used in real-time colorimetric biosensing applications [25–28]. Colorimetric sensors are attractive because they do not require expensive instruments. DNA-functionalized AuNP probes were among the first colorimetric assays to take advantage of the plasmon shift of AuNP aggregation from red to blue in the presence a target analyte [25, 27]. While this application utilizes simple spheres, nanoparticles of various sizes, shapes, and surface conjugation can be separated on a gel [29–33]. The mobility of NPs in gel electrophoresis is influenced by size and charge [29, 32]. As demonstrated by Hanauer, et. al., spherical AuNPs have size-dependent mobility, and the aspect ratio of gold nanorods influences their separation [29]. Xu, et. al. also demonstrated the separation and purification of a mixed sample of AuNPs (spheres and rods) based on size and shape using a preparative agarose gel column [30].
This is, to our knowledge, the first application of NPs in visualizing EMSAs. This NP-EMSA technique is convenient, fast, and cost-effective and it does not require blotting, the use of radioisotopes, or a gel imager. We expect that this approach could be adapted to other aptamers and for the use of a mixture of aptamers or different types of nanoparticles with distinct optical properties. In addition, we have shown that this aptamer binds specifically to mCRP and not pCRP as had been previously described [19, 20]. Since mCRP is shown to possess pro-inflammatory and pro-atherogenic effects [8–10], the availability of an mCRP sensor will have significant value in clinical assays to more accurately predict risk of CVD.
Supplementary Material
Acknowledgments
The authors wish to acknowledge support from the NIH 1R15GM088960-01 (SMR), NSF CBET-1033161 (SMR), and AHA 10POST4070019 (MSW). They thank Dr. Marilyn Mackiewicz for proof-reading the manuscript and technical discussion, the UCD Biology department for use of the Odyssey imager and the University of Denver, Department of Physics for use of the AFM.
Abbreviations
- AuNP
gold nanoparticle
- CRP
C-reactive protein
- CVD
cardiovascular disease
- mCRP
monomeric CRP′
- NP-EMSA
nanoparticle-based electrophoresis;mobility shift assay
- ON
oligonucleotide
- pCRP
pentameric CRP
Footnotes
The authors have declared no conflict of interest.
References
- 1.Black S, Kushner I, Samols D. J Biol Chem. 2004;279:48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- 2.Ji SR, Wu Y, Zhu L, Potempa LA, Sheng FL, Lu W, Zhao J. FASEB J. 2007;21:284–294. doi: 10.1096/fj.06-6722com. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Suresh MV, Hammond DJ, Rusinol AE, Potempa LA, Agrawal A. Clin Chim Acta. 2009;406:151–155. doi: 10.1016/j.cca.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang MK, Binder CJ, Torzewski M, Witztum JL. Proc Natl Acad Sci USA. 2002;99:13043–13048. doi: 10.1073/pnas.192399699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber J, Vales A, Mitulovic G, Blumer M, Schmid R, Witztum JL, Binder BR, Leitinger N. Arterioscl Thromb Vasc Biol. 2002;22:101–107. doi: 10.1161/hq0102.101525. [DOI] [PubMed] [Google Scholar]
- 6.van der Zee PM, Biro E, Trouw LA, Ko Y, de Winter RJ, Hack CE, Sturk A, Nieuwland R. Clin Immunol. 2010;135:490–495. doi: 10.1016/j.clim.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Potempa LA, Siegel JN, Fiedel BA, Potempa RT, Gewurz H. Mol Immunol. 1987;24:531–541. doi: 10.1016/0161-5890(87)90028-9. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhardt SU, Thiele JR, Bannasch H, Stark GB, Peter K. Cell Cycle. 2009;8:3885–3892. doi: 10.4161/cc.8.23.10068. [DOI] [PubMed] [Google Scholar]
- 9.Eisenhardt SU, Habersberger J, Murphy A, Chen YC, Woollard KJ, Bassler N, Qian HW, von zur Muhlen C, Hagemeyer CE, Ahrens I, Chin-Dusting J, Bobik A, Peter K. Circ Res. 2009;105:128–U155. doi: 10.1161/CIRCRESAHA.108.190611. [DOI] [PubMed] [Google Scholar]
- 10.Khreiss T, Jozsef L, Potempa LA, Filep JG. Circulation. 2004;109:2016–2022. doi: 10.1161/01.CIR.0000125527.41598.68. [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 13.Juskowiak B. Anal Chim Acta. 2006;568:171–180. doi: 10.1016/j.aca.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 14.Wang KM, Li W, Tan WH, Ma CB, Yang XH. Analyst. 2007;132:107–113. doi: 10.1039/b614138b. [DOI] [PubMed] [Google Scholar]
- 15.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem Int Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 17.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 18.Kim SD, Ryu JS, Yi H-K, Kim S-C, Zhang B-T. Preliminary Proceedings of the Tenth International Meeting on DNA Computing (DNA10); 2004. pp. 334–343. [Google Scholar]
- 19.Huang CJ, Lin HI, Shiesh SC, Lee GB. Biosens Bioelectron. 2010;25:1761–1766. doi: 10.1016/j.bios.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 20.Bini A, Centi S, Tombelli S, Minunni M, Mascini M. Anal Bioanal Chem. 2008;390:1077–1086. doi: 10.1007/s00216-007-1736-7. [DOI] [PubMed] [Google Scholar]
- 21.Ryder SP, Recht MI, Williamson JR. In: RNA-Protein Interaction Protocols. Lin R-J, editor. Humana Press Inc; Totowa: 2008. pp. 99–115. [Google Scholar]
- 22.Fried MG, Hellman LM. Nat Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdanov A, Zhang S, Metelev V, Tabatadze D, Zamecnik PC. Proc Natl Acad Sci USA. 2008;105:4156–4161. doi: 10.1073/pnas.0800162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang MS, Black JC, Knowles MK, Reed SM. Anal Bioanal Chem. 2011 doi: 10.1007/s00216-011-5174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai CS, Yu TB, Chen CT. Chem Comm. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Lu Y. J Am Chem Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Lu Y. Nat Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 28.Zhao WA, Chiuman W, Brook MA, Li YF. Chembiochem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 29.Hanauer M, Pierrat S, Zins I, Lotz A, Sonnichsen C. Nano Lett. 2007;7:2881–2885. doi: 10.1021/nl071615y. [DOI] [PubMed] [Google Scholar]
- 30.Xu XY, Caswell KK, Tucker E, Kabisatpathy S, Brodhacker KL, Scrivens WA. J Chromatogr A. 2007;1167:35–41. doi: 10.1016/j.chroma.2007.07.056. [DOI] [PubMed] [Google Scholar]
- 31.Zanchet D, Micheel CM, Parak WJ, Gerion D, Alivisatos AP. Nano Lett. 2001;1:32–35. [Google Scholar]
- 32.Patra HK, GuhaSarkar D, Dasgupta AK. Anal Chim Acta. 2009;649:128–134. doi: 10.1016/j.aca.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Pellegrino T, Sperling RA, Alivisatos AP, Parak WJ. J Biomed Biotechnol. 2007;26796:9. doi: 10.1155/2007/26796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.