Abstract
A significant number of macromolecular structures solved by electron cryo-microscopy and X-ray crystallography obtain resolutions of 3.5–6Å, at which direct atomistic interpretation is difficult. To address this, we developed pathwalking, a semi-automated protocol to enumerate reasonable Cα models from near-atomic resolution density maps without a structural template or sequence-structure correspondence. Pathwalking uses a novel approach derived from the Traveling Salesman Problem to rapidly generate an ensemble of initial models for individual proteins, which can later be optimized to produce full atomic models. Pathwalking can also be used to validate and identify potential structural ambiguities in models generated from near-atomic resolution density maps. In this work, examples from the EMDB and PDB are used to assess the broad applicability and accuracy of our method. With the growing number of near-atomic resolution density maps from cryo-EM and X-ray crystallography, pathwalking can become an important tool in modeling protein structures.
INTRODUCTION
Macromolecular assemblies are critical for nearly every biological process, and thus extremely important in discovering targets for disease prevention, as well as increasing our knowledge of basic cellular events (Sali et al., 2003; Sali and Kuriyan, 1999). The most common techniques for imaging macromolecular assemblies are X-ray crystallography and electron cryo-microscopy (cryo-EM) (Chiu, Baker and Almo, 2006). While X-ray crystallography is capable of resolving macromolecular assemblies, it is often difficult to obtain well-diffracting crystals and construct atomic models for larger or less stable assemblies. As such, it is typically used to solve the structures of single proteins or small, stable protein complexes. In cryo-EM, a sample does not need to be crystallized; rather, thousands of individual particle images from a solution environment are combined to generate a 3-D density map for very large (200+ kDa) and often transitory complexes (Baumeister and Steven, 2000; Frank, 2002).
Both X-ray crystallography and cryo-EM encounter frequent difficulties in obtaining structures of large assemblies at atomic resolution (better than 3 Å). Nearly one-third of all the macromolecular assemblies (>300 kDa) solved by X-ray crystallography have resolutions worse than 3.5 Å. While cryo-EM has resulted in several near-atomic resolution (3.5–4.7 Å) density maps (Zhou, 2008; Grigorieff and Harrison, 2011; Baker et al., 2010; Hryc et al., 2011), the vast majority of cryo-EM maps have resolutions between 5 and 20 Å. For such cryo-EM maps, fitting atomic models of known components, typically from X-ray crystallography, is a relatively common approach for building models of entire assemblies. However, fitting individual structures may not accurately reflect the structure of the component in the context of the assembly or in solution.
Typically in analyzing macromolecular assemblies, a density map is examined for visible features (Baker et al., 2010). At low resolutions (worse than 10 Å), this may describe the overall size and shape of the assembly and locations of individual components. At subnanometer resolutions, secondary structure elements (SSE) become visible, with α-helices appearing as cylinders and β-sheets appearing as thin surfaces (Baker, Ju and Chiu, 2007; Jiang et al., 2001). At near-atomic resolutions (3.5–4.7 Å), additional features become discernable in a density map such as the pitch of helices, separation of individual strands in β-sheets and even some bulky sidechains (Ludtke et al., 2008; Zhang et al., 2008). However, it is presumed that the polypeptide chain may not be confidently resolved until ~3.5 Å resolution (Blundell and Johnson, 1976), limiting direct model building at non-atomic resolutions.
Despite the ambiguity in intermediate resolution density maps, model building is still possible. In cryo-EM, de novo modeling techniques (Baker et al., 2011) have been used to construct models for a variety of samples at resolutions better than 5 Å (Chen et al., 2011; Liu et al., 2010; Zhang et al., 2010; Cong et al., 2010; Jiang et al., 2008; Ludtke et al., 2008). In these examples, SSEs were used to infer topological information when coupled with a density skeleton (Ju, Baker and Chiu, 2007; Abeysinghe et al., 2008b; Abeysinghe and Ju, 2009).
De novo modeling relies heavily on visual interpretation, clearly defined SSEs in both the sequence and density map and manual structure assignment. Registration of SSEs in the sequence and structure, combined with geometric information, can then be used to anchor an initial protein backbone trace in the density map (Abeysinghe et al., 2008a). Without reliably detectable SSEs, no or possibly wrong correspondences between sequence and structure can be determined. In these cases, without a priori knowledge, accurate models cannot be constructed. As such, the accurate localization of SSEs in the sequence and density is critical for de novo modeling. This type of modeling can be extremely time consuming and is susceptible to human bias; few methods for assessing model quality from non-atomic resolution density maps exist.
In an effort to streamline the model building and validation process, we have created a new set of utilities to automatically enumerate putative configurations of protein structure models in subnanometer resolution density maps. Pathwalking utilizes combinatorial optimization strategies from the Traveling Salesman Problem (TSP) (Lawler, 1985) paradigm to compute possible cyclical paths through the density map using pseudoatoms, free of any sequence or structure constraints. In this work, we present a complete set of tools, methodology and examples of pathwalking. Authentic and simulated density maps from the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) at a broad range of resolutions (3.5–8 Å) illustrate the ability of pathwalking to quickly produce first-approach Cα models.
METHOD: ALGORITHM
Pathwalking is based on a set of novel computational tools that builds upon our de novo modeling approaches at near-atomic resolutions (Baker et al., 2011). It has the unique advantage of being sequence and template “free”, meaning that the primary sequence or a related structural template is not required in the construction of the initial model. This can be advantageous for structure determination in difficult-to-model proteins. Overall, pathwalking can be broken down into several discrete steps (Figure 1, Figure S1). First, a set of nodes (pseudoatoms) is populated within the density map. Next, a set of potential paths through these points is calculated. These represent “first-approach” models, which are “topologically-correct” but not fully stereochemically or density-refined. Finally, a path is refined and the sequence is threaded on to the model.
Figure 1.
The pathwalking protocol. The basic pathwalking protocol is shown. The five basic steps in pathwalking are shown in the box, along with a brief description and/or diagram. Corresponding utilities at each of these stages is shown in green. The overall procedure is shown as a flow-chart. Steps connected with a dotted line are optional and shown in grey. Figure S1 provides an alternative view of the protocol. Details on pseudoatom placement and the TSP-based solver can be found in the supplemental experimental procedures and depicted in Figure S2 and S3, respectively.
Pseudoatom placement
One caveat in pathwalking is that a subunit/domain must be extracted from the entire macromolecular assembly. Several semi-automated tools, such as EMAN2’s e2segment3d.py (Tang et al., 2007) and Segger (Pintilie et al., 2010), are available to segment out the density. In our examples, manual segmentation using UCSF’s Chimera (Pettersen et al., 2004) was performed.
Once segmented, pseudoatoms (Cα atoms) are computationally placed within the density map. The number of pseudoatoms placed corresponds to the number of Cα atoms in the protein, as defined by the primary sequence. Here, we use a k-means clustering routine to identify N number of segments from the density map, where N represents the number of pseudoatoms to be placed. The center of each segment is assigned a pseudoatom. To maintain cluster sizes approximating a residue, the routine is modified to enforce minimum and maximum separation distances (user-tunable parameters).
Alternatively, an undetermined number of pseudoatoms can be placed in a density map at a given threshold based purely on minimum and maximum distance criteria. This does not require the user to specify the number of clusters as in the case of the k-means approach, only minimum and maximum distances criteria. As the number of pseudoatoms is not directly enforced, varying the density threshold and distance parameters may be necessary to achieve the desired number of pseudoatoms.
In lower resolution density maps (5–8 Å), placement of the Cαs can be augmented by identifying secondary structure elements. In this context, pseudoatoms are first placed along detected α-helices. The remaining pseudoatoms can then be placed using either aforementioned approach. Examples of pseudoatom placement can be seen in Figure S2 and further detailed in the Supplemental Experimental Procedures.
Path detection
Next, pseudoatoms must be connected to form a “reasonable” structural model, satisfying a set of polypeptide constraints: every pseudoatom is connected to two other pseudoatoms (except the N- and C-terminus), all pseudoatoms must be included and deviation from the observed 3.8 Å Cα-Cα bond distance must be minimized. Generating the best possible path is a computationally complex NP-hard organizational problem. A naive approach based on exhaustive search of all possible models quickly becomes intractable, as there are (n-1)!/2 possible solutions, where n is the number of amino acids in the polypeptide. While this can be simplified, typical proteins sizes are still far too complex to solve.
Fortunately, this problem is analogous to the Traveling Salesman Problem (TSP), where the goal is to find the shortest cyclic path that visits each node exactly once (Applegate, 2006). This is a foundational problem and many successful algorithms, both heuristic methods and exact-solution optimizations, have been developed. Software implementing these methods is widely available, including Concorde (Applegate, 2006), which uses a cutting-plane method to find exact solutions, and LKH (Helsgaun, 2009), a flexible implementation of the Lin-Kernighan heuristic method to quickly find near-optimal solutions.
In tracing a protein backbone, pseudoatoms are nodes in a complete undirected graph, while the potential connections between nodes are modeled as edges. While the TSP solvers attempt to find the shortest distance between nodes, the distance along a protein backbone trace is not necessarily the minimal path length. Rather, we express the distance between nodes as a weighted deviation from 3.8 Å, the prototypical Cα-Cα distance, and minimize the total path deviation. Specifically, a pairwise matrix of edge weights based for all pseudoatoms is calculated using a weighted distance function: (3.8 Å - distance(i1, i2))^2. This weighted distance matrix can be passed directly to an “off-the-shelf” TSP solver, such as Concorde and LKH. The weighted distance function allows some flexibility in the distance between pseudoatoms in a path, reflecting the uncertainty in pseudoatom placement, while helping to eliminate outliers.
In pathwalking, we utilize unmodified distributions of both the Concorde and LKH solvers, called directly from e2pathwalker.py (Supplemental Experimental Procedures). Both TSP solvers work quickly and produce good initial Cα models. Model construction may result in a number of potential paths, which can be assessed for their structural plausibility (e.g. incorporating known SSEs and stereochemistry sanity checks) and subsequently refined with other programs such as Rosetta (Bradley et al., 2005; DiMaio et al., 2009).
METHOD: IMPLEMENTATION
The pathwalking procedure is implemented in three separate utilities in EMAN2 (Tang et al., 2007), a freely available image processing toolkit for cryo-EM. Each of these tools is written in Python and utilizes EMAN2 dependencies and the aforementioned TSP solvers. SSE detection is optional for pathwalking.
Pseudoatom placement
Placing pseudoatoms for initial model building is accomplished with EMAN2’s e2segment3d.py (Tang et al., 2007), implementing both the k-means and simple distance algorithms. When using the modified k-means clustering routine (Figure S2, cyan spheres), the user defines the number of clusters as the number of pseudoatoms (nseg) to be placed, along with a density threshold (thr) and minimum and maximum separation in Ångstroms (maxsegsize, minsegsep). Based on empirical observations from hundreds of protein structures, a range of 3.5 Å to 4.2 Å covered all Cα-Cα distances. This range was our starting criteria for pseudoatom placement and connectivity (described later). The density threshold corresponds to the value at which the user can begin to resolve density features, such as separation of β-strands, the pitch of an α-helix or large sidechains, while maintaining connectivity. Each instance of pseudoatom placement is unique, and multiple runs of this program with the same parameters may result in similar but non-identical pseudoatom placement.
e2segment3d.py target.mrc --process=segment.kmeans:ampweight=0:nseg=100:thr=1:maxsegsize=4.2:minsegsep=3.5:verbose=1 --pdbout=pa-out.pdb
For the distance-based pseudoatom placement routine (Figure S2, orange spheres), the user does not specify the number of clusters (pseudoatoms), only a density threshold (thr) and minimum and maximum pseudoatom distances in pixels (maxsegsize, minsegsep). As the number of pseudoatoms is not directly enforced, varying the threshold and distance parameters will be necessary to achieve the desired number of pseudoatoms.
e2segment3d.py target.mrc --process=segment.distance:minsegsep=3:maxsegsep=4:thr=0.5:verbose=1 --pdbout=pa-out.pdb
Pseudoatom placement can be carried out on the density map or a density skeleton. In either case, the user is required to visually inspect the pseudoatom placement. In a noisy or poorly segmented density map, spurious pseudoatoms may be placed outside the main protein density. Manual adjustment of pseudoatom positions may be necessary to correct outliers and can be accomplished by moving pseudoatoms with molecular modeling tools such as Chimera, Coot or Gorgon (Pettersen et al., 2004; Emsley et al., 2010; Baker et al., 2011). Low pass filtering will remove some high-resolution features, like side chain densities, and may improve pseudoatom placement in higher resolution maps.
Pathwalking
e2pathwalker.py in EMAN2 calculates paths through the pseudoatoms. This program requires the user to provide a set of pseudoatoms in PDB format. The user may specify options such as minimum and maximum pseudoatom path lengths (dmin, dmax). One or both termini can be given as arguments to the program (start, end). Specifying the termini is potentially useful during model refinement if the termini are close to each other or buried in the core of the protein.
Support is provided for two high-performance TSP solvers (solver): LKH (Helsgaun, 2009), an approximate solver based on a modified Lin-Kernighan heuristic, and Concorde (Applegate, 2006), an exact solver utilizing the cutting-plane method. Both solvers are called as sub-processes and produce the same high quality paths, usually within seconds. With the LKH solver, the ordering of pseudoatoms can be specified (fix) (i.e. the user can enforce pairwise connections between pseudoatoms, such as those in helices).
e2pathwalker.py contains an option to iteratively run the routine (iterations) with a specified amount of Gaussian noise applied to pseudoatom coordinates (noise). This type of perturbation is useful in producing alternate paths. Statistics are generated on the resulting ensemble of models including an “occupancy” for each edge.
e2pathwalker.py pseudoatoms.pdb --solver=lkh --start=1 --end=523 --dmin=3.5 --dmax=4.2 --fix=fixed.txt --noise=0.2 --iterations=100
e2pathwalker.py produces an ordered set of pseudoatoms. An initial path can be refined by making small adjustments to atom placement and enforcing certain connectivities. For assessing model quality, e2pathwalker.py produces a Cα Ramachandran plot (Cα−Cα−Cα vs. Cα−Cα−Cα−Cα) (Kleywegt, 1997) and a table listing bond distances and angles. These measures can be used in combination with visual inspection to identify regions of the model with poor geometry or fit to density.
Sequence assignment
After the pseudoatoms have been ordered, the sequence is threaded onto the pseudoatoms to generate a structural model. e2seq2pdb.py reads a text file containing the primary sequence of the target protein. The sequence is threaded both forward and reverse through the pseudoatoms; correlation with known structural information and/or secondary structure prediction can be used to help determine the correct direction of the sequence assignment. Two structural models are written out as a Cα-only PDB files.
e2seq2pdb.py path.pdb seq.txt model-out.pdb
RESULTS
Initial evaluation of the pathwalking protocol was broken into three phases. First, we examined the use of a TSP solver in finding the correct path through an optimal set of pseudoatoms. Second, we evaluated the ability to place suitable pseudoatoms within a density map. Finally, we assessed the pathwalking protocol to find and evaluate paths through density maps at subnanometer resolution. It should be noted that this procedure and its utilities can be applied to any density map at near-atomic resolution and, in some favorable cases, at subnanometer resolution.
We created two benchmarks from PDB structures to evaluate our tools. The first set contained 737 non-redundant protein structures of various sizes and fold types (single chain, contiguous backbone without gaps from all fold classes). For this benchmark, the Cα atoms represented the pseudoatom positions; density maps were not simulated for these structures. A second benchmark was created from a subset of the first containing a representative structure from each of the 40 unique CATH architectures. For all these structures, simulated density maps were constructed using EMAN’s pdb2mrc program at 5 Å resolution (1 Å/pixel). Furthermore, one structure from each of the four fold classes was simulated at 6, 7 and 8 Å resolution.
Finding paths with a TSP solver
To test the TSP solvers’ capabilities of obtaining the correct path through a set of pseudoatoms, Cαs from each of the 737 structures in the first benchmark were processed with e2pathwalker.py (using LKH and Concorde solvers) enforcing minimum and maximum distances (3.5 Å and 4.2 Å, respectively). An example of the pathwalking results derived from the Cα coordinates of the GroEL X-ray structure (PDB ID: 1SS8) (Chaudhry et al., 2004) is shown in Figure S3A–C. In all 737 cases, both TSP solvers in e2pathwalker.py were able to identify the correct path through the pseudoatoms, though the directionality of the path was undetermined.
Next, we added Gaussian noise to the pseudoatom positions of the first benchmark where the mean of the Gaussian distribution was defined as 3.8 Å. The standard deviation of the function was varied from 0.1 to 1 σ. The correct path was determined in greater than 95% of models in which σ was at or below 0.2, corresponding to normally distributed Cα-Cα distances ranging from 2.95 to 4.6 Å (Figure S3D). Once past 0.2 σ, breaks were introduced in the models, and either partial or incorrect folds were found.
Pseudoatom placement
Both pseudoatom placement routines in e2segment3d.py were used to define pseudoatoms in simulated density maps from the second benchmark such that the total number of pseudoatoms corresponded to the total number of amino acids in the protein. Placement of the pseudoatoms with both routines roughly corresponded to the positions of the Cα atoms in the atomic model (Figure S2A, B). In all example structures, the average deviation from the known Cα positions was less than 2 Å.
Evaluating the Pathwalking protocol
Pathwalking with simulated data
After establishing that the TSP solvers could be used to accurately find proper backbone traces through a set of ideally spaced pseudoatoms and that we could reliably place pseudoatoms in a density map, we ran the complete pathwalking protocol on the 40 simulated density maps from the second benchmark data set, including both pseudoatom generation and path identification. For each pathwalking model, we used five parameters to measure the extent of structural agreement between the model and the known structure: Cα RMS deviation, percent of total Cαs within a 3 and 5 Å radius when compared to the corresponding Cα position in the known structure, percent of correctly registered Cαs and the topology score from the CLICK webserver (Nguyen et al., 2011). Cα RMSD describes the overall model error, while the 3 and 5 Å radii percentage and the percent of correctly registered Cαs reflect the quality of atom placement. With CLICK, a topology independent alignment between the model and known structure is computed. From this superposition, a topology score is calculated and reported from 0–1, where 1 indicates an identical topology between the known and query models. Particularly important is the fact that CLICK is tolerant of model conformational variations that do not disrupt topology.
Results using the LKH-TSP solver are summarized in Table 1, and nearly identical results were obtained with the Concorde TSP search method. For the 5 Å resolution benchmark, the mean RMS deviation was 3.32 Å with a standard deviation of 1.52 Å. The mean percentage of Cα atoms within 3 Å and 5 Å of their true position was 54.67±25.94 and 80.05±22.28. The mean percentage of correctly registered Cαs was 39.03±23.96. The CLICK score varied less with an average of 0.96 and a standard deviation of 0.06. Normalized based on sequence length, the mean RMS deviation was 3.89 Å, the mean percentage of Cαs within 3 Å and 5 Å was 44.87 and 71.8, respectively, the mean percentage of correctly registered residues was 30.6, and the mean CLICK topology score was 0.94.
Table 1.
Pathwalking results on simulated density maps. A summarized table of pathwalking on the benchmark of 40 5 Å simulated density maps. Reported resolution and number of amino acids for each protein is shown. Additionally, the Cα RMS deviation, the percent of total Cαs within a 3 and 5 Å radius when compared to the corresponding Cα position in the known structure, the percent of correctly registered Cαs and the topology score from the CLICK webserver (0–1 where 1 corresponds to an identical topology between the known and pathwalker structures) are reported. The results summarized in the table were from the LKH-TSP solver, though nearly identical results were obtained with the Concorde TSP search method. Images of all 40 pathwalking models can be found in Figure S4.
| PDB ID |
Length (aa) |
RMSD (Å) |
Cα within 3Å radius (%) |
Cα within 5Å radius (%) |
Correctly registered Cαs (%) |
Topology score |
|---|---|---|---|---|---|---|
| Alpha | ||||||
| 1h12 | 404 | 5.66 | 20.8 | 46 | 12.1 | 0.84 |
| 1oai | 59 | 1.67 | 84.8 | 100 | 78 | 1.00 |
| 1ppr | 155 | 2.73 | 60.7 | 96.1 | 32.3 | 0.82 |
| 1qsa | 363 | 6.28 | 3.6 | 28.7 | 3 | 0.89 |
| 2erl | 40 | 1.70 | 85 | 100 | 77.5 | 1.00 |
| Alpha/Beta | ||||||
| 1b25 | 209 | 2.36 | 66.5 | 97.1 | 48.3 | 0.88 |
| 1ejd | 207 | 2.14 | 70.5 | 99.5 | 60.4 | 1.00 |
| 1ewf | 180 | 5.18 | 24.4 | 54.4 | 20.6 | 1.00 |
| 1h70 | 255 | 2.15 | 76.1 | 98.0 | 54.1 | 1.00 |
| 1igd | 61 | 2.42 | 57.4 | 100 | 50.8 | 1.00 |
| 1j0p | 108 | 3.96 | 29.6 | 69.4 | 13 | 1.00 |
| 1ogq | 313 | 3.88 | 39.6 | 70.9 | 14.1 | 0.82 |
| 1plq | 258 | 2.97 | 54.7 | 84.5 | 42.6 | 0.94 |
| 1vbw | 68 | 1.36 | 98.5 | 100 | 86.8 | 1.00 |
| 1vq8 | 115 | 5.37 | 34.8 | 48.7 | 27.8 | 1.00 |
| 1wru | 88 | 2.04 | 80.7 | 100 | 58 | 1.00 |
| 2eiy | 164 | 3.93 | 31.1 | 75 | 23.8 | 0.96 |
| 2hba | 52 | 3.69 | 44.2 | 84.6 | 7.7 | 1.00 |
| 3hms | 91 | 1.49 | 89.0 | 95.6 | 84.6 | 1.00 |
| Beta | ||||||
| 1a1x | 106 | 3.03 | 64.2 | 89.6 | 17 | 0.95 |
| 1auu | 55 | 2.62 | 65.5 | 87.3 | 56.4 | 1.00 |
| 1h8p | 44 | 1.92 | 88.6 | 100 | 50 | 1.00 |
| 1i5p | 198 | 3.04 | 56.1 | 83.8 | 43.9 | 0.93 |
| 1iwm | 177 | 5.68 | 23.2 | 45.8 | 11.3 | 1.00 |
| 1lkt | 104 | 3.03 | 39.4 | 98.1 | 26.9 | 1.00 |
| 1m3y | 188 | 5.99 | 13.3 | 38.3 | 7.4 | 0.96 |
| 1mkn | 59 | 1.50 | 86.4 | 100 | 78 | 1.00 |
| 1n7v | 177 | 6.54 | 17.5 | 40.1 | 10.7 | 1.00 |
| 1p9h | 159 | 4.59 | 37.1 | 56 | 22 | 0.91 |
| 1rg8 | 141 | 3.01 | 51.1 | 87.2 | 36.9 | 1.00 |
| 1tl2 | 235 | 2.20 | 77 | 99.6 | 44.7 | 0.90 |
| 1v3w | 173 | 4.12 | 32.4 | 56.1 | 26.6 | 1.00 |
| 1w6s | 595 | 5.76 | 21.3 | 47.2 | 11.4 | 0.95 |
| 1yfq | 342 | 4.53 | 30.1 | 66.4 | 17.8 | 0.95 |
| 2bf6 | 383 | 3.66 | 41.5 | 73.1 | 23 | 1.00 |
| 2bmo | 137 | 2.15 | 75.9 | 97.1 | 54 | 0.91 |
| 2dpf | 111 | 2.33 | 67.6 | 97.3 | 49.5 | 1.00 |
| 2hnu | 81 | 2.09 | 75.3 | 100 | 58 | 1.00 |
| 3c7x | 196 | 2.35 | 73 | 90.3 | 59.7 | 0.83 |
| Irregular | ||||||
| 1ba3 | 53 | 1.73 | 98.1 | 100 | 60.4 | 1.00 |
In all instances, pathwalking on simulated density maps was able to produce topologically correct models (CLICK score close to 1) even in instances where the RMSD was relatively high and the number of correctly registered Cα atoms was low. In examining models with high RMS deviations or low CLICK scores, the major source of error was in maintaining correct helical geometry; pseudoatom placement was non-optimal and produced “back-tracing” that resulted in distorted helices (Figure S2C, D). A secondary source of error came from the CLICK alignment routine, which occasionally misaligned repeated structural elements, such as blades in multi-bladed β-propeller proteins. Overall, pathwalking performed well, identifying the correct protein topology for all structures in the simulated data set at 5 Å resolution (Figure S4).
Further benchmarking of the pathwalking protocol was done on representative structures from each of the four CATH classes at 6, 7, and 8 Å resolution. Results from this benchmark were evaluated as described above. At 6 Å resolution, pathwalking produced correct paths, though the reported statistics were generally worse than the 5 Å resolution data. At 7 Å and 8 Å resolutions, only models from two of the four density maps had the correct topology. Interestingly, the correct models at 7 Å and 8 Å differed. In all cases, RMS deviations increased and the percent of correctly registered Cα atoms decreased with resolution. Thus, we infer the boundaries of our protocol to accurately and reliably identify correct models to vary according to SSEs and resolvability of features. Results are summarized in Table 2 and Figure S5.
Table 2.
Pathwalking results at subnanometer resolutions. The table summarizes pathwalking results for four representative structures from the 40 protein benchmark set simulated at 6, 7 and 8 Å resolution. As in Table 1, the number of amino acids for each protein, resolution, Cα RMS deviation, the percent of total Cαs within a 3 and 5 Å radius when compared to the corresponding Cα position in the known structure, the percent of correctly registered Cαs and the topology score from the CLICK webserver (0–1 where 1 corresponds to an identical topology between the known and pathwalker structures) are reported. Images of all resulting pathwalking models can be found in Figure S5.
| PDB ID |
Length (aa) |
Resolution (Å) |
RMSD (Å) |
Cα within 3Å radius (%) |
Cα within 5Å radius (%) |
Correctly registered Cαs (%) |
Topology score |
|---|---|---|---|---|---|---|---|
| Alpha | |||||||
| 1ppr | 155 | 6 | 3.11 | 50.3 | 92.9 | 27.7 | 0.72 |
| 7 | 4.31 | 32.3 | 70.3 | 18.7 | 0.80 | ||
| 8 | 5.36 | 23.9 | 52.9 | 14.2 | 0.76 | ||
| Alpha/Beta | |||||||
| 3hms | 91 | 6 | 3.38 | 39.6 | 84.6 | 19.8 | 1.00 |
| 7 | 9.33 | 13.2 | 29.7 | 3.3 | 0.45 | ||
| 8 | 10.78 | 8.8 | 17.6 | 5.5 | 0.73 | ||
| Beta | |||||||
| 2hnu | 81 | 6 | 2.34 | 71.6 | 100 | 40.7 | 1.00 |
| 7 | 7.99 | 18.5 | 34.6 | 8.6 | 0.00 | ||
| 8 | - | - | - | - | 0.00 | ||
| Irregular | |||||||
| 1ba3 | 53 | 6 | 2.01 | 86.8 | 100 | 54.7 | 1.00 |
| 7 | 4.90 | 13.2 | 60.4 | 7.5 | 1.00 | ||
| 8 | 6.43 | 9.4 | 47.2 | 5.7 | 0.00 | ||
Pathwalking on authentic density maps
After evaluation with simulated data, real cryo-EM density maps at subnanometer resolutions were selected from the EMDB for testing the pathwalking protocol. For each data set selected, a structural model was previously determined and deposited in the PDB: vp6 from the 3.88 Å structure of rotavirus (EMDB ID: 1461 PDB ID:1QHD) (Mathieu et al., 2001; Zhang et al., 2008), GroEL monomer at 4.0 Å resolution (EMDB ID: 5001 PDB ID:1SS8) (Chaudhry et al., 2004; Ludtke et al., 2008), Aquaporin-1 at 3.8 Å resolution by electron crystallography (PDB ID:1IH5) (Murata et al., 2000), several protein chains from the 6.4 Å resolution structure of T. thermophilus 70S ribosome (EMDB ID: 5030 PDB ID:3FIN,3FIC) (Schuette et al., 2009) and P8 capsid protein from the 7.9 Å resolution rice dwarf virus (EMDB ID: 1375 PDB ID: 1UF2) (Liu et al., 2007; Nakagawa et al., 2003). The resolution definition was different among these maps and map quality/resolvability differed considerably though similar resolutions may be reported.
A single subunit was first isolated from the entire density map manually using UCSF Chimera, normalized with EMAN’s proc3d utility and SSE localization was done with SSEHunter. Pseudoatoms, with the total number corresponding to protein length, were placed in density maps using the k-means option from e2segment3d.py with spacing intervals from 3.5 to 4.2 Å. Path determination was carried out using the LKH-TSP solver in e2pathwalker.py with minimum and maximum distances of 3.2 and 4.5 Å, respectively.
Initial paths through the pseudoatoms were examined in context of the density map and detected SSEs. Adjustments of pseudoatom positions were performed to improve path geometry and eliminate density outliers. Three to five rounds of iterative path determination and optimization, beginning with pseudoatoms in SSEs followed by loops, were required to generate reasonable models and improve agreement to the density map. An example of an initial pathwalking model with and without SSE constraints is shown in Figure S6. In final models, the corresponding primary sequence was threaded onto the model for further evaluation. Model construction and evaluation with the pathwalking protocol took approximately one-half to a full day per data set by an intermediate-level user for proteins up to 500 amino acids.
The final pathwalking model for Aquaporin-1, rotavirus vp6 and GroEL monomer matched the fold of the known protein structure, though deviations in the assignment of some amino acids can be seen (Figure 2). Pathwalking on authentic density maps were evaluated as described for the simulated data (Table 3). The Cα RMS deviations were 4.63 Å for Aquaporin-1, 7.4 Å for rotavirus vp6 and 7.51 Å for GroEL; the fold for each of these proteins appeared to be nearly identical to that of the known structure with CLICK topology scores of 1, 0.93 and 0.79, respectively. This suggests that the pathwalking models are topologically equivalent to the known structure. As our test data set represents different protein folds, our results show that pathwalking is insensitive to protein fold as, at this resolution, helices, loops and strands were relatively well-resolved.
Figure 2.
Pathwalking on near-atomic resolution density maps. The pathwalking protocol was performed on Aquaporin at 3.8 Å (PDB ID:1IH5), rotavirus vp6 at 3.88 Å (EMDB ID: 1461 PDB ID:1QHD) and a GroEL monomer at 4.0 Å (EMDB ID: 5001 PDB ID:1SS8). The left column shows the density maps; the middle column shows the pathwalking model in the density; the right column shows the RMS deviation from the known structure. For the structures in the right column, relative error is shown in two ways: colored from blue to red based on RMS deviation at each Cα (blue: no deviation, white: RMS deviation of the model versus the corresponding known structure, red: maximum) and ribbon thickness from lowest RMS (thinnest) to highest RMS (thickest) deviation. See Figure S6 for an example of unsupervised pathwalking on rotavirus vp6.
Table 3.
Pathwalking results on authentic density maps. A summarized table of all pathwalking generated models is presented. As in Table 1, the number of amino acids for each protein, resolution, the Cα RMS deviation, the percent of total Cαs within a 3 and 5 Å radius when compared to the corresponding Cα position in the known structure, the percent of correctly registered Cαs and the topology score from the CLICK webserver (0–1 where 1 corresponds to an identical topology between the known and pathwalker structures) are reported. The results summarized in this table were from the LKH-TSP solver. For RDV P8 and MM-cpn, only the results for the first pathwalker model are reported.
| Data set | Reported Resolution (Å) |
Number of Amino Acids |
Cα RMSD (Å) |
Cα within 3Å radius (%) |
Cα within 5Å radius (%) |
Correctly registered Cαs (%) |
Topology score |
|---|---|---|---|---|---|---|---|
| Aquaporin-1 | 3.8 | 220 | 4.63 | 27.3 | 57.7 | 25.5 | 1.00 |
| Rotavirus VP6 | 3.8 | 397 | 3.99 | 49.6 | 72.5 | 40.8 | 0.93 |
| GroEL | 4.0 | 524 | 7.51 | 12 | 35.8 | 10.3 | 0.79 |
| GroEL-Rosetta | 4.0 | 524 | 6.31 | 25.8 | 45.5 | 25.4 | 0.98 |
| Ribosome chain P | 6.4 | 84 | 3.08 | 4.51.8 | 83.1 | 45.8 | 0.88 |
| Ribosome chain Q | 6.4 | 100 | 4.42 | 36.4 | 69.7 | 31.3 | 1.00 |
| Ribosome chain H | 6.4 | 139 | 8.09 | 18.8 | 35.5 | 15.2 | 0.93 |
| Ribosome chain B | 6.4 | 220 | 9.86 | 3.2 | 15.4 | 4.1 | 0.96 |
| Ribosome chain G | 6.4 | 156 | 8.75 | 20 | 27.1 | 19.4 | 1.00 |
| Ribosome chain N | 6.4 | 61 | 4.89 | 21.7 | 47.7 | 15 | 1.00 |
| RDV P8* | 7.9 | 421 | 15.49 | 3.1 | 12.4 | 1 | 0.27 |
| MM-cpn* | 4.3 | 508 | 3.13 | 56.2 | 84.3 | 51.9 | 1.00 |
| Epsilon 15 gp7 | 4.5 | 335 | 9.02 | 4.2 | 14 | 5.1 | 0.75 |
In the 6.4 Å resolution structure of the 70S T. thermophilus, density corresponding to six chains from the 30S ribosome were extracted using UCSF’s Chimera and modeled via pathwalking as described above (Figures 3, S7 and Table 3). At this resolution, β-strands are not visible, though loops and helices are generally well-resolved.
Figure 3.
Pathwalking on the 6.4 Å resolution ribosome density map. A. The 30S subunit from the cryo-EM structure of the 70S ribosome (EMDB ID: 5030 PDB ID:3FIN, 3FIC) is shown with the X-ray structures of protein chains P (B, green), Q (C, yellow), H (D, cyan), B (E, red), G (F, blue) and N (G, purple). The pathwalking models for chains P, Q, H, B, G and N are shown clockwise in panels B through G, respectively. High relative error is depicted in the enlarged red regions, and the thin, blue regions indicate relatively low error. A more detailed view of the models can be seen in Figure S7.
In chains B and H, the correct structural model was determined without user intervention. While the RMS deviation in chains H and B were among the largest (9.86 and 8.09 Å), CLICK scores of 0.96 and 0.93 suggest that the models are topologically equivalent to their known structures. In chains G, N and P, the correct fold was determined but required three to five iterations of pseudoatom optimization and path determination. Typically, paths through SSEs contained non-protein like features (like jumps between β-strands) and required manual adjustment. Models for chains G and N had perfect CLICK scores (1.0), while the chain P model had a CLICK score of 0.88, suggesting that these models were topologically equivalent to their known structures. For the final chain (Q), pathwalking resulted in a reasonable but incorrect structural model (CLICK score of 0.75). In examining the differences, the pathwalking model exhibited swapped strands in the central β-sheet domain. A single round of pseudoatom optimization and path determination produced a model that was consistent with the reported secondary structure, with a CLICK score of 1.0 (Figure S7, row 6). Unfortunately, no automated model checking is currently available in pathwalking; evaluation of possible models must be done visually and checked against any known structural information.
The pathwalking protocol was also applied to the 7.9 Å structure of the rice dwarf virus P8 capsid protein (Figure 4A). In P8, the well-resolved lower domain is nearly all α-helical, while the upper domain is nearly entirely β-sheet, with characteristic flat surfaces. In the lower domain, α-helices were detected using SSEHunter; potential connections between helices were visible when examining SSEHunter’s density skeleton (Figure 4B). 421 pseudoatoms were assigned and an initial path was calculated using the above protocol (Figure 4C). The path contained correct connectivity in the lower helical domain, but no reasonable path through the β-sheet domain was identified. A Cα RMS deviation of 15.49 Å and a CLICK score of 0.27 over the entire protein indicated a poor trace. The CLICK topology score for the lower domain was 1.0. An additional 100 models were calculated by adding Gaussian noise (0.2 σ) to the P8 pseudoatom coordinate positions (Figure 4D). In the resulting models, the lower domain paths were all similar, agreeing with the X-ray structure. In the upper domain, the paths deviated significantly from each other and no model agreed with the X-ray structure. Simply put, the upper domain β-sheets did not have enough structural features to accurately place and connect pseudoatoms. As in the simulated density maps at this resolution range, the lack of resolvable structural features is prohibitive in finding good paths through a density map with pathwalking.
Figure 4.
Pathwalking on the 7.9 Å resolution rice dwarf virus P8 density map. (A) The cryo-EM density map for P8 is shown with the X-ray structure superimposed and is rainbow colored N- to C- terminus (blue to red) (EMDB ID: 1375 PDB ID: 1UF2). (B) At this resolution the density skeleton (red) shows connectivity in the lower helical domain but is ambiguous in the upper β-sheet domain. (C) The initial pathwalking model is shown rainbow colored N- to C- terminus (blue to red). (D) A gallery of pathwalking models run with added Gaussian noise is shown. The lower helical domain is well resolved in nearly all of the 100 models (lower panel), while the upper β-sheet domain varied considerably in all of the models (upper panel).
Overall, the mean RMS deviation was 6.86 Å with a standard deviation of 3.48 Å for models from authentic density maps. The mean percentage of Cα atoms within 3 Å and 5 Å of their true position was 21.75±17.39 and 46.21±25.67, respectively. The mean percentage of correctly registered Cαs was 22.37±16.35. The mean CLICK topology score was 0.88 with a standard deviation of 0.2. These results are comparable to the simulated data and exhibit similar trends in the reported metrics; topologically correct models were generally obtained for density maps better than 7 Å resolution. Beyond this resolution, correct topological models could be obtained but were generally less accurate.
Modeling errors
Examining the pathwalking models when compared to the known structure revealed that the overall protein topology was accurate though some register shifts (shifted sequence assignments on the pseudoatom level) were apparent in the final models (Figure 5A). Most of the register shifts were on the order of 1–3 residues, though the position of the pseudoatom was generally close (~2 Å) to a Cα atom in the known structure.
Figure 5.
Modeling Errors. Typical model errors consist of: register shifts (A, rotavirus vp6), cross-overs in β-sheets (B, ε15 gp7), missing or under-populated pseudoatoms in the density (C, GroEL) and pseudoatoms (red) over-populated in β-sheets at subnanometer resolutions (D, rice dwarf virus P8). In A, the pathwalking model is shown in rainbow color with black labels and the X-ray structure (PDB ID:1QHD) is shown in gray with red labels. In (C), the pathwalking model for GroEL is shown in rainbow color and the X-ray structure (PDB ID:1SS8) is shown in grey.
In the near-atomic resolution density maps, a common error in modeling was crossovers in β-sheets; rather than producing parallel/anti-parallel strands in β-sheets, the path jumped between strands, making a “zig-zag” pattern (Figure 5B). These jumps often occurred in pairs, compensating for the alterations in the path, and were typically found in regions containing long, multi-stranded β-sheets. In practice, minimal low-pass filtering of the density map and/or manual manipulation of the pseudoatoms can correct this error.
Pseudoatom placement was another source of model error. Variations in density, or regions that were either excluded or improperly segmented resulted in poor pseudoatom placement, thereby affecting the overall structure of the protein. An example of this can be seen in the GroEL apical domain, in which a density protrusion was missed during pseudoatom placement (Figure 5C). Such errors account for relatively high RMS deviations but allow for correct protein topology.
Model optimization
Pathwalking models, calculated in a few seconds, are intended to be initial models with correct topology, but they are not constrained for optimal protein stereochemistry or refined fit to density features (i.e. visible sidechains). As shown earlier, a topologically correct model of GroEL was constructed from the 4.0 Å density map (CLICK score 0.79), though the overall RMS deviation of the model compared to 1SS8 was 7.51 Å. Examining the differences at the amino acid level indicated that the majority of model deviations were due to register shifts. Similar types of errors have been reported in de novo cryo-EM modeling (Jiang et al., 2008; Ludtke et al., 2008). While this difference is larger than what was reported previously (Ludtke et al., 2008), the original de novo model was manually optimized using the density map features. Starting with the GroEL pathwalking model, we carried out an initial optimization step using Rosetta (DiMaio et al., 2009) to improve both fit to the density map and stereochemistry (Figure 6). In this approach, the GroEL pathwalking model was broken up into three domains, the equatorial, intermediate and apical domains. Each domain was subjected to only one round of refinement with Rosetta. After this optimization step, the top models for each domain were concatenated and compared to 1SS8 and the original pathwalking model.
Figure 6.
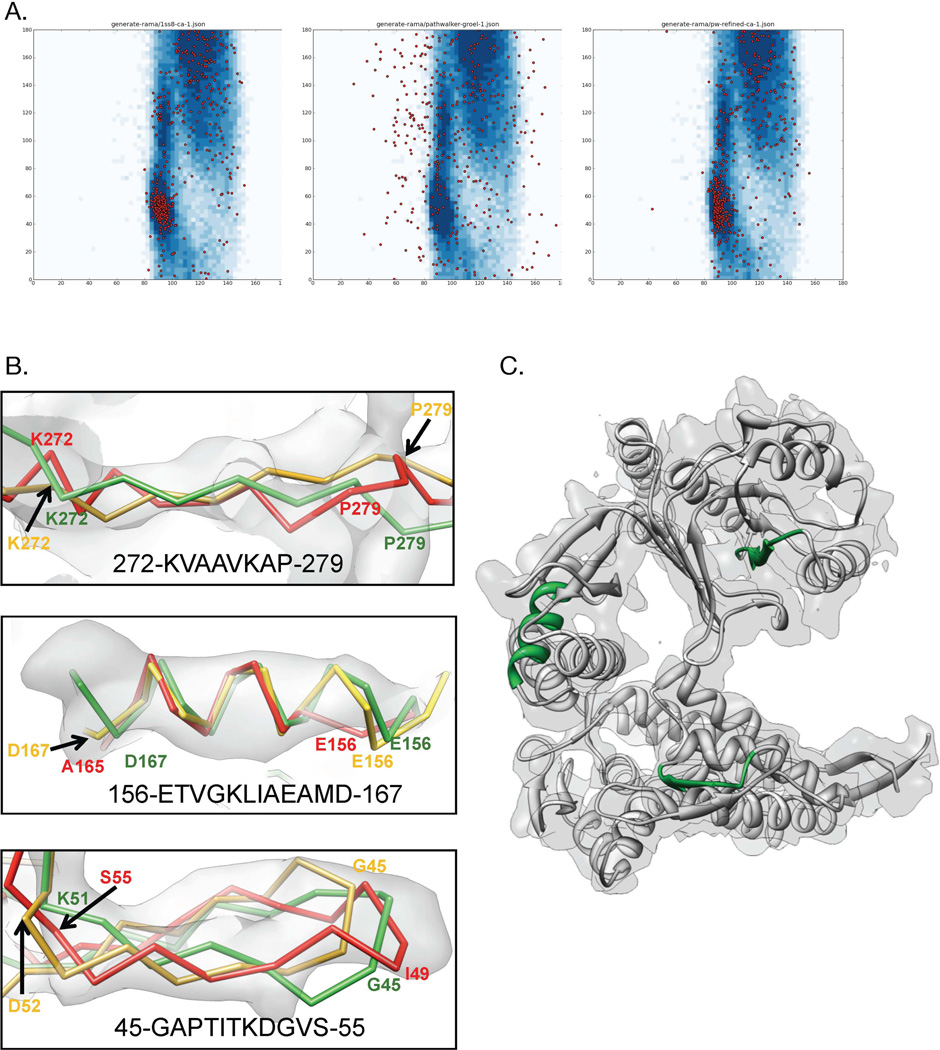
Model Refinement. The initial pathwalking model for GroEL was refined using Rosetta. A plot of a Cα Ramachandran angles for the GroEL X-ray structure (PDB ID:1SS8), initial pathwalking model and the Rosetta refined pathwalking model are shown in (A), from left to right. Three selected regions from the refined GroEL model, highlighted in green in (C), are shown in (B). In (B), the X-ray structure is shown in green, initial pathwalking model is shown in red, and the refined pathwalking model is shown in yellow. The corresponding sequence is shown for each region.
After one iteration of density-constrained refinement with Rosetta, the RMS deviation dropped to 6.45 Å (~16.4% improvement) and the CLICK score improved to 0.98. Additionally, the Cα Ramachandran plot for the optimized model improved significantly (Figure 6A). As no stereochemical constraints were enforced during the pathwalking procedure, this type of optimization step is essential in producing accurate protein structures. In addition to improved stereochemistry and geometry, the optimized model fit the density better and sequence registration errors were fixed in most locations (Figure 6B, C). Small register shifts were generally alleviated, though larger shifts (4+ amino acids) were typically only partially corrected. Further iterations of Rosetta refinement protocol will continue to improve the model and fix larger errors.
Validating an existing model
Pathwalking can also be used to assess reliability and accuracy of all types of de novo backbone models, whereby Gaussian noise can be added to pseudoatom positions and new paths calculated. Adding positional noise allows the pathwalking utilities to explore alternate paths through the density.
A model for the entire Mm-cpn assembly (EMDB ID: 5137, PDB ID: 3LOS) (Zhang et al., 2010), determined from the 4.3 Å resolution cryo-EM density map using our de novo modeling protocol (Zhang et al., 2010) and refined by Direx (Schröder, Brunger and Levitt, 2007), was used to investigate possible alternative paths with pathwalking. Using the Cαs from the de novo model, an initial path was generated with e2pathwalker.py (Figure S8A). The pathwalking model had an RMS deviation of 3.58 Å when compared to the X-ray structure (PDB ID: 3KFB) (Pereira et al., 2010). Gaussian noise was added in increments of 0.1 σ and new paths were calculated using both TSP routines (Figure S8B). The path for Mm-cpn was determined correctly >95% of the time at or below a noise-level of 0.5 σ in pseudoatom positions (Cα-Cα distances between 1.92 and 5.67 Å). Crossovers in β-sheets began to occur at noise levels of 0.3 σ but did not affect the overall fold of the protein as compensating crossovers occurred nearby.
Like Mm-cpn, a de novo model for ε15 gp7 was built manually from the 4.5 Å resolution density map prior to the availability our pathwalking procedures (Jiang et al., 2008). However, computational refinement of the model was not carried out. No atomic model is currently available for ε15 gp7. Running the full TSP pathwalking protocol on the ε15 gp7 density map resulted in an initial model with a relatively high RMS deviation (9.03 Å) but topologically equivalent model when compared to the hand-built de novo model (Figure 7A). Running 100 iterations of e2pathwalker.py with Gaussian noise (0.2 σ) on both the hand-built de novo and pathwalking initial model generated several alternate models (Figure 7B) comprised of the basic HK97 bacteriophage coat protein structure (Helgstrand et al., 2003). In the alternate models, swaps in the ordering of loops and β-strands in the A-domain were seen (Figure 7B–D). Nearly all of these models could be ruled out once sequence was threaded onto the model due to differences in secondary structure. Interestingly, at least one alternative model agreed with both the secondary structure predictions and density map suggesting a possible alternative fold for gp7 (Figure 7C, D).
Figure 7.
Model Validation. The pathwalking protocol was performed on one subunit of the ε15 gp7 density map at 4.5 Å (EMDB ID: 5003 PDB ID:3C5B). In (A), the left column shows the density maps; the middle column shows the pathwalking model, rainbow colored from N- to C- terminus, in the density map; the right column shows the RMS deviation from the previously reported de novo model. High relative error is depicted in the enlarged red regions, and the thin, blue regions indicate relatively low error. (B) The pathwalking models for ε15 gp7 (EMDB ID: 5003), varied considerably even with small amounts of noise, particularly in the highly β-sheet A-domain (inset). The two possible paths for ε15 gp7 are shown in (C); zoomed-in views of the A-domain in the two possible gp7 models are shown in (D). The models are rainbow colored from N- to C- terminus (blue to red). An additional example with Mm-cpn is shown in Figure S8.
DISCUSSION
Current de novo model building procedures generally rely on the presence of structural landmarksfrom which manual or semi-automated model building is initiated. Pathwalking rapidly constructs first-approach models, represented as Cα backbone traces that are topologically equivalent to the protein’s tertiary structure, without requiring a priori knowledge. Such models serve as initial starting points for further refinement with software such as Rosetta, Modeller or Direx (Alber et al., 2007; Bradley et al., 2005; Schröder, Brunger and Levitt, 2007).
Pathwalking is unique in that it is completely de novo, sequence-free, template free, semi-automated and suitable for use on maps from 3 to 7 Å resolution. Unlike most of the modeling tools in cryo-EM, pathwalking does not use a structural template for model building, refinement or evaluation. Furthermore, pathwalking minimizes user intervention, unlike interactive modeling tools like Gorgon, O or Coot (Baker et al., 2011; Emsley et al., 2010; Jones et al., 1991). X-ray crystallographic tools exist for (semi-) automatic model building, however these utilities are targeted to higher resolution density maps, though some can potentially be applied to 3–4 Å resolution density maps (Cohen et al., 2004; Cowtan, 2006).
While pathwalking is almost completely automated, many control points have been added to allow for user input regarding potential paths. Evaluated visually, a good path should: connect all pseudoatoms such that each is visited only once, contains no intersecting path segments, have reasonable connectivity (bond distances and angles) and have connections within/bounded by the density map. Additionally, the model is expected to have “realistic” structural features. Regions in the density map shown to have helices should have pseudoatoms and a path arranged helically; regions containing β-sheets should have parallel/anti-parallel strands. Threading the primary sequence on to a path and evaluating it in the context of SSEs and sidechain density can also be used in the evaluation a model. If the user perceives a problem with the path or wishes to evaluate alternate paths, pathwalking can be run multiple times simply by varying the parameters for pseudoatom placement and/or path searching, adding constraints or manually adjusting “bad” regions of the trace. Such interventions may improve registration of SSEs and sidechains in the density map, which are not explicitly considered in pathwalking.
Pathwalking Accuracy
For evaluating pathwalking, we created a large enchmark data set. In the initial test, we examined the TSP-solvers for pathwalking in a set of 737 non-redundant protein structures. In this data set, we used the position of the known Cα atoms as the pseudoatom inputs to e2pathwalker.py. This test showed that a correct path could be identified given reasonably spaced pseudoatoms. In the second benchmark, we considered not only the problem of path tracing but also the problem of placing pseudoatoms in simulated density maps. Our pathwalking approach produced correct topological models in all the examples, though some non-protein like geometries were observed. In the final set of tests, we examined the entire pathwalking procedure on authentic density maps ranging from ~4–8 Å resolution. This benchmark covered a wide range of fold-types and was representative of maps deposited in the EMDB and PDB. While in the higher resolution data sets paths through the density maps contained a limited number of ambiguities, lower resolution density maps, like the ribosome density map, did not have unambiguous paths and were considerably harder targets. It should also be noted that some of the higher resolution density maps were not uniformly resolved and contained regions where the density was considerably more difficult to evaluate (apical domain of GroEL). Overall, the set of simulated and authentic density maps provide a realistic baseline for what users should expect with density maps in the “near-atomic” resolution range.
In nearly all of our test cases, pathwalking produced topologically correct models (CLICK score close to 1), though the exact amino acid assignment was often out of register, resulting in relatively high RMS deviations when compared to the known structure (Tables 1–3). The emphasis in pathwalking is that models can be built directly from the density map with correct topologies, despite errors in amino acid assignments. As demonstrated, this level of error can be corrected with additional optimization steps (DiMaio et al., 2009). In GroEL, a single iteration of density-based refinement using Rosetta resulted in improved stereochemistry and geometry, and also repaired a vast majority of the sequence shifts, lowering the RMS deviation by 16.4% (Figure 6). Additional rounds of refinement would likely further improve model quality.
In the cases where pathwalking did not give the correct fold on the first iteration, models typically did not agree with the secondary structure predictions. In chain Q from the ribosome density map, several strands and loops were transposed (Figure S7). The model visually appeared to agree with the density map, however it did not agree with the secondary structure, indicating a bad topological path. In this case, it was possible to constrain well-defined regions and calculate an alternate path (Figure S7, row 6).
Pathwalking Limitations
Our approach requires that a single subunit be accurately segmented from the entire density map. Missing portions or extra density will result in poor pseudoatom placement (Figure 5C). Depending on the level of mis-segmentation, pathwalking may not yield the correct protein fold. Therefore, it is imperative that segmentation be as accurate as possible. In practice, segmentation and model building at subnanometer resolutions are usually coupled and, as such, the pathwalking protocol may need to be run iteratively as subunit boundaries are defined.
With pathwalking, it is possible that the connections between pseudoatoms could be adversely effected by non-optimal pseudoatom placement. The TSP solvers do not consider this uncertainty. By adding random perturbations of varying strength to the pseudoatom coordinates and running e2pathwalker.py many times, alternative models can be computed. In most cases, the ensemble of the models will agree topologically, though differences may be seen in poorly resolved regions. Degenerate paths in a “fuzzy” loop may connect the same pseudoatoms in different orders yet still maintain the protein fold. Conversely, the same path may be achieved with a different set of pseudoatoms. In these cases, the user is required to judge which order of connectivity is best based on features in the density map, path geometry and a priori information. Additionally, a user can explicitly add or remove connections based on other biochemical information and/or visual interpretation. In all cases, the best model can generally be selected visually such that it meets basic protein structure requirements.
Map resolution is also a factor in model accuracy. From our benchmarks, it was possible to construct first–approach models even at 7–8 Å resolution with our pathwalking tools. As all density maps vary in composition, quality and resolution, it is difficult to assign hard limits for pathwalking. This is in part due to the various resolution definitions, variability in resolvability of density maps and the SSE content in the protein. The accuracy of pathwalking is a direct reflection of the resolvability of features in a density map. At subnanometer resolutions, α-helices tend to be better resolved than loops and β-sheets, making it possible to construct models for all helical proteins at lower resolutions (Figures S2–S5). A well-defined map containing mostly helices at 7 Å resolution will undoubtedly yield better results than a poorly resolved density map of an all-β protein at 4.5 Å resolution. Ultimately, the resolvability of structural features dictates the limitations of our approach. Therefore, we cannot specify an absolute resolution range for pathwalking.
Model Validation with Pathwalking
Beyond model construction, our pathwalking procedures can be used to assess de novo model validity and report potential alternative topologies. As in the case of ε15 gp7, alternate models using the pathwalking procedure can highlight potential areas of structural ambiguity. This can be particularly useful when dealing with models where resolvability is limited.
Pathwalking represent the first step in sequence and template-free modeling in near-atomic resolution density maps. This process is capable of rapidly computing first-approach models for individual subunits in large macromolecular complexes. Additionally, the same utilities can be used to validate models and display alternate topologies. We believe our pathwalking tools will become an important part of model building and validation for the growing number of near-atomic resolution density maps by cryo-EM and X-ray crystallography.
Supplementary Material
ACKNOWLEDGEMENTS
This research is supported by grants from NIH through the National Center for Research Resources (P41RR002250), National Institute of General Medical Science (R01GM079429 and R01GM080139), Common Fund (PN2EY016525) and National Science Foundation (IIS-0705644, IIS-0705474). M. R. Baker and I. Rees are supported by a postdoctoral and predoctoral training fellowship respectively from the National Library of Medicine Training Program in Computational Biology and Biomedical Informatics provided by the Keck Center and Gulf Coast Consortia (T15LM007093).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abeysinghe S, Ju T, Baker ML, Chiu W. Shape modeling and matching in identifying 3D protein structures. Comput Aided Design. 2008a;40:708–720. [Google Scholar]
- Abeysinghe SS, Ju T. Interactive skeletonization of intensity volumes. The Visual Compter. 2009;25:627–635. [Google Scholar]
- Abeysinghe SS, Baker ML, Chiu W, Ju T. Segmentation-free skeletonization of grayscale volumes for shape understanding; IEEE International Conference on Shape Modeling and Applications; 2008b. pp. 63–71. [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Applegate DL. The traveling salesman problem: a computational study. Princeton: Princeton University Press; 2006. [Google Scholar]
- Baker ML, Abeysinghe SS, Schuh S, Coleman RA, Abrams A, Marsh MP, Hryc CF, Ruths T, Chiu W, Ju T. Modeling protein structure at near atomic resolutions with Gorgon. J Struct Biol. 2011;174:360–373. doi: 10.1016/j.jsb.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Baker MR, Hryc CF, Dimaio F. Analyses of subnanometer resolution cryo-EM density maps. Methods Enzymol. 2010;483:1–29. doi: 10.1016/S0076-6879(10)83001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner DW, Bloomer A, Petsko GA, Phillips DC, Wilson IA. Atomic coordinates for triose phosphate isomerase from chicken muscle. Biochem Biophys Res Commun. 1976;72:146–155. doi: 10.1016/0006-291x(76)90972-4. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Steven AC. Macromolecular electron microscopy in the era of structural genomics. Trends Biochem Sci. 2000;25:624–631. doi: 10.1016/s0968-0004(00)01720-5. [DOI] [PubMed] [Google Scholar]
- Blundell TL, Johnson LN. Protein crystallography. New York: Academic Press; 1976. [Google Scholar]
- Bradley P, Malmström L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KMS, Baker D. Free modeling with Rosetta in CASP6. Proteins. 2005;61 Suppl 7:128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- Chaudhry C, Horwich AL, Brunger AT, Adams PD. Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J Mol Biol. 2004;342:229–245. doi: 10.1016/j.jmb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, Dimaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Almo SC. Structural biology of cellular machines. Trends Cell Biol. 2006;16:144–150. doi: 10.1016/j.tcb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, Lamzin VS, Kleywegt GJ, Perrakis A. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J, Basak AK, Roy P, Stuart D. The crystal structure of bluetongue virus VP7. Nature. 1995;373:167–170. doi: 10.1038/373167a0. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Helsgaun K. General k-opt submoves for the Lin-Kernighan TSP heuristic. Mathematical Programming Computation. 2009;1:119–163. [Google Scholar]
- Hryc CF, Chen DH, Chiu W. Near-atomic resolution cryo-EM for molecular virology. Current Opinion in Virology. 2011;1:110–117. doi: 10.1016/j.coviro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Joh NH, Oberai A, Yang D, Whitelegge JP, Bowie JU. Similar energetic contributions of packing in the core of membrane and water-soluble proteins. J Am Chem Soc. 2009;131:10846–10847. doi: 10.1021/ja904711k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Ju T, Baker ML, Chiu W. Computing a family of skeletons of volumetric models for shape description. Comput Aided Des. 2007;39:352–360. doi: 10.1016/j.cad.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ. Validation of protein models from Calpha coordinates alone. J Mol Biol. 1997;273:371–376. doi: 10.1006/jmbi.1997.1309. [DOI] [PubMed] [Google Scholar]
- Lawler EL. The Traveling salesman problem: a guided tour of combinatorial optimization. New York: Wiley; 1985. [Google Scholar]
- Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen D-HH, Song J-LL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 2001;20:1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Nguyen MN, Tan KP, Madhusudhan MS. CLICK--topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res. 2011;39:W24–W28. doi: 10.1093/nar/gkr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, Goulet DR, King JA, Frydman J, Adams PD. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem. 2010;285:27958–27966. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Kuriyan J. Challenges at the frontiers of structural biology. Trends Cell Biol. 1999;9:M20–M24. [PubMed] [Google Scholar]
- Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.