Abstract
Hepcidin is an antimicrobial peptide hormone involved in the metabolism of iron, encoded for by the HAMP gene mainly in hepatocytes. It’s expressed at lower levels in other cells such as the macrophages. The mechanisms that determine tissue-specific expression of hepcidin remain unclear. GATA- and its co-factor Friend of GATA (FOG) modulate the tissue-specific transcription of other genes involved in the metabolism of iron. GATA proteins are group of evolutionary conserved transcriptional regulators that bind to the consensus motif –WGATAR– in the promoter. We characterized a 1·3 kb fragment of the 5′-flanking sequence of the HAMP gene in Huh7 cells, which express HAMP. Transfection of 5′-deletions of the HAMP promoter in Huh7 cells revealed two regions, −932/−878 and −155/−96, that when deleted decreased promoter activity. Using site-directed mutations in the HAMP promoter region −155/−96 we identified two subregions, −138/−125 and −103/−98, which when mutated suppressed promoter activity by 70 and 90% respectively. Site −103/−98 with a sequence –TTATCT– to which endogenous GATA proteins 4 and 6 bind and transactivate HAMP is a GATA-regulatory element (RE). Mutation of the GATA-RE abrogated binding of GATA proteins 4 and 6 to the promoter and blunted the GATA transactivation of HAMP. FOG proteins 1 and 2 suppressed the endogenous and exogenous GATA activation of the HAMP promoter. We concluded that the GATA-RE, –TTATCT– in the HAMP promoter region −103/−98 is crucial for the GATA-4 and GATA-6 driven transcription of hepcidin in Huh7 cells and that FOG proteins moderate the transcription by suppressing the GATA transactivation of HAMP.
Introduction
The concentration of iron in the body is regulated by an antimicrobial peptide hormone, hepcidin, which is encoded by the HAMP gene that is expressed mainly in the liver by hepatocytes (Roetto et al. 2003, Pietrangelo 2006). Hepcidin is also expressed at lower levels in other organs such as the heart, brain, spinal cord, stomach, and pancreas and in cells such as adipocytes and macrophages (Merle et al. 2007, Kulaksiz et al. 2008, Hanninen et al. 2009, Isoda et al. 2010). Increases in the concentration of iron and inflammation upregulate the transcription of hepcidin, while iron deficiency and hypoxia downregulate it (Nicolas et al. 2002). At present, it is unclear as to what the molecular mechanisms involved in the tissue-specific expression of hepcidin are.
Studies on the HAMP promoter indicate that the region −235/+50 bp is crucial for the basal expression of hepcidin in hepatocytes, since mutation of the CCAAT/enhancer-binding protein (C/EBP) site −231/−222 (Courselaud et al. 2002), cyclic AMP response element-binding protein (CREB) site −138/−131 (Vecchi et al. 2009), bone morphogenetic protein-response element (BMP-RE) site −84/−79 (Verga Falzacappa et al. 2008), and signal transducer and activator of transcription 3-response element (STAT3-RE) site −72/−64 (Pietrangelo et al. 2007, Verga Falzacappa et al. 2007) in this region abrogate the basal transcription of HAMP.
In hepatocytes, GATA- and its co-factor Friend of GATA (FOG) modulate the tissue-specific transcription of other genes involved in the metabolism of iron, in particular: GATA-1 is a transcriptional enhancer of hemochromatosis (HFE; Mura et al. 2004) and erythropoietin receptor (EPOR; Chiba et al. 1991); GATA proteins 2 and 3 are transcriptional repressors of EPO; while GATA-4 is a transcriptional activator of EPO (Imagawa et al. 1997, Dame et al. 2004, Obara et al. 2008a).
GATA proteins are evolutionarily conserved cell-specific transcriptional regulators that bind to the consensus motif –WGATAR– in the promoter region of several genes (Ko & Engel 1993). Six GATA-factors with a highly conserved zinc finger DNA-binding domain have been identified (Weiss & Orkin 1995, Molkentin 2000). In hepatocytes, GATA-1, GATA-4, and GATA-6 are expressed in the prenatal period; while GATA-2, GATA-3, and GATA-6 in the postnatal period (Dame et al. 2004, Divine et al. 2004). In mice, the disruption of either the GATA-4 or GATA-6 gene results in embryonic death, due to defects in formation of either the heart tube or extra-embryonic endoderm respectively (Kuo et al. 1997, Molkentin et al. 1997, Zhao et al. 2005). GATA proteins regulate the expression of other genes through functional interactions with FOG-proteins (Robert et al. 2002, Obara et al. 2008a).
At present, two FOG proteins have been identified: FOG-1, mainly expressed in erythroid and megakaryocytic cells (Tsang et al. 1997) and FOG-2 in the heart, brain, and gonads (Lu et al. 1999, Svensson et al. 1999, Tevosian et al. 1999) and at low levels in the liver (Lu et al. 1999). FOG-1 knockout mice die in mid-gestation from anemia and cessation in erythroid maturation (Tsang et al. 1998); while FOG-2 knockout mice exhibit cardiovascular defects leading to embryonic death (Tevosian et al. 1999, Svensson et al. 2000b).
In this study, we identified a GATA-RE, –TTATCT– in the HAMP promoter region −103/−98 that is crucial for the FOG (1 and 2) repression of the endogenous and exogenous GATA (4 and 6) transactivation of HAMP.
Materials and methods
Cell culture
HepG2 cells were cultured as recommended by American Type Culture Collection (Manassas, VA, USA); while Huh7 cells (gift from Dr K Pantopoulos) were cultured in DMEM essential medium (WISENT, Inc., Montreal, QC, Canada) supplemented with 10% heat inactivated fetal bovine serum (WISENT, Inc.), 1% non-essential amino acids (WISENT, Inc.), 50 μg/ml of penicillin–streptomycin (WISENT, Inc.), and 0·5 μg/ml amphotericin B (WISENT, Inc.). Huh7 and HepG2 cells were incubated at 37 °C under 5% CO2.
RNA isolation, reverse transcription, PCR, and real-time PCR
Total RNA from hepatocytes was isolated using TRizol as recommended by the manufacturer (Invitrogen Canada). The RNA (2 μg) was used as a template to synthesize the first-strand cDNA using Omniscript reverse transcriptase-PCT (RT-PCR) system (Qiagen). The first-strand cDNA synthesized (20 μl) was diluted to 120 μl with RNase-free water, 5 μl of which was used as a template in quantitative RT-PCRs. Adult human liver total RNA was purchased from Clontech Laboratories, Inc. Quantification of gene expression was performed by a Rotor Gene 3000 Real-time DNA Detection System (Montreal Biotech, Kirkland, QC, Canada) with QuantiTect SYBRGreen I PCR kits (Qiagen), as described previously (Constante et al. 2007). The oligonucleotides used to amplify β-actin, HAMP, GATA-2, GATA-3, GATA-4, GATA-6, FOG-1, and FOG-2 genes are listed in Table 1. The PCR products were confirmed by agarose gel electrophoresis and sequencing (University McGill and Génome Québec Innovation Centre, Montréal QC). Gene expression levels in the liver tissue or hepatoma cells were represented relative to the expression of the housekeeping gene β-actin.
Table 1.
Oligonucleotides used in this study for PCRs and electromobility shift assays
| Application
|
Description
|
Sequencea
|
|---|---|---|
| Real-time PCR | HAMP | Forward: 5′-ACCAGAGCAAGCTCAAGACC-3′ |
| Reverse: 5′-CTCTGCAAGTTGTCCCGTCT-3′ | ||
| β-Actin | Forward: 5′-CTGAACCCCAAGGCCAAC-3′ | |
| Reverse: 5′-AGAGGCGTACAGGGATAGCA-3′ | ||
| GATA-2 | Forward: 5′-GCAACCCCTACTATGCCAAC-3′ | |
| Reverse: 5′-GTCTTGGAGAAGGGGCTCAC-3′ | ||
| GATA-3 | Forward: 5′-CTCATTAAGCCCAAGCGAAG-3′ | |
| Reverse: 5′-TTTTTCGGTTTCTGGTCTGG-3′ | ||
| GATA-4 | Forward: 5′-TGGCCTGTCATCTCACTACG-3′ | |
| Reverse: 5′-AAGACCAGGCTGTTCCAAGA-3′ | ||
| GATA-6 | Forward: 5′-GCCAACTGTCACACCACAAC-3′ | |
| Reverse: 5′-TGGAGTCATGGGAATGGAAT-3′ | ||
| FOG-1 | Forward: 5′-ACAGACGAGAAGCCCAAAGA-3′ | |
| Reverse: 5′-CCCTTGTGGTGGAGATGAAG-3′ | ||
| FOG-2 | Forward: 5′-GCTTCTATTTTGCCCACAGC-3′ | |
| Reverse: 5′-ACAGGCTGGAAATCTGATGG-3′ | ||
| HAMP promoter | −1234 bp | Forward: 5′-GCGGATCCAGGCTAGAATCTCAGCTCTG-3′ |
| −1117 bp | Forward: 5′-TCCGACGGGGAGGAGGaTCCTAAAAGGGAGCAG-3′ | |
| −932 bp | Forward: 5′-AAATGTACTCATCGGATCCTAGATGTTAGCTATTA-3′ | |
| −878 bp | Forward: 5′-GTTTTATAGACAAGATCTCACTCTGTCACCCA-3′ | |
| −816 bp | Forward: 5′-GCGGATCCTGGGCTTAAGCGATCTGCCTCAG-3′ | |
| −155 bp | Forward: 5′-AGCGGATCCTCTGCCGGCTGAGGGTGACACAA-3′ | |
| −96 bp | Forward: 5′-GCGGATCCCGCCTTTTCGGCGCCACCACCT-3′ | |
| +73 bp | Reverse: 5′-CCGGTACCATCGTGCCGTCTGTCTGGC-3′ | |
| Mutagenesis site I | M21 −928/−926 bp | S: 5′-TAAAATGTACTCATCGGACTacgGATGTTAGCTATTACTATTAC-3′ |
| M22 −923/−921 bp | S: 5′-TGTACTCATCGGACTGTAGAcacTAGCTATTACTATTACTGCT-3′ | |
| M23 −918/−916 bp | S: 5′-TCATCGGACTGTAGATGTTAatcATTACTATTACTGCTATTTT-3′ | |
| M24 −913/−911 bp | S: 5′-GGACTGTAGATGTTAGCTATcgtTATTACTGCTATTTTATGTT-3′ | |
| M25 −908/−906 bp | S: 5′-GTAGATGTTAGCTATTACTAccgCTGCTATTTTATGTTTTATA-3′ | |
| M26 −903/−901 bp | S: 5′-TGTTAGCTATTACTATTACTatcATTTTATGTTTTATAGACAG-3′ | |
| M27 −898/−896 bp | S: 5′-GCTATTACTATTACTGCTATcccATGTTTTATAGACAGGGTCT-3′ | |
| M28 −893/−891 bp | S: 5′-TACTATTACTGCTATTTTATaccTTATAGACAGGGTCTCACTC-3′ | |
| Mutagenesis site II | M2 −143/−141 bp | S: 5′-GAACACACCTCTGCCGGCTagaGGTGACACAACCCTGTTCC-3′ |
| M3 −138/−136 bp | S: 5′-CACCTCTGCCGGCTGAGGGcagCACAACCCTGTTCCCTGTC-3′ | |
| M4 −133/−131 bp | S: 5′-CTGCCGGCTGAGGGTGACAtggCCCTGTTCCCTGTCGCTCT-3′ | |
| M5 −128/−126 bp | S: 5′-CTGAGGGTGACACAACCtcaTTCCCTGTCGCTCTGTTC-3′ | |
| M6 −123/−121 bp | S: 5′-GGTGACACAACCCTGTTtttTGTCGCTCTGTTCCCGC-3′ | |
| M7 −118/−116 bp | S: 5′-GACACAACCCTGTTCCCTGctaCTCTGTTCCCGCTTATCTC-3′ | |
| M8 −113/−111 bp | S: 5′-CCCTGTTCCCTGTCGCTtcaTTCCCGCTTATCTCTCCC-3′ | |
| M9 −108/−106 bp | S: 5′-GTTCCCTGTCGCTCTGTTtttGCTTATCTCTCCCGCCTT-3′ | |
| M10 −103/−100 bp | S: 5′-CTGTCGCTCTGTTCCCGCccgTCTCTCCCGCCTTTTCGG-3′ | |
| M11 −98/−96 bp | S: 5′-TCGCTCTGTTCCCGCTTATCctcCCCGCCTTTTCGGCGCCACC-3′ | |
| EMSA | WT −121/−83 bp | S: 5′-CTGTCGCTCTGTTCCCGCTTATCTCTCCCGCCTTTTCGG-3′ |
| MT −102/−98 bp | S: 5′-CTGTCGCTCTGTTCCCGCTcgctcCTCCCGCCTTTTCGG-3′ | |
| ChIP | −155/+73 bp HAMP promoter | S: 5′-CCTCTGCCGGCTGAGGGTGACACAA-3′ |
| AS: 5′-CATCGTGCCGTCTGTCTGGCTGTCC-3′ |
S, sense sequence; AS, antisense.
Cloning sites are underlined while mutations are in lower case letters.
Plasmids
A 1307 bp fragment (−1234/+73) of the 5′-flanking end of the HAMP gene was generated from human genomic DNA by PCR using the primers listed in Table 1. Deletion constructs (−1117, −932, −878, −816, −155, and −96 bp) were generated using primers listed in Table 1 and the HAMP plasmid −1234/+73 bp as a template. All the promoter fragments were cloned into a modified pXP1 luciferase reporter plasmid obtained from Dr Jacques Tremblay (CHUQ, Laval University, Quebec, Canada; Tremblay & Viger 1999). Site-directed, serially arranged tri-nucleotide mutations were introduced in the HAMP promoter construct −1234/+73 bp using the Quik-Change II XL mutagenesis kit (Stratagene, La Jolla, CA, USA) and oligonucleotides listed in Table 1. All plasmids were verified by digestion with restriction enzymes and sequencing (McGill University and Génome Québec Innovation Centre). Expression vectors for GATA-1, GATA-3, GATA-4, GATA-6, FOG-1, and FOG-2 were kindly provided by Dr Robert Viger (CHUQ, Laval University) and have been described previously (Tremblay & Viger 1999, 2001, Tremblay et al. 2001, Robert et al. 2002).
Transfections and luciferase assays
Transfections of Huh7 cells were performed in 12-well plates using the Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Briefly, on the day before transfection 8×104 hepatocytes were seeded with 1000 μl of antibiotic-free media per well. Hepatocytes were co-transfected with 1580 ng/well of either wild-type (WT) or mutated (MT) HAMP promoter constructs along with 20 ng of phRL-TK (Renilla Luciferase). Five hours after transfection, the media was replaced with fresh antibiotic-free media. Hepatocytes were lysed 40 h following transfection and the Dual-Luciferase Assay System (Promega Corporation) used to determine Luciferase activities (firefly and Renilla) with a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA, USA). When evaluating the effect of either GATA (1, 3, 4, and 6) or FOG (1 and 2) protein on the HAMP promoter, Huh7 cells were co-transfected with increasing concentrations (0–787·5 ng/well) of the expression vectors encoding the full-length GATA or FOG protein in addition to the HAMP reporter constructs (787·5 ng/well). The total amount of DNA was maintained at 1·6 μg/well by adding an empty pcDNA3 vector (Invitrogen). In all experiments, data reported represent an average of at least three experiments, done in triplicate, using at least three different DNA preparations.
In silico analysis
Analysis for potential regulatory elements (RE) in the HAMP promoter was performed using Evolutionary Conserved Regions ECR Browser (http://ecrbrowser.dcode.org) and Transcription Factor SEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html; Heinemeyer et al. 1998).
Electromobility shift assays
Huh7 cells were cultured in petri dishes (60 cm2) and the nuclear protein extracts prepared using a kit (Active Motif, Carlsbad, CA, USA). The probe used in electromobility shift assay (EMSA) was obtained by enzymatic labeling the 3′-end of a double-stranded oligonucleotide corresponding to the promoter region −121/−83 (Table 1) of the HAMP promoter by incorporation of a single digoxigenin (DIG)-labeled ddUTP using the DIG kit (Roche Diagnostics). Briefly, 100 pmol of the WT oligonucleotide was added (on ice) to 4 μl 5× labeling buffer, 4 μl, 25 mM CoCl2, 1 μl 0·05 mM DIG-11-ddUTP and 400 U terminal transferase in a final volume of 20 μl and then incubated at 37 °C for 15 min. The efficiency of the labeling was tested in a dot blot assay as described by the manufacturer (Roche Diagnostics). The probe (0·8 ng) was incubated with 7·5 μg of nuclear protein extracts from Huh7 cells along with 1 μl poly [d(I-C)], 0·1 μg poly-L-lysine in 20 μl of binding buffer for 15 min at room temperature. In the competition experiments, specific binding of nuclear protein extracts from Huh7 cells to the probe was challenged with an excess (125× and 250× molar excess) of unlabeled oligonucleotides (Table 1). The unlabeled oligonucleotides carried a sequence that corresponded to either the WT promoter region −121/−83 or a MT (an oligonucleotide bearing a mutation of the GATA-binding site –TATCT– (Table 1)). In the super-shift experiments, the nuclear protein extracts that were used were prepared from Huh7 cells 40 h following their transfection with 24 μg of either an expression vector encoding the full-length WT GATA protein (4 or 6) or the empty plasmid vector, pcDNA3 (Invitrogen). Specific binding of the nuclear extracts harvested from the transfected Huh7 cells to the probe was evaluated in the presence of either an antibody raised against the overexpressed protein (either GATA-4 or GATA-6) or in the presence of normal goat IgG. The latter was used as a control for the goat anti-human GATA-4 polyclonal antibody (Part 965831; R&D Systems, Inc., Minneapolis, MN, USA) and goat anti-human GATA-6 polyclonal antibody (Part 965812; R&D Systems, Inc.) that was used against overexpressed GATA-4 and GATA-6 proteins respectively. Aliquots of nuclear extracts harvested from Huh7 cells that had been transfected with the empty vector were incubated with either goat anti-human GATA-4 polyclonal antibody (Part 965831; R&D Systems, Inc.) or goat anti-human GATA-6 polyclonal antibody (Part 965812; R&D Systems, Inc.). Normal goat IgG (Part 965806; R&D Systems, Inc.) was used as a control for both the antibodies used.
All the DNA–protein mixtures above were migrated through a 6% polyacrylamide gel in 0·5×TBE at 84 V then transferred at 400 mA to a positively charged nylon membrane which was then probed for the DIG-labeled oligonucleotide with specific antibodies as recommended by the DIG-kit manufacturer (Roche Diagnostics).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays for GATA-4 and GATA-6 were performed as specified in the ExactaChiP Human GATA-4 and GATA-6 ChIP Kits, Catalog numbers ECP2606 and ECP1700, respectively (R&D Systems, Inc.), with a few modifications. In experiments for endogenous binding of GATA proteins 4 and 6 to the HAMP promoter, Huh7 cells were grown to 90% confluence in cell culture media containing 10% FBS. However, when evaluating the effect of IL6 on the binding of GATA proteins 4 and 6 to the HAMP promoter, Huh7 cells were grown to 80% confluence in cell culture media containing 10% FBS, the latter (FBS) was then withdrawn from the culture media for 10 h (overnight). The cell culture media was then replenished with fresh media free of FBS with or without IL6 (2·5 ng/ml) for 6 h. Cells (~1×107 cells per 100 mm petri dish) from both experiments were fixed using 1% formaldehyde and the chromatin sheared (~300–1000 bp) using a Misonix Ultrasonic XL-2000 Liquid Processor, by pulsing seven times for 23 s at a power output of 7 W with rest intervals on ice of 45 s between each pulse. The cross-linked protein–DNA samples were pre-cleared with a slurry of protein A plus protein G agarose beads (Catalog number: IP05; EMD Biosciences, Inc., Darmstadt, Germany), 1 mg/ml salmon sperm DNA (Cat 15632-011; Sigma–Aldrich) and 1 mg/ml BSA (Sigma–Aldrich). The samples were then immunoprecipitated using either goat anti-human GATA-4 polyclonal antibody (Part 965831; R&D Systems, Inc.) or goat anti-human GATA-6 polyclonal antibody (Part 965812; R&D Systems, Inc.), overnight at 4 °C while being gently swirled with a rotor. Normal goat IgG (Part 965806; R&D Systems, Inc.) was used as the control for the antibodies in the ChIP assay. The protein–DNA complexes were pulled using streptavidin-conjugated beads from Sigma–Aldrich as a stationary phase. In the PCRs, either 1/20 of the input chromatin used for the immunoprecipitation experiments or 50% (25 μl) of the total immunoprecipitated DNA was used as templates. Water and 10 ng of the full-length HAMP promoter construct (−1234/+73) served as the PCR negative and positive controls respectively. Primers were utilized specific for the proximal region of the HAMP promoter region −155 to +73 bp (Table 1). PCRs were carried out using Phusion high-fidelity DNA Polymerase (New England Biolabs, Pickering, Ontario, Canada) at 95 °C for 5 min, followed by 32 cycles of 94 °C for 1 min, 61 °C for 1 min, and 72 °C for 30 s and completed by a final extension of 5 min at 72 °C. The PCR products were then analyzed by electrophoresis on a 1·5% ethidium bromide-stained agarose gel. When evaluating the effect of IL6 on the binding of GATA proteins to the HAMP promoter, the PCRs were analyzed by densitometry. The readings taken on a gray scale were normalized to the input signal for each treatment (with or without IL6) and then binding was then expressed in relative arbitrary units.
Statistical analysis
The data were analyzed by a one-way ANOVA (Sigma-Stat Version 2.0, Jadel Corporation, San Rafael, CA, USA). Multiple range comparisons of paired means were done using a Fishers LSD test or the Newman–Keuls test. Level of significance was set at P<0·05. Data are reported as mean±S.E.M.
Results
Expression of HAMP in the liver and hepatoma cell lines
Reverse quantitative PCR was performed to evaluate the expression of hepcidin mRNA in the adult human liver and the hepatoma cell lines (Huh7 and HepG2). Using β-actin as a reference gene, the relative mRNA expression of HAMP was higher in Huh7 cells compared with either HepG2 cells or adult human liver, 3·7±0·4, 1·33±0·4, and 2·83±0·12 respectively (P<0·05). The presence of HAMP mRNA in the hepatoma cell lines indicates that these cell lines have the capacity to transcribe HAMP.
Localization of the regulatory elements of the HAMP promoter
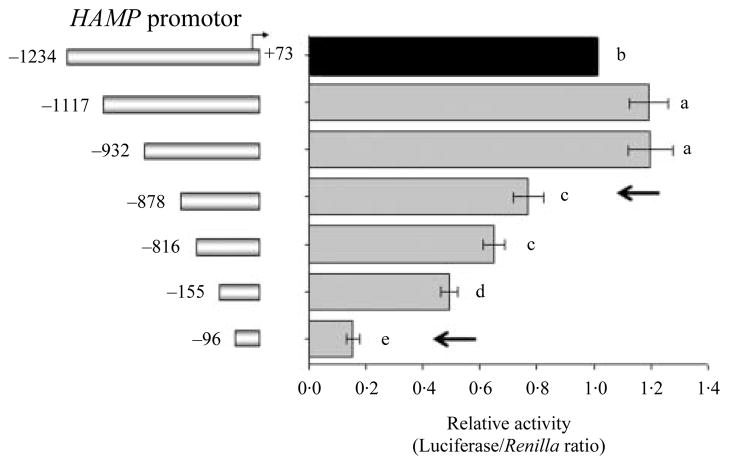
To locate the regulatory elements (RE) important for transcription hepcidin in hepatocytes, a 1307 bp (−1234/+73) HAMP promoter along with 5′-deletion constructs were generated by from human genomic DNA. Promoter fragments of <1000 bp were shown to be sufficient to evaluate basal transcription of HAMP (Courselaud et al. 2002, Bayele et al. 2006, Verga Falzacappa et al. 2007, Vecchi et al. 2009). Deletion of the HAMP promoter from −1234 to −1117 bp led to an increase in promoter activity in Huh7 cells (P<0·05; Fig. 1). Subsequent deletions from −932 to −878 bp and −816 to −155 bp decreased the HAMP promoter activity (P<0·05; Fig. 1). Promoter activity reached ebb following the deletion to −96 bp (P<0·05; Fig. 1). The deletion from −816 to −155 bp lead to the removal of a C/EBPα bind site −231/−222 bp that was previously reported to be important for the basal HAMP promoter activity (Courselaud et al. 2002, Verga Falzacappa et al. 2007). The drastic decrease in promoter activity that followed the deletion of the distal −932/−878 bp and proximal −155/−96 bp regions, indicated the removal of putative RE in the HAMP promoter that are crucial for high promoter activity under steady-state culture conditions.
Figure 1.
Distal and proximal sites of the HAMP promoter are crucial for its basal activity in hepatocytes. Huh7 cells in a 12-well plates were co-transfected with 1580 ng/well of the HAMP promoter constructs (the 5′-end point of each construct is shown to the left of the y-axis) along with 20 ng of phRL-TK (Renilla Luciferase) as an internal control. Total DNA was kept at 1·6 μg/well. Deletion of a distal (−932/−878) and proximal (−155/−96) region as indicated by arrows, severely blunted the HAMP promoter activity in Huh7 cells. Results are presented as mean±S.E.M. of the relative activity (Luciferase/Renilla ratio, gray bars) of each promoter fragment with respect to the −1234/+73 bp (black bar). At least three experiments were done in triplicate using three different DNA preparations for each construct. Different superscripts indicate significant differences between means (a–eP<0·05).
Fine mapping of the regulatory regions in the human HAMP promoter
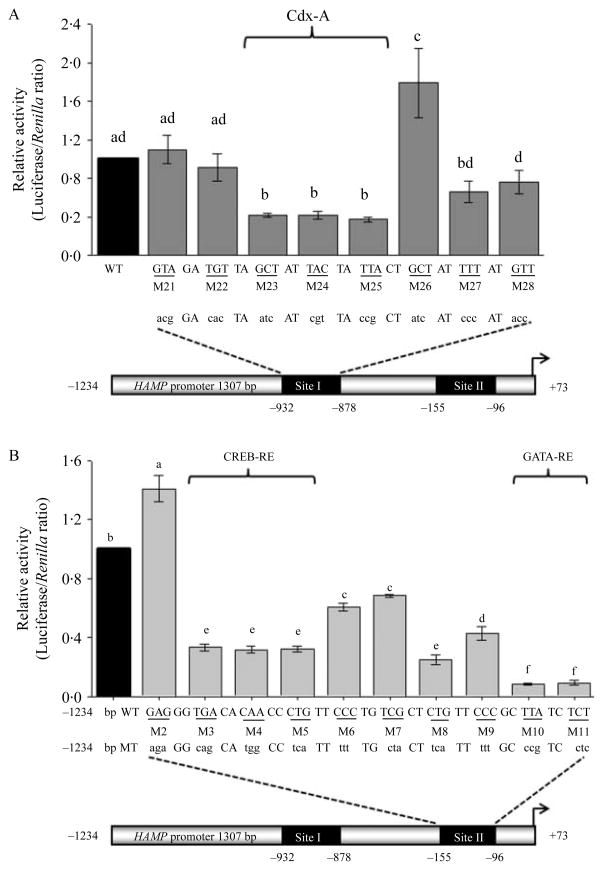
To better define the RE located within the distal (−932/−878) and proximal (−155/−96) promoter regions, sites I and II, respectively, we performed both in silico analysis and site-directed mutagenesis. Using the Transcription Factor SEARCH program to scan the region −932/−878 for potential regulatory elements, we found a A/T-rich region −918/−906 with the WT sequence –GCTATTACTATTA– is a potential CdxA-RE (Heinemeyer et al. 1998). To further investigate the site, eight serially linked scanning mutations were introduced in the distal (−932/−878) HAMP promoter region (Fig. 2A). Each construct harbored a different tri-nucleotide mutation in the context of the −1234/+73 construct. Tri-nucleotide mutations in constructs, M23, M24, and M25 decreased HAMP promoter activity compared with the WT construct (P<0·05; Fig. 2A). We did not investigate the effects of CdxA on HAMP promoter activity nor it’s binding to the A/T-rich sequence since CdxA is not expressed in the normal liver (Gautier-Stein et al. 2003).
Figure 2.
Distal and proximal regulatory elements in the HAMP promoter are crucial for its high basal activity in hepatocytes. Huh7 cells were transfected with 1580 ng/well of either a wild-type (WT, −1234/+73) or a mutated HAMP promoter constructs (mutants, M21–M28 in A and M2–M11 in B) along with 20 ng of phRL-TK (Renilla Luciferase) as an internal control. Tri-nucleotide mutations were introduced within the distal site −932/−878 (A) and proximal site −155/−96 (B) promoter regions of the full-length promoter (−1234/+73). In each case the WT HAMP promoter sequence is shown below the x-axis in upper case while the corresponding mutated tri-nucleotides are in lowercase. At least three experiments were done in triplicate using three different DNA preparations for each construct. Results are the relative activity of the constructs (mean±S.E.M., gray bars) with respect to the WT promoter fragment −1234/+73 (black bars). A different superscript indicates a difference between means (a–fP<0·05).
Ten mutants were generated by introducing serially arranged tri-nucleotide mutations in the proximal promoter region −143/−96 (site II) of the HAMP promoter, in the context of the −1234/+73 construct (Fig. 2B). Transfection of the HAMP promoter constructs in Huh7 cells revealed mutations M3–M11 decreased the promoter activity compared with the WT promoter (P<0·05; Fig. 2B). Site II, encompasses two subregions: a 13-nucleotide (−138/−125) and a 6-nucleotide A/T-rich (−103/−98). Using bioinformatics (ECR Browser and TFSEARCH) we established that the WT sequence –TGACACAACCCTG– in the subregion −138/−125, mutated in the promoter constructs M3–M5 was a CREB-RE (Vecchi et al. 2009); while the WT sequence –TTATCT– in the subregion −103/−98, mutated in the promoter in constructs M10 and M11 is a GATA-RE (Imagawa et al. 1997, Fox et al. 1999, Robert et al. 2002, Obara et al. 2008a,b). To see if the GATA-RE was crucial for the transcriptional activity of the HAMP promoter in HepG2 cells, M10, M11 and the WT constructs were transfected in the hepatoma cells. Mutations of either M10 or M11 decreased the promoter activity to 0·28±0·02 or 0·33±0·03, respectively, compared with the WT promoter activity (1·0, P<0·05).
GATA- and its co-factor FOG modulate the HAMP promoter activity
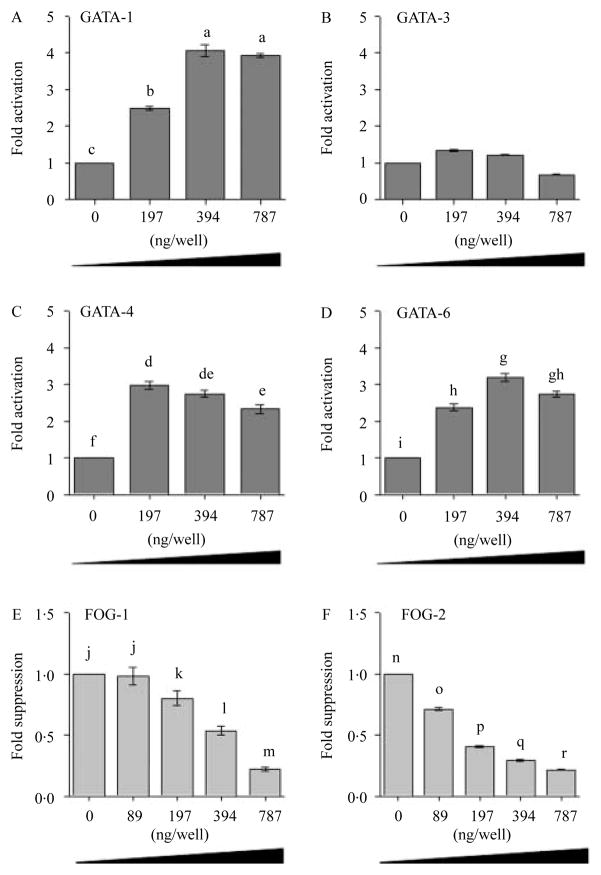
In silico analysis indicated that the GATA-RE in the proximal promoter site −103/−98 had a high probability of binding GATA proteins 1, 3, 4, and 6. We then determined if the GATA proteins 1, 3, 4, and 6 and its modulator protein FOG (1 and 2) were involved in the regulation of the HAMP promoter activity. Co-transfection of Huh7 cells with the HAMP promoter (−1234/+73) along with increasing concentrations of the expression vectors encoding GATA proteins 1, 4, and 6 led to a dose-dependent increase in the HAMP promoter activity (P>0·05; Fig. 3A–D), while the co-transfection of Huh7 cells with the HAMP promoter construct −1234/+73 bp along with increasing concentrations (0–787 ng) of either FOG-1 or FOG-2 expression vector led to a dose-dependent suppression of the HAMP promoter activity (P>0·05; Fig. 3E and F). We concluded that GATA proteins 1, 4, and 6 transactivate the HAMP promoter; while FOG proteins (1 and 2) suppress the activity.
Figure 3.
GATA- and its co-factor Friend of GATA (FOG) modulate the basal activity of the HAMP promoter. Huh7 cells were co-transfected with 787·5 ng/well of the full-length wild-type HAMP promoter construct (−1234/+73) along with increasing concentrations (0–787·5 ng/well) of the expression vectors encoding either GATA (1, 3, 4, and 6, A–D) or FOG (1 or 2, E and F) protein. The total DNA concentration transfected was kept at 1·6 μg/well in 12-well plates by adding an empty vector pcDNA3 (Invitrogen). At least three experiments were done in triplicate using three different DNA preparations for each construct. Results are the mean (±S.E.M.) relative change in luciferase activity (fold activation as dark gray bars or suppression as light gray bars) of the HAMP promoter construct with increase in concentrations of either GATA (1, 4, and 6) or FOG (1 and 2) with respect to the cells co-transfected with the promoter construct along with an empty pcDNA3 expression vector (Invitrogen). A different superscript indicates a statistical difference between means (a–rP<0·05).
FOG-1 and FOG-2 suppress the GATA-induced transactivation of the HAMP in Huh7 cells
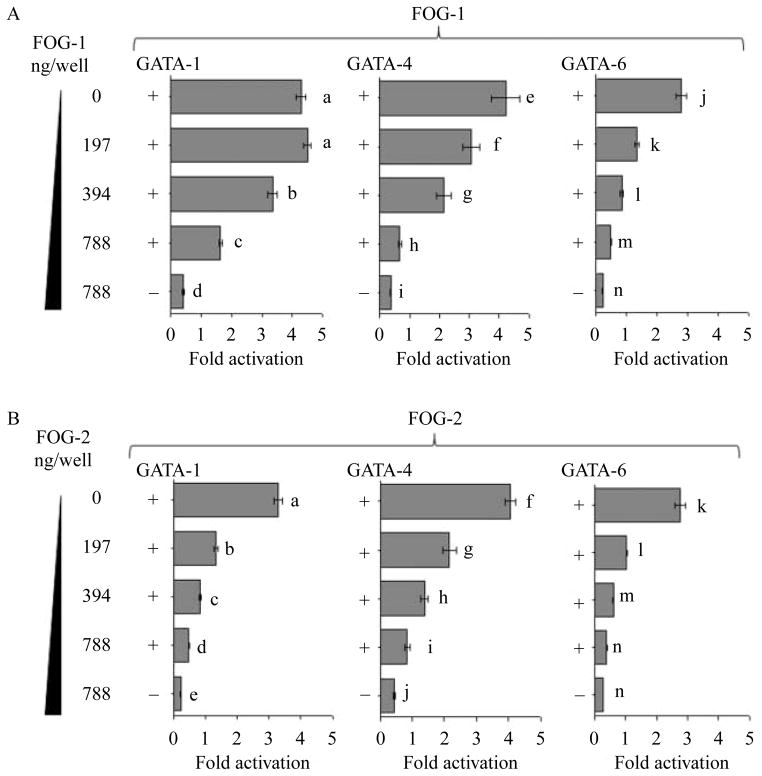
The transactivation capacity of GATA-factors is modulated by their co-factor FOG proteins 1 and 2 (Fox et al. 1999, Robert et al. 2002, Obara et al. 2008a). To establish if FOG-1 and FOG-2 suppressed the exogenous GATA-induced transactivation of HAMP we co-transfected Huh7 cells with the HAMP promoter construct −1234/+73 bp along with expression vectors of either GATA-1 (400 ng/well) or GATA-4 (200 ng/well) or GATA-6 (400 ng/well) in combination with increasing concentrations (0–787 ng/well) of expression vectors encoding FOG proteins (1 or 2). FOG-1 and FOG-2 suppressed the exogenous GATA proteins 1, 4, and 6 transactivation of the HAMP (P>0·05; Fig. 4A and B).
Figure 4.
FOG-1 and FOG-2 repress the exogenous GATA transactivation of the HAMP promoter. Huh7 cells were co-transfected with the full-length wild-type (WT) HAMP promoter construct (−1234/+73, 787·5 ng/well) along with increasing concentrations of expression plasmids encoding either FOG-1 or FOG-2 protein (left of the y-axis, A and B) in addition expression plasmids encoding either GATA-1 (400 ng/well) or GATA-4 (200 ng/well) or GATA-6 (400 ng/well, A and B). To evaluate the basal firefly luciferase activity for controls the WT HAMP promoter (−1234/+73) was co-transfected with the empty expression vector (pcDNA3; Invitrogen). Total DNA concentration transfected was kept at 1600 ng/well in 12-well plates by adding an empty vector (pcDNA3; Invitrogen). At least three experiments were done in triplicate using three different DNA preparations for each construct. The relative luciferase activities of the treatments were determined with respect to the basal firefly luciferase activity of the promoter fragment −1234/+73 (mean±S.E.M.). A different superscript indicates a significant difference in fold suppression (a–nP<0·05).
GATA transactivation of the HAMP is dependent on a GATA-RE in the proximal promoter region
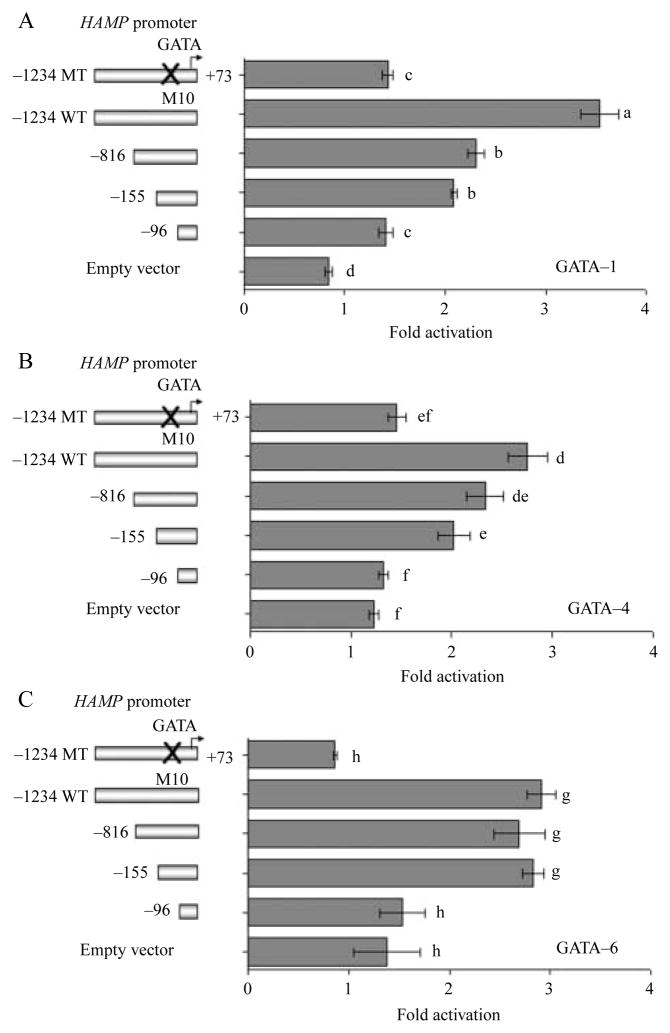
To evaluate whether the transactivation of the HAMP promoter was dependent on a GATA-RE in the promoter region −103/−98 we co-transfected Huh7 cells with different HAMP promoter constructs (M10 and deletion constructs −1234/+73, −816/+73, −155/+73, and −96/+73) along with expression vectors encoding GATA proteins 1, 4, and 6 (Fig. 5A–C). The GATA-RE in the proximal HAMP promoter region −103/−98 bp was mutated and deleted in constructs M10 and −96/+73 bp respectively. Mutation or deletion of the GATA-RE sequence blunted the exogenous GATA-1, GATA-4, and GATA-6 transactivation of the HAMP promoter in Huh7 cells (Fig. 5A–C). The transactivation of the HAMP by GATA proteins 1, 4, and 6 is therefore dependent on the presence of a GATA-RE in proximal promoter region (−103/−98).
Figure 5.
The GATA transactivation of HAMP is dependent on a GATA-regulatory element in the proximal promoter region. Huh7 cells were co-transfected with 787·5 ng/well of either full-length −1234/+73 bp wild-type or a mutant HAMP promoter (carrying a mutated GATA-RE) or 5′-deletion constructs as shown to the left of the y-axis along in addition with either expression plasmids encoding full-length GATA-1 (400 ng/well, A) or GATA-4 (200 ng/well, B) or GATA-6 (400 ng/well, C). To evaluate the control or basal firefly luciferase activity of every HAMP promoter construct, each promoter construct was co-transfected with the empty vector (pcDNA3; Invitrogen). Total DNA concentration transfected was kept at 1600 ng/well in 12-well plates by adding the empty vector (pcDNA3). At least three experiments were done in triplicate using three different DNA preparations for each construct. Results are the relative firefly luciferase activity of the co-transfected cells with respect to the same HAMP promoter co-transfected with only the empty vector (pcDNA3; mean ±S.E.M.). A different superscript indicates a significant difference between means (a–hP<0·05).
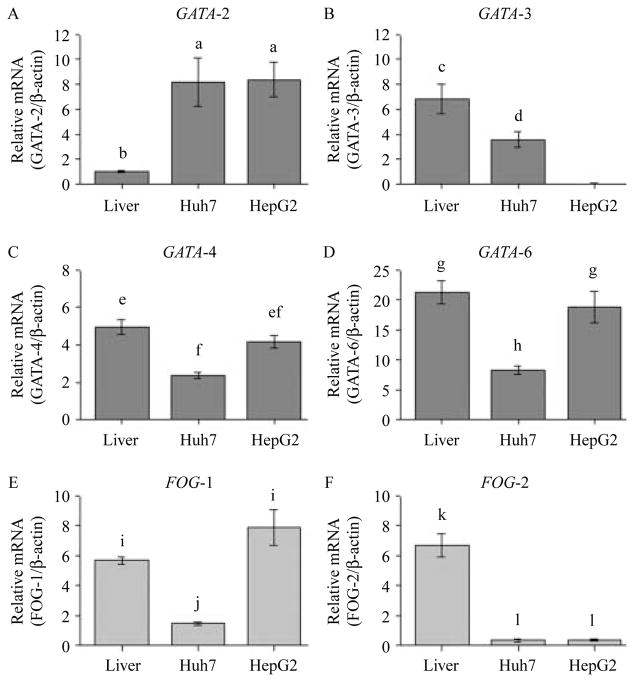
Expression of GATA-factors in adult human liver and hepatoma cell lines
To determine which of the GATA (1–6) and FOG (1 and 2) factors are relevant for the transcription of HAMP under steady cell culture conditions, we used quantitative RT-PCR to determine their expression in the adult human liver and hepatocytes. We established that mRNA for GATA-factors 2, 3, 4, and 6, as well as FOG-1 and FOG-2 are present in the adult human liver and Huh7 cells although the levels of FOG-2 mRNA were almost undetectable in Huh7 (Fig. 6). HepG2 cells expressed GATA-2, GATA-4, and GATA-6, as well as FOG-1 mRNA but GATA-3 and FOG-2 were undetectable in HepG2 cells (Fig. 6). We were unable to detect GATA-1 and GATA-5 mRNA in the adult human liver and the hepatoma cells.
Figure 6.
The expression of GATA (2, 3, 4, and 6; panels A–D) and FOG (1 and 2; panels E and F) in the human liver and hepatoma cell lines. Total RNA from hepatocytes was isolated using TRizol (Invitrogen Canada). An aliquot of the total RNA (2 μg) was used as a template to synthesize the first-strand cDNA using the Omniscript reverse transcriptase-PCR system (Qiagen), 5 μl of diluted cDNA (1:6) was then used as a template in the RT-PCRs. Quantification of gene expression was performed by a Rotor Gene 3000 Real-time DNA Detection System (Montreal Biotech, Kirkland, QC, Canada) with QuantiTect SYBRGreen I PCR kits (Qiagen). Amplifications were performed in duplicate using at least three different preparations of first-strand cDNAs for each of the three different RNA extractions while adult human liver total RNA was purchased from Clontech Laboratories, Inc. Expression levels for GATA (dark gray bars) and FOG (light gray bars) were normalized to the housekeeping gene β-actin. The results are presented as mean (±S.E.M.) of the relative mRNA expression. A different superscript indicates a statistically significant difference between means (a–lP<0·05).
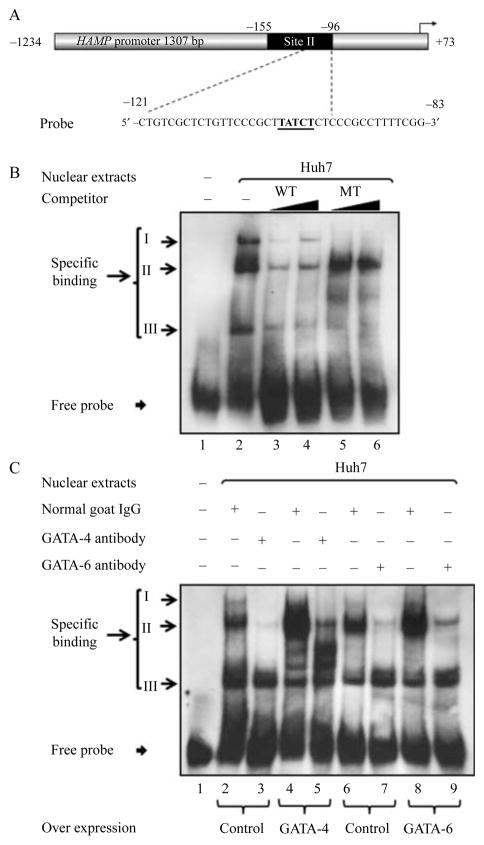
Several proteins bind to the proximal promoter region −121/−83 of the HAMP promoter
An EMSA was performed by nuclear extracts from Huh7 cells and a probe that spanned the region −121/−83 bp of the HAMP promoter to determine whether the DNA–protein binding was specific (Fig. 7A and Table 1). Three distinct probe–nuclear protein complexes were identified, complexes I–III (Figs 7B, lane 2 and 8B, lane 2). Increasing the molar concentrations (125× and 250×) of unlabeled oligonucleotide corresponding to the WT probe displaced all the protein–DNA complexes (Fig. 7B, lanes 3 and 4), while increased concentrations (125× and 250×) of oligonucleotide harboring the mutated GATA-RE at the site −103/−98 (Table 1), was unable to compete with the probe in the formation of complex II; however, it effectively out-competed the WT probe for nuclear protein binding in the formation of complexes I and III (Fig. 7B; lanes 5 and 6).
Figure 7.
Several proteins specifically bind to the −121/−83 bp region of the HAMP promoter. EMSA was used to assess the binding of nuclear protein extracts (7·5 μg) prepared from Huh7 cells to a double-stranded DIG-labeled oligonucleotide corresponding to the wild-type (WT) promoter region −121/−83 bp of the HAMP gene (A; the underlined sequence indicates the GATA-RE). Nuclear protein extracts from Huh7 cells formed specific complexes (I–III) with the probe as indicated to the left of the image (B). Protein binding was challenged by increasing molar concentrations (125× and 250×; black triangles) of unlabeled oligonucleotides corresponding to the WT probe and the mutant (MT), the sequences of the WT and mutant used are shown in Table 1. To evaluate binding of GATA proteins to the HAMP promoter (C) nuclear protein extracts 7·5 μg) were prepared from Huh7 cells 40 h following transfection with 24 μg of either an expression vector encoding the full-length WT GATA protein (GATA-4 (C, lanes 4 and 5) or GATA-6 (C, lanes 8 and 9)) or an empty plasmid vector (pcDNA3 (C, lanes 2, 3, 6, and 7)). Specific binding to a double-stranded DIG-labeled oligonucleotide corresponding to the WT promoter region −121/−83 bp of the HAMP gene (A) of nuclear extracts from Huh7 cells overexpressing either GATA-4 (C, lanes 4 and 5) or GATA-6 (C, lanes 8 and 9) protein was evaluated in the presence of either an antibody directed toward the overexpressed GATA protein (anti-GATA-4 (C, lane 5) or anti-GATA-6 (C, lane 9)) or normal goat serum as a control (C, lanes 4 and 8). Overexpressed GATA protein (4 or 6) incubated with the probe in presence normal goat serum showed a bigger complex II (C, lanes 4 and 6) than control nuclear extracts collected from Huh7 cells transfected with an empty vector [pCDNA3] that were incubated with the probe in presence of normal goat serum (C, lanes 2 and 6). Complex II was abrogated by incubation of the probe with nuclear extracts containing the overexpressed GATA protein along with an antibody directed against the corresponding overexpressed protein (goat anti-human GATA-4 polyclonal antibody or goat anti-human GATA-6 polyclonal antibody for GATA proteins 4 or 6, respectively), as shown in C, lane 5 (in the case of GATA-4) or lane 9 (in the case of GATA-6).
GATA-4 and GATA-6 bind to the proximal promoter region of the HAMP
Since the sequence –TTATCT– is a putative GATA-binding site for nuclear proteins isolated from Huh7 cells that when mutated compromises the ability of nuclear proteins to specifically bind to the DNA (Imagawa et al. 1997, Fox et al. 1999, Robert et al. 2002, Obara et al. 2008a,b), we decided to confirm if GATA proteins 4 and 6 bind to the GATA-RE at −103/−98 of the HAMP promoter. Binding of the probe was performed with nuclear extracts that were harvested from Huh7 cells following overexpression of either GATA-4 (Fig. 7C, lanes 4 and 5) or GATA-6 (Fig. 7C, lanes 8 and 9) protein in the presence of either an antibody directed toward the overexpressed GATA protein (anti-GATA-4 (Fig. 7C, lane 5) or anti-GATA-6 (Fig. 7C, lane 9)) or normal goat serum, the latter was a control for each of the antibodies used (Fig. 7C, lanes 4 and 8). Nuclear extracts collected from Huh7 cells that overexpressed either GATA-4 or GATA-6 incubated with the probe in presence normal goat serum showed a bigger complex II (Fig. 7C, lanes 4 and 6) than the control nuclear extracts that were collected form Huh7 cells transfected with the empty vector pCDNA3 (Invitrogen) and then incubated with the probe in presence of normal goat serum (Fig. 7C, lanes 2 and 6). Incubation with an antibody directed against the overexpressed GATA protein (goat anti-human GATA-4 polyclonal antibody (Part 965831; R&D Systems, Inc.) in case of GATA-4 or goat anti-human GATA-6 polyclonal antibody (Part 965812; R&D Systems, Inc.) in the case of GATA-6) abrogated complexes I and II while having no effect on complex III, as shown in Fig. 7C, lane 5 (in the case of GATA-4) and lane 9 (in the case of GATA-6) compared with when normal goat IgG was used in place of the antibody, Fig. 7C, lane 4 (anti-GATA-4) and lane 8 (anti-GATA-6). The data indicate that complex II corresponds to GATA-4 and GATA-6, since mutation of the GATA-RE failed to displace complex II that was subsequently abrogated by anti-GATA-4 and anti-GATA-6 antibodies respectively (Fig. 7B and C).
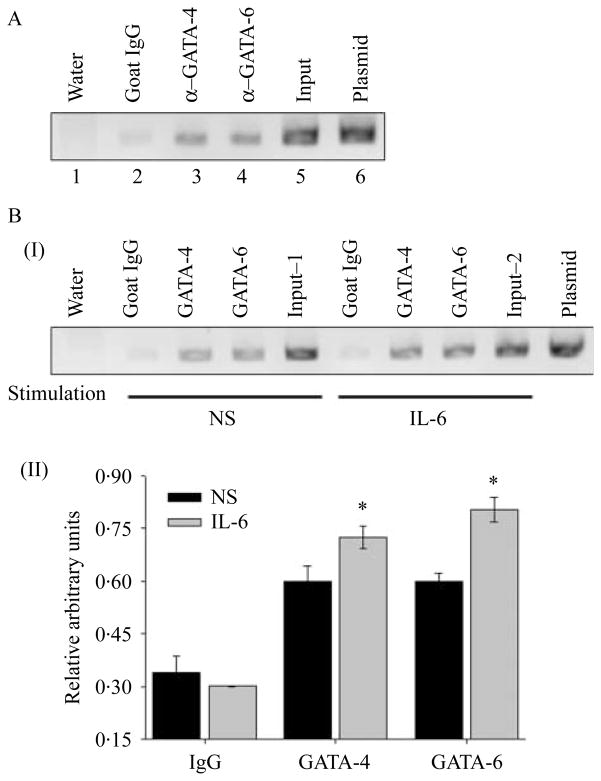
To confirm the recruitment of GATA-4 and GATA-6 to the proximal HAMP promoter in vivo, a ChIP assays were performed with antisera for GATA-4 and GATA-6. In Fig. 8A, a band of the expected size (~230 bp) corresponding to the proximal HAMP promoter was detected in the samples immunoprecipitated with the antisera against GATA-4 (Fig. 8A, lane 3) and GATA-6 (Fig. 8A, lane 4) which indicated that both GATA-4 and GATA-6 are associated with the proximal HAMP promoter in Huh7 cells. Treatment of Huh7 cells with 2·5 ng/ml of IL6 for 6 h increased the binding of GATA proteins 4 and 6 to the proximal HAMP promoter (Fig. 8B).
Figure 8.
GATA proteins 4 and 6 specifically bind to the GATA-RE at −103/−98 of the HAMP promoter. In vivo recruitment of GATA proteins (4 and 6) to the proximal HAMP promoter was determined by ChIP assays in Huh7 cells that were cultured under either steady-state culture conditions (A) or in media free of fetal bovine serum following treatment with or without IL6 (2·5 ng/ml) (B). An aliquot of chromatin (1/20) preparation before immunoprecipitation (input) was used as a positive control for the chromatin that was immunoprecipitated with antisera against GATA-4 (goat anti-human GATA-4 polyclonal antibody; A, lane 3) and GATA-6 (goat anti-human GATA-6 polyclonal antibody, lane 4). Normal goat serum (IgG; A, lane 2) was used as a negative control for the ChIP. A 230 bp DNA fragment containing the GATA-4 and GATA-6 binding sites (GATA-RE) within the HAMP promoter was amplified by PCR. A plasmid containing the HAMP promoter served as a positive control for the PCR (C, lane 6) while water served as a negative control (C, lane 1). When evaluating binding of GATA proteins to the HAMP promoter following treatment with IL6 for 6 h, the PCRs were analyzed by densitometry following electrophoresis on a 1·5% ethidium bromide-stained agarose gel (B, I), two readings for GATA-4 or GATA-6 binding were taken on a gray scale for each experiment and normalized to the input signal, input 1 or 2 for treatment with no stimulation (NS) or with IL6 stimulation respectively. The results are presented as the mean relative arbitrary units of three experiments (B, II; *P<0·05).
We concluded that in Huh7 cells, endogenous GATA-factors 4 and 6 are the cell-specific transcription factors involved in the transcriptional regulation of HAMP; while FOG-1 modulates the GATA-4 and GATA-6 induced transcription of HAMP through the GATA-RE in the proximal HAMP promoter region.
Discussion
In this study, HAMP mRNA was detected in both Huh7 and HepG2 cells, which indicated that the cell lines had the capacity to transcribe hepcidin. Promoter analysis in Huh7 cells revealed an increase in promoter activity when the 5′-end of the HAMP promoter fragment was deleted from −1234 to −1117 bp. The increase in activity may have been due to elimination of repressive elements upstream of −1117 bp. The presence of alternating d(TG)n-type purine–pyrimidine di-nucleotide repeats in the deleted region could have suppressed the HAMP promoter activity (Naylor & Clark 1990, Bergeron et al. 2011); however, in silico analysis of the deleted region revealed a potential CP-2 transcription factor binding site −1191/−1181 with a WT sequence –GGCCATGCCCT– (Heinemeyer et al. 1998). CP-2 is expressed in hepatocytes and acts either as a transcription enhancer or repressor (Veljkovic & Hansen 2004, Jang et al. 2010). CP-2 binds to the transferrin promoter and upregulates its transcription (Jang et al. 2010). High levels of transferrin and CP-2 occur in concert with the downregulation of hepcidin during iron deficiency anemia (Macedo & de Sousa 2008). However, during inflammation high levels of hepcidin are coincident with the downregulation of transferrin and CP-2 (Ritchie et al. 1999) and as such, CP-2 might be a transcriptional repressor of HAMP.
In this study, deletion from −816 to −155 bp decreased HAMP promoter activity in Huh7 cells, consistent with the elimination of the oxygen-sensitive transcription factor C/EBPα binding site, −231/−222 (Courselaud et al. 2002, Verga Falzacappa et al. 2007). C/EBPα was shown to increase the basal expression of hepcidin in hepatocytes (Courselaud et al. 2002, Verga Falzacappa et al. 2007). The introduction of site-directed mutations within the site −156/−96 of the promoter construct −1234/+73 revealed two regulatory subregions, −138/−125 and −103/−98 which when mutated blunted the HAMP basal activity by 70 and 90% respectively. The upstream element in the promoter subregion −138/−125 is a CREB-RE to which the liver-specific transcription factor CREB-H binds and increases the basal expression of hepcidin (Vecchi et al. 2009). CREB-H is also involved in the upregulation of hepcidin during inflammation (Vecchi et al. 2009). Mutation of the downstream promoter subregion (−103/−98) with the WT sequence –TTATC– severely blunted the promoter activity in both Huh7 and HepG2 cells, as was recently reported in HepG2 cells, while the current manuscript was under review (Island et al. 2011). We established that the WT sequence –TTATCT– located at position −103/−98 of the HAMP promoter is a GATA-RE (Imagawa et al. 1997, Fox et al. 1999, Tremblay et al. 2001, Robert et al. 2002, Obara et al. 2008a, Island et al. 2011). In our study, GATA proteins (4 and 6) were able to bind to the GATA-RE and activate the transcription of hepcidin in Huh7 cells. However, Island et al. (2011), using protein extracts from HepG2 cells were unable to demonstrate the binding of GATA-6. In this study, using nuclear extracts from Huh7 cell in which GATA-6 was over-expressed, we showed GATA-6 was able to bind to the GATA-RE in the HAMP promoter, likewise, endogenous GATA-6 was able to bind to the HAMP promoter in vivo. The disparity between both studies could have been due to the difference in cell types or may indicate differences in affinity with which the GATA proteins 4 and 6 bind to the GATA-RE, with the former having a higher affinity than the latter. A higher GATA-4 binding affinity than GATA-6 for the GATA-RE may explain why we detected the same endogenous GATA protein binding intensities to the HAMP promoter although the expression levels of GATA-6 were higher than those of GATA-4; in addition, GATA-4 had a higher trans-activation potency than GATA-6. Binding affinity is dependent on the nucleotide sequences flanking the GATA-RE or the interactions of GATA proteins with other co-transcription factors (Molkentin 2000). Despite the differential affinities, GATA-4 was reported to facilitate the binding of GATA-6 to the GATA-RE and functionally acting in concert with it (Molkentin 2000). In postnatal cardiomyocytes, GATA-4 was shown to have a higher binding affinity than GATA-6 for the GATA-RE in the promoter region of the atrial natriuretic factor (ANF) and B-type atrial natriuretic peptide (BNP) genes (Charron et al. 1999); however, the two GATA proteins (4 and 6) co-localize and interact functionally and physically to provide cooperative activation of the ANF and BNP promoters (Charron et al. 1999). In this study, GATA proteins 4 and 6 were able to act in synergy in the transactivation of HAMP (data not shown). Although GATA-factors have similar DNA-binding properties, they possess distinct spatial–temporal expression patterns and play different physiological functions (Matsuda et al. 1994, Tremblay et al. 2001, Cantor et al. 2002, Dame et al. 2004, Divine et al. 2004). In hepatocytes, the expression of GATA-1 and GATA-4 is limited to the prenatal period while that of GATA-6 to both the pre- and postnatal period (Dame et al. 2004, Divine et al. 2004). Spatial–temporal distribution therefore places GATA-1, GATA-4, and GATA-6 as mediators of basal transcription of hepcidin in fetal hepatocytes while GATA-6 as the mediator in postnatal hepatocytes. GATA proteins regulate other genes through functional interactions with FOG-1 and FOG-2 (Crispino et al. 1999, Vyas et al. 1999, Cantor et al. 2002, Katz et al. 2002, Obara et al. 2008a).
In all vertebrates, FOG is capable of interacting with all GATA proteins since the N-terminal zinc fingers of GATA proteins that forms a physical and functional bridge with FOG is highly conserved (Crispino et al. 1999, Vyas et al. 1999, Cantor et al. 2002, Katz et al. 2002, Obara et al. 2008a). FOG-1 and FOG-2 can act either as enhancers or repressor of the GATA-mediated transcription (Tremblay et al. 2001, Robert et al. 2002, Dame et al. 2004, Obara et al. 2008a). In this study, FOG-1 and FOG-2 suppressed the endogenous or exogenous GATA-dependent activation of HAMP. FOG-2 was a more potent suppressor of the GATA-dependent activation of the HAMP promoter. The apparent differential potencies of FOG proteins could be a mechanism for fine-tuning the suppression of HAMP in hepatocytes (Robert et al. 2002). The repressive nature of FOG proteins is in part due to their association with the potent transcriptional co-repressor, C-terminal-binding protein-2 (Holmes et al. 1999, Svensson et al. 2000a, Robert et al. 2002). Therefore, it is likely that FOG proteins serve a crucial physiological role as modulators of GATA-dependent transcription of HAMP given the in vivo roles it plays in the regulation of other genes (Matsuda et al. 1994, Tremblay et al. 2001, Cantor et al. 2002, Dame et al. 2004, Divine et al. 2004).
At present, promoter studies have shown that increases in the concentration of iron and inflammation upregulate the transcription of hepcidin through BMP-RE site −84/−79 (Verga Falzacappa et al. 2008) and STAT3-RE site −72/−64 (Pietrangelo et al. 2007, Verga Falzacappa et al. 2007), respectively, while iron deficiency and hypoxia downregulate it through the C/EBP site −231/−222 (Nicolas et al. 2002). It is not clear as to how the physiological changes in concentration of iron or oxygen tension in blood affect either FOG or GATA and subsequently hepcidin. In HepG2 cells, the concentration of GATA-4 was neither affected by proinflammatory cytokines (IL1β and tumor necrosis factor-α) nor hypoxia (Dame et al. 2004). In this study, we noted an increased binding of GATA proteins (4 and 6) to the HAMP promoter in Huh7 cells following treatment with IL6 as such it is likely that during inflammation the increased recruitment of GATA proteins to the HAMP promoter contributes to the higher expression of hepcidin in hepatocytes. IL6 is a proinflammatory cytokine responsible for the increased expression of HAMP during inflammation or following treatment with lipopolysaccharide (Andrews 2004). While the current manuscript was under review, Island et al. (2011) reported that in HepG2 cells mutation of the GATA-RE impaired the IL6 induction of hepcidin gene expression, but did not prevent the response to BMP6. They suggested that GATA-4 could be involved in the control of hepcidin expression and that alteration of its expression may contribute to the development of iron-related disorders (Island et al. 2011). Incidentally, GATA-4 was shown to be absent in hepatocytes that were isolated from normal mice and humans during the postnatal period (Dame et al. 2004, Divine et al. 2004). The absence of GATA-4 in hepatocytes during the postnatal period was also cited as the reason for the switch in the expression of the EPO gene from fetal hepatocytes to the adult kidney in mice and humans (Dame et al. 2004). The changes in concentrations of the proinflammatory cytokines (IL6) in the body during the postnatal period may therefore affect levels of either GATA-6 or FOG protein (1 and 2) in hepatocytes and the nature of the response being dependent on the type and strength of the input signal: the increase in GATA-6 with or without a decrease in FOG upregulating the transcription of HAMP; while the increase in concentration of FOG proteins with or without a decrease in GATA-6 concentrations downregulating the transcription of HAMP. In mice, inactivation of either GATA (4 and 6) or FOG (1 and 2) causes embryonic death, hindering in vivo assessment of the role of transcriptional factors in the expression of hepcidin in hepatocytes (Kuo et al. 1997, Molkentin et al. 1997, Zhao et al. 2005).
In this study, we concluded that the GATA regulatory region in the proximal HAMP promoter is important for the basal expression of hepcidin in hepatocytes, GATA-factors 4 and 6 bind to the GATA-RE –TTATCT–in the promoter region −103/−98 and upregulate the transcription of HAMP, while FOG through its ability to suppress both the endogenous and exogenous GATA-induced transcription of HAMP acts as an alternative port for the regulation of hepcidin.
Acknowledgments
Funding
This work was supported by grants from the Canadian Institutes of Health Research (CIHR, grant no. MOP44045) and Natural Sciences and Engineering Research Council of Canada (NSERC, grant number 298515-2012) to M M S. M M S is a recipient of a Research Scholarship (Senior) from the FRSQ (Fonds de la recherche en santé du Québec). E T B is a recipient of postdoctoral fellowships from the Institut du Cancer de Montréal and Centre de Recherche du CHUM, Université de Montréal, QC, Canada.
We are grateful to Dr Robert Viger for the GATA (1, 3, 4, and 6) and FOG (1 and 2) expression vectors, Dr Jacques Tremblay for the pXP1 luciferase reporter plasmid, and Dr Rejean Lapointe for human tissue cDNA. We also thank Annie Calvé, Drs Muhammad Zafarullah and Richard Betrand for their technical help and advice.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution statement
E T B developed, designed, and executed the experiments, collected, analyzed, and interpreted the data and then wrote the manuscript. M M S assisted in developing the study, discussing the data, and writing the manuscript.
References
- Andrews NC. Anemia of inflammation: the cytokine-hepcidin link. Journal of Clinical Investigation. 2004;113:1251–1253. doi: 10.1172/JCI21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayele HK, McArdle H, Srai SK. Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood. 2006;108:4237–4245. doi: 10.1182/blood-2005-07-027037. [DOI] [PubMed] [Google Scholar]
- Bergeron F, Bagu ET, Tremblay JJ. Transcription of platelet-derived growth factor receptor {alpha} in Leydig cells involves specificity protein 1 and 3. Journal of Molecular Endocrinology. 2011;46:125–138. doi: 10.1530/JME-10-0145. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakar-yocytic maturation. Molecular and Cellular Biology. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Molecular and Cellular Biology. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Ikawa Y, Todokoro K. GATA-1 transactivates erythropoietin receptor gene, and erythropoietin receptor-mediated signals enhance GATA-1 gene expression. Nucleic Acids Research. 1991;19:3843–3848. doi: 10.1093/nar/19.14.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constante M, Wang D, Raymond VA, Bilodeau M, Santos MM. Repression of repulsive guidance molecule C during inflammation is independent of Hfe and involves tumor necrosis factor-alpha. American Journal of Pathology. 2007;170:497–504. doi: 10.2353/ajpath.2007.060437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, et al. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Crosstalk between C/EBP pathway and iron metabolism. Journal of Biological Chemistry. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Molecular Cell. 1999;3:219–228. doi: 10.1016/S1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Dame C, Sola MC, Lim KC, Leach KM, Fandrey J, Ma Y, Knopfle G, Engel JD, Bungert J. Hepatic erythropoietin gene regulation by GATA-4. Journal of Biological Chemistry. 2004;279:2955–2961. doi: 10.1074/jbc.M310404200. [DOI] [PubMed] [Google Scholar]
- Divine JK, Staloch LJ, Haveri H, Jacobsen CM, Wilson DB, Heikinheimo M, Simon TC. GATA-4, GATA-5, and GATA-6 activate the rat liver fatty acid binding protein gene in concert with HNF-1alpha. American Journal of Physiology Gastrointestinal and Liver Physiology. 2004;287:G1086–G1099. doi: 10.1152/ajpgi.00421.2003. [DOI] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO Journal. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier-Stein A, Domon-Dell C, Calon A, Bady I, Freund JN, Mithieux G, Rajas F. Differential regulation of the glucose-6-phosphatase TATA box by intestine-specific homeodomain proteins CDX1 and CDX2. Nucleic Acids Research. 2003;31:5238–5246. doi: 10.1093/nar/gkg747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanninen MM, Haapasalo J, Haapasalo H, Fleming RE, Britton RS, Bacon BR, Parkkila S. Expression of iron-related genes in human brain and brain tumors. BioMed Central Neuroscience. 2009;10:36. doi: 10.1186/1471-2202-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Research. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes M, Turner J, Fox A, Chisholm O, Crossley M, Chong B. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. Journal of Biological Chemistry. 1999;274:23491–23498. doi: 10.1074/jbc.274.33.23491. [DOI] [PubMed] [Google Scholar]
- Imagawa S, Yamamoto M, Miura Y. Negative regulation of the erythropoietin gene expression by the GATA transcription factors. Blood. 1997;89:1430–1439. [PubMed] [Google Scholar]
- Island ML, Fatih N, Leroyer P, Brissot P, Loreal O. GATA-4 transcription factor regulates hepatic hepcidin expression. Biochemical Journal. 2011;437:477–482. doi: 10.1042/BJ20110225. [DOI] [PubMed] [Google Scholar]
- Isoda M, Hanawa H, Watanabe R, Yoshida T, Toba K, Yoshida K, Kojima M, Otaki K, Hao K, Ding L, et al. Expression of the peptide hormone hepcidin increases in cardiomyocytes under myocarditis and myocardial infarction. Journal of Nutritional Biochemistry. 2010;21:749–756. doi: 10.1016/j.jnutbio.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Jang SM, Kim JW, Kim CH, An JH, Kang EJ, Kim CG, Kim HJ, Choi KH. Control of transferrin expression by beta-amyloid through the CP2 transcription factor. FEBS Journal. 2010;277:4054–4065. doi: 10.1111/j.1742-4658.2010.07801.x. [DOI] [PubMed] [Google Scholar]
- Katz SG, Cantor AB, Orkin SH. Interaction between FOG-1 and the corepressor C-terminal binding protein is dispensable for normal erythropoiesis in vivo. Molecular and Cellular Biology. 2002;22:3121–3128. doi: 10.1128/MCB.22.9.3121-3128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Molecular and Cellular Biology. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaksiz H, Fein E, Redecker P, Stremmel W, Adler G, Cetin Y. Pancreatic beta-cells express hepcidin, an iron-uptake regulatory peptide. Journal of Endocrinology. 2008;197:241–249. doi: 10.1677/JOE-07-0528. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes and Development. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Molecular and Cellular Biology. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo MF, de Sousa M. Transferrin and the transferrin receptor: of magic bullets and other concerns. Inflammation and Allergy Drug Targets. 2008;7:41–52. doi: 10.2174/187152808784165162. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kobune Y, Noda C, Ichihara A. Expression of GATA-binding transcription factors in rat hepatocytes. FEBS Letters. 1994;353:269–272. doi: 10.1016/0014-5793(94)01062-5. [DOI] [PubMed] [Google Scholar]
- Merle U, Fein E, Gehrke SG, Stremmel W, Kulaksiz H. The iron regulatory peptide hepcidin is expressed in the heart and regulated by hypoxia and inflammation. Endocrinology. 2007;148:2663–2668. doi: 10.1210/en.2006-1331. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. Journal of Biological Chemistry. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes and Development. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Mura C, Le Gac G, Jacolot S, Ferec C. Transcriptional regulation of the human HFE gene indicates high liver expression and erythropoiesis coregulation. FASEB Journal. 2004;18:1922–1924. doi: 10.1096/fj.04-2520fje. [DOI] [PubMed] [Google Scholar]
- Naylor LH, Clark EM. d(TG)n.d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Research. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Viatte L, Bennoun M, Beaumont C, Kahn A, Vaulont S. Hepcidin, a new iron regulatory peptide. Blood Cells Molecules and Diseases. 2002;29:327–335. doi: 10.1006/bcmd.2002.0573. [DOI] [PubMed] [Google Scholar]
- Obara N, Suzuki N, Kim K, Nagasawa T, Imagawa S, Yamamoto M. Repression via the GATA box is essential for tissue-specific erythropoietin gene expression. Blood. 2008a;111:5223–5232. doi: 10.1182/blood-2007-10-115857. [DOI] [PubMed] [Google Scholar]
- Obara N, Ueno Y, Fukushima K, Nakagome Y, Kakazu E, Kimura O, Wakui Y, Kido O, Ninomiya M, Kogure T, et al. Transient elastography for measurement of liver stiffness measurement can detect early significant hepatic fibrosis in Japanese patients with viral and nonviral liver diseases. Journal of Gastroenterology. 2008b;43:720–728. doi: 10.1007/s00535-008-2225-2. [DOI] [PubMed] [Google Scholar]
- Pietrangelo A. Molecular insights into the pathogenesis of hereditary haemochromatosis. Gut. 2006;55:564–568. doi: 10.1136/gut.2005.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O. Reference distributions for the negative acute-phase proteins, albumin, transferrin, and transthyretin: a comparison of a large cohort to the world’s literature. Journal of Clinical Laboratory Analysis. 1999;13:280–286. doi: 10.1002/(SICI)1098-2825(1999)13:6<280::AID-JCLA5>3.0.CO;2-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert NM, Tremblay JJ, Viger RS. Friend of GATA (FOG)-1 and FOG-2 differentially repress the GATA-dependent activity of multiple gonadal promoters. Endocrinology. 2002;143:3963–3973. doi: 10.1210/en.2002-220280. [DOI] [PubMed] [Google Scholar]
- Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nature Genetics. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. PNAS. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson EC, Huggins GS, Dardik FB, Polk CE, Leiden JM. A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. Journal of Biological Chemistry. 2000a;275:20762–20769. doi: 10.1074/jbc.M001522200. [DOI] [PubMed] [Google Scholar]
- Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, Dardik FB, Leiden JM. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nature Genetics. 2000b;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. PNAS. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Viger RS. Transcription factor GATA-4 enhances Mullerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Molecular Endocrinology. 1999;13:1388–1401. doi: 10.1210/me.13.8.1388. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Viger RS. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology. 2001;142:977–986. doi: 10.1210/en.142.3.977. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Robert NM, Viger RS. Modulation of endogenous GATA-4 activity reveals its dual contribution to Mullerian inhibiting substance gene transcription in Sertoli cells. Molecular Endocrinology. 2001;15:1636–1650. doi: 10.1210/me.15.9.1636. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/S0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes and Development. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40. doi: 10.1016/j.gene.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. Journal of Molecular Medicine. 2008;86:531–540. doi: 10.1007/s00109-008-0313-7. [DOI] [PubMed] [Google Scholar]
- Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Experimental Hematology. 1995;23:99–107. [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Molecular and Cellular Biology. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]