Abstract
Radial spokes are ubiquitous components of motile cilia and flagella and play an essential role in transmitting signals that regulate the activity of the dynein motors, and thus ciliary and flagellar motility. In some organisms the 96 nm axonemal repeat unit contains only a pair of two spokes, RS1 and RS2, while most organisms have spoke triplets with an additional spoke RS3. The spoke pair in Chlamydomonas flagella has been well characterized, while spoke triplets have received less attention. Here, we used cryo-electron tomography and subtomogram averaging to visualize the 3D structure of spoke triplets in Strongylocentrotus purpuratus (sea urchin) sperm flagella in unprecedented detail. Only small differences were observed between RS1 and RS2, but the structure of RS3 was surprisingly unique and structurally different from the other two spokes. We observed novel doublet specific features that connect RS2, RS3 and the nexin-dynein regulatory complex, three key ciliary and flagellar structures. The distribution of these doublet specific structures suggests that they could be important for establishing the asymmetry of dynein activity required for the oscillatory movement of cilia and flagella. Surprisingly, a comparison with other organisms demonstrated both that this considerable radial spoke heterogeneity is conserved and that organisms with radial spoke pairs contain the basal part of RS3. This conserved radial spoke heterogeneity may also reflect functional differences between the spokes and their involvement in regulating ciliary and flagellar motility.
Keywords: cryo-electron tomography, axoneme, radial spoke triplet, RS3, RS3S
INTRODUCTION
The general architecture of motile cilia and flagella is highly conserved, such as the nearly universal “9+2” arrangement of the microtubules in the axonemal core. Each of the peripheral 9 doublet microtubules (DMTs) that surround the two central single microtubules of the central pair complex (CPC) is built from a 96 nm long unit that repeats along the length of the axoneme. The dynein motors are arranged in two rows, the inner and outer dynein arms, along the length of the A-tubule of each DMT and can “walk” along the B-tubule of the neighboring DMT to generate sliding motion between the doublets. The nexin-dynein regulatory complex (N-DRC), which connects neighboring DMTs [Heuser et al., 2009; Nicastro et al., 2006; Porter and Sale, 2000], is thought to restrict this sliding motion and, thus, to convert sliding into the bending motion of the axoneme [Satir, 1968; Summers and Gibbons, 1971]. For effective beating and to generate the propagating, oscillatory motions typical of beating cilia and flagella, let alone to vary beating patterns and waveforms in response to environmental signals, the action of the tens of thousands of dynein motors in a single organelle must be precisely controlled. Genetic, structural and biochemical studies of Chlamydomonas flagella suggest that several axonemal components are involved in this regulation. The CPC, the radial spokes (RSs) [Warner and Satir, 1974; Piperno et al., 1981; Diener et al., 1993; Smith and Lefebvre, 1997; Mitchell and Sale, 1999; Smith and Yang, 2004], the I1 inner dynein arm [Porter and Sale, 2000; Wirschell et al., 2007; Wirschell et al., 2009] and the N-DRC [Heuser et al., 2009; Rupp and Porter, 2003] are all thought to transmit mechanical and/or chemical signals that control dynein activity and, thus, ciliary and flagellar bending movement.
The RSs are anchored to the DMT and project toward the CPC, where the spoke heads transiently interact with the CPC projections [Goodenough and Heuser, 1985; Mitchell and Sale, 1999; Omoto et al., 1999; Smith and Lefebvre, 1997; Warner and Satir, 1974]. Depending on the species, the RSs repeat in pairs (e.g., Chlamydomonas, Sarcophaga) or triplets (e.g., Tetrahymena, Trypanosoma, sea urchins, mammals) every 96 nm along the DMTs, with highly conserved intervals of 32 nm between RS1-RS2, and then either 64 nm between RS2-RS1 for RS pairs, or 24 nm between RS2-RS3 and 40 nm between RS3-RS1 for RS triplets [Warner, 1970; Dentler and Cunningham, 1977; Goodenough and Heuser, 1985; Koyfman et al., 2011; Nicastro et al., 2005; Nicastro et al., 2006; Olson and Linck, 1977]. In Chlamydomonas and other organisms, mutations that cause RS defects result usually in ciliary and flagellar paralysis [Afzelius, 1985; Afzelius and Eliasson, 1979; Witman et al., 1978]. Several studies of human respiratory disease or Primary Ciliary Dyskinesia (also called immotile cilia syndrome) showed that the RSs were shorter or missing in the patient cilia [Afzelius, 2004; Plesec et al., 2008; Sturgess et al., 1979]. In addition, a recent study linked mutations of human spoke head proteins to Primary Ciliary Dyskinesia [Castleman et al., 2009]. Therefore, it is important to understanding how RSs are involved in the regulation of dynein activity and ciliary motility, and how RS malfunction relates to human disease.
Biochemical studies have identified at least 23 radial spoke proteins in Chlamydomonas flagella, including proteins that play essential roles in signal transduction, such as calmodulin, the EF hand-containing proteins and the A-kinase anchor protein [Patel-King et al., 2004; Yang et al., 2006]. Several lines of evidence suggest that the RSs serve as mechano-chemical transducers that transmit signals from the CPC to the dynein arms to regulate dynein-driven movement, which generates the cilia and flagella waveform [Smith and Yang, 2004]. However, a complete understanding of the mechanism by which the RSs control this motility will require detailed knowledge of both the 3D structure and the subunit organization of the RSs and an understanding of how these components interact with other axonemal complexes to facilitate regulatory signal transduction.
Previous classical electron microscopy studies described the RSs as T-shaped structures composed of a thin stalk that attaches to the A-tubule of the DMTs and a “T-bar”-shaped head that is adjacent to the central pair projections [Chasey, 1972; Chasey, 1974; Curry and Rosenbaum, 1993; Goodenough and Heuser, 1985; Warner, 1970; Warner and Satir, 1974]. Our recent cryo-electron tomography (cryo-ET) study of the RS pair in Chlamydomonas flagella revealed the detailed 3D structure of RS1 and RS2 [Barber et al., 2011]. Major findings of this investigation included: i) the visualization of direct connections between the RS bases and inner dynein arms (IDAs) and the N-DRC, respectively, ii) the ultrastructure of the two spoke heads and interactions between them, and iii) the observation of a short structure, termed the Radial Spoke 3 Stand-in (RS3S), that occupies the site where a third spoke, RS3, would be found in organisms with radial spoke triplets. RS3S, however, did not resemble any part of the two radial spokes, RS1 and RS2 [Barber et al., 2011]. It is unknown why some organisms have motile cilia and flagella with only two radial spokes, such as Chlamydomonas, while most have triplets, including mammalian cilia [Olson and Linck, 1977], but because most of our knowledge about the RSs originates from studying Chlamydomonas flagella, little is known about RS3.
In this study, we used cryo-ET and image processing to analyze the ultrastructure of RS triplets. Our 3D subtomographic averages of sea urchin sperm flagella show unprecedented detail and allowed two surprising discoveries. First, they revealed significant structural heterogeneity between RS3 and the other two RSs, not only in sea urchin, but also in protists. Second, our results showed structural similarities between RS3 and the RS3S complex in Chlamydomonas flagella, suggesting that RS3S is the basal part of RS3 in an organism that was thought to contain only a radial spoke pair. RS3 also shows doublet specific features, indicating that RS3 may play a unique role in regulating ciliary and flagellar bending.
RESULTS
Cryo-ET reveals the overall arrangement of the RS triplets in sea urchin flagella
We used cryo-ET to determine the 3D structure of RS triplets from flagella of the sea urchin Strongylocentrotus purpuratus. Eight tomograms of intact, frozen-hydrated sea urchin sperm flagella were selected for subtomographic averaging (for sample tomogram see Supporting Information Fig. S1A and S1B). We aligned and averaged 1300 axonemal repeats from all nine DMTs, resulting in a subtomographic average with a relatively uniform resolution of 3.6 nm in the center of the 96 nm repeat unit and corrected missing wedge (Supporting Information Fig. S1C and S1D). To obtain the best possible 3D images of each RS, local alignments with smaller subtomographic volumes were calculated centered on each RS individually. The final resolutions for locally aligned and averaged structures of RS1, RS2 and RS3 were 3.8 nm, 3.6 nm and 3.8 nm, respectively (Fig. 1E–1P; Supporting Information Fig. S1D).
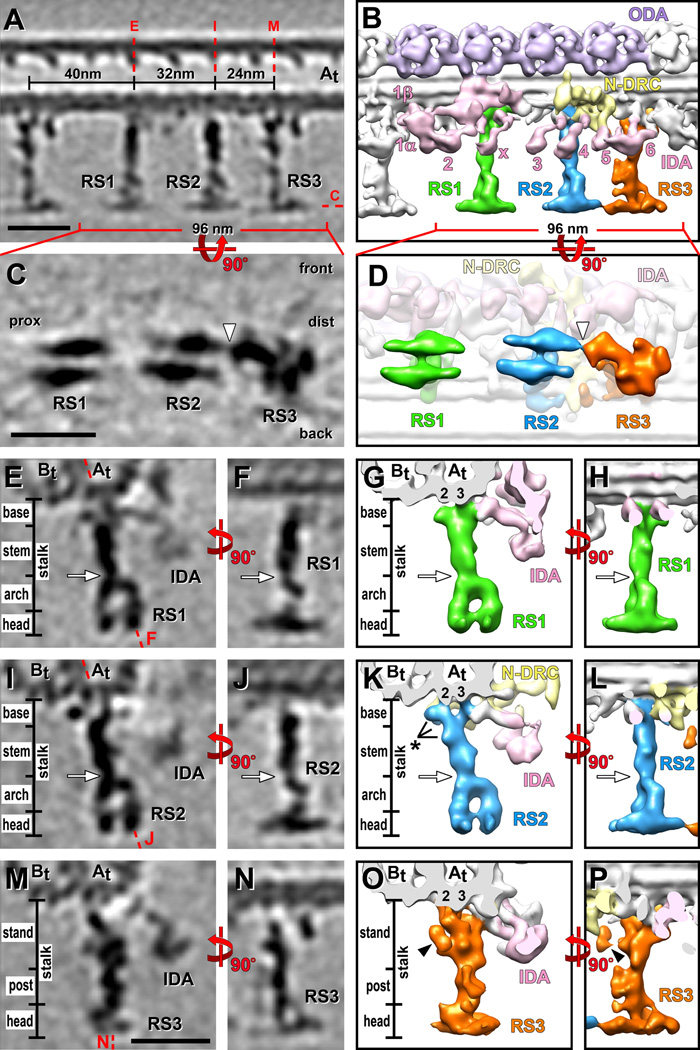
Figure 1. 3D structure of sea urchin RSs.
Tomographic slices (A, C, E, F, I, J, M and N) and isosurface renderings (B, D, G, H, K, L, O and P) show the averaged axonemal repeat in longitudinal views (A–D, F, H, J, L, N and P), from the front (A–B, F, H, J, L, N and P), the bottom (C and D) (looking from the central pair towards the DMT) and in cross-section (E, G, I, K, M and O). (A–D) Overview of the RS arrangement. Red dotted lines in (A) indicate the locations of the tomographic slices shown in (C, E, I and M). Note that both heads of RS1 and RS2 consist of two rotationally symmetric halves, while the RS3 head has an irregular shape (C and D); the narrow part of the RS3 head points proximal and connects to the front half of the RS2 head (white arrowhead in C and D). The front, back, proximal (prox) and distal (dist) sides of the average are indicated in (C) for easier orientation in relation to the front view shown in (A). (E–P) Details of the RS structures. Both RS1 (green) and RS2 (blue) can be separated into four regions: the base, the stem, the arch and the head domain closest to the central pair (E–L). RS1 and RS2 have a very similar overall structure, but also some small differences (black asterisk in K). RS1 has a single attachment site to protofilament A3 of the A-tubule (At 3) (E and G), while the two-pronged RS2 base is attached to protofilaments, A2 and A3 (At 2, 3) (I and K). The RS3 structure (orange) is distinct from RS1 and RS2 and can be separated into three regions: stand, post and head; none of these regions resemble parts of the RS1 or RS2 structure. Note the peanut shaped density (O and P; black arrowhead); a doublet specific analysis (see Figure 3) revealed that this density, the radial spoke joist (RSJ), is only present in some DMTs (Figure 3K) and thus is in the here shown average of all doublets only partially visible. RS3 is also attached to the A-tubule protofilaments A2 and A3 (At 2, 3), but differently orientated compared to RS2 (compare K and O). Red dotted lines in (E, I and M) indicate the locations of the tomographic slices shown in (F, J and N), respectively. Base, stem and arch together, or stand and post together respectively, represent the portion of the RS called ”stalk” in previous publications. White arrows indicate the waist, a narrow region that separates the stem and arch (E–L). Other labels: B-tubule (Bt), inner dynein arm (IDA 1α, 1β, 2–6 and x; rose) [Nicastro et al., 2009], Nexin-Dynein Regulatory Complex (N-DRC, yellow) [Heuser et al., 2009], outer dynein arm (ODA, purple). Structure color coding is preserved in all following figures. Scale bars are 25 nm; scale bar in (M) is applicable to (E, F, I, J, M and N).
The subtomographic average of the 96 nm repeat unit allows the visualization of all major axonemal complexes, such as the DMTs, the rows of inner and outer dynein arms, the N-DRC and RSs at – for sea urchin flagella – unprecedented detail (Figs. 1 and 2). As previously reported [Goodenough and Heuser, 1985; Nicastro et al., 2005], the spokes RS1-RS3 were arranged in triplets with the typical intervals of 32, 24 and 40 nm distance between RSs (Fig. 1A and 1B).
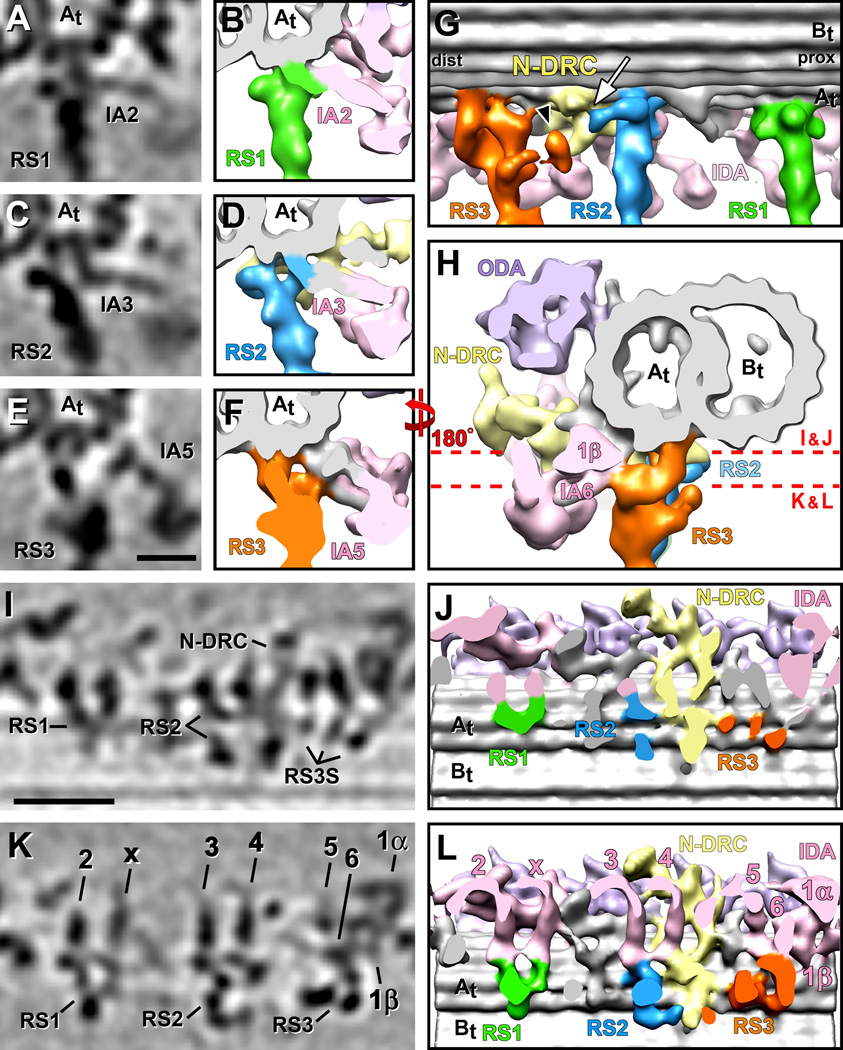
Figure 2. Connections between the RSs and neighboring structures.
(A–L) Tomographic slices (A, C, E, I and K) and isosurface renderings (B, D, F–H, J and L) of the averaged axonemal repeat from sea urchin flagella show several connections originating from the RSs’ bases. Cross-sections from proximal (A–F) and distal (H), and longitudinal bottom views (I–L; cut through the base of the RSs as indicated by red dotted lines in H) reveal connections between IDA tails and the RSs: dynein IA2 and IdaX connect with the front of the RS1 base (A, B and I–L); dynein IA3 connects to the front of the RS2 base (C, D and I–L); and the dynein arms IA5, IA6 and I1β connect with the front region of the RS3 stand (E, F and H–L). (G) A longitudinal view from the back reveals the three docking points of RS3 to the A-tubule and a connection between RS2 and the N-DRC (white arrow). Also note the protrusion termed RSJ (black arrowhead), which is only present in some DMTs and connects to the N-DRC (see Fig. 3 for more details). The proximal (prox) and distal (dist) sides of the average, as well as the A- and B-tubules (At, Bt) are indicated for easier orientation. (I and J) The DMT attachments are different between the three RSs: RS1 has one attachment site to the A-tubule; RS2 has two attachment sites, and RS3 docks at least at three points to the A-tubule. Scale bars are 12.5 nm in (E), applicable for (A, C and E), and 25 nm in (I), applicable for (I and K).
Three-dimensional structure of RS1 and RS2
The overall shapes of RS1 and RS2 were very similar, with a length of 42 nm measured from the A-tubule towards the CPC and a width that varied from 3–7 nm along the stalk to a maximum of 24 nm across each RS head measured along the DMT axis (Fig. 1A–1D). Based on their electron densities in the 3D averages, RS1 and RS2 each have an estimated mass of ~2.5 MDa. Their stalks can be divided into three distinct subregions: an 8 nm long spoke base that anchors the RSs to the DMT, a 15 nm stem in the middle, and a 12 nm arch that connects to the 7 nm thick spoke head (Fig. 1E–1L; Supporting Information Movie S1). A constriction, termed the waist, was present at the stem-arch intersection (Fig. 1E–L, white arrows).
The head and arch regions of RS1 and RS2 appeared structurally identical and had estimated masses of 900 KDa for the head and 600 KDa for the arch. Each head of RS1 and RS2 consists of two apparently identical halves that are connected in the head center (Fig. 1C–1D; Supporting Information Movie S1). The two pillars of the arch link the two halves of each RS head to the stem (Fig. 1E–1L; Supporting Information Movie S1). In longitudinal views of the DMTs, the pillars line up behind each other, so that the stalk and head give RS1 and RS2 their typical T-shaped appearance (Fig. 1F, 1H, 1J and 1L), but in cross-sectional view the spokes are shaped like stirrups due to the arch (Fig. 1E, 1G, 1I and 1K). We observed no connection between the heads of RS1 and RS2.
While the overall structures of RS1 and RS2 was very similar, we also observed small structural differences between these two RSs, especially in their basal regions (Supporting Information Movie S1). RS2 has two small additional densities attached to the stem region and the back prong of the base; latter density connects the RS2 base to the N-DRC (Figs. 1K and 2G). RS1 has a U-shaped attachment to only one protofilament, A3* of the A-tubule, while the base of RS2 has two prongs that connect to protofilaments A2 and A3 (Figs. 1G, 1K; 2I and 2J; Supporting Information Movie S2).
Three-dimensional structure of RS3 and conserved RS heterogeneity
RS3 bears no structural resemblance to RS1 and RS2. The length of RS3 is 43 nm, only slightly longer than RS1 and RS2, but its width varied from 3–16 nm along the stalk to a maximum of 26 nm across the RS3 head when viewed longitudinally (Fig. 1A–1D). Although the estimated mass of RS3 is with 2.8 MDa only 300 kDa larger than the other spokes, RS3 is structurally unique. The wide RS head still conveys an overall T-shape appearance (Fig. 1N and 1P), explaining why previous studies with lower resolution could not detect this RS heterogeneity. We refer to this widest part of RS3 still as the RS head region, despite of its structural differences compared to the heads of RS1 and RS2 (see below). However, based on the 3D structure we could not correlate the subregions of the RS3 stalk to the base, stem and arch region of RS1 and RS2. To reflect this structural uniqueness of the RS3 stalk we gave its two subregions new names: first, the RS3 stand, which encompasses a 1.3 MDa large, 21 nm long, complex density that reaches from the docking site at the DMT to a constriction at about half the length of the RS; second, the RS3 post, which connects between the RS3 stand and head (Fig. 1M–1P; Supporting Information Movie S1). There are several small, wing-like protrusions along the RS3 post (Fig. 1P). The head of RS3 is asymmetrically shaped and the narrower, proximal part seems to connect to the front half of the RS2 head (Fig. 1C and 1D).
The RS3 stand is attached to protofilaments A2 and A3 of the A-tubule through three connections that align along the longitudinal axis of the DMT (Fig. 2I and 2J). This is in contrast to RS2, which attaches to the same protofilaments, but the two prongs of the RS2 base align in a plane perpendicular to the DMT axis (Fig. 2I and 2J). The RS3 stand contains three major domains: 1) a small bow-shaped density links the two proximal DMT docking sites along the DMT length (Figs. 1A and 2G), 2) followed by a bulbous core that extends to the central constriction of the stalk (Figs. 1O, 1P and 2G), and 3) a large arm that starts at the most distal docking side of the RS3 and projects forward to connect to the core and the DMT docking site of the tail domain of dynein inner arm IA6 (dynein a/d) (Fig. 2G, 2H and 2L).
This RS heterogeneity was also observed in Tetrahymena (Supporting Information Fig. S2), a protist with cilia that contain radial spoke triplets. Cryo-ET and subtomogram averaging of Tetrahymena axonemes revealed RS structures that were very similar to those observed in sea urchin flagella, including the distinct structure of RS3, i.e. the small bow-shaped density which docks the spoke to the DMT and the bulbous core extending away from the microtubule (Supporting Information Fig. S2).
RS3 contains or connects to doublet specific structures
To generate the mainly planar waveform of undulating sea urchin sperm flagella [Woolley and Vernon, 2001], dynein activity and interdoublet sliding must be limited to subsets of DMTs, rather than all nine DMTs [Hayashi and Shingyoji, 2008]. Therefore, structural heterogeneity between complexes on different (subsets of) DMTs, so called doublet specific structures, are of great interest, because they could contribute to the non-uniform, asymmetric distribution of dynein activity and microtubule sliding. With the RSs being key regulators of dynein activity we therefore asked the question whether the spokes contained or connected to doublet specific features. We calculated doublet specific averages for each DMT by first determining the doublet numbering in each tomogram based on the location of the 5–6 bridge that forms permanent links between doublets 5 and 6 [Afzelius, 1959]; then we averaged the same DMTs from each tomogram into doublet specific averages (DMT 1–9 in Supporting Information Fig. S3). These doublet specific averages confirmed the overall architecture of the RSs that is present on all nine DMTs, but also revealed striking doublet specific differences in structures connecting to RS3 (Fig. 3; Supporting Information Fig. S3).
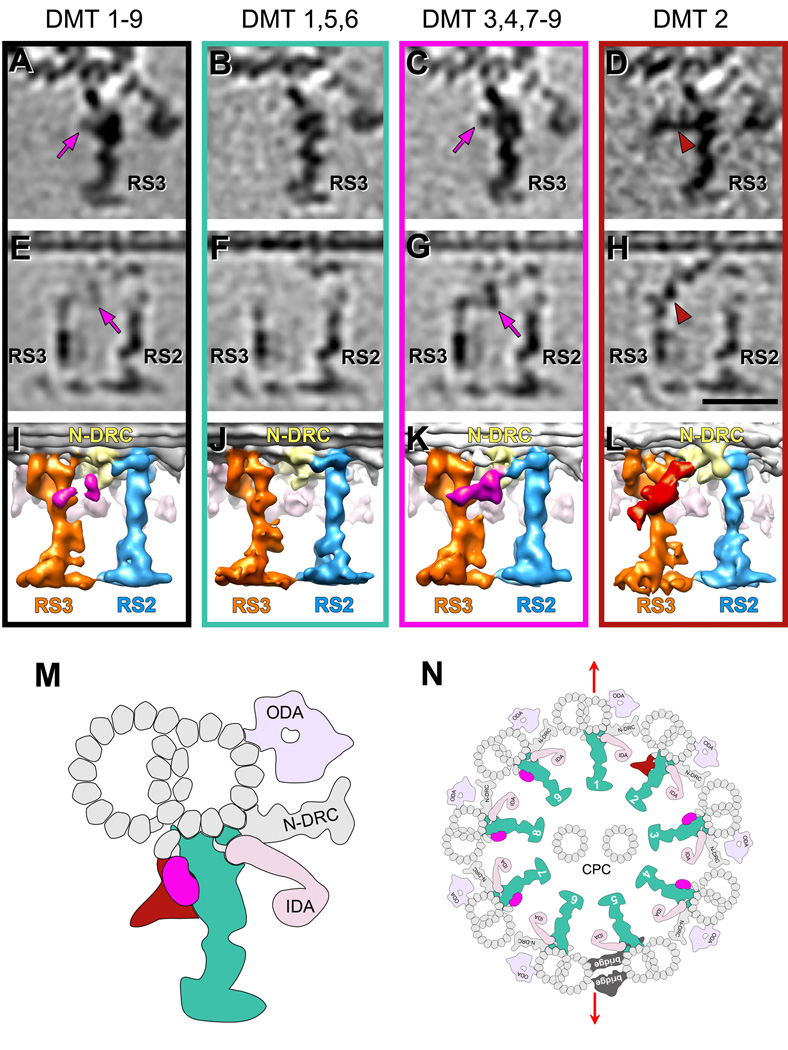
Figure 3. Doublet specific differences in the RS3 structure.
(A–L) Tomographic slices (A–H) and isosurface renderings (I–L) of averaged axonemal repeats from sea urchin flagella viewed from the proximal (A–D) and back side (E–L) show doublet specific features that connect RS3 to RS2 and the N-DRC. Averages of repeats from all nine DMTs (A, E and I) are compared to class averages that include the following groups of doublets: DMTs 1, 5 and 6 (B, F and J), DMTs 3, 4, 7–9 (C, G and K) and DMT 2 (D, H and L). Magenta arrows in (A, C, E and G) point to a density called RSJ (colored in magenta in (I, K)). Brown arrowheads in (D and H) highlight another density called spur (colored in brown in (L)). Both densities protrude from the back of RS3 and connect with the N-DRC but are distinct from each other. Averages of each individual doublet are shown in Fig. S3. Scale bar: 25 nm (valid for A–H). (M and N) Schematic models of RS3 (M) and an entire axoneme (N) from sea urchin flagella viewed in cross-section from proximal. The consensus part of RS3 that seems consistent among all individual doublets is colored in turquoise; the RSJ present on DMTs 3, 4 and 7–9 is colored magenta; the spur present only on DMT 2 is colored brown. Note the position of the 5–6 bridge between DMTs 5 and 6. Red arrows indicate the flagellar beating plane. Previous studies showed that the bending is perpendicular to the plane that contains both microtubules of the central pair complex (CPC); DMTs 2–4 and 7–9 are thought to be critical for flagella motility [Lindemann and Lesich, 2010]. These are the same doublets that have the doublet specific structures, the RSJ and the spur, connecting RS3 to RS2 and N-DRC.
All observed doublet specific differences related to RS3 are limited to a region that reaches from the back of the RS3 stand to the base of RS2. The comparison of doublet specific averages revealed a ~450 kDa mass that is only present on DMTs 3,4,7–9 and that is suspended between RS3 and RS2 like a “joist” (termed here Radial Spoke Joist (RSJ); magenta arrows and density in Fig. 3C, 3G and 3K; Supporting Information Fig. S3). String-like densities connect the central mass to the back of RS3 close to the tip of the bulbous core and to the base of RS2, as well as to the base plate of the N-DRC (Fig. 3G and 3K). The same region around RS3 on DMT 2 shows a much larger, “spur”-like structure with an estimated mass of ~800 kDa and the following two major parts: a connection between the back of RS3 and the N-DRC base plate – similar to the RSJ, but located closer to the DMT and without the central globular mass, and second a cone-shaped projection that starts in the same region like the RSJ density, but extends ~20 nm backwards perpendicular to the RS3 stalk (Fig. 3D, 3H and 3L; Supporting Information Figs. S3 and S4). Neither the RSJ nor the spur structures were detected on DMTs 1, 5 and 6 (Fig. 3B, 3F and 3J; Supporting Information Figs. S3). Therefore, in an average of all 9 DMTs only a weak density of the RSJ remains visible of the doublet specific structures (Fig. 3A, 3E and 3I).
RSs interact directly with IDAs and the N-DRC
The mechanism how RS signals reach the dyneins and regulate the motor proteins’ activity is not clear. Shedding light on this, our cryo-ET averages revealed two interesting types of connections of the RSs with other axonemal complexes: for one several IDAs connect with their tail domains directly to the bases of RS1 and RS2, and the stand of RS3; in addition, both RS2 and RS3 seem to connect directly to the base plate of the N-DRC, a known, major regulatory hub that coordinates dynein activity (Figs. 2 and 3; Supporting Information Movie S2). Specifically the tails of the dynein arm IA2 (dynein a/d) and the doublet specific arm IdaX (dynein b/g) [Nicastro, 2009; Bui et al., 2009] were anchored to the front of RS1 (Figs. 1G, 2A, 2B and 2I-2L), and IA3 (dynein c) was attached to the front prong of the base of RS2 (Fig. 2C, 2D, 2I–2L), while the back prong extended distally to connect to the N-DRC (Fig. 2G). We also observed 3 connections between inner dyneins and the arm domain of the RS3 stand. Two dynein arms, IA5 (dynein b/g) and IA6 (dynein a/d), contacted the RS3 stand through their tails, but in case of the I1 β heavy chain (f dynein) the connections were directly to the dynein motor domain (Fig. 2E, 2F, 2H, 2K and 2L). The doublet specific connections of RS3 to the N-DRC through the RSJ (Fig. 3C, 3G and 3K) or spur structure (Fig. 3D, 3H and 3L) have been described above. The borders between the dynein tails and the RS structures are only approximations based on previously published images showing the size and shape of the inner dynein heavy chain c with full length tail domain (IA3) [Burgess et al., 2004].
DISCUSSION
The comparison between Strongylocentrotus, Tetrahymena and Chlamydomonas cilia and flagella shows conserved RS heterogeneity and the presence of the basal part of RS3 in Chlamydomonas
Previous EM studies have described all RSs uniformly as T-shaped structures [Chasey, 1972; Chasey, 1974; Curry and Rosenbaum, 1993; Dentler and Cunningham, 1977; Goodenough and Heuser, 1985; Hopkins, 1970; Warner, 1970; Warner and Satir, 1974; Nicastro et al., 2005]. In addition, classical EM studies of Chlamydomonas mutants indicated that mutations in RS genes caused the same structural defects in both RS1 and RS2 [Curry and Rosenbaum, 1993; Diener et al., 1993; Huang et al., 1981]. Even though the distance between RS1 and RS2 is different from the distance between RS2 and RS3 in organisms with RS triplets, and despite the fact that studies of RSs in mussel gill cilia showed different tilting of RS3 compared to RS1 and RS2 [Warner and Satir, 1974], RSs were generally thought of as homogeneous structures.
Our recent comparative study of Chlamydomonas WT flagella and several RNA-interference mutants lacking parts of the conserved Calmodulin and Spoke-associated Complex (CSC) [Dymek and Smith, 2007] showed that in the knockdowns many of the axonemal repeat units were specifically missing RS2, while RS1 was always present [Dymek et al., 2011]. These results strongly suggested heterogeneity among spokes and that the CSC is located at or near the base of RS2 [Dymek et al., 2011]. Our most recent cryo-ET study of the 3D structures of RSs in WT and RS mutant axonemes of Chlamydomonas revealed small structural differences between the basal regions of RS1 and RS2, and thus confirmed minor spoke heterogeneity between RS1 and RS2 [Barber et al., 2011]. These structural differences between Chlamydomonas RS1 and RS2 were very similar to differences observed in the present study for RS1 and RS2 in sea urchin flagella. However, in sea urchin flagella we found two additional differences between RS1 and RS2: RS1 has a single docking site to the A-tubule, but forms connections with two IDAs, while the RS2 attachment is 2-pronged with one IDA connecting to the front prong (Fig. 2I–2L), which is very similar to the arrangement of RS2 in Chlamydomonas [Barber et al., 2011].
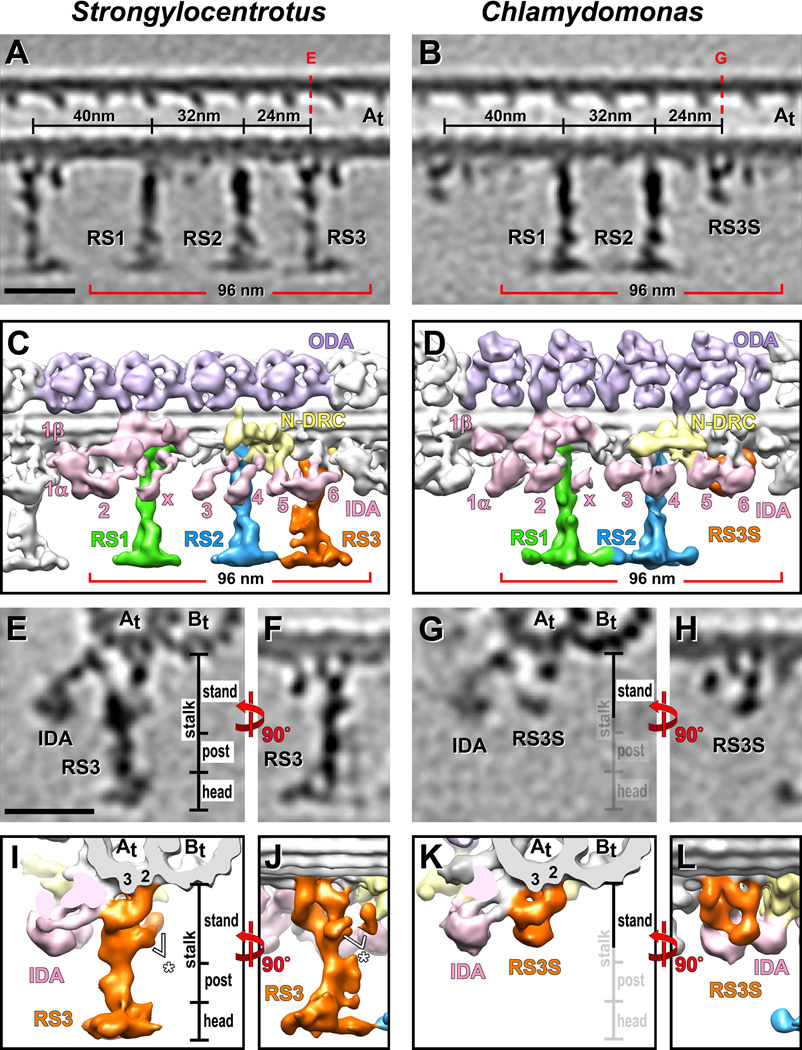
A comparison of the subtomogram averages of Strongylocentrotus (sea urchin) flagella and Tetrahymena cilia revealed a highly similar organization of the spoke triplets in these two species, including the unique structure of RS3. In place of RS3, Chlamydomonas flagella contain a shorter structure [Bui et al., 2008], termed Radial Spoke 3 Stand-in (RS3S), that lacks structural similarities with spokes RS1 and RS2 and therefore its relationship to RSs had remained unclear [Barber et al., 2011]. Here we found that the 3D structure of the Chlamydomonas ~18 nm long RS3S [Barber et al., 2011] correlates remarkably well with the structure of the basal half of RS3, the 21 nm long RS3 stand, in sea urchin and Tetrahymena axonemes (Fig. 4; Supporting Information Fig. S2). Both RS3 and RS3S are attached to the protofilaments A2 and A3 of the A-tubule through 3 connections and have contact with the tail domain of the inner dynein arm IA6 (dynein a or d). The specific locations of RS3 and RS3S within the 96 nm axonemal repeat, their highly similar, yet complex and unique structures in organisms reaching from protists to sea urchins suggests both that RS3S is the basal part of RS3, and that radial spoke heterogeneity of RS3 vs. RS1 and RS2 is highly conserved across species.
Figure 4. Comparison of the Strongylocentrotus RS triplet and Chlamydomonas RS pair with special emphasis on RS3 and RS3S.
(A–L) Tomographic slices (A, B and E–H) and isosurface renderings (C, D and I–L) from averaged axonemal repeats of Strongylocentrotus (sea urchin) (A, C, E, F, I and J) and Chlamydomonas (algae) (B, D, G, H, K and L), viewed from the front (A–D), back (F, H, J and L) and in cross-section from distal (E, G, I and K), show that the structure of RS3S in Chlamydomonas closely resembles the RS3 stand in Strongylocentrotus. Red dotted lines in (A and B) indicate the locations of the tomographic slices shown in (E and G). The white asterisks in (I and J) highlight parts of the RSJ structure that is attached to the back of RS3. The post and head region of RS3, as well as the RSJ structure are not present in Chlamydomonas. The labels for these regions are faded in (G and K) for easier comparison. Panels A and C are the same as Figure 1A and 1B, and panel D is reproduced from Barber et al. [2011] Figure 3A with minor modifications. Scale bars are 25 nm (scale bar in A valid for B; scale bar in E valid for F–H).
The proteome of the RS triplet
RSs are large, multi-subunit complexes with at least 23 radial spoke proteins (RSP1–23) in Chlamydomonas [Patel-King et al., 2004; Piperno et al., 1977; Piperno et al., 1981; Yang et al., 2001; Yang et al., 2006]. RSP1–17 were identified by 2D gel analysis of the Chlamydomonas WT compared to the pf14 mutant, which contains a mutation in RSP3 and lacks any identifiable RS structure in classical EM images [Piperno et al., 1977; Piperno et al., 1981]. RSP18–23 were later identified from purified Chlamydomonas RS fractions [Yang et al., 2001; Yang et al., 2006; Patel-King et al., 2004]. Although Chlamydomonas is the only organism in which comprehensive proteomic studies of the RS proteins have been conducted, evidence exists that several of the spoke proteins are conserved, e.g., members of the ‘RSP3’ superfamily have been identified in 14 species [Ralston et al., 2006; Marchler-Bauer et al., 2009]. However, with the mounting evidence for radial spoke heterogeneity, to some degree between RS1 and RS2 [Dymek et al., 2011; Barber et al., 2011], but especially between RS3/RS3S and the remaining spokes, the question arises as to how complete our current understanding of the proteome of the RS triplets is. The observed structural RS heterogeneity will likely be reflected in a different protein composition of the RSs. Therefore most known RS proteins probably correspond to the proteome of RS1 and RS2, because Chlamydomonas flagella lack a full length RS3 and our preliminary structural data of the spokeless Chlamydomonas pf14 mutant indicates that RS3S is still present in these mutant axonemes [data not shown].
While no proteomics data are available for RSs in sea urchin flagella, studies of two organisms with cilia and flagella containing RS triplets, Trypanosoma and Ciona, have been conducted. Recent studies of Ciona sperm flagella have detected 15 RS proteins, of which 11 proteins were identified by mass spectrometry [Padma et al., 2003; Satouh and Inaba, 2009; Satouh et al., 2005]. However, in these studies, spokes were isolated by immunoprecipitation with an anti-RSP3 antibody. It is possible that in contrast to RS1 and RS2, RS3 might not contain RSP3 protein, in which case RS3 would not have been immunoprecipitated and RS proteins unique to RS3 not been detected in this study. RSP3 knockdown mutants in Trypanosoma, have reduced numbers of spokes, although the number of remaining RSs seen in EM cross-sections and the degree of rudimentary flagellar motility were higher than expected based on the dramatic reduction of rsp3 transcripts [Ralston et al., 2006]. While the presence of two-thirds of the RSs could be due to incomplete knockdown of RSP3, it is also possible that RSP3 is not essential for assembly and DMT docking of RS3.
The structure of RS1 and RS2 seemed overall well conserved between Chlamydomonas and sea urchin flagella (Fig. 4A–4D). The most obvious differences in the RS1 and RS2 structure between these two species were seen in the RS heads. Although they all consist of two rotationally symmetric halves, the RS halves in sea urchin flagella were considerably smaller in size (~50% based on our mass estimations) and lacked the RS1-RS2 connection between the RS heads (Figs. 1C, 1D and 4A–4D), that was observed in Chlamydomonas flagella [Barber et al., 2011]. The Chlamydomonas spoke heads contain at least 5 RSPs (RSP1, 4, 6, 9 and10) [Yang et al., 2006]. In Ciona flagella only 3 homologues of Chlamydomonas head proteins were identified so far [Padma et al., 2003; Satouh and Inaba, 2009; Satouh et al., 2005]. The sizes of the RS1 and RS2 heads in Tetrahymena axonemes appear similar to that of the Chlamydomonas RS heads, but at the current resolution it is unclear if the RS1-RS2 connection is also present [data not shown]. Future studies that can correlate current and new proteomics data with RS structure, are required for a more comprehensive understanding of the RS triplet proteome.
The role of the doublet specific structures connected to RS3
For effective beating and to generate the complex oscillatory motions typical of beating cilia and flagella the action of the tens of thousands of dynein motors in a single organelle must be spatiotemporally controlled. In most cilia and flagella, including those of sea urchins, the CPC is positioned so that the plane that includes both central microtubules is perpendicular to the plane of the principal bend (Fig. 3N) [Sale, 1986]. Using the standard numbering convention for sea urchin flagella, doublets 5 and 6 are permanently linked by the 5–6 bridge, which is thought to prevent significant interdoublet sliding between these two DMTs [Afzelius, 1959]. In a typical axoneme, doublets 5 and 6 are opposite to doublet 1, and a plane running through DMT 1 and the 5–6 bridge is usually perpendicular to the plane running through both CPC microtubules. Doublets 2–4 and 7–9 are arranged on opposite sides of the beating plane and are believed to contribute most of the force required for the bending motion [Brokaw, 1989; Holwill and Satir, 1994; Lindemann and Lesich, 2010; Nakano et al., 2003; Shingyoji et al., 1977; Wargo et al., 2004]. Dyneins are minus-end-directed motor proteins [Sale and Satir, 1977; Vale and Toyoshima, 1988], and this unidirectionality means that the cyclical bending in alternating directions requires alternating the activity of dynein arms between the two sides left and right of the beating plane. A well accepted model for asynchronous dynein arm activity, known as the switch point hypothesis, explains the cyclic beating in the light of unidirectional inter-doublet sliding [Lindemann and Lesich, 2010; Satir, 1989; Satir and Matsuoka, 1989].
Remarkably, the here observed doublet specific features that connect RS3 to both RS2 and the N-DRC, are limited to exactly those subsets of doublets, DMTs 2–4 and 7–9, that are crucial for generating the driving forces for axoneme bending [Brokaw, 1989; Holwill and Satir, 1994; Lindemann and Lesich, 2010; Nakano et al., 2003; Shingyoji et al., 1977; Wargo et al., 2004]. This distribution in two DMTs groups on opposing sides would agree well with the switch point hypothesis. The RSJ structure was observed on doublets 3, 4 and 7–9, while a larger, spur-like structure locates to roughly the same region on doublet 2 (Fig. 3). These structures could be involved in transmitting signals that are important for establishing dynein activity localized to specific DMTs in the axoneme. They are also currently the only known structures that form a triad junction between RS3, RS2 and the N-DRC, possibly allowing signal integration, feedback or cross talk between these three major complexes in cilia and flagella. The complex “spur” structure on doublet 2 seems to break the otherwise symmetrical distribution of doublet specific structures into two opposing groups of DMTs.
Here we established that the presence of a third RS, as full-length RS3 or remnant RS3S, as well as RS heterogeneity are highly conserved features of cilia and flagella. This lays a crucial foundation for future studies that will determine a more comprehensive understanding about the subunit composition of individual RSs and their functions for regulating ciliary and flagellar motility.
MATERIALS AND METHODS
Specimen Preparation
Live sea urchins (Strongylocentrotus purpuratus purchased from Monterey Abalone Co. Monterey, California) were kept in the laboratory at 4°C for up to 3 weeks in artificial seawater. Spawning was induced by the injection of 1 to 2 ml of 0.5 M KCl into the perivisceral cavity [Gibbons, 1982]. Sperm samples were collected and kept on ice without dilution in artificial seawater to prevent sperm activation [Gatti and Christen, 1985]
Tetrahymena thermophila (strain B2086) cells were grown to a density of approximately 3×105/ml in 250 ml of SPP medium (1% proteose peptone, 0.1% yeast extract and 0.2% glucose) at 30°C. The cilia were isolated as previously described [Wloga et al., 2008], with minimal modification. Briefly, cells were collected by centrifugation, washed with 10 mM Tris pH 7.5 and resuspended in 20 ml of 10 mM Tris, 50 mM sucrose, 10 mM CaCl2 with protease inhibitors, pH 7.5. Cilia were detached from the cells by adding 350 µl of 0.5 M acetic acid. After 1 min, 180 µl of KOH were added. The cilia-containing supernatant was separated from the cell bodies and debris by two centrifugation steps at 1,860 × g for 5 min. Then the cilia were collected by centrifugation at 10,000 × g for 15 min, and were demembranated during a 30 min incubation in HMEEK buffer (30 mM HEPES, 25 mM KCl, 5 mM MgSO4, 0.1 mM EDTA, 1.0 mM EGTA, pH 7.4) with 0.2% Triton X-100. The axonemes were collected by centrifugation at 10,000 × g for 10 min and were resuspended in HMEEK buffer without Triton X-100.
Quantifoil holey carbon grids (Quantifoil Micro Tools GmbH, Jena, Germany) were glow discharged and then coated with 10 nm colloidal gold (Sigma-Aldrich, St. Louis, MO). Grids were loaded in a self made plunge freezer. 3 µl of sea urchin sperm diluted in artificial seawater or 3 µl of Tetrahymena axonemes were added to the grid followed by 1 µl of a tenfold concentrated 10 nm colloidal gold solution. The grid was blotted from the front side with a filter paper for 2 s and immediately plunge frozen in liquid ethane. The vitrified sample was then stored in liquid nitrogen until data collection.
Cryo-electron tomography
EM images were collected using a Tecnai F30 transmission electron microscope (FEI, Eindhoven, NL) equipped with a field emission gun and a postcolumn energy filter (Gatan, Pleasanton, CA). Cryo-samples were transferred into the microscope with a cryo-holder (Gatan) and kept below the devitrification temperature (~ −140°C) at all times. Samples were imaged at 300 keV, with −8 µm defocus, under low dose conditions and in the zero-loss mode of the energy filter (20 eV slit width). Several tilt-series of images were recorded automatically from approximately −65 to +65° with 1.5 – 2.5° angular increments, using the SerialEM image acquisition software [Mastronarde, 2005]. The cumulative electron dose was limited to ~100 e/Å2. All images were recorded on a 2k × 2k charge-coupled device camera (Gatan) at a magnification of 13,500×, resulting in a pixel size of ~1 nm.
Image processing
Cryo-electron tomograms were reconstructed using the IMOD software package [Kremer et al., 1996] with fiducial marker alignment and weighted back projection. Only tomograms of intact and non- or only mildly compressed flagella and axonemes were used for further image processing. Subtomograms containing the 96 nm long axonemal repeats were extracted from eight tomograms of sea urchin sperm flagella and three tomograms of Tetrahymena axonemes, aligned and averaged in 3D using the PEET (Particle Estimation for Electron Tomography) software [Nicastro et al., 2006]. In addition, subtomogram averages from five tomograms of Chlamydomonas axonemes that were previously published in [Heuser et al., 2009; Barber et al., 2011] were used for comparison. Averaging repeats from DMTs with different orientations relative to the tilt axis allowed for the compensation of the missing wedge, resulting in relatively uniform resolution [Nicastro et al., 2006]. Two different aligning strategies were employed: a) to optimize the alignment accuracy either for the entire 96 nm axonemal repeat using global alignment with relatively large particle volumes (Figs. 1A, 2 and 4A; Supporting Information Fig. S2); or b) to resolve the structure of a specific complex of interest at the highest possible detail; in this case we used smaller subtomographic volumes for local alignments that were calculated centered on the structure of interest, e.g. on RS1–RS3. The resulting averages of each RS local alignment resolved the RSs at a slightly higher level of detail (Figs. 1B–1P, 3, 4C, 4E, 4F, 4I and 4J). For the doublet specific analysis of axonemal structures in sea urchin flagella first the DMT numbers were identified in all tomograms based on the location of the 5–6 bridge, a permanent link between doublets 5 and 6 [Afzelius, 1959]. Then doublet specific averages were generated by combining all axonemal repeats from the same doublet number from all tomograms (Supporting Information Fig. S3). When a doublet specific feature was present on a subset of doublets, these doublet specific averages could be combined into a class average to increase the signal-to-noise ratio and thus resolution of the specific feature (Fig. 3). The UCSF Chimera package [Pettersen et al., 2004] was used for 3D visualization by isosurface rendering and for volume size measurements. Mass estimations were calculated using an average protein density of 1.43 g/cm3 [Quillin and Matthews, 2000]. The resolution was estimated in a (20 nm)3 sub-volume in the center of all averages using the Fourier shell correlation method [Harauz and Van Heel, 1986]. All aligned repeat units were divided into two groups (odd and even numbers) and the Fourier shell at which a correlation value of 0.5 was reached was used to estimate the resolution (Supporting Information Fig. S1D).
Supplementary Material
Acknowledgements
We thank Dr. Daniel T. N. Chen for providing Strongylocentrotus purpuratus sperm and Dr. Chen Xu for outstanding management of the Brandeis EM facility. We are grateful to Dr. Xiaofeng Fu and Mikael Garabedian for technical assistance with generating the Tetrahymena subtomogram averages. This work was supported by grants from the National Institutes of Health (GM083122), from the W. M. Keck Foundation, and by a Pew Biomedical Scholars Award (to D. Nicastro).
Footnotes
The protofilaments were numbered according to the most widely accepted protofilament numbering system [Linck and Stephens, 2007] (Fig. 1G, 1K and 1O).
Supporting Information is available online.
REFERENCES
- Afzelius B. Electron microscopy of the sperm tail; results obtained with a new fixative. J Biophys Biochem Cytol. 1959;5(2):269–278. doi: 10.1083/jcb.5.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius BA. The immotile-cilia syndrome: a microtubule-associated defect. CRC Crit Rev Biochem. 1985;19(1):63–87. doi: 10.3109/10409238509086788. [DOI] [PubMed] [Google Scholar]
- Afzelius BA. Cilia-related diseases. J Pathol. 2004;204(4):470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius BA, Eliasson R. Flagellar mutants in man: on the heterogeneity of the immotile-cilia syndrome. J Ultrastruct Res. 1979;69(1):43–52. doi: 10.1016/s0022-5320(79)80041-6. [DOI] [PubMed] [Google Scholar]
- Barber CF, Heuser T, Carbajal-González BI, Botchkarev VV, Nicastro D. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-08-0692. mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ. Direct measurements of sliding between outer doublet microtubules in swimming sperm flagella. Science. 1989;243(4898):1593–1596. doi: 10.1126/science.2928796. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183(5):923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186(3):437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Oiwa K, Knight PJ. The structure of dynein-c by negative stain electron microscopy. J Struct Biol. 2004;146(1–2):205–216. doi: 10.1016/j.jsb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SC, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84(2):197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasey D. Further observations on the ultrastructure of cilia from Tetrahymena pyriformis. Exp Cell Res. 1972;74(2):471–479. doi: 10.1016/0014-4827(72)90403-x. [DOI] [PubMed] [Google Scholar]
- Chasey D. The three-dimensional arrangement of radial spokes in the flagella of Chlamydomonas reinhardii. Exp Cell Res. 1974;84(1):374–380. doi: 10.1016/0014-4827(74)90418-2. [DOI] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24(4):224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Dentler WL, Cunningham WP. Structure and organization of radial spokes in cilia of Tetrahymena pyriformis. J Morphol. 1977;153(1):143–151. doi: 10.1002/jmor.1051530110. [DOI] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123(1):183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22(14):2520–2531. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179(3):515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti JL, Christen R. Regulation of internal pH of sea urchin sperm. A role for the Na/K pump. J Biol Chem. 1985;260(12):7599–7602. [PubMed] [Google Scholar]
- Gibbons IR. Sliding and bending in sea urchin sperm flagella. Symp Soc Exp Biol. 1982;35:225–287. [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100(6):2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Hayashi S, Shingyoji C. Mechanism of flagellar oscillation-bending-induced switching of dynein activity in elastase-treated axonemes of sea urchin sperm. J Cell Sci. 2008;121:2833–2843. doi: 10.1242/jcs.031195. [DOI] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187(6):921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwill ME, Satir P. Physical model of axonemal splitting. Cell Motil Cytoskeleton. 1994;27(4):287–298. doi: 10.1002/cm.970270402. [DOI] [PubMed] [Google Scholar]
- Hopkins JM. Subsidiary components of the flagella of Chlamydomonas reinhardii. J Cell Sci. 1970;7(3):823–839. doi: 10.1242/jcs.7.3.823. [DOI] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88(1):80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyfman AY, Schmid MF, Gheiratmand L, Fu CJ, Khant HA, Huang D, He CY, Chiu W. Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc Natl Acad Sci USA. 2011;108(27):11105–11108. doi: 10.1073/pnas.1103634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lindemann CB, Lesich KA. Flagellar and ciliary beating: the proven and the possible. J Cell Sci. 2010;123(Pt 4):519–528. doi: 10.1242/jcs.051326. [DOI] [PubMed] [Google Scholar]
- Linck RW, Stephens RE. Functional protofilament numbering of ciliary, flagellar, and centriolar microtubules. Cell Motil Cytoskeleton. 2007;64:489–495. doi: 10.1002/cm.20202. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Sale WS. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol. 1999;144(2):293–304. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano I, Kobayashi T, Yoshimura M, Shingyoji C. Central-pair-linked regulation of microtubule sliding by calcium in flagellar axonemes. J Cell Sci. 2003;116(Pt 8):1627–1636. doi: 10.1242/jcs.00336. [DOI] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102(44):15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nicastro D. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol. 2009;91:1–39. doi: 10.1016/S0091-679X(08)91001-3. [DOI] [PubMed] [Google Scholar]
- Olson GE, Linck RW. Observations of the structural components of flagellar axonemes and central pair microtubules from rat sperm. J Ultrastruct Res. 1977;61(1):21–43. doi: 10.1016/s0022-5320(77)90004-1. [DOI] [PubMed] [Google Scholar]
- Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. Rotation of the central pair microtubules in eukaryotic flagella. Mol Biol Cell. 1999;10(1):1–4. doi: 10.1091/mbc.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma P, Satouh Y, Wakabayashi K, Hozumi A, Ushimaru Y, Kamiya R, Inaba K. Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the Ascidian Ciona intestinalis. Mol Biol Cell. 2003;14(2):774–785. doi: 10.1091/mbc.02-06-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, Gorbatyuk O, Takebe S, King SM. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol Biol Cell. 2004;15(8):3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJ. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74(4):1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88(1):73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesec TP, Ruiz A, McMahon JT, Prayson RA. Ultrastructural abnormalities of respiratory cilia: a 25-year experience. Arch Pathol Lab Med. 2008;132(11):1786–1791. doi: 10.5858/132.11.1786. [DOI] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151(5):F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 7):791–794. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- Ralston KS, Lerner AG, Diener DR, Hill KL. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell. 2006;5(4):696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, Porter ME. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol. 2003;162(1):47–57. doi: 10.1083/jcb.200303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci USA. 1977;74(5):2045–2049. doi: 10.1073/pnas.74.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS. The axonemal axis and Ca2+-induced asymmetry of active microtubule sliding in sea urchin sperm tails. J Cell Biol. 1986;102(6):2042–2052. doi: 10.1083/jcb.102.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. Studies on cilia. 3. Further studies on the cilium tip and a "sliding filament" model of ciliary motility. J Cell Biol. 1968;39(1):77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. The role of axonemal components in ciliary motility. Comp Biochem Physiol Comp Physiol. 1989;94(2):351–357. doi: 10.1016/0300-9629(89)90558-6. [DOI] [PubMed] [Google Scholar]
- Satir P, Matsuoka T. Splitting the ciliary axoneme: implications for a "switch-point" model of dynein arm activity in ciliary motion. Cell Motil Cytoskeleton. 1989;14(3):345–358. doi: 10.1002/cm.970140305. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Inaba K. Proteomic characterization of sperm radial spokes identifies a novel spoke protein with an ubiquitin domain. FEBS Lett. 2009;583(13):2201–2207. doi: 10.1016/j.febslet.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Padma P, Toda T, Satoh N, Ide H, Inaba K. Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol Biol Cell. 2005;16(2):626–636. doi: 10.1091/mbc.E04-09-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingyoji C, Murakami A, Takahashi K. Local reactivation of Triton-extracted flagella by iontophoretic application of ATP. Nature. 1977;265(5591):269–270. doi: 10.1038/265269a0. [DOI] [PubMed] [Google Scholar]
- Smith EF, Lefebvre PA. The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil Cytoskeleton. 1997;38(1):1–8. doi: 10.1002/(SICI)1097-0169(1997)38:1<1::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57(1):8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgess JM, Chao J, Wong J, Aspin N, Turner JA. Cilia with defective radial spokes: a cause of human respiratory disease. N Engl J Med. 1979;300(2):53–56. doi: 10.1056/NEJM197901113000201. [DOI] [PubMed] [Google Scholar]
- Summers KE, Gibbons IR. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc Natl Acad Sci USA. 1971;68(12):3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Toyoshima YY. Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell. 1988;52(3):459–469. doi: 10.1016/s0092-8674(88)80038-2. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, McPeek MA, Smith EF. Analysis of microtubule sliding patterns in Chlamydomonas flagellar axonemes reveals dynein activity on specific doublet microtubules. J Cell Sci. 2004;117(Pt 12):2533–2544. doi: 10.1242/jcs.01082. [DOI] [PubMed] [Google Scholar]
- Warner FD. New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J Cell Biol. 1970;47(1):159–182. doi: 10.1083/jcb.47.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner FD, Satir P. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol. 1974;63(1):35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Hendrickson T, Sale WS. Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil Cytoskeleton. 2007;64(8):569–579. doi: 10.1002/cm.20211. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yang C, Yang P, Fox L, Yanagisawa HA, Kamiya R, Witman GB, Porter ME, Sale WS. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol Biol Cell. 2009;20(13):3044–3054. doi: 10.1091/mbc.E09-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB, Plummer J, Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J Cell Biol. 1978;76(3):729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Eddé B, Bré MH, Levilliers N, Redeker V, Duan J, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7(8):1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley DM, Vernon GG. A study of helical and planar waves on sea urchin sperm flagella, with a theory of how they are generated. J Exp Biol. 2001;204:1333–1345. doi: 10.1242/jeb.204.7.1333. [DOI] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153(6):1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119(Pt 6):1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.