Abstract
We describe a case of proven donor transmission of carbapenem-resistant Acinetobacter baumannii, which resulted in severe infectious complications after lung transplantation. A single blaOXA-23 positive strain, belonging to a new multilocus sequence type (ST231), was isolated from donor and recipient, who died 65 days after transplantation. This report highlights the current challenges associated with the potential transmission of multidrug-resistant infections through organ transplantation.
Keywords: carbapenem resistance, Acinetobacter baumannii, lung transplant, donor-transmitted infection
Carbapenem-resistant Acinetobacter baumannii (CRAB) is an emerging pathogen, frequently associated with nosocomial outbreaks of infection (1). Although healthcare-associated infections are commonly seen among lung transplant (LT) recipients (2), data are scarce on the epidemiology and outcome of CRAB infections in these patients (3, 4). Most cases reported to date were typical postoperatively acquired infections, occurring several weeks after admission to an intensive care unit (ICU), and associated with mechanical ventilation or exposure to broad-spectrum antibiotics. In the present report we describe, for the first time to our knowledge, confirmed donor-recipient transmission of CRAB causing severe infectious complication after LT. The study was approved by the institutional review board of the hospital.
Case report
On April 22, 2007, right lung transplantation was performed on a 50-year-old female patient with idiopathic pulmonary fibrosis. The patient was diagnosed with idiopathic pulmonary fibrosis in 2005, and was admitted to the hospital for respiratory insufficiency for 3 months in 2006, when pulmonary transplantation was indicated. About a month before lung transplantation, she was hospitalized for oxygen therapy support, because of difficulties with home care assistance. The donor, who had been admitted to the emergency room of another hospital 2 days before, was evaluated according to the LT protocol of the Federal University of Rio de Janeiro. The partial pressure of oxygen/fraction of inspired oxygen (PO2/FIO2) was >300, chest x-ray was normal, with no sign of bronchial aspiration by bronchoscopy. The perioperative antimicrobial prophylaxis consisted of vancomycin plus cefepime.
On April 23, 2007, postoperative day (POD) 1, the patient was afebrile and hemodynamically stable. She was weaned off mechanical ventilation and extubated. Post-transplant immunosuppressive medication consisted of tacrolimus, azathioprine, and methylprednisolone, and Pneumocystis jirovecii pneumonia prophylaxis of trimethoprim-sulfamethoxazole. On the following day, however, she presented with fever (38°C), arterial hypotension, and respiratory failure. A chest x-ray showed an infiltrate in the lower third of the right hemithorax. The patient was reintubated and returned to mechanical ventilation. PO2/FiO2 just after reintubation was 164. Additionally, norepinephrine infusion was started, and meropenem was substituted for cefepime. On the same day, the results of donor’s bronchoalveolar lavage (BAL) culture became available, yielding 105 colony-forming units (CFU)/mL of A. baumannii, susceptible to ampicillin-sulbactam, meropenem, imipenem, and amicacin. Unfortunately, this result for meropenem was later shown to be wrong (see “Microbiologic investigation” below). On POD 6, vasopressor therapy was stopped and PO2/FiO2 was 240. However, she was persistently febrile and a partial dehiscence of the surgical wound was noticed in association with purulent discharge draining from the wound and thoracic drain. A computerized tomography of the thorax detected bilateral consolidations with air bronchogram and small amount of liquid in the right pleural space. On POD 9, CRAB was isolated from BAL (105 CFU/mL) and from the surgical wound specimen. The isolate was susceptible only to amikacin and polymyxin B. Therefore, intravenous (IV) polymyxin B was substituted for meropenem and tacrolimus dosage was reduced.
By POD 29, after 19 days of polymyxin B therapy, the patient had been afebrile for 10 days. Although she could not tolerate weaning off mechanical ventilation, the PO2/FiO2 ratio was 260. The surgical wound presented granulation tissue and no purulent secretion in the wound or through the thoracic drain. However, her serum creatinine had risen to 2.1 mg/dL leading to the decision to stop polymyxin B therapy. On the following days, the serum creatinine progressively returned to baseline level (1.0 mg/dL). However, on POD 46, polymyxin B had to be restarted in association with inhaled amikacin therapy. At that time, the patient presented recurrence of clinical and radiological evidences of pneumonia as well as recrudescence of the purulent drainage at the surgical wound. CRAB had been again isolated from BAL (105 CFU/mL) and surgical site specimens collected 2 days earlier. As a transbronchial lung biopsy showed the coexistence of cytomegalovirus pneumonia, IV ganciclovir therapy was also prescribed. After a transient improvement during the first few days of the second course of polymyxin B therapy, fever returned and the respiratory function progressively worsened. On POD 53, inhaled polymyxin B replaced inhaled amikacin. No organisms were isolated in the microbiological investigation performed at this time. On POD 57, as the patient’s clinical status continued to deteriorate, and empiric amphotericin B therapy was started. On POD 61, immunosuppressive therapy with tacrolimus and azathioprine was stopped, but she died on POD 65.
Microbiological investigation
All A. baumannii isolates obtained from BAL, tracheal aspirates, and surgical wound were saved for further analyses at a reference laboratory. Biochemical tests and rpoB gene sequencing (5) confirmed the species identification of all isolates. Antimicrobial susceptibility was determined by disk-diffusion; polymyxin B minimum inhibitory concentration (MIC) was determined by broth microdilution in triplicate (6). The presence of the blaOXA23 gene, one of the major determinants of carbapenem resistance in A. baumannii, was investigated by polymerase chain reaction assay (7). Strain typing was performed for all isolates by random amplification of polymorphic DNA (RAPD) with primer M13, and for 2 representative isolates by pulsed field gel electrophoresis (PFGE) (8). Cluster analysis was performed with GelComparII, version 4.01 (Applied Maths) using Dice index and unweighted pair group method with arithmetic averages. Isolates with indistinguishable band profiles were included in a single genotype. Study isolates were compared to a collection of strains from a surveillance system established in Curitiba, Brazil (8). Two isolates from the study and one from the surveillance collection were subjected to multilocus sequence typing (9).
All isolates were resistant to cefepime, ceftazidime, ciprofloxacin, gentamicin, and imipenem. The isolate from the donor BAL was susceptible to ampicillin/sulbactam and intermediate to meropenem while all others were resistant to both. All were susceptible to polymyxin (MIC 0.5–2.0 mg/mL). Most isolates were susceptible to amikacin except for 3 isolates from tracheal aspirates that were resistant. All A. baumannii harbored the blaOXA23 gene and belonged to a single RAPD genotype. This strain belongs to a new sequence type (ST), ST231, and was indistinguishable from PFGE genotype D that caused fatal infections in 2 patients (one case of meningitis and a vascular catheter-related infection) in Curitiba (8). Characteristics of isolates are described in Table 1, and RAPD cluster analysis is shown in Figure 1. ST231 belongs to the same clonal complex as other STs already described in Argentina and Japan (ST109) and Italy (ST197) (http://pubmlst.org).
Table 1.
Characteristics of Acinetobacter baumannii isolates studied
| Isolate | Date | Specimen | Meropenem | Amikacin | Amoxicillin- sulbactam | Imipenem | RAPD | ST | Reference |
|---|---|---|---|---|---|---|---|---|---|
| I04* | Apr 22, 2007 | BAL** | S | S | S | S | D | NT | PR |
| I | S | S | R | D | NT | PR | |||
| I05 | Apr 28, 2007 | Wound secretion | R | S | S | R | D | NT | PR |
| I06 | Apr 28, 2007 | BAL | R | S | R | R | D*** | 231 | PR |
| I16 | Jun 5, 2007 | Wound secretion | R | S | R | R | D | NT | PR |
| I18 | Jun 5, 2007 | BAL | R | S | R | R | D | 231 | PR |
| I20 | Jun 7, 2007 | BAL | R | S | R | R | D | NT | PR |
| 1198 | May 2003 | Tracheal secretion | D*** | 231 | (8) |
Isolate obtained from donor.
Results on top are for isolate tested at the microbiology laboratory of the hospital, and below are for test in our research laboratory.
Genotypes confirmed by PFGE. All isolates were resistant to cefepime, ciprofloxacin, ceftazidime, piperacillin-tazobactam, tobramicin, gentamicin, trimethoprim-sulfamethoxazole.
RAPD, random amplification of polymorphic DNA; ST, sequence type; PR, present report; BAL, bronchoalveolar lavage; S, susceptible; NT, not typed; I, intermediate; R, resistant; PFGE, pulsed field gel electrophoresis.
Fig. 1.
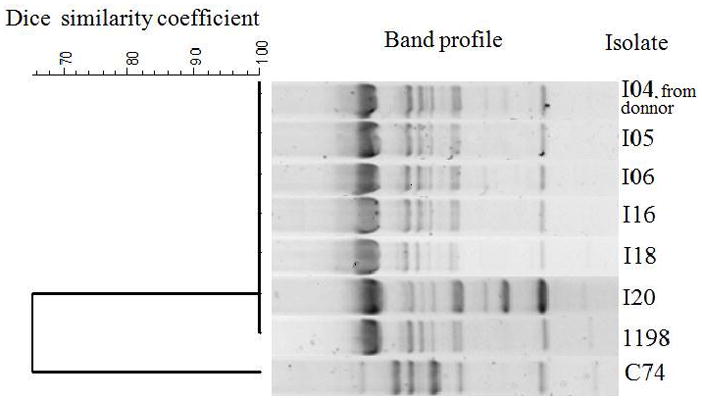
Dendrogram of RAPD band profiles obtained with Acinetobacter baumannii study isolates. C74 is a control isolate from laboratory collection. RAPD, random amplification of polymorphic DNA.
Discussion
Although donor lungs are frequently colonized with microorganisms, recent studies have reported a low incidence of donor-transmitted bacterial infections with no significant influence on the outcome of LT recipients (2, 10, 11). However, none of these studies assessed specifically the impact of donor colonization with multidrug-resistant (MDR) pathogens. The documented experience of preoperative colonization of LT candidates with MDR bacteria has shown that outcome varies according to the colonizing species. Survival after LT is not influenced by recipient’s preoperative colonization with MDR Pseudomonas aeruginosa. However, preoperative colonization with Burkholderia cenocepacia is associated with a much lower survival, compared with other species of the Burkholderia cepacia complex (12). The influence of recipient’s preoperative colonization with CRAB remains undefined.
The present report shows that donor lung colonization with CRAB may result in early and severe infection in recipients. Donor-to-recipient transmission was confirmed by strain typing that showed that donor and recipient isolates belonged to a single genotype. An additional possibility is that the patient acquired the strain from the hospital environment or other patients; however, Acinetobacter had not been obtained in any cultures from patients admitted to the ICU in the preceding month.
The scarce available data on the outcome of CRAB infections in organ transplant recipients suggest an increased mortality compared with non-transplant patients (3, 13). A single-center study showed that LT recipients were more likely to have persistent and fatal CRAB infection compared with non-immunosuppressed patients (3). These data are in line with the relapsing course and fatal outcome of the present case. Such observations raise concern about the clinical impact of donor-transmitted CRAB infection among LT recipients, which warrant further studies.
We must recognize, however, that other factors might have influenced the outcome in this case. The possible contribution of cytomegalovirus pneumonia or other undetected coinfections cannot be excluded. Secondly, the delay in starting polymyxin B because of the initial incorrect results of susceptibility tests of the donor’s isolate may have contributed to the patient’s deterioration. The first antibiogram showed the isolate was susceptible to carbapenems, but repeat testing in our reference laboratory gave an intermediate result for meropenem and showed resistance for imipenem. In fact, small differences in the inhibition halo may occur and are inherent to the technique. The isolate was most likely resistant due to the presence of the blaOXA23. Finally, we cannot rule out the influence of putative virulence traits of the infecting strain. Little is known about the virulence of A. baumannii, but experimental data suggest this is a highly variable feature among strains (14). Interestingly, the strain isolated in this case belonged to a newly described sequence type also found in a few patients from another Brazilian city with death attributed to CRAB infection. As this genotype belongs to a clonal group that has widespread geographic distribution, this may be a matter of concern for transplant centers elsewhere.
Should further studies confirm that donor lung colonization with CRAB portends a worse prognosis for LT recipients, the development of a faster microbiologic method for its detection would be very helpful for screening potential donors in highly endemic centers. Unfortunately, because of the relatively long processing time of cultures, most cases of donor lung colonization become evident only after the organ has already been harvested. A timely diagnosis of CRAB colonization would probably assist in the decision to accept organ donation and in adjusting the perioperative prophylaxis.
Acknowledgments
Support: This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)/Comissão Fullbright-Brasil, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) of Brazil and Fogarty International Program in Global Infectious Diseases (TW006563) of the National Institute of Health.
Footnotes
Author contributions: N.M. supervised all microbiological analysis, which was conducted with the help of V.G. (susceptibility tests), T.S. and R.D. (molecular analysis to provide species identification), and F.P. (molecular typing). W.F. and S.N. followed the medical case and provided important insights for the interpretation of data collected. J.M. and A.C.M. conducted the surveillance for colonization and infection for Acinetobacter species in the ICU. L.D.C. investigates Acinetobacter species in Brazil and provided strains for comparisons, helped interpret data related to the strains in Rio de Janeiro, and ran some of the molecular typing analyses. L.W.R. provided advice in the molecular epidemiology of the infection. I.S.M., B.M.M., G.S.-L., and C.H.R.B. organized the whole manuscript.
References
- 1.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42 (5):692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 2.Mattner F, Fischer S, Weissbrodt H, et al. Post-operative nosocomial infections after lung and heart transplantation. J Heart Lung Transplant. 2007;26 (3):241–249. doi: 10.1016/j.healun.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Nunley DR, Bauldoff GS, Mangino JE, Pope-Harman AL. Mortality associated with Acinetobacter baumannii infections experienced by lung transplant recipients. Lung. 2010;188 (5):381–385. doi: 10.1007/s00408-010-9250-7. [DOI] [PubMed] [Google Scholar]
- 4.Sopirala MM, Pope-Harman A, Nunley DR, Moffatt-Bruce S, Ross P, Martin SI. Multidrug-resistant Acinetobacter baumannii pneumonia in lung transplant recipients. J Heart Lung Transplant. 2008;27 (7):804–807. doi: 10.1016/j.healun.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 5.La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006;44 (3):827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Document M100-S19: Clinical and Laboratory Standards Institute. Jan, 2009. Performance Standards for Antimicrobial Susceptibility Tests: Nineteenth Informational Supplement. [Google Scholar]
- 7.Jeon BC, Jeong SH, Bae IK, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in Korea. J Clin Microbiol. 2005;43 (5):2241–2245. doi: 10.1128/JCM.43.5.2241-2245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schimith Bier KE, Luiz SO, Scheffer MC, et al. Temporal evolution of carbapenem-resistant Acinetobacter baumannii in Curitiba, southern Brazil. Am J Infect Control. 2010;38 (4):308–314. doi: 10.1016/j.ajic.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43 (9):4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos S, Caramori M, Teixeira R, et al. Bacterial and fungal pneumonias after lung transplantation. Transplant Proc. 2008;40 (3):822–824. doi: 10.1016/j.transproceed.2008.02.049. [DOI] [PubMed] [Google Scholar]
- 11.Bonde PN, Patel ND, Borja MC, et al. Impact of donor lung organisms on post-lung transplant pneumonia. J Heart Lung Transplant. 2006;25 (1):99–105. doi: 10.1016/j.healun.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 12.van Delden C, Blumberg EA. Multidrug resistant gram-negative bacteria in solid organ transplant recipients. Am J Transplant. 2009;9 (Suppl 4):S27–34. doi: 10.1111/j.1600-6143.2009.02890.x. [DOI] [PubMed] [Google Scholar]
- 13.Trottier V, Namias N, Pust DG, Jr, et al. Outcomes of Acinetobacter baumannii infection in critically ill surgical patients. Surg Infect (Larchmt) 2007;8 (4):437–443. doi: 10.1089/sur.2006.029. [DOI] [PubMed] [Google Scholar]
- 14.de Breij A, Dijkshoorn L, Lagendijk E, et al. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One. 2010;5 (5):e10732. doi: 10.1371/journal.pone.0010732. [DOI] [PMC free article] [PubMed] [Google Scholar]


