Abstract
Major breakthroughs at the beginning of this century in high-throughput technologies have profoundly transformed biological research. Significant knowledge has been gained regarding our biological system and its disease such as malignant transformation. In this review, we summarize leading discoveries in prostate cancer research derived from the use of high-throughput approaches powered by microarrays and massively parallel next-generation sequencing (NGS). These include the seminal discovery of chromosomal translocations such as TMPRSS2-ERG gene fusions as well as the identification of critical oncogenes exemplified by the polycomb group protein EZH2. We then demonstrate the power of interrogating genomic and epigenomic data in understanding the plethora of mechanisms of transcriptional regulation. As an example, we review how androgen receptor (AR) binding events are mediated at multiple levels through protein-DNA interaction, histone and DNA modifications, as well as high-order chromatin structural changes.
Keywords: integrative genomics, FoxA1, EZH2, androgen receptor, nucleosome positioning, transcriptional regulation, gene fusion
1. Introduction
Prostate cancer is the most commonly diagnosed non-skin cancer in American men. It was estimated that 1 out of 6 men will be diagnosed with prostate cancer in their life time, while 1 in 35 will succumb to the disease. Like many other human diseases, a wide spectrum of genetic aberrations has been associated with prostate cancer [1]. Elevated androgen signaling is a major pathway to prostate cancer initiation and progression. Androgen is a hormonal growth factor that activates androgen receptor (AR), a hormonal transcription factor. In the classical model, cytoplasmic AR, once bound by the ligand androgen, translocates to the nucleus to bind cis-regulating DNA elements and turn on prostate-specific gene expression. The dependence of prostate cancer on androgen signaling stems from the early work of Charles Huggins over 60 years ago [2]. PSA, a prototype androgen-induced gene, has been used for early prostate cancer detection for decades until its recent suspension. In addition, the understanding of prostate cancer dependence on androgen signaling has also laid the foundation for androgen-deprivation therapy, which remains the mainstay treatment of advanced prostate cancer today. Despite these successes, prostate cancer remains largely an incurable disease.
The lethality of prostate cancer arises primarily from castration-resistant prostate cancer (CRPC). With the ultimate goal to treat CRPC, there are two major challenges in prostate cancer research. First, highly sensitive and specific biomarkers are needed to distinguish aggressive from indolent prostate cancer, so that intensive treatment may be applied early on only to selected patients. Molecular classification based on genetic biomarkers that allows personalized medicine is particularly important for prostate cancer due to its high heterogeneity and variable progressive potentials. The second challenge is to develop novel therapeutic strategies to cure CRPC. To meet these challenges, it is essential to first understand the molecular mechanisms controlling prostate cancer progression and castration resistance. This has become increasingly achievable in the exciting era of technological breakthroughs with the sequencing of the human genome. The use of high-throughput approaches including microarrays and massively parallel next-generation sequencing (NGS) techniques in the last decade has led to the identification of numerous oncogenes and tumor suppressor genes in an unprecedented speed. In addition, while cellular behavior is determined by the set of genes a cell expresses, gene expression in turn is regulated at multiple levels through various genetic and epigenetic mechanisms, which can now be readily probed at the global scale. Integrating these various levels of information is critical for revealing the underlying regulatory network and for effectively perturbing this regulation in order to correct aberrant gene expression for cancer treatment.
In this review, we provide an overview of how technological advances have transformed prostate cancer research and also of achievements in prostate cancer genomics in the past decade. In particular, we describe how a number of high-throughput approaches have been utilized to monitor global gene expression patterns and to detect the various mechanisms of transcriptional regulation by transcription factors, DNA methylation, histone methylation and high-order chromatin structures (Figure 1 and Table 1). Bioinformatics and integrative analytical approaches have been essential in consolidating the large volume of genomic data to generate testable hypothesis. We also discuss new frontiers in prostate cancer research to achieve further understanding of prostate cancer etiology and progression. This knowledge of prostate cancer biology will ultimately change how we think about and treat prostate cancer.
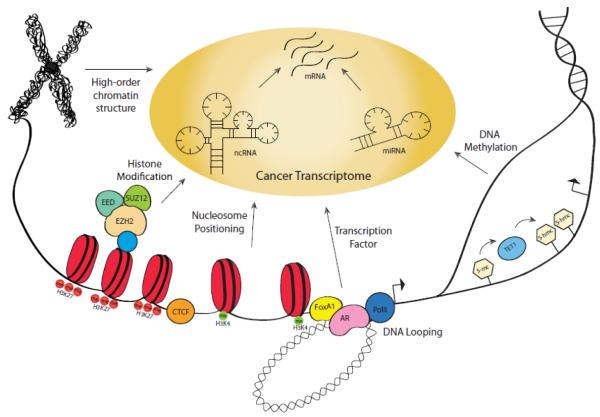
Figure 1. The multi-tier regulation of prostate cancer transcriptome.
The transcriptome of a cell includes the expression of thousands of genes/mRNA, hundreds of miRNAs, as well as an ever-growing number of ncRNAs. The expression of genes are tightly regulated by multiple mechanisms including small RNAs, transcription factors, DNA and histone modifications, as well as high-order chromatin structures including nucleosome positioning and DNA looping.
Table 1.
Summary of recent genomic technologies powered by deep sequencing.
| Name | Description | Usage | Reference |
|---|---|---|---|
| ChIP-Seq | ChIP enrichment followed by sequencing |
- Whole-genome mapping of protein-DNA interaction |
[56, 57, 78] |
| - Genome-wide Histone modification patterns |
|||
| mRNA-Seq | Sequencing of all mRNA |
- Gene fusion | [45-48] |
| - Expression Profiling | |||
| - non-coding RNA expression | |||
| - Novel transcript | |||
| - Alternative splicing | |||
| Small RNA-seq | Sequencing of small RNAs such as miRNA |
- small RNA discovery | [142, 143] |
| - small RNA expression profiling |
|||
| DNase-Seq | Sequencing of DNase- accessible DNA fragments |
- Identify nucleosome- depleted regions |
[125] |
| - Determine active gene regulatory elements |
|||
| - Nucleosome positioning | |||
| FAIRE-Seq | Sequencing of free DNA that is not crosslinked to any protein by formaldehyde |
- Genome-wide mapping of protein-free DNA |
[126] |
| - Active gene regulatory elements |
|||
| - Nucleosome positioning | |||
| MNase-Seq | Sequencing of nucleosome-protected DNA |
- Genome-wide mapping of nucleosome-protected DNA |
[132, 144] |
| - Nucleosome positioning | |||
| Hi-C | Chromatin proximity ligation and sequencing |
- Genome-wide spatial chromatin proximity maps |
[138] |
| - 3-dimensional protein-DNA interaction |
|||
| - DNA looping | |||
| ChIA-PET | ChIP-enrichment followed by chromatin proximity ligation and sequencing |
- Chromatin interaction analysis |
[140, 141] |
| - 3D chromosomal structure around a transcription factor |
|||
| - DNA looping |
2. Analysis of Prostate Cancer Transcriptome
The search of prostate cancer-associated genes and mutations has been ongoing for several decades. However, before the sequencing of the human genome, disease genes were discovered by examining one gene at a time which was cumbersome and inefficient. Therefore, only few genes have been shown to play important roles in prostate cancer. These include loss and mutation of tumor suppressor genes such as Nkx3.1, PTEN, and p53, amplification and/or overexpression of oncogenes such as AR [3], and promoter hypermethylation of tumor suppressors such as GSTP1 [4]. With the completion of the human genome project at the end of the last century, the advent of microarray technologies and, most recently, massively parallel NGS technologies have revolutionized the way fundamental biological questions are addressed [5, 6]. The power of these high-throughput approaches lies in their ability to comparatively analyze gene expression and regulatory patterns at the genome-wide scale.
2.1 Expression microarray profiling of prostate cancer
The behavior of a cell is dictated by the set of genes and proteins that it expresses. Microarrays containing thousands of DNA fragments complementary to the coding regions of all known genes provide a valuable tool to simultaneously monitor the expression of all genes in a cell. In the early days, microarrays contain excessive cDNAs that are isolated from cDNA libraries and printed on glass slides, which are hybridized to two samples labeled with distinct dyes, commonly Cy 3 and Cy5 [7, 8]. The ratio of the two dye signals for each cDNA spot on the array is used as a measure of differential gene expression between the samples for the corresponding gene. Following the completion of the human genome project, microarray technologies have evolved rapidly to contain chemically synthesized short-oligonucleotide DNAs as probes for genes. The most commonly used microarrays today include Agilent microarrays, Affymetrix chips, and Illumina bead arrays.
By comparing gene expression profiles between normal and diseased samples, one will be able to identify differentially expressed genes, which would be strong candidates of disease genes. Using human cDNA microarrays of less than 10,000 elements, several groups have profiled tens of normal and neoplastic prostatic specimens and developed signature gene expression that is characteristic of localized and metastatic prostate cancer tissues [9-11]. For example, in 2002 three studies concurrently reported AMACR as a new molecular marker of prostate cancer [12-14]. AMACR up-regulation at the protein level was confirmed in nearly all prostate cancers by tissue-microarray staining [15]. This knowledge was rapidly translated to the bedside; immunohistochemical staining of AMACR in prostate biopsies has been used as a prognostic tool in clinical pathology [16]. Another important gene discovered through microarray profiling of prostate cancer is the Polycomb group (PcG) protein Enhancer of Zeste homolog 2 (EZH2), which highly specifically catalyzes the epigenetic silencer Histone H3 Lysine 27 trimethylation (H3K27me3) [17]. EZH2 was found overexpressed in metastatic prostate cancer and its high expression level in clinically localized prostate cancer predicts poor prognosis. Since its first implication in prostate cancer, more than 300 papers have been published in less than a decade reporting EZH2 function in a variety of cancer types [18]. Many other genes such as Hepsin, IGFBP3, and CXCL2 have also been associated with prostate cancer by various cancer profiling microarray studies [9-11, 19-21]. However, it is important to note that cancer profiling studies do not always agree with one another due to variability in experimental procedures, microarray platforms, analytical methods, and patient cohorts.
Besides disease gene discovery, expression profiling of a large number of primary tumors with clinical follow-up has also enabled the identification of novel diagnostic and prognostic markers or gene signatures. For example, using Affymetrix chips Glinsky et al. (2004) and Stephenson et al. (2005) each profiled an independent set of 79 prostate cancer tissues from patients with distinct clinical outcomes and identified gene signatures that could predict biochemical failure following radical therapy with high accuracy [22, 23]. In another study, Yu et al. (2004) profiled the expression pattern of 66 tumors and derived a 70-gene signature to distinguish metastatic prostate cancer from nonaggressive ones [20]. Moreover, the expression levels of numerous individual genes such as EZH2, SLIT2, and UBE2C have also been associated with clinical outcome [24, 25].
Understanding the characteristics of hundreds of prostate tumors at the molecular level holds the promise for molecular sub-classification. This is particularly important as prostate cancer is a highly heterogeneous disease, for which further stratification may better define a specific disease and guide improved targeted therapy. In 2005, through microarray profiling of primary tissues followed by Cancer Outlier Profile Analysis, Tomlins et al. made the seminal discovery of recurrent genomic translocation fusing the androgen-sensitive TMPRSS2 promoter with the coding regions of the oncogenic ETS family transcript factor ERG in prostate cancer [21]. Multiple studies have rapidly confirmed this gene fusion in independent patient cohorts. TMPRSS2-ERG gene fusion was detected in 40-80% of localized and metastatic prostate tumors but not in any benign samples [26-28]. This was the first major recurrent gene fusion identified in epithelial tumors and represented a paradigm shift in our investigation of the mechanisms underlying solid tumor initiation and progression. This study has drawn much attention from the cancer research community and has provoked the search of gene fusions in other epithelial tumors [29]. Within a short period of time, a number of new translocations were found in prostate cancer although with much lower rate of recurrence [30, 31]. A majority of these genomic rearrangements lead to the fusion of an androgen-regulated promoter, such as those of SLC45A3, CANT1, and KLK2 with an ETS family transcription factor, including ERG, ETV1, ETV4, and ETV5. However, it is important to note that some gene fusions may occur through mechanisms other than translocation, such as trans-splicing and direct transcript read-through as is the case of SLC45A3-ELK4 [32]. In addition, not all 5′ fusion partners are androgen-responsive genes as is the case of HNRPA2B1-ETV1 fusion [33]. Due to the specificity of these gene fusions in prostate cancer but not in normal tissues, they become great candidates for prostate cancer diagnosis. A recent report by Tomlins et al. indeed demonstrated that urine TMPRSS2-ERG transcript is associated with indicators of clinically significant prostate cancer at biopsy and prostatectomy [34].
2.2 mRNA-Seq to detect gene fusions in epithelial tumors
Lately, the search of gene fusions was made significantly more efficient due to the ability to directly sequence the entire cancer transcriptome. The genome, epigenome, transcriptome, and interactome of a given sample can now be readily determined through deep sequencing using various NGS platforms such as the Illumina Genome Analyzer/Hi-Seq, ABI SOLiD, Roche 454 Genome Sequencer, and the Helicos HeliScope single molecule sequencer [35-41]. RNA sequencing (specifically mRNA-Seq) of several prostate cancer samples confirmed TMRPSS2-ERG gene fusions and also uncovered new recurrent chimeric transcripts [42]. The power of transcriptome sequencing in detecting chimeric transcripts was further enhanced by paired-end sequencing technology coupled with bioinformatics analysis [31]. Applying this technique to a large number of tumor samples should lead to the discovery of predominant chimeric sequences and the major recurrent gene fusions in a given cancer type. Indeed, Palanisamy et al. used paired-end transcriptome sequencing to search for translocations in ETS rearrangement-negative prostate cancers and identified gene fusions to BRAF and CRAF in 1-2% of prostate carcinomas [43]. These RAF pathway gene rearrangements were also found in other solid tumors such as gastric cancer and melanoma. Similarly, Pflueger et al. performed mRNA-Seq analysis of 25 human prostate cancer samples for the presence of chimeric fusion transcripts [44]. In addition to confirming gene fusions to ERG and ETV1 they also discovered four novel genomic rearrangements involving non-ETS genes. While mRNA-Seq is surely a useful tool for gene fusion discovery, many artificial gene fusions may be generated during library preparation steps. Sophisticated software tools are often required to filter out the biological fusions which should be further confirmed using conventional approaches.
2.3 Next-generation sequencing of prostate cancer transcriptome
Besides discovering chromosomal translocations, mRNA-Seq has also been using in monitoring gene expression, detecting lowly-expressed genes, and uncovering alternative splicing and novel transcripts [45-47]. However, for studies in species with sequence information fully decoded, such as human and mouse, microarrays are still the method of choice for gene expression profiling. Besides the higher cost, mRNA-Seq data suffers a number of limitations including the cumbersome steps involved in data analysis, biases caused by sample prep protocols, and the difficulty in reliably measuring lowly-expressed genes as a majority of the sequencing reads are consumed by the abundant genes. However, this may soon change with the technology rapidly evolving and the price for NGS continuously dropping. The mRNA-Seq data does offer several leverages for gene expression analysis such as its independence on pre-designed probes and the potential to identify novel transcripts. For the analysis of large samples, target exome capture followed by massively parallel sequencing has become an efficient strategy to selectively sequence the portion of the coding genome, which constitutes a majority of known disease-causing mutations [48]. Whole exome-sequencing of 23 prostate cancers has recently revealed a spectrum of mutation frequencies in lethal prostate cancers [49]. The mRNA-Seq results have also been utilized to analyze long non-coding RNA (ncRNA) transcripts [50]. For example, Prensner et al. discovered 121 unannotated prostate cancer-associated ncRNA transcripts by ab initio assembly of mRNA-Seq data of a cohort of 102 prostate tissues and cell lines. In particular, one of these ncRNA transcripts is a prostate-specific regulator of cell proliferation and a target of the Polycomb Repressive Complex [51]. By contrast, small RNA sequencing, which uses DNA libraries that are prepared distinctly from mRNA-Seq, has been more frequently used to infer small RNA expression levels and to identify novel ones. For example, the miRNAs expressed in androgen-dependent and -independent prostate cancer cell lines have recently been determined [52]. At least 83 miRNAs are significantly differentially expressed between androgen-dependent and castration-resistant prostate cancers. Therefore, microarrays and NGS have rapidly transformed our understanding of prostate cancer genomics and transcriptome.
3. Transcription factor regulation of gene expression
High-throughput genomic approaches including microarrays and NGS have revealed hundreds of genes that are differentially expressed between normal and cancerous samples. This brings up an equally challenging problem as the lack of disease genes in terms of identifying the central genes for therapeutic intervention. It has thus become increasingly important to determine the converging points of the many disease genes/pathways and to identify the key disease-causing events. A systematic understanding of the regulatory network or interactome that controls cancer process is thus critical. While the set of genes and proteins a cell expresses determine its cellular behavior, gene expression in turn is tightly regulated by multiple mechanisms, including transcription factor binding, epigenomic modifications, as well as high-order chromosomal structures (Figure 1).
3.1 ChIP to determine protein-DNA interactions
Chromatin Immunopreciptation (ChIP) assay has been very useful in identifying the genomic loci that a specific protein binds [53]. It captures protein-DNA interaction in living cells and determines the genomic regions that are bound or occupied by a protein of interest [54, 55]. When coupled with genome-wide DNA microarrays (ChIP-chip) or massively parallel sequencing (ChIP-Seq), ChIP provides a whole-genome view of DNA binding events of a given protein [54, 56, 57]. Using Agilent 60-mer oligonucleotide microarrays spanning the proximal promoter (−0.8 to +0.2kb around TSS) regions of over 17,000 genes we later identified 1,165 PcG- or H3K27me3-occupied genes in three metastatic prostate cancer tissues and revealed remarkable similarity of PcG targets between poorly differentiated cancer and pluripotent embryonic stem cells [57]. Similarly, using customized high-density tiling arrays, Gal-Yam et al. reported frequent switching of PcG occupancy and promoter DNA hypermethylation in PC3 prostate cancer cells [58]. Over the years, EZH2 has been shown to regulate a large number of tumor suppressor genes such as CNR1, DAB2IP, RUNX3, and SLIT2 [25, 59-62],
Due to their high prevalence in prostate cancer, TMPRSS2-ERG gene fusions and their protein products have also been an area of extensive investigation. Using ChIP-Seq we mapped the genomic landscape of ERG in the TMPRSS2-ERG fusion positive VCaP prostate cancer cell line as well as in a fusion-positive primary prostate cancer tissue [63]. Analysis of the ChIP-Seq data revealed approximately 38,000 ERG binding events, approximately 50% of which locate around the TSS of known genes.
The transcription factor that has been the most extensively studied in prostate cancer is probably the androgen receptor (AR). Following technological advances the genomic landscape of AR has been repeatedly analyzed by combining ChIP with various platforms as they became available. In 2007, coupling AR ChIP with ENCODE tiling arrays Takayama et al. identified 10 AR binding sites and validated AR recruitment to these genes[64], while using proximal promoter microarrays Massie et al. revealed AR binding to 1532 genomic sites and, for the first time, suggested novel pathways of androgen signaling cooperated by ETS-family transcription factor ETS1 [65]. Using custom NimbleGen oligonucleotide tiling arrays covering approximately 104 kb genomic regions centered on the TSS of 548 pre-selected androgen-regulated genes, another report showed that a large proportion of AR binding sites are not within the proximal promoter regions [66]. Wang et al. performed ChIP-on-chip using chromosome 21 and 22 tiling arrays and reported 89 AR binding sites in the LNCaP prostate cancer cells and further confirmed that AR functions primarily through distal enhancers [67]. Recently, using ChIP-Seq technologies the genomic landscape of AR was determined at high resolution [35, 56]. For example, ChIP-Seq analysis of PC3 cells expressing ectopic AR demonstrated that AR directly induces genes of the growth-inhibition response program [68]. We and others have also mapped the genomic landscape of AR in a number of androgen-dependent prostate cancer cell lines such as LNCaP and VCaP as well as in primary prostate cancer tissues [63, 69, 70].
3.2 ChIP-Seq to analyze transcription factor co-regulation
In addition to target gene identification genome-wide analysis of protein-DNA interaction also provides an excellent tool for the investigation of co-localization and co-regulation of two or more transcription factors over the entire genome. In 2002, Shang et al. revealed the androgen receptor transcription complex including a variety of coactivators such as GRIP1 and CBP and corepressors such as NCoR and SMT [71]. The assembly of the AR complexes involves chromosomal looping of the promoter and enhancer of the target gene such as PSA [72]. In 2007, the same group mapped the cis-regulatory sites of AR on Chromosomes 21 and 22 by coupling ChIP with oligonucleotide tiling microarrays and reported AR interaction with co-factors FoxA1, GATA2 and OCT1 [67]. The transcription factor FoxA1 was thought to act as a pioneer factor binding to nucleosomal DNA to pry open the chromatin at cell type-specific enhancers and subsequently recruit lineage-specific hormonal receptors such as AR and estrogen receptor (ER) [73]. Using a fine breast cancer cell line model that expresses AR but not ER, Robinson et al. revealed an AR cistrome in breast cancer cells that resembles that of ER [74]. This alteration of AR specificity is dictated by and dependent on the breast cell-specific FoxA1 binding. Knockdown of FoxA1 reduces chromatin-associated AR and inhibits AR binding to target loci in breast cancer cells. This functionality of AR in occupying the ER cistrome and regulating ER-mediated transcriptional program further confirmed FoxA1 as a pioneer factor for AR binding. However, in prostate cancer studies have shown that loss of FoxA1 in androgen-sensitive LNCaP prostate cancer cells does not prevent androgen induction of target genes such as PSA and TMPRSS2 [24, 67]. In addition, although there is a significant overlap between AR and FoxA1 binding events, ChIP-Seq studies have also revealed that a large set of AR-bound loci are not simultaneously bound by FoxA1, and vice versa [63]. These results suggest that (1) not all AR binding is dependent on FoxA1 and (2) FoxA1 may have more complicated roles than simply being pioneering factor for AR. Recently, two groups attempted to further untangle the interaction between FoxA1 and AR cistrome [69, 70]. Through ChIP followed by deep sequencing, they revealed markedly increased, rather than decreased, AR binding events upon FoxA1 knockdown in LNCaP cells. They reported 3 categories of AR binding events that vary in their response to FoxA1 knockdown: (1) lost AR loci that are dependent on FoxA1 expression, (2) conserved AR loci that are independent of FoxA1, and (3) gained AR loci upon FoxA1 knockdown. Obviously, a majority of previous studies have focused mainly on the first category of AR target genes that are pioneered by FoxA1 with only a few studies touched upon the second category, however, without mechanistic details. These current studies, however, clearly demonstrated an AR transcriptional program in the absence of FoxA1. In fact, there are far more AR binding sites gained than lost upon FoxA1 depletion. Furthermore, in contrast to previous studies showing invariant AR expression upon FoxA1 over-expression or knockdown in prostate cancer cells [24], these studies reported increased AR expression [69] and binding to target genes including PSA upon FoxA1 depletion, suggesting an inhibitory role of FoxA1 on AR signaling, at least to some target genes [74]. This is groundbreaking as it seems under certain context FoxA1 may have an inhibitory role on AR signaling, which is opposing to what was believed in the field for years. Interestingly, while one of these two studies indeed demonstrated an association of FoxA1 with good prognosis, the other study showed that high FoxA1 is predictive of poor clinical outcome. This inconsistency may be due to varied roles of FoxA1 at different stages of prostate cancer. Supporting the former study, in breast cancer FoxA1 has been shown as a significant marker of good prognosis [75]. Despite of this controversy, the consensus is that FoxA1 might contribute significantly to human malignancy [76]. Disappointingly, neither studies investigated the function of FoxA1 itself nor did they examine how AR is recruited to genomic loci in the absence of FoxA1. A recent study has just begun to examine the role of FoxA1 in castration resistant prostate cancer [77]. Further investigation of AR and FoxA1 cistrome and function in the context of each other is much in need and should help further clarify the regulatory network involving these two essential transcription factors of prostate cancer. We believe some of the inconsistent results may be easily explained by cell type-specific and disease stage-specific effects of FoxA1 in prostate cancer.
3.3 Interrogating ChIP-Seq and expression profiling data
ChIP-Seq has been extremely useful for a comprehensive view of protein-DNA interaction over the chromatin. However, it is obvious that not all genes that are bound by a transcription factor respond at the expression level upon dys-regulation of this factor. On the other hand, genes that are differentially regulated by a transcription factor are not necessary directly bound by the factor. Interrogating genome-wide protein-DNA interaction data with gene expression profiling data has thus been very useful in narrowing down to the direct target genes for further functional characterization. We performed integrative analysis of PcG target genes through ChIP-chip analysis of EZH2, SUZ12, and H3K27me3 and expression microarray profiling of prostate cells with EZH2 overexpression or knockdown. We found that PcG suppresses a large set of tumor suppressor genes, most of which are developmental regulators that are silenced by EZH2 in embryonic stem cells in order to maintain pluripotency [57]. We and others have characterized a number of of direct EZH2 target genes including ADRB2 [78], CDH1 [79], SLIT2 [25], and DAB2IP [60, 61]. The suppression of these tumor suppressor genes contribute to EZH2-mediated prostate cancer tumorigenesis such as cell proliferation, transformation, migration, invasion, and/or metastasis using in vitro cell lines and in vivo mouse models.
Similarly, several groups have employed integrative genomics analysis to understand the downstream pathways of the TMPRSS2-ERG gene fusions. Using ChIP-Seq we mapped the genomic landscape of AR and ERG in the TMPRSS2-ERG fusion-positive VCaP prostate cancer cells. Comparative analysis of their binding sites revealed that ERG binds to genomic regions that are also bound by AR [63]. Considering that ERG is positively regulated by AR, we initially hypothesized that ERG facilitates AR signaling thus forming a positive-feedback loop. Surprisingly, integration of expression microarray data revealed that AR-induced genes are down-regulated in TMPRSS2-ERG fusion-positive prostate cancers. This finding was further confirmed at individual gene level showing that ERG directly binds to a large number of AR-induced genes such as PSA and TMPRSS2 to suppress their expression [63]. In addition, bioinformatics analysis of gene set enrichment revealed that ERG-regulated genes are enriched for Polycomb target genes. Integration with ChIP-Seq data revealed direct ERG binding at the EZH2 promoter. This observation led to further experimentation demonstrating that ERG directly induces EZH2 expression in prostate cancer cells. Concurrently, through bioinformatics analysis of prostate cancer microarray profiling data another group reported EZH2 up-regulation in ERGhigh tumors. Through PCR analysis of ERG ChIP-enriched DNA they subsequently demonstrated that ERG directly induces EZH2 expression [80]. Interestingly, they also showed that ESE3, another ETS family transcription factor, directly inhibits EZH2 expression.
Integrative analyses of genome-wide protein-DNA interaction and gene expression data have also been very useful in the study of AR-mediated transcriptional regulation (Figure 2). A number of studies have employed expression microarrays to study androgen-regulated gene expression and ChIP-based assays to identify direct AR target genes [63, 67]. Coupling ChIP with genome-wide tiling arrays, Wang et al. compared the AR cistrome in LNCaP cells with that in its androgen-independent derivative LNCaP-abl cells [24]. This study led to the identification of cell cycle-related genes such as UBE2C that are bound and regulated by AR only in LNCaP-abl cells, thus providing a novel mechanism to androgen-independent cell growth. Recently, Massie et al. integrated genomic data with metabolomic profiling to reveal that AR regulates aerobic glycolysis and anabolism [81]. By combining AR genomic studies with gene expression data in clinical samples, they identified CAMKK2, the calcium/calmodulin-dependent kinase kinase 2, as a metabolic master regulator downstream of androgen signaling in prostate cancer. This study thus illustrated the power of unbiased integrative approaches in identifying novel disease pathways and promising therapeutic targets.
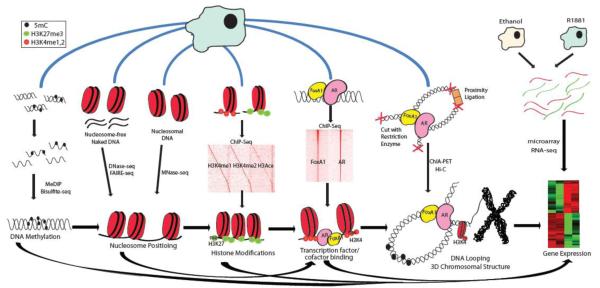
Figure 2. Integrative genomic analysis to understand AR regulation in prostate cancer.
The genes that are differentially expressed upon androgen treatment were determined using microarray profiling or RNA-Seq. To identify the subset of the differentially expressed genes that is directly regulated by AR, ChIP-Seq was performed to map the genomic landscape of AR. ChIP-Seq analysis of AR cofactors confirmed AR and FoxA1 colocalization, while ChIP-Seq analysis of histone modifications demonstrated that H3K4me1 and H3K4me2 marks FoxA1 and AR binding sites. Analysis of nucleosome positioning using DNase-Seq, FAIRE-Seq, or lately MNase-Seq suggested that H3K4 methylations pry open nucleosomes to allow FoxA1 and AR binding. Nucleosomes positioning in turn is largely influenced by DNA sequence preferences. In addition, genome-wide DNA methylation patterns which can be determined using MeDIP-chip, MeDIP-Seq or direct bisulfite-sequencing also have substantial effect on gene expression. Furthermore, ChIA-PET and Hi-C assays are able to reveal intra- and inter-chromosomal DNA loopings that are essential for the AR enhancer function.
However, it is of interest to note that although androgen-repressed genes can be observed in a majority of the datasets derived from these studies, in most cases, they are largely overlooked and have not been selected for further characterization. Only very few studies have examined genes that are down-regulated upon androgen treatment [82]. For example, activated AR has been shown to cooperate with HDAC in down-regulating E-cadherin expression in prostate cancer, thus promoting tumor metastasis [83]. AR has also been shown to inhibit the expression of c-Met and Smad3 by inhibiting Sp1-induced transcription [84, 85]. Some studies reported that AR inhibits cyclin D1, TGF-β1 and CDK2 via binding to negative androgen-response elements [86, 87], while others demonstrated that AR suppresses α-subunit and Runx2 genes through mechanisms independent of AR binding domain [88, 89]. Although a majority of these previous studies proposed indirect mechanisms for AR-mediated repression, with genome-wide AR binding events readily mappable and the understanding of AR acting primarily through distal enhancers, it has recently begun to unveil that AR may directly function as a transcriptional repressor. The lack of direct AR-repressed genes to date may be in part due to the difficulties in mapping AR binding elements at distal enhancers in the past. Indeed, a recent study has shown that AR directly suppresses itself by recruiting H3K4 demethylase LSD1 [90]. It will be of great interest to decipher the full spectrum of AR function and characterize direct AR-repressed genes, which are often tumor suppressors of critical functionality.
4. Epigenomic modifications regulate prostate cancer
In the recent years evidence has emerged that gene expression is also controlled by the epigenome including DNA methylation, histone modifications, nucleosomal structure, and RNA-associated regulation such as miRNA, non-coding RNA and pseudo-genes [91] (Figure 1). Following the technological advances much has been achieved lately in understanding the epigenome and its function in prostate cancer.
4.1 DNA methylation and prostate cancer
Alterations of methylation patterns at the 5-position of cytosine (5mC) residues within CpG dinucleotides, including global hypomethylation within the intergenic or intragenic regions and promoter hypermethylation at gene-specific loci, have been associated with prostate cancer [91, 92]. Promoter hypermethylation is a common mechanism for silencing tumor suppressor genes in cancer. To date, more than 160 genes have been frequently analyzed for promoter hypermethylation and potentially contributing to prostate cancer, including GSTP1, APC, CCND2, CDH1, RASSF1, and PTGS2. A comprehensive list of the commonly hypermethylated genes can be found in several recent reviews [92, 93]. Out of these genes, promoter hypermethylation of GSTP1 occurs in a majority of prostate cancer and has been developed into epigenomic biomarker for clinical tests [94]. By examining target genes of PcG protein EZH2, we have reported SLIT2 promoter hypermethylation in aggressive prostate cancer [25]. To evaluate aberrant DNA methylation as biomarkers for prostate cancer diagnosis and/or prognosis, another study recently analyzed absolute methylation levels of 28 candidate genes in 48 prostate cancer samples and confirmed the prognostic potential of several genes including SLIT2 promoter hypermethylation [95].
In addition to locus-specific DNA methylation analysis, genome-wide DNA methylation profiling is increasingly possible due to the development of high-throughput technologies in the last decade. The plethora of techniques including array-based and deep sequencing-based assays that are available for methylation analysis have been recently reviewed with a fine comparison of their strengths and weaknesses [96]. While singe-base-pair resolution of bisulphite sequencing may soon become possible for complex genomes as sequencing costs drop, to date the most commonly used techniques are array- or deep sequencing-analysis of selected regions. Multiple approaches have been utilized to enrich highly methylated DNA, including chromatin immunoprecipitation using antibodies specific for 5-methylcytosine (termed MeDIP) or methyl-CpG binding domain (MBD) proteins [97]. Meissner et al. has developed methods to analyze methylation pattern using reduced representation bisulfite sequencing of selected regions of the genome through MspI or BglII digestion [98]. Many studies, however, have utilized commercially available arrays such as the Illumina Infinium HumanMethylation BeadChip that contains over 26,000 CpG sites at the promoters of more than 14,000 genes. By analyzing methylation profiles of hundreds of prostate cancer samples, novel prognostic and diagnostic methylation biomarkers such as EFEMP1 has been identified [99, 100]. Similarly, we have used MeDIP and MethylPlex (Rubicon Genomics, Inc) coupled with array hybridization or deep-sequencing to study global CpG island methylation in prostate cancer [99]. This study confirmed an overall increase in promoter methylation during prostate cancer progression and identified 2,481 cancer-specific methylation regions. In addition, differentiation methylation in LINE-1 between TMPRSS2-ERG fusion-positive and –negative prostate tumors was observed, although the mechanism for this difference remains to be determined. Despite these important findings, genome-wide methylation data to date are largely biased due to technical limitations [101]. The use of whole-genome bisulfite sequencing and in particular single-molecule sequencing approaches in the future shall shed light on DNA methylation at single-base-pair resolution across the genome, which will very likely lead to important discoveries.
The regulation of DNA methylation has recently become an area of intense focus since the seminal discovery of TET1 as an enzyme that converts 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC) [102]. TET1 and 5-hmC may play critical roles in DNA de-methylation and thus gene regulation. A number of studies have already begun to investigate the important function of TET1 in embryonic stem cells and during differentiation [103-106]. The 5-hmC is highly abundant in embryonic stem cells and its abundance decreases upon differentiation. While TET1 has been investigated in acute myeloid leukemia wherein it is fused to MLL [102, 107, 108], only very recently have the expression pattern of TET1 and the genomic distribution of 5-hmC come to be explored in other cancer types. For example, two studies have recently demonstrated decreased 5-hmC in colorectal and prostate cancer tissues [109, 110]. It will be exceedingly intriguing to determine the genomic landscape of TET1, 5-hmC and 5-mC in cancers such as prostate cancer. Global mapping of TET1 and 5-hmC have been proven achievable through ChIP-Seq using anti-TET1 or anti-5hmC antibodies in mouse embryonic stem cells [104, 105, 111]. Furthermore, Song et al. has recently developed a method to determine genome-wide distribution of 5-hmC by transferring chemically modified glucose moiety onto the hydroxyl group of 5-hmC for highly specific biotin pull down and affinity enrichment [103, 112]. These advances should enable comparative analysis of 5-hmC genomic landscapes among various tissues and cancer types in the near future. It is to be determined whether 5-hmC will provide innovative disease-specific methylation signatures beyond those derived from 5-mC.
4.2 Histone modifications and transcriptional regulation
Chromatin modification at histone tails can alter chromatin structure and is a potent cellular mechanism in regulating gene expression. Through immunohistochemical staining of primary prostatectomy tissues, Seligson et al. first determined the levels of several histone acetylation and methylation markers and reported that histone modification patterns predict clinical outcome [113]. Using ChIP-chip assays regulatory enhancers were found to be marked with cell-type-specific histone modification patterns, which seem to subsequently dictate cell-type-specific transcriptional programs, which in prostate cancer is largely mediated by androgen receptor [114]. Through ChIP-chip analysis of FoxA1 binding sites in breast and prostate cancer cells, Lupien et al. demonstrated that cell type-specific histone signatures denoted by H3K4me1/2 but not H3K9me2 first recruits FoxA1 to distal enhancers, which subsequently recruits lineage-specific hormonal factors such as AR to dictate cell type-specific transcriptional program [73]. ChIP-Seq analyses have confirmed substantial overlap between the binding events of AR, FoxA1 and H3K4me/2 methylations [63, 69]. In androgen-independent prostate cancer, altered histone H3K4 methylation directs AR binding to novel target genes [24]. In addition, recent studies showed that chromatins containing distinct H3K4me1 and H3K27ac profiles direct AR in binding to new genomic loci upon FoxA1 loss in prostate cancer [69]. Moreover, AR is able to suppress its own gene expression by recruiting LSD1 for repressive chromatin remodeling [90].
Over the years, a large amount of transcriptional co-regulators have been found to interact with AR and modulate its activity, many of which such as CBP, p300 and HDAC are known to function by altering histone modification patterns [115]. HDACs, which removes histone acetylation and promotes histone methylation, is commonly considered as co-repressors that antagonize AR activity. Paradoxically, Welsbie et al. recently demonstrated that HDACs are required for AR-mediated activation of target genes such as PSA and KLK2 [116]. In addition, LSD1, the first histone lysine demethylase identified, is also an AR co-factor [117]. LSD1 was initially found to specifically demethylate histone H3K4 as a component of co-repressor complexes [118, 119]. In the context of AR biology, LSD1, however, demethylates repressive histone H3K9 methylation to promote AR-dependent transcription. This altered specificity in substrates is believed due to phosphorylation of histone H3 at threonine 6 by protein kinase C beta I, which prevents LSD1 from demethylating H3K4 during AR-dependent gene activation [120]. This model, however, was recently challenged by the finding that AR recruits LSD1 to suppress the expression of AR itself and other genes through H3K4 demethylation [90]. The interplay between AR, LSD1 and histone demethylation may be much more complicated and context-dependent.
4.3 Nucleosome positioning regulates transcription
Besides DNA and histone modifications, gene expression is also regulated by high-order chromatin structure that affects the accessibility of cis-regulatory elements by transcription factors. Nucleosomes are the basic repeating unit of chromatin, composed of ~147 bp DNA wrapped around a histone octamer core [121]. Nucleosomes are not evenly spaced across the genome. Their positioning is regulated by multiple mechanisms such as DNA sequence preference and transcription factor competition. Compared to the nucleosomal DNA, the “naked” DNA between two nucleosomes are much more accessible by DNA-binding proteins such as transcription factors [122]. Therefore, nucleosome positioning is a potent mechanism in regulating DNA accessibility and consequently transcriptional regulation. There are a number of techniques to determine open chromatin regions, many of which have lately been coupled with DNA microarrays and deep-sequencing for genome-wide discoveries. DNase-Seq utilizes DNase I to selectively digest nucleosome-free therefore DNase I-hypersensitive sites to uncover regulatory elements including promoters, enhancers and silencers that are presumably occupied by transcription factors [123]. It has been used to map all active regulatory elements in a cell under a specific condition, although these active elements may be occupied by either transcription activators or repressors [124, 125]. Similarly, FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) uses formaldehyde crosslinking and phenol-chloroform extraction to isolate nucleosome-depleted DNA from DNA fragments that are bound by any proteins, a majority of which are histones [126]. Analysis of seven human cell lines demonstrated high concordance in the regulatory regions revealed by DNase-Seq and FAIRE-Seq [112]. However, the combination of both methods is more effective than either one alone in identifying potential regulatory DNA elements. Using FAIRE followed by tiling array hybridization, Eeckhoute et al. reported that a significant portion of FoxA1-bound sites are in closed chromatin and marked by repressive histone marks, thus suggesting that FoxA1 alone is required but insufficient for enhancer activity [127]. On the other hand, this also indicates that FoxA1 may have unreported roles within repressive chromatin that are distinct from its pioneering effect for AR-mediated transcriptional activation.
Although DNase-Seq and FAIRE-Seq are suggestive of nucleosome-free regions, it was not until recently has it become possible to more precisely infer genome-wide nucleosome positioning. Using nucleosome-resolution histone modification ChIP-Seq data, bioinformatics tools such as NPS have lately been developed to determine nucleosome positioning based on the location of histones and their modifications [128]. In prostate cancer cells, analysis of H3K4 methylation ChIP-Seq data revealed the nucleosome dynamics underlying AR function [129]. For at least some androgen-responsive loci, central nucleosomes that were present at the AR binding sites were dismissed to the flanking regions upon androgen treatment, thus opening up chromatin for subsequent transcriptional activation. However, it is yet to understand how nucleosome dynamics regulate transcriptional repression following androgen treatment. As AR was recently shown to possess inhibitory function through repressive chromatin remodeling [90], it will be important for future studies to delineate the different nucleosomal dynamics that may be involved.
Technological advances in last few years have also made it increasingly possible and affordable to determine genome-wide nucleosome positioning through deep sequencing. MNase-Seq utilizes micrococcal nuclease (MNase) to digest accessible “naked” DNA and then isolate DNA fragments that are protected by nucleosomes from MNase digestion for subsequent deep sequencing. This will ultimately produce a genome-wide map of positioned nucleosomes (Figure 2). Unlike DNase-Seq and FAIRE-Seq that sequence the small percentage of open chromatin DNA, MNase-Seq requires sequencing of the majority of the genome that is wrapped around histone cores of nucleosomes. This has imposed major challenges in terms of cost and massive data handling until recently. The first few genome-wide patterns of nucleosome positioning were thus mapped in small-genome model organisms such as yeast. Segal et al. have shown that the genome sequence is highly predictive of the in vivo nucleosome positions and nucleosomes are depleted from the yeast promoters which often contain tracts of adenosine-thymidine base pairs that are unfavorable to nuclosomes [122, 130]. While promoters and transcription factor binding sites in yeast typically have low intrinsic nucleosome occupancy, regulatory elements in human have high GC content that are favorable to nucleosomes [131]. This was thought important in restricting DNA accessibility to transcription factors in more complex genomes. Several groups have pioneered in mapping genome-wide nucleosome positioning in human cells [132, 133]. In prostate cancer, several studies have started to implicate a role of nucleosome occupancy in epigenetic silencing and AR activation, although at current stage most of these studies are limited to few genes/regions [134, 135]. In the next few years, the dynamics of nucleosome positioning at the global scale during different stages of prostate cancer will likely be determined.
4.3 High-order chromatin structural regulation of gene expression
Besides nucleosome positioning, three-dimensional (3D) chromosomal architecture also play important roles in transcriptional regulation. For example, DNA looping between enhancer and promoter is an important regulatory mechanism for enhancer-bound transcription factors such as AR and FoxA1 [134, 136]. Chromosome conformation capture (3C)-based assays are often used to analyze chromosomal interaction and spatial organization. Coupling ChIP with 3C, previous studies have demonstrated AR recruitment of co-activators to create chromosomal loops between the enhancer and promoter of AR-bound genes such as PSA and TMPRSS2 [72]. Chromosomal proximity mediated by AR binding and DNA looping, when coupled with DNA double-stranded breaks, has also been appealed guilty in causing chromosomal translocations such as TMPRSS2:ERG gene fusions in prostate cancer [137].
With emerging evidence showing that a majority of the AR binding events occur at distal enhancers, the DNA looping architecture around AR binding sites becomes critical for the understanding of AR-mediated transcriptional regulation. A majority of current studies of AR cistrome assumed that AR binding events regulate the nearest genes. However, as DNA looping brings distal chromosomal regions or even different chromosomes to local proximity, it is possible for an AR binding event to regulate the expression of distant genes or even genes located on different chromosomes. This complexity of transcriptional regulation through intrachromosomal or interchromosomal DNA looping has just begun to be unveiled. Notably, Lieberman-Aiden et al. has reported a method termed Hi-C to probe genome-wide 3D DNA-protein interaction by coupling 3C-like proximity-based ligation with NGS [138]. Using Hi-C they were able to reverse-engineer the spatial proximity maps of the human genome at 1 megabase resolution. While this study surely demonstrated the potential power of Hi-C in studying the dynamic conformations of whole genome, the complexity of the data is overwhelming and might be too intimidating for most biologists to handle. Indeed, more success has been gained using Hi-C in reconstructing the 3-D architecture of smaller genomes such as those of bacteria [139]. Besides Hi-C, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) provides another effective strategy to study global chromatin interactions. Unlike Hi-C which probes all protein-DNA interactions in a cell, ChIA-PET first uses ChIP to enrich chromatin-DNA complexes involving a particular protein of interest before proximity-based DNA ligation and paired-end sequencing [140]. ChIA-PET is thus able to determine specifically the 3D chromosomal structure around a transcription factor of interest such as hormonal receptors (Figure 2). For example, using ChIA-PET the majority of estrogen receptor (ER)-bound enhancers was found anchored at gene promoters through long-range DNA looping or chromatin interactions. Computational software packages such as ChIA-PET tool have been developed to provide automatic processing of ChIA-PET sequence data [141]. As the technology matures, it will be of high importance to study the genomic architecture around AR binding events and to determine how this architecture may be altered during genetic perturbation and cancer progression.
5. Concluding remarks and perspectives
Using high-throughput technologies, we have gained substantial understanding of prostate cancer transcriptome and its complex mechanisms of regulation (Figure 2). Like many other diseases, prostate cancer is marked by an array of aberrant genetic and epigenetic events ultimately leading to abnormal gene expression patterns and uncontrolled cellular behavior. Employing DNA microarrays and NGS platforms, the community has begun to understand prostate cancer in much more details and at the genome scale. Integrative analysis of genomic and epigenomic data has never been more important in our investigation of cancer and has led to many seminal discovery. Despite these important achievements, advanced prostate cancer and many of other late-stage cancers remain essentially incurable. The key pathway in prostate cancer, the AR signaling, seems much more complicated than previously thought and remains the lead pathway for therapeutic interaction. Studies on this disease pathway have shown controversial results but also bring critical insights into the mechanisms underlying prostate cancer. Future investigation using ever-improving high-throughput technologies shall reveal novel aspects that are critical for prostate cancer initiation and progression.
Acknowledgements
We thank Dr. Hongjian Jin for helpful discussion. This work was supported in part by the NIH P50CA090386 Career Development Award (to J.Y.), U54CA143869 pilot project (to J.Y.), R00CA129565 (to J.Y.), and the U.S. Department of Defense PC080665 grant (to J.Y.), and the Research Scholar Grant #RSG-12-085-01 from the American Cancer Society (to J.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on Prostatic Cancer. I. The Effect of Castration, of Estrogen and of Androgen Injection on Serum Phosphatases in Metastatic Carcinoma of the Prostate. Cancer Res. 1941;1(4):293–97. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 3.Ruijter E, et al. Molecular genetics and epidemiology of prostate carcinoma. Endocr Rev. 1999;20(1):22–45. doi: 10.1210/edrv.20.1.0356. [DOI] [PubMed] [Google Scholar]
- 4.Kopper L, Timar J. Genomics of prostate cancer: is there anything to “translate”? Pathol Oncol Res. 2005;11(4):197–203. doi: 10.1007/BF02893851. [DOI] [PubMed] [Google Scholar]
- 5.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 6.Schena M, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 7.Hughes TR, et al. Microarray analysis of RNA processing and modification. Methods Enzymol. 2006;410:300–16. doi: 10.1016/S0076-6879(06)10014-2. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, et al. Evaluation and optimization of procedures for target labeling and hybridization of cDNA microarrays. Mol Vis. 2002;8:130–7. [PubMed] [Google Scholar]
- 9.Dhanasekaran SM, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–6. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, et al. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61(12):4683–8. [PubMed] [Google Scholar]
- 11.Welsh JB, et al. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61(16):5974–8. [PubMed] [Google Scholar]
- 12.Kuefer R, et al. alpha-Methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161(3):841–8. doi: 10.1016/s0002-9440(10)64244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J, et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62(8):2220–6. [PubMed] [Google Scholar]
- 14.Zhou M, et al. Alpha-Methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002;26(7):926–31. doi: 10.1097/00000478-200207000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Rubin MA, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287(13):1662–70. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 16.Carswell BM, et al. Detection of prostate cancer by alpha-methylacyl CoA racemase (P504S) in needle biopsy specimens previously reported as negative for malignancy. Histopathology. 2006;48(6):668–73. doi: 10.1111/j.1365-2559.2006.02409.x. [DOI] [PubMed] [Google Scholar]
- 17.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 18.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 19.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101(3):811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 21.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 22.Glinsky GV, et al. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–23. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson AJ, et al. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104(2):290–8. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Cao Q, Yu J, Wu L, Dallol A, Li J, Latif F, Wu J, Chinnaiyan AM. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene. 2010;29(39):5370–80. doi: 10.1038/onc.2010.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, et al. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66(17):8347–51. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 27.Soller MJ, et al. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45(7):717–9. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 28.Clark J, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26(18):2667–73. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 29.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 30.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106(30):12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69(7):2734–8. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 34.Tomlins SA, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3(94):94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marguerat S, Wilhelm BT, Bahler J. Next-generation sequencing: applications beyond genomes. Biochem Soc Trans. 2008;36(Pt 5):1091–6. doi: 10.1042/BST0361091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 40.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 41.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 42.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pflueger D, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21(1):56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 46.Marioni JC, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48(3):249–57. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108(41):17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu G, et al. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS One. 2010;5(11):e15519. doi: 10.1371/journal.pone.0015519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25(3):99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 54.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83(3):349–60. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1(2):729–48. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 57.Yu J, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67(22):10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 58.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105(35):12979–84. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18(13):1592–605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280(23):22437–44. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 61.Min J, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16(3):286–94. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujii S, et al. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283(25):17324–32. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takayama K, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26(30):4453–63. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- 65.Massie CE, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8(9):871–8. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolton EC, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21(16):2005–17. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin B, et al. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS One. 2009;4(8):e6589. doi: 10.1371/journal.pone.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(3):601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19(5):631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 73.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson JL, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30(15):3019–27. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehta RJ, et al. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- 76.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30(19):3885–94. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang C, et al. Definition of a FoxA1 Cistrome that is crucial for G1 to S-phase cell-cycle transit in castration-resistant prostate cancer. Cancer Res. 2011;71(21):6738–48. doi: 10.1158/0008-5472.CAN-11-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu J, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12(5):419–31. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 79.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kunderfranco P, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5(5):e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Massie CE, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–33. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grosse A, Bartsch S, Baniahmad A. Androgen receptor-mediated gene repression. Mol Cell Endocrinol. 2011 doi: 10.1016/j.mce.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 83.Liu YN, et al. Activated androgen receptor downregulates E-cadherin gene expression and promotes tumor metastasis. Mol Cell Biol. 2008;28(23):7096–108. doi: 10.1128/MCB.00449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verras M, et al. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007;67(3):967–75. doi: 10.1158/0008-5472.CAN-06-3552. [DOI] [PubMed] [Google Scholar]
- 85.Song K, et al. DHT selectively reverses Smad3-mediated/TGF-beta-induced responses through transcriptional down-regulation of Smad3 in prostate epithelial cells. Mol Endocrinol. 2010;24(10):2019–29. doi: 10.1210/me.2010-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lanzino M, et al. Inhibition of cyclin D1 expression by androgen receptor in breast cancer cells--identification of a novel androgen response element. Nucleic Acids Res. 2010;38(16):5351–65. doi: 10.1093/nar/gkq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi W, et al. Negative Androgen-Response Elements Mediate Androgen-Dependent Transcriptional Inhibition of TGF-{beta}1 and CDK2 Promoters in the Prostate Gland. J Androl. 2011 doi: 10.2164/jandrol.110.011999. [DOI] [PubMed] [Google Scholar]
- 88.Heckert LL, Wilson EM, Nilson JH. Transcriptional repression of the alpha-subunit gene by androgen receptor occurs independently of DNA binding but requires the DNA-binding and ligand-binding domains of the receptor. Mol Endocrinol. 1997;11(10):1497–506. doi: 10.1210/mend.11.10.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baniwal SK, et al. Repression of Runx2 by androgen receptor (AR) in osteoblasts and prostate cancer cells: AR binds Runx2 and abrogates its recruitment to DNA. Mol Endocrinol. 2009;23(8):1203–14. doi: 10.1210/me.2008-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai C, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20(4):457–71. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perry AS, et al. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 2010;7(12):668–80. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 93.Park JY. Promoter hypermethylation in prostate cancer. Cancer Control. 2010;17(4):245–55. doi: 10.1177/107327481001700405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150(9):3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vasiljevic N, et al. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis Markers. 2011;30(4):151–61. doi: 10.3233/DMA-2011-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 97.Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques. 2008;44(1):35, 37, 39 passim. doi: 10.2144/000112708. [DOI] [PubMed] [Google Scholar]
- 98.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim JH, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21(7):1028–41. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kobayashi Y, et al. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21(7):1017–27. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hinoue T, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2011 doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szulwach KE, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7(6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–93. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 106.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–75. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62(14):4075–80. [PubMed] [Google Scholar]
- 108.Langemeijer SM, Aslanyan MG, Jansen JH. TET proteins in malignant hematopoiesis. Cell Cycle. 2009;8(24):4044–8. doi: 10.4161/cc.8.24.10239. [DOI] [PubMed] [Google Scholar]
- 109.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haffner MC, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2(8):627–37. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 114.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 116.Welsbie DS, et al. Histone deacetylases are required for androgen receptor function in hormone-sensitive and castrate-resistant prostate cancer. Cancer Res. 2009;69(3):958–66. doi: 10.1158/0008-5472.CAN-08-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 118.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 119.Lee MG, et al. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 120.Metzger E, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464(7289):792–6. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 121.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 122.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boeger H, et al. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11(6):1587–98. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 124.Shibata Y, Crawford GE. Mapping regulatory elements by DNaseI hypersensitivity chip (DNase-Chip) Methods Mol Biol. 2009;556:177–90. doi: 10.1007/978-1-60327-192-9_13. [DOI] [PubMed] [Google Scholar]
- 125.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giresi PG, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eeckhoute J, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19(3):372–80. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y, et al. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458(7236):362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tillo D, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5(2):e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474(7352):516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen Z, et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30(12):2405–19. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andreu-Vieyra C, et al. Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol Cell Biol. 2011;31(23):4648–62. doi: 10.1128/MCB.05934-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu D, et al. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol Metab. 2011 doi: 10.1016/j.tem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139(6):1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Umbarger MA, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44(2):252–64. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li G, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11(2):R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liang RQ, et al. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33(2):e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu C, et al. Elucidation of the small RNA component of the transcriptome. Science. 2005;309(5740):1567–9. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 144.Fu Y, et al. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4(7):e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]