Abstract
New chemotherapeutics active against multidrug-resistant Mycobacterium tuberculosis (M. tb) are urgently needed. We report on the identification of an adamantyl urea compound displaying potent bactericidal activity against M. tb and a unique mode of action, namely the abolition of the translocation of mycolic acids from the cytoplasm where they are synthesized to the periplasmic side of the plasma membrane where they are transferred onto cell wall arabinogalactan or used in the formation of virulence-associated outer membrane trehalose-containing glycolipids. Whole genome sequencing of spontaneous resistant mutants of M. tb selected in vitro followed by genetic validation experiments revealed that our prototype inhibitor targets the inner membrane transporter, MmpL3. Conditional gene expression of mmpL3 in mycobacteria and analysis of inhibitor-treated cells validate MmpL3 as essential for mycobacterial growth and support the involvement of this transporter in the translocation of trehalose monomycolate across the plasma membrane.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB) in humans, currently infects one-third of the world’s population in its latent form and claims about 1.7 million lives annually making it a leader among the causes of mortality in developing countries1. Infections by M. tb have been estimated by the World Health Organization at 9.4 million new cases during 2009 and the global number of TB cases is still rising at a rate of 0.6% per year fueled by the HIV/AIDS pandemic, poverty and the emergence of multidrug and extensively-drug-resistant strains of M. tb2–3. In this context, continued research aimed at developing new effective anti-TB drugs with bactericidal mechanisms different from those of presently available agents is urgently needed.
The structure and composition of the cell envelope of mycobacteria differentiate these microorganisms from other prokaryotes. With this in mind, considerable effort has been placed on investigating the cell envelope structure and the biosynthesis of its hallmark entities in order to identify attractive drug targets. The interest in drugs targeting the biosynthesis of mycolic acids, in particular, is clearly demonstrated by the therapeutic efficacy of several key anti-TB agents, such as isoniazid and ethionamide, and various other compounds, including isoxyl, thiolactomycin and thiacetazone4–6. Mycolic acids are very long chain (C60-C90) α-branched β-hydroxylated fatty acids found covalently attached to the major cell wall polysaccharide, arabinogalactan (AG), or esterifying outer membrane glycolipids such as trehalose monomycolate (TMM), trehalose dimycolate (TDM; cord factor), glucose monomycolate or glycerol monomycolate. Mycolic acids occur as a mixture of structurally-related molecules that differ primarily from one another by the nature of the chemical groups at the proximal and distal positions of their main meromycolic acid chains. They represent major constituents of the cell envelope of mycobacteria (and 40 to 60% of the cell’s dry weight) and are known to play essential roles in the formation of the outer membrane of M. tb as well as in its interactions with the host7–13. The pathway leading to their biosynthesis is for the most part unique to mycobacteria (for a review4–5,8,14). In M. tb, the multifunctional type I fatty acid synthase (FAS-I) synthesizes C16-C18 and C24-C26 fatty acids in a bimodal fashion. A type II multi-enzyme complex (FAS-II) then catalyzes the processive addition of multiple malonate units onto the C16-C18 products of FAS-I to generate the long-chain meromycolic acid (C48-C64). The pre-formed mero chain is further functionalized with cis or trans double bonds, cis or trans cyclopropane rings and polar functional groups such as methyl ethers, ketones, esters and epoxides. Condensation of the C23-C27α-branch with the meromycolic chain in a process involving the acyl-AMP ligase FadD32 and the polyketide synthase Pks13 yields the full-size α-alkyl β-ketoacyl derivatives15 which, upon release from Pks13, undergo a final reduction step catalyzed by the reductase CmrA, yielding the final mycolic acid products16–17. Although three decades of studies have allowed most aspects of the biosynthesis of mycolates to be elucidated some important steps remain undefined. Among these is the fate of the completed mycolic acids upon their release from Pks13 and reduction by CmrA. How are mycolic acids translocated from their cytoplasmic production site to the periplasm where they are then transferred to their final cell envelope acceptors? Under what form are they translocated across the inner membrane? What is the nature of the export system involved?
According to the generally accepted model, the presence of trehalose in the cells under normal growth conditions is a prerequisite for the successful transfer of mycolates onto their cell envelope acceptors14,18–21. Thus, it may be hypothesized that the conjugation of mycolic acids and trehalose to form TMM in M. tb occurs in the cytoplasm where both compounds are synthesized or imported19,21, by means of an as yet unknown enzymatic activity. Subsequently, TMM or another intermediate mycolic acid acceptor14,22 may be translocated across the inner membrane by an unknown transporter before serving as the substrate for the mycolyltransferases of the antigen 85 complex (FbpA, FbpB and FbpC) whose role is to transfer the mycolic acid chain from TMM onto either AG or another molecule of TMM, yielding TDM23–27.
Random screening of a compound library against whole M. tb bacilli in culture revealed a highly active adamantyl 2,3,4 trifluorophenyl urea with an apparent unique mechanism of action on cell envelope biosynthesis. We report here on the use of this small chemical compound to identify the long sought transporter responsible for the translocation of mycolic acids across the plasma membrane.
RESULTS
Identification of an adamantyl urea inhibitor of M. tb
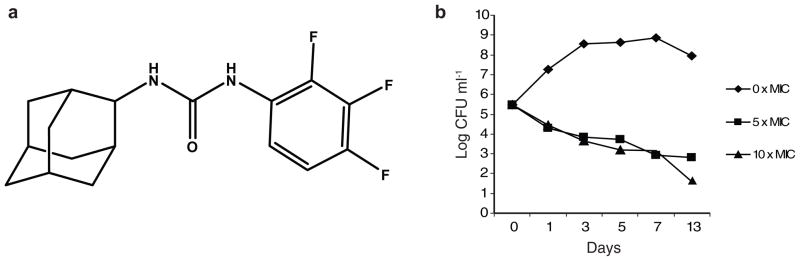
The screening of some 12,000 compounds from LeadScreen™ (preformatted screening set commercialized by Tripos; St Louis, MO) at a concentration of 10 μg ml−1 against 7H9-OADC-Tween 80 grown M. tb H37Rv led to the identification of a novel urea derivative [1-(2-adamantyl)-3-(2,3,4-trifluorophenyl)urea; compound # AU1235 (1)] (Fig. 1A) with a minimum inhibitory concentration (MIC) of 0.1 μg ml−1 (0.3 μM) (minimum bactericidal concentration of 0.1 μg ml−1)28 (see Supplementary Results, Supplementary Table 1). Importantly, AU1235 was similarly active against MDR clinical isolates of M. tb displaying resistance to isoniazid, rifampicin, and pyrazinamide in addition to streptomycin, fluoroquinolones and/or ethambutol (Table 1). This lack of cross-resistance with currently used anti-TB drugs suggested that AU1235 targets a novel biological function. AU1235 also inhibited Mycobacterium smegmatis and Mycobacterium fortuitum although the MICs (3.2 to 6.4 μg ml−1) were significantly higher than against M. tb and M. bovis BCG (Table 1). Exposure of M. tb H37Rv in log-phase growth (Abs600 nm = 0.2) to 0.5 μg ml−1 AU1235 (corresponding to 5 times MIC) resulted in a reduction in viable CFUs of about 2 log units after 5 days indicating that this compound is bactericidal in vitro (Fig. 1B). A higher concentration of AU1235 (10 times MIC) did not increase the killing effect within the first 7 days indicating that the killing was time-dependent rather than concentration-dependent (Fig. 1B). In an anaerobic model involving non-replicating M. tb H37Rv bacilli, AU1235 at 10 μg ml−1 showed no detectable activity suggesting that it acts on a biosynthetic pathway required for active bacterial multiplication.
Figure 1. Structure and bactericidal activity of AU1235.
(a) Structure of AU1235 (1) [1-(2-adamantyl)-3-(2,3,4-trifluorophenyl)urea] (b) Kill-kinetics showing decrease in viability of M. tb H37Rv in 7H9-OADC-Tween 80 medium containing AU1235 at 0.5 and 1 μg ml−1 (5 and 10 × MIC). The results for each drug concentration are representative of at least two independent experiments.
Table 1.
AU1235 susceptibilities for drug-susceptible and MDR isolates of M. tb and different other Mycobacterium spp.
| Mycobacterium species | MIC of AU1235 (μg ml−1) |
|---|---|
| M. tb H37Rv TMC102 | 0.1 |
| M. tb resistant to STR, INH, RIF, PZA | < 0.12 |
| M. tb resistant to STR, INH, RIF, PZA, FQ (isolate # 1) | < 0.12 |
| M. tb resistant to STR, INH, RIF, PZA, FQ (isolate # 2) | < 0.12 |
| M. tb resistant to EMB, INH, RIF, PZA, FQ | < 0.12 |
| M. tb H37Ra ATCC 25177 | 0.1 – 0.2 |
| M. bovis BCG Pasteur | 0.1 |
| M. avium 104 | > 12.8 |
| M. smegmatis mc2155 | 3.2 |
| M. fortuitum ATCC 6841 | 3.2 – 6.4 |
| M. chelonae ATCC 35752 | > 12.8 |
| M. abscessus ATCC 19977 | > 12.8 |
MIC of AU1235 that inhibited 100% of the growth of mycobacteria was determined in 7H9-S-OADC-Tween 80 broth at 37°C as described under the Methods section. DMSO was present in the medium at a final concentration of 2%. INH, isoniazid, STR, streptomycin; RIF, rifampicin; PZA, pyrazinamide; FQ, fluoroquinolones; EMB, ethambutol.
AU1235 was tested against a panel of Gram negative and Gram positive bacteria, including other Corynebacterineae such as Corynebacterium glutamicum, and was completely inactive (see Supplementary Table 2). Thus, this compound is specific for mycobacteria and shows best activity against M. tb, including MDR M. tb strains.
Macromolecular effects of AU1235 on M. tb and M. smegmatis
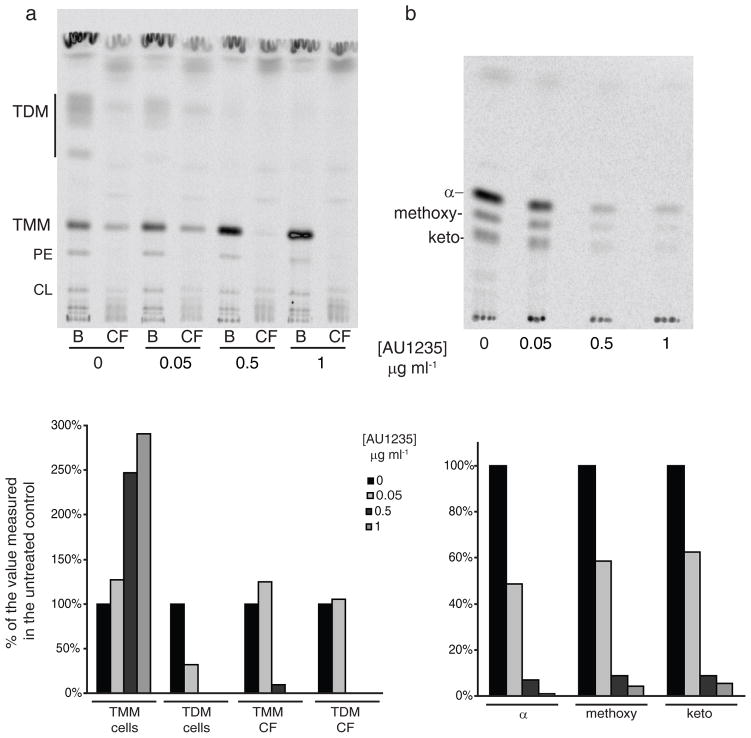
The mechanism of inhibition of M. tb by AU1235 was studied through metabolic labeling of untreated and AU1235-treated M. tb cells with various radiolabeled precursors including [5,6-3H]uracil, [8-3H]guanine, [35S]protein-labeling mix, [U-14C]glucose and [1,2-14C]acetic acid to monitor RNA, DNA, protein, polysaccharide and fatty acid/(glyco)lipid synthesis, respectively. Whereas no obvious inhibitory effect of the compound on RNA, DNA, protein and polysaccharide synthesis was detected (see Supplementary Fig. 1), a clear inhibitor concentration-dependent decrease in trehalose dimycolates (TDM) and all three forms of cell wall-bound mycolic acids (α-, methoxy- and keto-) was visible in the treated cells (Fig. 2A–2B). Interestingly and further supporting the idea that our inhibitor targeted a novel biological process in the biosynthesis of mycolic acids, TMM synthesis was apparently unaffected in the adamantyl urea-treated cells. AU1235, however, did substantially affect the sub-cellular distribution of TMM in that, as the concentration of AU1235 increased in the medium, less TMM was shed by the bacilli in the culture filtrate and more remained associated to the cells (Fig. 2A). In the presence of Tween 80 in the culture medium, M. tb indeed sheds some of its outermost components allowing an easier inventory of its outer membrane composition29. The extraction of surface-exposed lipids with water-saturated butanol further supported this conclusion in that the proportion of TMM found in the butanol fraction of untreated M. tb bacilli was 3 times greater than that found in the butanol fraction of M. tb cells treated with AU1235 at 5xMIC for 5 hr (see Supplementary Fig. 2a). Further sub-fractionation of untreated and AU1235-treated M. tb H37Ra followed by analysis of the sub-cellular fractions by TLC and autoradiography, although imperfect due to the notorious difficulty of separating outer from inner membranes in mycobacteria, revealed some accumulation of TMM in the inner membrane of the treated cells (see Supplementary Fig. 2b–2c).
Figure 2. Effect of AU1235 on mycolic acid biosynthesis and transfer in M. tb.
M. tb H37Ra cultured in 7H9-OADC-Tween 80 broth was treated for 5 hr at 37°C with either no inhibitor or with AU1235 at a concentration of 0.05 μg ml−1, 0.5 μg ml−1, or 1 μg ml−1 (0.5x to 10x MIC). [14C]-acetate was added to the cultures at the same time as the inhibitor. (a) Bacterial cells (B) and culture filtrates (CF) were collected and the lipids contained in each of these fractions extracted as described under the Methods section. The same volume of samples was loaded per lane. The TLC was developed in the solvent system [chloroform:methanol:water] (20:4:0.5, by vol.) and revealed by autoradiography. PE, phosphatidylethanolamine; CL, cardiolipin. (b) Mycolic acid methyl esters were prepared from delipidated cells as described48. α–, methoxy- and keto- denote the three forms of mycolic acids produced by M. tb. The same volume of samples was loaded per lane. The TLC was developed thrice in the solvent system [n-hexanes:ethyl acetate] (95:5, by vol.) and revealed by autoradiography. The amount of radioactivity incorporated in the products of interest was semi-quantified using a PhosphoImager and the results (expressed as a % of the value measured in the untreated control) are presented as histograms under their corresponding autoradiograms.
Altogether, our results thus indicate that AU1235 affects the translocation of TMM to the outer membrane of M. tb. Additional TLC analyses of radiolabeled lipid products and LC/MS analyses of the lipidomes of non-radiolabeled treated and untreated M. tb H37Ra otherwise failed to reveal any accumulating free mycolic acids or mycolate-containing glycolipids in the cells (data not shown).
Similar labeling experiments with [1,2-14C]acetic acid in M. smegmatis revealed identical effects of the inhibitor on the synthesis of TDM and cell wall-bound mycolates in this species (see Supplementary Fig. 3).
We conclude from these experiments that AU1235 does not inhibit the biosynthesis of mycolic acids per se but specifically their ability to be transferred onto their cell wall and outer membrane acceptors, possibly as a result of altered TMM translocation across the plasma membrane. In this regard, the mechanism of inhibition of AU1235 differs considerably from those of isoniazid, isoxyl, ethionamide and thiolactomycin that all act at earlier stages of the biosynthetic pathway and lead to a complete shutdown of mycolic acid biosynthesis4–5 (see Supplementary Fig. 4).
In search of the lethal target(s) of AU1235 in M. tb
Because the abolition of TDM formation and mycolic acid transfer onto AG in treated cells might also have resulted from a general inhibition of the antigen 85 complex, we first assessed whether AU1235 had any inhibitory effect on the purified FbpA, FbpB and FbpC enzymes in vitro. The assay used to monitor mycolyltransferase activity was similar to the one described in ref. 24 and thus used TMM purified from M. tb as the mycolic acid donor and [U-14C]trehalose as the acceptor substrate. No inhibitory effect of AU1235 on any of the activities of the three mycolyltransferases was detected in this assay (see Supplementary Fig. 5).
The observation that adamantyl ureas are potent inhibitors of soluble α/β-hydrolase fold epoxide hydrolases (EHs) (E.C.3.3.2.3) and the known involvement of EHs in the metabolism of various lipid substrates30, had originally led to our interest in these enzymes as the potential lethal targets of AU1235 in the mycolic acid pathway. The genome of M. tb is indeed peculiar in encoding an unusually large number of potential EHs with six genes of as yet unknown function related by sequence to α/β-hydrolase fold EHs (ephA, ephB, ephC, ephD, ephE, and ephF). While our early results indicated that AU1235 clearly inhibits the activity of some M. tb EH enzymes in vitro28, our preliminary genetic and biochemical analyses of M. tb’s six putative EH genes also revealed that the inhibition of this family of enzymes is unlikely to account for the effects of our adamantyl urea on the mycolic acid pathway given that (i) all six EH genes are dispensable for the growth of M. tb in vitro and (ii) that their individual inactivation has no detectable effect on TMM and TDM contents (see Supplementary Fig. 6).
We thus set out to search for the target of our inhibitor using a more comprehensive approach involving the selection of spontaneous AU1235-resistant mutants of M. tb followed by whole genome sequencing. Resistant mutants were selected by plating M. tb H37Rv and H37Ra cultures on 7H11-OADC agar plates containing 0.2 to 0.4 μg ml−1 AU1235 (2 to 4x MIC). Spontaneous mutants of both M. tb isolates were isolated at a frequency of 4 × 10−9 (4xMIC) to 1 × 10−7 (2xMIC). Five M. tb H37Rv mutants arising from the plating of a single culture were selected for further studies and checked for susceptibility to anti-TB agents. The MIC of isoniazid, ethambutol, rifampicin, ciprofloxacin, amikacin and streptomycin against M. tb H37Ra and M. tb H37Rv AU1235-resistant mutants were similar to those measured against the M. tb parent strains (see Supplementary Table 3). Whole genome sequencing followed by comparative analysis of the sequences of the five AU1235-resistant isolates and parent M. tb H37Rv strain identified one single gene commonly affected in all resistant isolates. This gene encodes the putative inner membrane transporter MmpL3. Targeted sequencing of the mmpL3 gene in 17 M. tb H37Ra spontaneous-resistant mutants revealed the presence of the same mutation in all of these isolates as well. In all cases, the exact same SNP (G758→A) was found, resulting in an amino acid change at position 253 of the protein (G253E). G253 maps at the end of one of the predicted membrane spanning domains of MmpL3 and is conserved in all known mycobacterial MmpL3 orthologs, with the only exception of M. abscessus which displays a serine residue at this position.
Compared to overexpressing the wild-type version of mmpL3tb, expression of the mutated version, mmpL3tb-G253E, in M. smegmatis mc2155 increased 2-fold the resistance of this strain to AU1235 (Table 2). This increase in resistance was significantly more marked in an M. smegmatis mc2155 recombinant strain in which the endogenous mmpL3 gene had been disrupted and replaced by mmpL3tb-G253E as its only copy of the gene (Table 2) (Fig. 3A). It was indeed to be anticipated when working with putative multimeric proteins such as MmpL3 that the co-expression of a wild-type and mutated versions of the encoding gene might lead to the formation of hybrid forms of the protein that retained their susceptibility to AU1235. The M. smegmatis mmpL3 knock-out mutant expressing mmpL3tb-G253E thus provided a much cleaner genetic background for this study. Compared to the same knock-out strain expressing a wild-type copy of the mmpL3tb gene (mc2155ΔmmpL3/pMVGH1-mmpL3tb), mc2155ΔmmpL3/pMVGH1-mmpL3tb-G253E displayed an 8- to 16-fold increased resistance to AU1235 (Table 2). This effect was specific to the adamantyl urea as the MICs of other drugs against the same two recombinant strains were identical (see Supplementary Table 4). That the expression of mmpL3tb-G253E protected M. smegmatis from the effect of AU1235 on mycolic acids was verified by comparing the effect of treating the two M. smegmatis recombinant strains with increasing concentrations of the inhibitor (see Supplementary Fig. 7). MIC values thus correlate with the level of inhibition of mycolic acid transfer onto their cell envelope acceptors and both phenotypes are related to the presence of the G→E mutation at position 253 of MmpL3tb.
Table 2.
Effect of expressing the point-mutated mmpL3tb-G253E gene on the susceptibility of M. smegmatis to AU1235.
| Mycobacterial strain | MIC AU1235 |
|---|---|
| M. smegmatis mc2155 | 3.2 |
| mc2155/pMVGH1 | 3.2 |
| mc2155/pMVGH1-mmpL3tb | 1.6 |
| mc2155/pMVGH1-mmpL3tb-G253E | 3.2 |
| mc2155ΔmmpL3/pMVGH1-mmpL3tb | 0.4 – 0.8 |
| mc2155ΔmmpL3/pMVGH1-mmpL3tb-G253E | 3.2 – 6.4 |
MIC of AU1235 that inhibited 100% of the growth of M. smegmatis was determined in 7H9-S-OADC-Tween 80 at 37°C as described under the Methods section. MIC values are given in μg ml−1.
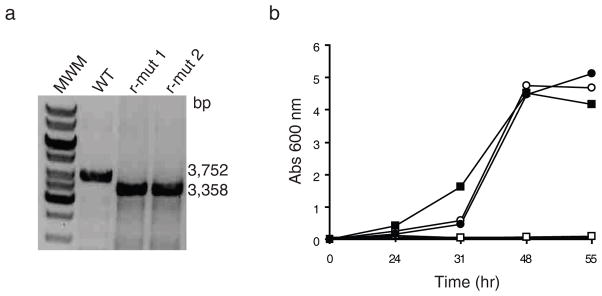
Figure 3. mmpL3 is an essential gene of M. smegmatis.
(a) Evidence for allelic replacement at the mmpL3 locus of M. smegmatis mc2155 in the presence of a rescue copy of mmpL3tb expressed from an episomal plasmid. Allelic exchange mutants were rescued with the mmpL3tb gene from M. tb expressed from either pMVGH1-mmpL3tb, pMVGH1-mmpL3tb-G253E or pSETetR-mmpL3tb. Allelic replacement was confirmed by PCR as described under the Methods section. The wild-type (WT) 3,752-bp amplification signal is replaced by a 3,358-bp fragment in the rescued mutants (r-mut) due to the 1,594-bp NotI deletion in the mmpL3 gene and insertion of a 1.2 kb-kanamycin resistance cassette. Only the profiles of two mc2155ΔmmpL3/pMVGH1-mmpL3tb strains are shown here as identical profiles were obtained will all rescued mutant forms. See Supplementary Figure 9 for the image of the full uncut gel.
(b) Growth characteristics of wild-type mc2155 (circles) and the conditional mutant, mc2155ΔmmpL3/pSETetR-mmpL3tb (squares), in 7H9-OADC-Tween 80 broth containing either no inducer (open symbols) or 50 ng ml−1 of anhydro-tetracycline (ATc) (full symbols) at 37°C. Reducing ATc concentration in the medium leads to the repression of mmpL3tb in the conditional mutant.
While these results strongly supported the assumption that MmpL3 served as the target of AU1235, the nature of this protein also made it possible that it acted as an efflux pump for this compound with the G→E mutation at position 253 increasing its effectiveness. To distinguish between these two hypotheses, further experiments were thus designed to analyze the effect of the point mutation on the accumulation of AU1235 in M. tb cells and functionally characterize this conserved mycobacterial transporter.
Accumulation of AU1235 in M. tb H37Ra cells
To determine whether the expression of mmpL3tbG253E led to the reduced accumulation of AU1235 in M. tb cells, M. tb H37Ra cultures either wild-type or expressing the G253E mutated version of MmpL3tb (spontaneous resistant isolates # 5 and 25) were treated for 4 hr at 37°C with 5 μM of the inhibitor. Bacterial pellets were then sub-fractionated into cell wall, membrane and cytosol, and the amount of AU1235 present in each of the compartments of the 3 strains quantified by LC/MS. No significant differences were noted between strains indicating that the increased resistance of the spontaneous isolates was unlikely to be the result of increased inhibitor efflux (see Supplementary Fig. 8).
Validation of the essentiality of MmpL3 in mycobacteria
Of the 13 to 14 MmpL-type proteins encoded by the genome of M. tb, mmpL3 is with mmpL11 the only mmpL gene conserved across the Mycobacterium genus, including M. leprae31. Further supporting the involvement of this gene in an important physiological process, mmpL3 was the only mmpL gene of M. tb H37Rv that couldn’t be knocked-out by allelic replacement in a systematic study aimed at studying the contribution of MmpL proteins to virulence and drug resistance32.
Similar to the situation in M. tb, we found that the disruption of mmpL3 by allelic replacement in M. smegmatis was not achievable unless a wild-type rescue copy of this gene was provided to the knock-out strain (Fig. 3A). Construction of conditional mutants of M. smegmatis in which the rescue copy of mmpL3tb was placed under control of a tetracycline-inducible promoter revealed that the merodiploid strains rapidly ceased growing when placed under non-permissive culture conditions where the expression of the rescue copy of mmpL3 was lost (Fig. 3B). Thus, expression of mmpL3 is essential for the growth of M. smegmatis. The fact that mmpL3tb could rescue the viability of the M. smegmatis mmpL3 knock-out mutant further indicates that the two mmpL3 orthologs have analogous functions.
Phenotypic effects of modulating mmpL3 expression
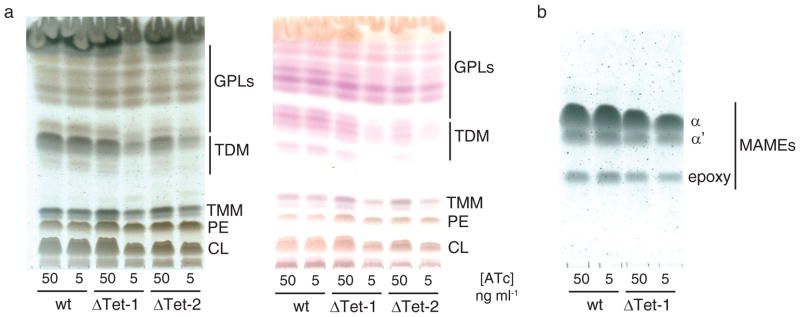
Loss of expression of mmpL3tb in the M. smegmatis conditional mutants grown under non-permissive conditions led to significant decreases in TDM formation (Fig. 4A) and mycolic transfer onto AG (Fig. 4B), thereby reproducing the effects of AU1235 on whole mycobacterial cells. LC/MS analyses of the lipidomes of the conditional mutants grown under non-permissive conditions otherwise failed to reveal any accumulating free mycolic acids, mycolyl-mannosylphosphopolyprenol22 or other mycolate-containing glycolipids in the cells (data not shown).
Figure 4. Effect of repressing mmpL3 expression on the mycolic acid content of M. smegmatis.
(a) Analysis of the extractable lipids from wild-type M. smegmatis mc2155 and two independent mc2155ΔmmpL3/pSETetR-mmpL3tb conditional mutants (ΔTet-1 and ΔTet-2) grown on 7H11-OADC agar in the presence of different concentrations of anhydro-tetracycline (ATc). Equal amounts of total cellular lipids from wild-type M. smegmatis mc2155 and the conditional mutants were run in the solvent system [chloroform:methanol:water] (20:4:0.5, by vol.). Left TLC plate: The plate was revealed with cupric sulfate to reveal all organic compounds. Right TLC plate: Samples were run similarly and the plate sprayed with α-naphthol to more specifically reveal glycolipids. Pink/purple-stained compounds correspond to glycolipids; yellow/brown-stained compounds migrating close to the origin are phospholipids.
The diffuse or doublet bands at the level of TDM and TMM correspond to various forms of these glycolipids esterified with different types of mycolic acid chains.
(b) Analysis of the cell wall-bound mycolates from wild-type M. smegmatis mc2155 and a mc2155ΔmmpL3/pSETetR-mmpL3tb conditional mutant (ΔTet-1). Cell wall-bound mycolic acid methyl esters (MAMEs) prepared from the same amount of wild-type and conditional mutant cells and loaded volume to volume are shown. A decrease in MAME content is seen in the conditional mutant grown at the lowest concentration of ATc. The TLC was developed thrice in the solvent system [n-hexanes:ethyl acetate] (95:5, by vol.) and the plates revealed with cupric sulfate. TMM, trehalose monomycolates; TDM, trehalose dimycolates; PE, phosphatidylethanolamine; CL, cardiolipin; GPLs, glycopeptidolipids.
Also worthy of note is the observation from the analysis of the cell wall-bound mycolates shown in Fig. 4B that the mmpL3 gene from M. tb was capable of complementing the transfer of all forms of mycolic acids produced by M. smegmatis, including the species-specific α′- and epoxy-forms not normally produced by the tubercle bacillus.
Altogether, the unchanged accumulation of AU1235 in the mmpL3-G253E-expressing M. tb cells and the fact that decreased mmpL3 expression mimicked the phenotypic effects of treating mycobacteria with AU1235 refute the hypothesis that MmpL3 serves as an efflux pump for the adamantyl urea and, instead, confirms this transporter as the direct target of our prototype inhibitor.
DISCUSSION
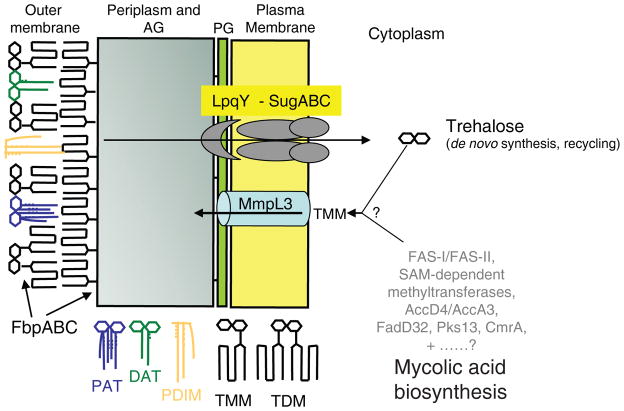
Whereas most steps of the mycolic acid biosynthetic pathway of M. tb have now been defined, the stage at which the transfer of mycolic acids to the periplasmic side of the plasma membrane occurs, the form under which they are translocated and the nature of the transporter involved have long remained enigmatic and the object of much speculation8,14,20,22. Yet, the fact that M. tb and M. smegmatis require trehalose for growth18–19, the recent identification of an ABC-transport system dedicated to the recycling and import of trehalose inside M. tb cells21 and the characterization of TMM as the direct substrate of the FbpABC mycolyltransferases on the periplasmic side of the plasma membrane23–24,27 supported the concept that TMM synthesis occurs on the cytoplasmic face of the plasma membrane and that this glycolipid represents the indispensable esterified form under which mycolates are then translocated across the inner membrane of mycobacteria. Consistent with this hypothesis, our results indicated a clear effect of chemically inhibiting the inner membrane transporter MmpL3 on the translocation of TMM in M. tb. This effect resulted in turn in a complete abolition of TDM formation and mycolic acid transfer onto AG, most likely reflecting the inaccessibility of the TMM generated intracellularly to the mycolyltransferases FbpABC located on the periplasmic side of the plasma membrane (Fig. 5). That TMM represents the direct substrate of the MmpL3 transporter is supported by the fact that this glycolipid was the only mycolic acid-containing compound accumulating in AU1235-treated M. tb cells. Given that TMM is the mycolic acid donor used in the mycolyltransferase reactions catalyzed by FbpABC, it is unlikely that a further downstream product of TMM serves as the substrate for the MmpL3 transporter. Interestingly and indicative of the relatively broad substrate specificity of MmpL3 in terms of mycolic acid forms, the expression of the M. tb ortholog of mmpL3 in an M. smegmatis mmpL3 knock-out mutant not only rescued the viability of this strain but also its ability to transfer all forms of mycolic acids onto AG, including M. smegmatis-specific forms not naturally produced by M. tb.
Figure 5. A model for mycolic acid biosynthesis and transport across the inner membrane.
Mycolic acid chains are shown in red. DAT, diacyltrehaloses; PAT, polyacyltrehaloses; PDIM, phthiocerol dimycocerosates. The trehalose released upon transfer of mycolic acids from TMM either onto AG or another molecule of TMM to form TDM is recycled and re-imported inside the cells by the ABC-transporter SugABC and the lipoprotein LpqY21; FbpABC are the mycolyltransferases responsible for the transfer of mycolic acids onto their cell envelope acceptors.
The apparent confinement of the MmpL family of proteins to mycobacteria31,33–34 is originally what has led to their ‘MmpL’ designation for ‘mycobacterial membrane proteins, large’. Although mycobacterial MmpL family proteins share low overall sequence similarity with other transporters, they are in fact a subset of the larger Resistance, Nodulation, and Division (RND) superfamily of inner membrane transporters33,35, the most well studied prokaryotic members of which are the major multidrug efflux pumps AcrB of E. coli and MexB of Pseudomonas aeruginosa36–37. Members of the RND family typically share topological features consisting of twelve membrane-spanning α-helices with two predicted soluble periplasmic loops between transmembrane domains 1 and 2, and between domains 7 and 8 that dictate the substrate specificity of the protein. These features are conserved in MmpL3 although the topology prediction and number of transmembrane domains (11 to 12) may vary with the secondary structure prediction algorithm. How AU1235 inhibits at the molecular level the transport activity of MmpL3tb is at present not known, but the localization of the resistance-conferring G253E mutation within a conserved charged region of RND transporters (D251, R259) suggests that the inhibitor may interfere with the transmembrane electrochemical proton gradient that provides energy for substrate translocation32, 35–37. The genome of M. tb encodes up to 14 MmpL proteins33, the function of only 4 of them (including MmpL3) is presently known. MmpL7 was shown to translocate major lipid constituents of the outer membrane known as the PDIM across the plasma membrane. mmpL7 mutants of M. tb are highly attenuated for virulence and accumulate PDIM inside the cells29,38. mmpL8 mutants of M. tb, which are also attenuated for virulence, fail to synthesize mature sulfolipid-I (SL-I) and, instead, accumulate inside the cytoplasm diacylated sulfolipid precursors which have the ability to stimulate CD1-restricted T cells39–41. MmpL11 was recently involved in heme iron acquisition in M. tb and, on the basis of its sequence homology to MmpL11 (25% identity) and ability of its predicted periplasmic domains to bind heme in vitro, MmpL3 was hypothesized to display a similar function42. Whether MmpL3 actually has this function in whole M. tb cells is at present not known but this finding raises the intriguing possibility that MmpL transporters may simultaneously act as exporters and importers and be involved in more than one physiological process in mycobacteria. In M. smegmatis, two MmpL transporters lacking clear orthologs in M. tb, TmtpB and TmtpC, participate in the synthesis of GPLs, key surface virulence factors produced by non-tuberculous mycobacteria31. Overall, MmpL transporters are thus emerging as key players in the building of the cell envelope and subsequent interactions of M. tb with the host. Sulfolipids, like TMM, are acyltrehaloses. So are diacyltrehaloses (DATs) and polyacyltrehaloses (PATs), other outer membrane glycolipids of M. tb involved in host-pathogen interactions whose biosynthetic polyketide synthase gene pks3/4 maps in close vicinity to an as yet uncharacterized mmpL gene, mmpL10 (for a review31,33,43). Importantly, PDIM, SL-I, DAT and PAT are restricted to pathogenic slow-growing Mycobacterium species43. Altogether, these findings thus indicate that several MmpL proteins of M. tb have specialized in the translocation of physiologically essential and/or biologically active (glyco)lipids of which many are acyltrehaloses. To this date, however, it is unclear how MmpLs function to translocate relatively large lipophilic molecules to the periplasm and how this process is driven energetically. The two large soluble periplasmic or cytosolic domains of MmpLs may carry the substrate specificity of these proteins. Alternatively, since MmpL proteins are likely to function as part of coupled biosynthesis/export machineries31,39–40,44, it was proposed that their ability to interact with some particular cytosolic biosynthetic enzymes dictated, at least in part, their substrate specificity31. The availability of a potent adamantyl urea inhibitor of MmpL3 provides unprecedented opportunities to investigate the molecular mechanisms of this unique family of transporters.
That adamantyl ureas may have more than one target in M. tb is supported by our earlier work showing a clear inhibition of some of M. tb’s putative EH enzymes in vitro28. The dispensable character of all six EH-like proteins of M. tb for growth and the phenotypic analysis of EH knock-out mutants showing an apparent lack of participation of these enzymes in TDM formation strongly argue, however, against these proteins being the bactericidal targets of AU1235. Likewise, in the event AU1235 inhibited other MmpL proteins than MmpL3 (an hypothesis that our preliminary analyses of the subcellular distribution of PDIM in AU1235-treated M. tb H37Rv cells does not currently support), such inhibition would be unlikely to lead to cell death given the non-essential character of these transporters32 and lack of demonstrated involvement in the mycolic acid pathway. Clearly, that MmpL3 is the bona fide primary target of our inhibitor is validated by the facts that mmpL3 was the only common mutated gene found in all spontaneous-resistant mutants of M. tb H37Rv and H37Ra (no mutations mapped to any of the EH genes or any other mmpL gene) and that the MIC of AU1235 correlates with the level of inhibition of TDM synthesis and mycolic acid transfer onto AG, two phenotypes that our analyses of AU1235-treated and recombinant M. smegmatis strains have related to the essential transporter, MmpL3.
The identification of a novel adamantyl urea-based compound showing high potency against M. tb in culture (including MDR strains) and with a novel mechanism of inhibition of the mycolic acid pathway opens new avenues for anti-TB drug development. Consistent with its mode of action, AU1235 is highly specific for mycobacteria. Important efforts to synthesize key structural analogs of AU1235 were undertaken and their testing against M. tb has revealed a clear structure activity relationship for this series28. Efforts are now underway to develop derivatives of these compounds efficacious in vivo.
METHODS
Strains and growth conditions
M. tb H37Rv TMC102 and ATCC 25618 and M. tb H37Ra ATCC 25177 were grown in Middlebrook 7H9-OADC broth (Difco) supplemented with 0.05% Tween 80 and on 7H11-OADC agar (Difco) at 37°C. M. smegmatis mc2155 was grown on LB agar, in 7H9-OADC-Tween 80 or in LB-Tween 80 broth at 37°C. Where required, kanamycin (25 μg ml−1), hygromycin (50 μg ml−1), sucrose (10%) and anhydro-tetracycline (0.1 to 50 ng ml−1) were added to the culture medium.
Generation of conditional mutants and complemented strains
A two-step procedure employing the counterselectable marker sacB45 was used to achieve allelic replacement at the mmpL3 (MSMEG_0250) locus of M. smegmatis mc2155. Briefly, the M. smegmatis mmpL3 gene and flanking regions was PCR-amplified from M. smegmatis mc2155 genomic DNA using the primers mmpL3smg.1 (5′-GGTCTAGACGCACTGCGCAGACGTGAGGG-3′) and mmpL3smg.2 (5′-GGTCTAGACAGGCGGCGGTGTGCTCGCG-3′) and a disrupted allele, mmpL3::kan, was obtained by replacing 1,594 bp of the coding sequence of this gene flanked by two NotI restriction sites by the kanamycin resistance cassette from pUC4K (GE Healthcare). mmpL3::kan was then cloned into the XbaI-cut pPR27-xylE to obtain pPR27mmpL3KX, the construct used for allelic replacement. pSETetR-mmpL3tb, an episomal rescue plasmid in which mmpL3tb was placed under control of a tetracycline-inducible promoter, was constructed by inserting successively into pSE10046, the tet repressor (tetR) from pMC2m46 and the entire coding sequence of mmpL3tb. Primer sequences for the conditional rescue construct are available upon request.
pMVGH1-mmpL3tb was obtained by cloning the entire coding sequence of mmpL3 from M. tb H37Rv, PCR-amplified using primers mmpL3tb.1 (5′-GGGCGCGCATATGTTCGCCTGGTGGGGTCGAAC-3′) and mmpL3tb.2 (5′-GGGAAGCTTAAGGCGTCCTTCGCGGCGAAG-3′), into the NdeI and HindIII restriction sites of pMVGH1, a derivative of the expression plasmid pVV1647 devoid of kanamycin resistance gene. pMVGH1-mmpL3tb-G253E was constructed similarly using genomic DNA from one of the AU1235 spontaneous resistant mutants of M. tb H37Rv as the template for PCR amplification. Allelic replacement at the mmpL3 locus of M. smegmatis was confirmed by PCR using primers DC1 (5′-GAGGCGACGGTCTTCACCAACAT-3′) and DC2 (5′-GCCTGGTCTACGCGCTGATCAC-3′) both located outside the mmpL3 region cloned into pPR27-xylE.
Selection of spontaneous AU1235-resistant mutants of M. tb
AU1235-resistant mutants were selected at 37°C (M. tb H37Rv) or 30°C (M. tb H37Ra) on 7H11 plates supplemented with OADC and 0.2 to 0.4 μg ml−1 of the inhibitor (2 to 4 × MIC) and scored for resistance 3 to 7 weeks post-inoculation.
Whole genome sequencing
Genomic DNA was isolated from freshly cultured M. tb H37Rv resistant isolates. Barcoded fragment libraries were constructed for each of the strains according to the SOLiD fragment library protocol (i.e. 50 bp fragment library) (Applied Biosystems). Libraries were pooled in equimolar ratios and sequencing was carried out using SOLiD V3 chemistry (Applied Biosystems). The data was reduced to sequence and reports about SNPs and in-dels are generated using the NextGen Software.
Whole cell radiolabeling experiments
Radiolabeling of whole mycobacterial cells with [1,2-14C]acetic acid (0.5 μCi ml−1; specific activity, 57 Ci mol−1, NEN Radiochemicals) was performed in 7H9-OADC-Tween 80 broth for 16 hr with shaking. The radiochemical was added to the cultures at the same time as the inhibitor.
Analytical procedures
Total lipids extraction from bacterial cells and culture filtrates, and preparation of fatty acid and mycolic acid methyl esters from extractable lipids and delipidated cells followed earlier procedures48. Cold and radiolabeled lipids and fatty acid/mycolic acid methyl esters were analyzed by TLC on aluminum-backed silica gel 60-precoated plates F254 (E. Merck). TLC plates were revealed by spraying with cupric sulfate (10% in a 8% phosphoric acid solution) or α-naphthol (1% in ethanol) and heating. Radiolabeled products were visualized by exposure of the TLC plates to Kodak X-Omat AR films at –80°C and quantified using a PhosphorImager (Typhoon, GE Healthcare). Alternatively, total lipids were run in both positive and negative mode and the released fatty acids/mycolic acids in negative mode only, on a high resolution Agilent 6220 TOF mass spectrometer interfaced to a LC. See Supplementary Methods for details.
Mycolyltransferase assays
The TMM transesterification assay described in ref. 24 was used to measure the mycolyltransferase activity of the purified FbpA FbpB and FbpC proteins in the presence of cold TMM purified from M. tb, [U-14C]trehalose and different concentrations of AU1235 (see Supplementary Fig. 5). Purified FbpA and FbpB proteins were obtained through the NIH - TB Vaccine Testing and Research Materials Contract and NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Ag85A, purified native protein from M. tb H37Rv (NR-14856); Ag85B, purified native protein from M. tb H37Rv (NR-14857). Purified recombinant FbpC (Ag85C) was kindly provided by Dr. K. Dobos (Colorado State University, Fort Collins, CO).
Drug susceptibility testing
MIC values of various antibiotics against Mycobacterium clinical isolates and recombinant strains were determined in 7H9-OADC-Tween 80 or 7H9-S-OADC-Tween 80 broth at 37°C in 96-well microtiter plates using the colorimetric resazurin microtiter assay and by visual readout for growth. Low-oxygen tension experiments were performed using the Rapid Anaerobic Dormancy (RAD) model as described in ref. 49. Control tubes contained methylene blue dye (1.5 μg ml−1) as an indicator of oxygen depletion. Different concentrations of AU1235 and control drugs (isoniazid, rifampicin, ethambutol and metronidazole) were injected through the septa of oxygen-depleted cultures and the cultures allowed to grow at 37o C with stirring for another 4 days, at which point the septa were removed and cultures serially diluted in saline and plated onto 7H11-OADC agar plates for enumeration of CFU.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID)/National Institutes of Health (NIH) grants AI085992, AI063054 and AI057836, the American Lebanese Syrian Associated Charities (ALSAC) and the Slovak Research and Development Agency under contract No. APVV-0441-10. We thank Dr. R. Dhiman for helpful scientific discussions and D. Dick for LC/MS analyses.
Footnotes
Author contributions
AEG, AJL, JK, REL, MM and MJ designed experiments. EJN and CM synthesized compounds. AEG, HP, VG, MSS, VJ, TH, SSC, JK, SEB and CM selected resistant mutants, constructed recombinant mycobacterial strains and performed MIC testing, enzymatic assays, metabolic labeling, lipid/mycolic acid analyses and genomic analyses. TH performed compound accumulation studies in M. tb. MM, REL and MJ wrote the paper.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.WHO. WHO Report 2009 - Global Tuberculosis Control. Epidemiology, Strategy, Financing. 2009. [Google Scholar]
- 2.Global Alliance for TB Drug Development. Scientific Blueprint for TB Drug Development. Tuberculosis. 2001;81:1–52. doi: 10.1054/tube.2001.0288. [DOI] [PubMed] [Google Scholar]
- 3.Center for Disease Control and Prevention (CDC) Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 4.Schroeder EK, de Souza ON, Santos DS, Blanchard JS, Basso LA. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis. Curr Pharm Biotechnol. 2002;3:197–225. doi: 10.2174/1389201023378328. [DOI] [PubMed] [Google Scholar]
- 5.Barry CE, Crick DC, McNeil MR. Targeting the formation of the cell wall core of Mycobacterium tuberculosis. Infectious Disorders Drug Targets. 2007;7:182–202. doi: 10.2174/187152607781001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alahari A, et al. Thiacetazone, an antitubercular drug that inhibits cyclopropanation of cell wall mycolic acids in mycobacteria. PLoS ONE. 2007;2:e1343. doi: 10.1371/journal.pone.0001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daffé M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Advances in microbial physiology. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 8.Barry CE, III, et al. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau E, et al. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol. 2000;36:630–637. doi: 10.1046/j.1365-2958.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 10.Glickman MS. Cording, cord factors and trehalose dimycolate. in. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. ASM Press; Washington DC: 2008. pp. 63–73. [Google Scholar]
- 11.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber B, et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bact. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Beken S, et al. Molecular structure of the Mycobacterium tuberculosis virulence factor, mycolic acid, determines the elicited inflammatory pattern. Eur J Immunol. 2011;41:450–60. doi: 10.1002/eji.201040719. [DOI] [PubMed] [Google Scholar]
- 14.Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavalda S, et al. The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J Biol Chem. 2009;284:19255–64. doi: 10.1074/jbc.M109.006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lea-Smith DJ, et al. The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J Biol Chem. 2007;282:11000–8. doi: 10.1074/jbc.M608686200. [DOI] [PubMed] [Google Scholar]
- 17.Bhatt A, Brown AK, Singh A, Minnikin DE, Besra GS. Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta-oxo-mycolic acid analogs and accumulation of ketones. Chem Biol. 2008;15:930–9. doi: 10.1016/j.chembiol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodruff PJ, et al. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem. 2004;279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 19.Murphy HN, et al. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2005;280:14524–14529. doi: 10.1074/jbc.M414232200. [DOI] [PubMed] [Google Scholar]
- 20.Tropis M, et al. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J Biol Chem. 2005;280:26573–85. doi: 10.1074/jbc.M502104200. [DOI] [PubMed] [Google Scholar]
- 21.Kalscheuer R, Weinrick B, Veeraraghavan U, Besra GS, Jacobs WR., Jr Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;107:21761–6. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besra GS, et al. Identification of the apparent carrier in mycolic acid synthesis. Proc Natl Acad Sci USA. 1994;91:12735–12739. doi: 10.1073/pnas.91.26.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sathyamoorthy N, Takayama K. Purification and characterization of a novel mycolic acid exchange enzyme from Mycobacterium tuberculosis. J Biol Chem. 1987;262:13417–13423. [PubMed] [Google Scholar]
- 24.Belisle JT, et al. Role of the major antigen of Mycobacterium tuberculosis in the cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 25.Jackson M, et al. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999;31:1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 26.Puech V, et al. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Mol Microbiol. 2002;44:1109–22. doi: 10.1046/j.1365-2958.2002.02953.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanki AK, Boucau J, Ronning DR, Sucheck SJ. Antigen 85C-mediated acyl-transfer between synthetic acyl donors and fragments of the arabinan. Glycoconj J. 2009;26:589–96. doi: 10.1007/s10719-008-9211-z. [DOI] [PubMed] [Google Scholar]
- 28.Brown JR, et al. The structure activity relationship of urea derivatives as anti-tuberculosis agents. Bioorganic and Medicinal Chemistry. 2011 doi: 10.1016/j.bmc.2011.07.034. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho LR, et al. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 30.Morisseau C, Hammock BD. Epoxide hydrolases: Mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 31.Jain M, Chow ED, Cox JS. The MmpL protein family. In: Daffé M, Reyrat J-M, editors. The mycobacterial cell envelope. ASM Press; Washington, DC: 2008. pp. 201–210. [Google Scholar]
- 32.Domenech P, Reed MB, Barry CE., III Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tekaia F, et al. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tubercle and Lung Disease. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 34.De Rossi E, Ainsa JA, Riccardi G. Role of mycobacterial efflux transporters in drug resistance: an unsolved question. FEMS Microbiol Rev. 2006;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- 35.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–13. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 36.Murakami S. Multidrug efflux transporter, AcrB-the pumping mechanism. Curr Opin Struct Biol. 2008;18:459–65. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Sennhauser G, Bukowska MA, Briand C, Grutter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol. 2009;389:134–45. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 39.Converse SE, et al. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc Natl Acad Sci USA. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domenech P, et al. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J Biol Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 41.Gilleron M, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tullius MV, et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci USA. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson M, Stadthagen G, Gicquel B. Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis: biosynthesis, transport, regulation and biological activities. Tuberculosis. 2007;87:78–86. doi: 10.1016/j.tube.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathogens. 2005;1:12–19. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson M, Camacho LR, Gicquel B, Guilhot C. Gene replacement and transposon delivery using the negative selection marker sacB. In: Parish T, Stocker NG, editors. Mycobacterium tuberculosis protocols. Vol. 54. Humana Press; Totowa N J: 2001. pp. 59–75. [DOI] [PubMed] [Google Scholar]
- 46.Guo XV, et al. Silencing essential protein secretion in Mycobacterium smegmatis by using tetracyclin repressors. J Bacteriol. 2007;189:4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korduláková J, et al. Definition of the first mannosylation step in phosphatidylinositol synthesis: PimA is essential for growth of mycobacteria. J Biol Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- 48.Stadthagen G, et al. p-hydroxybenzoic acid synthesis in Mycobacterium tuberculosis. J Biol Chem. 2005;280:40699–40706. doi: 10.1074/jbc.M508332200. [DOI] [PubMed] [Google Scholar]
- 49.Leistikow RL, et al. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 2010;192:1662–70. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.