Abstract
Glioblastoma Multiforme (GBM) is the most malignant and lethal form of astrocytoma. The GBM patient survival time of approximately one year necessitates the identification of novel molecular targets and more effective therapeutics. Cadherin-11, a calcium-dependent cell-cell adhesion molecule and mesenchymal marker, plays a role in both normal tissue development and in cancer cell migration. The functional significance of cadherin-11 in GBM has not been investigated. Here, we show that cadherin-11 is expressed in human GBM tumors and human glioma stem-like cells by immunohistochemical labeling. Additionally, we show that cadherin-11 is expressed in human glioma cell lines by immunoblotting. shRNA mediated knockdown of cadherin-11 expression in human glioma cell lines results in decreased migration and growth factor-independent cell survival in vitro. More importantly, knockdown of cadherin-11 inhibits glioma cell survival in heterotopic and orthotopic mouse xenograft models. Together, our results demonstrate the functional significance of cadherin-11 expression in GBM and provide evidence for a novel role of cadherin-11 in promoting glioma cell survival in an in vivo environment. Thus, our studies suggest cadherin-11 is a viable molecular target for therapeutic intervention in GBM.
Keywords: Cadherin-11, glioma, cell migration, cell survival, mesenchymal lineage
Introduction
Glioblastoma Multiforme (GBM), classified as Grade IV astrocytoma by the World Health Organization, constitutes the most common and highly malignant form of intracranial glioma tumor (1, 2). The primary treatment option for GBM is surgery followed by chemotherapy and/or radiation (1). In spite of advances in therapeutic and diagnostic research, GBM patients have a survival time of approximately one year post diagnosis (1). GBM cells are highly dispersive, can proliferate extensively and migrate throughout the brain parenchyma contributing to the lethality of this disease (1, 2). Therefore, identification of novel molecular targets regulating migration and survival pathways in GBM is essential for development of targeted therapeutics.
The heterogeneity of GBM at the cellular level hinders our understanding of the disease rendering the design and development of effective therapeutic strategies difficult (1, 3, 4). To understand the molecular changes in GBM, several groups have studied the global gene expression profiles and genetic alterations that drive tumor aggressiveness in GBM (4–9). Recently, GBM has been classified into 4 subtypes: Proneural, Neural, Classical and Mesenchymal based on a comprehensive analysis of The Cancer Genome Atlas (TCGA) dataset (10). The mesenchymal phenotype is associated with the increased migration and invasion of GBM cells (11). Conversion of neural cells into cell types showing a mesenchymal phenotype has been demonstrated in brain neoplasms (4, 11). In addition, gene expression profiles from GBM patients with a poor prognosis showed overexpression of genes associated with the mesenchymal lineage (4). Multiple glioma cell lines derived from human GBM, including U-87 MG, express genes associated with a mesenchymal phenotype (11).
During tumor progression, cadherin switching occurs: a process wherein expression of epithelial E-cadherin is switched to mesenchymal-associated cadherins like N-cadherin and cadherin-11 (12, 13). Cadherins are a major class of cell-cell adhesion molecules that interact with catenins to regulate adhesion and migration (14–16). Thus, the cadherin-catenin complex plays important roles in regulating biological processes like the rearrangement of actin cytoskeleton and gene transcription to control tissue morphogenesis (14–18). Cadherin-11 (also called OB-cadherin or CDH11) was initially identified in mouse osteoblasts (19) and is expressed in normal mesoderm-derived tissues during development, including mesenchymal cells in the kidney and brain (20). Additionally, cadherin-11 is expressed in multiple cancers including breast, prostate, colon, gastric, renal cell and osteosarcoma (21–28). Cadherin-11 plays a role in tumor invasion and progression (17, 29, 30). Cadherin-11 expression in malignant breast and prostate cancer is associated with increased invasiveness and poor prognosis (24, 27, 28). In addition, cadherin-11 expression is suggested to promote homing and metastasis of breast and prostate cancer cells to bone in experimental models (31, 32). Recently, cadherin-11 has been implicated in regulating migration in glioma cells in vitro (33). The functional importance of cadherin-11 in regulating glioma migration and cell survival in vivo has not yet been investigated.
In this study, we show that cadherin-11 is expressed in the center and edge of human GBM tumors, human glioma progenitor-like cells and in two commonly used human glioma cell lines, U-87 MG and LN-229. shRNA-mediated knockdown of cadherin-11 in U-87 MG and LN-229 glioma cells decreased cell migration in vitro. Additionally, shRNA mediated reduction of cadherin-11 expression in U-87 MG and LN-229 glioma cells significantly decreased survival in vitro in growth factor-independent colony formation assays. Remarkably, shRNA-mediated knockdown of cadherin-11 in U-87 MG cells significantly reduced tumor size while it completely inhibited survival of LN-229 glioma cells in tumor xenografts in vivo. Our data demonstrate a novel function for cadherin-11 in regulating glioma cell survival in vitro and in vivo, implicating cadherin-11 as a potential therapeutic target in GBM.
Materials and Methods
Cell culture
The human glioma cell lines, U-87 MG and LN-229 were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in a humidified atmosphere of 5% CO2 at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with fetal bovine serum (10% for U-87 MG and 5% for LN-229) (HyClone, Logan, UT) and antibiotic-antimycotic (Invitrogen, Carlsbad, CA). The cell lines are authenticated by ATCC using tests like short tandem repeat (STR) profiling.
Human brain tissue extraction
Cortical surface and tumor tissue from human patients was resected as described previously (34) using approved protocols from the University Hospitals Case Medical Center and Case Comprehensive Cancer Center’s Institutional Review Board.
Lentiviral infection
Lentiviral shRNA construct TRCN0000054336 (Sigma-Aldrich, St. Louis, MO) that targets a region in the extracellular domain of human cadherin-11 mRNA was used to suppress cadherin-11 expression. pLKO.1 (Sigma-Aldrich, St. Louis, MO), a lentiviral control shRNA construct was used as a control for TRCN0000054336. A lentiviral construct encoding green fluorescent protein (GFP) was synthesized by inserting the GFP coding sequence (35) into the pCDH1-MCS2 Cloning and Expression Vector (System Biosciences, Mountain View, CA). Production of lentivirus and infection of U-87 MG and LN-229 glioma cells was performed as previously described (36). RETRO-tek HIV-1 p24 Antigen ELISA (ZeptoMetrix Corporation, Buffalo, NY) was used to titer lentiviruses. A protocol from Sigma-Aldrich (St. Louis, MO) was used to compute the concentration of HIV-1 p24 antigen and transducing units per milliliter (TU/ml) for every picogram (pg) of p24 antigen. U-87 MG and LN-229 cells plated at a density of 2 × 105 were infected with 2.07 × 106 TU of lentivirus encoding control or cadherin-11 shRNA. Reduction in cadherin-11 protein levels following infection with lentivirus encoding cadherin-11 shRNA was examined by immunoblotting.
Immunoblotting
Cell lysates were prepared by incubating the cells in a buffer containing 100 mM Tris pH 7.5, 1% Triton-X-100, 0.1% sodium dodecyl sulfate (SDS), 140 mM NaCl, 0.5% sodium deoxycholate, 5 mM EDTA, protease inhibitors (1 mM benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 μg/ml pepstatin A) and phosphatase inhibitors [(1 mM sodium fluoride (NaF), 1 mM sodium orthovanadate (NaVO3)] on ice for 30 min. After centrifugation at 10,000 rpm for 3 min, the supernatant was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membrane. Cadherin-11 expression was analyzed by immunoblotting using monoclonal antibodies to cadherin-11 (Invitrogen, Carlsbad, CA), which recognize the intracellular portion of human cadherin-11 protein. Anti-vinculin antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Primary antibodies were detected using goat anti-mouse secondary antibodies conjugated to HRP (Jackson Immuno Research Laboratories, Inc., West Grove, PA). Secondary antibodies were detected by chemiluminescence (GE Healthcare, Piscataway, NJ) using an Image Quant™ LAS 4000 digital imaging system (GE Healthcare, Piscataway, NJ). Densitometry analysis was performed using Image J software (37).
Scratch wound assay
U-87 MG and LN-229 glioma cells were infected with lentivirus encoding control or cadherin-11 shRNA. 3 days post infection, the monolayer of cells was scratched with a pipette tip to form a wound. Cells were imaged at time 0 and 24 hr post scratching. The data was analyzed to quantify cell migration as previously described (36).
Spot assay
We developed a new quantitative method for analyzing cell migration in vitro. U-87 MG and LN-229 glioma cells infected with lentivirus encoding control or cadherin-11 shRNA were trypsinized and resuspended at a concentration of 2.5 × 104 cells per microliter (μl) in DMEM (Invitrogen, Carlsbad, CA). 2 μl of cells resuspended in DMEM and diluted 1:1 in 4 mg/ml Growth Factor Reduced (GFR) BD Matrigel™ Matrix (BD Biosciences, Franklin Lakes, NJ) were pipetted as a spot in a tissue-culture dish and incubated at 37°C for 5 min to gel. Media was added and the spot was imaged as day 0 image using a 20x objective on a Leica DMI 6000 B automated inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) using a Retiga EXi camera (QImaging, Surrey, BC, Canada). Media was changed each day to ensure a constant supply of nutrients and to prevent growth-arrest of cells. The spot was re-imaged after 3 days (for U-87 MG) and 4 days (for LN-229) to assess the migration of cells. Each spot was imaged by taking multiple, automated, sequential images using Scan Slide feature in MetaMorph software (Molecular Devices, Downington, PA) to image the entire area covered by the spot at day 0 and also the area covered by the migrating cells away from the spot in 3 and 4 days. These multiple images were stitched together using MetaMorph software to give one complete image of the spot.
To quantitate cell migration away from the spot, MetaMorph software was used. Briefly, images taken at the 0 and 3 or 4 day time points were pseudo-colored in 2 different colors and overlayed to outline the boundary of the day 0 spot. The extent of cell migration was analyzed by measuring the distance migrated by the cells away from the initial spot in micrometers. 10 measurements per spot and a minimum of 3 spots per condition were averaged. The data was plotted using Microsoft Excel to determine the distance migrated by the leading edge of the cells at the 3 or 4 day time point.
Colony formation assay
U-87 MG and LN-229 glioma cells were infected with lentivirus encoding control or cadherin-11 shRNA. 3 days post infection, cells were trypsinized and plated at low density. Cells were allowed to form colonies for 14 days under growth factor-deprived conditions after which plates were stained with crystal violet to visualize colonies as previously described (38).
Heterotopic xenograft tumors implanted in mouse flank
NIH athymic nude female mice (NCr-nu/+, NCr-nu/nu) were maintained according to institutional policies in the Animal Core Facility at Case Western Reserve University. Approved protocols from the Institutional Animal Care and Use Committee were followed for all animal procedures. U-87 MG and LN-229 cells co-infected with lentivirus encoding either control or cadherin-11 shRNA and GFP for 3 days were trypsinized and resuspended in phosphate buffered saline (PBS) at a concentration of 2 × 104 cells per μl. Cells resuspended in PBS were diluted in BD Matrigel™ Matrix (BD Biosciences, Franklin Lakes, NJ). 250–280 μl of the cell and Matrigel mixture (0.8 × 106 and 1.5 × 106 total number of cells for U-87 MG and LN-229 cells, respectively) was injected subcutaneously into the right flank region of athymic nude mice (6 to 8 weeks old). Live anaesthetized animals were imaged for brightfield and GFP fluorescence on 1, 5, 15, 10 and 20 days post tumor implantation to assess tumor growth in vivo using the Maestro FLEX In Vivo Imaging System with appropriate filters for GFP (tumor; excitation = 445–490 nm, emission = 515 nm long-pass filter, acquisition settings = 500–720 in 10 nm steps). Acquisition settings were 120 ms for GFP and brightfield images. The background autofluorescence spectra from non-tumor regions of the flank were collected. The multispectral fluorescence images were background subtracted and unmixed using Maestro software (Cambridge Research and Instrumentation, Inc., Woburn, MA) to detect the GFP signal from the fluorescent tumor cells. Three weeks after injection of the tumor cells, animals were sacrificed and imaged as described above. The tumors were then excised and measured with calipers (Fine Science Tools, Inc., Foster City, CA). Tumor volume was computed from the length, width and height measurements. Excised flank tumors were then snap-frozen on dry ice for transport and frozen for sectioning to perform immunohistochemistry.
Immunohistochemistry
Excised flank tumors or brains injected with GFP expressing tumor cells were fixed in 4% paraformaldehyde in PEM buffer (80 mM Pipes, 1 mM magnesium chloride, 3% sucrose) pH 7.4. The fixative was washed off using PBS and the flank tumors and brains were incubated sequentially in 10% sucrose in PBS, 25% sucrose in PBS and then embedded in Tissue Freezing Medium (TFMTM, Electron Microscopy Sciences, Hatfield PA). Flank tumors were then sectioned at 7 μm intervals and labeled with anti-human vimentin rabbit monoclonal antibody (NeoMarkers, Fremont, CA) to detect human tumor cells. After washing off TFMTM with water, the sections were incubated in block buffer containing PBS, 2% goat serum, 0.25% saponin and 0.5% BSA to permeabilize the cells and block non-specific binding. Tissue sections were then incubated with anti-human vimentin primary antibodies overnight at 4°C that were detected by incubating with AlexaFluor® 568 conjugated goat anti-rabbit (Invitrogen, Carlsbad, CA) secondary antibodies for 1.5 hr at room temperature. Sections were then coverslipped with Citifluor Antifadent Mounting Medium, AF1 (Electron Microscopy Sciences, Hatfield, PA) and imaged. Fluorescent images were taken with a 20x objective using a Leica DMI 6000B automated inverted microscope (Leica Microsystems Inc., Buffalo Grove, IL) fitted with a Retiga EXi camera (QImaging, Surrey, BC, Canada). Formalin fixed paraffin embedded human tumor tissue and normal cortical surface tissue sections were immunolabeled with anti-human cadherin-11 (LifeSpan BioSciences, Inc., Seattle, WA). Briefly, sections were deparaffinized by sequential washes in Xylene and ethanol and then subjected to antigen retrieval by heating in sodium citrate buffer (pH 6.0) at 100°C for 18 min. The sections were allowed to cool for 20 min and incubated in block buffer as mentioned above. Subsequently, sections were labeled with anti-human cadherin-11 primary antibody overnight at 4°C and detected using AlexaFluor® 568 conjugated goat anti-rabbit (Invitrogen, Carlsbad, CA) secondary antibody by incubation for 1.5 hr at room temperature. Fluorescent images were taken using a 20x objective as described above.
Sequential 20 μm thick frozen sections of mouse xenograft tumors formed from CD133+ enriched neurospheres of human GBM tumors were immunolabeled with anti-human vimentin and cadherin-11 antibodies as described above following antigen retrieval.
Orthotopic xenograft tumors implanted intracranially in mice
U-87 MG and LN-229 cells infected with lentivirus encoding control or cadherin-11 shRNA and GFP were injected intracranially into athymic nude mice as described previously (34). For implantation, infected cells were resuspended in PBS at a concentration of 12,500 cells per μl. 3 weeks post implantation, the animals were sacrificed, whole brains were excised and imaged using the Maestro FLEX In Vivo Imaging System. The acquisition settings used for U-87 MG intracranial tumors were 10 ms exposure for GFP fluorescent images. The acquisition settings used for LN-229 intracranial tumors were 75 ms exposure for GFP fluorescent images. The position of the injection site was determined by comparing sections containing GFP fluorescence to the brightfield images. The images were analyzed as described above.
Xenograft tumors of primary human glioma stem-like cells (CW 421) implanted intracranially in mice
Cultures enriched in glioma stem-like cells (GSCs) were isolated from a patient with newly diagnosed glioblastoma multiforme as previously described (39). The patient provided informed consent under a protocol approved by University Hospital Case Medical Center Institutional Review Board. The tumor was dissociated by papain dissociation system (Worthington Biochemical Corp., Lakewood, NJ) and filtered with a 70 μm cell strainer to remove undissociated tissue according to the manufacturer’s instructions. Cells were cultured in stem-cell culture medium supplemented with B27 (without vitamin A) (Invitrogen, Carlsbad, CA), EGF (10 ng/ml) and bFGF (10 ng/ml). Cells were then cultured for 24 or 48 hr to recover surface antigens. CD133+ cells were derived by separation using magnetic cell sorting columns using microbead-conjugated CD133 antibodies (Miltenyi Biotec Inc., Auburn, CA). The stem-like nature of the CD133+ cells was confirmed based on the criterion of proliferation, replication, limited dilution assay, serum induced differentiation, and tumorigenicity of CD133+ enriched neurospheres in nude mice. For this study, an alliquot of 300,000 neurospheres was spun down and washed twice with PBS and implanted intracranially into nude mice at a concentration of 1×105 cells/μl for a total of 3×105 cells in accordance with Case Western Reserve University Institutional Animal Care and Use Committee approved protocol to Andrew E. Sloan. Mice were maintained until animals exhibited morbidity or neurological symptoms. Brains of euthanized mice were collected and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and cryoprotected in ascending amounts of sucrose. The brains were embedded in OCT and sectioned into 20 μm sections and subjected to immunohistochemical labeling as described above.
Statistical analyses
Data was collected from at least 3 independent experiments. For in vivo experiments, data was analyzed from at least 3 animals injected per condition. Error bars denote standard error and statistical significance was assessed by an unpaired Student’s t-test.
Results
Analysis of Cadherin-11 expression
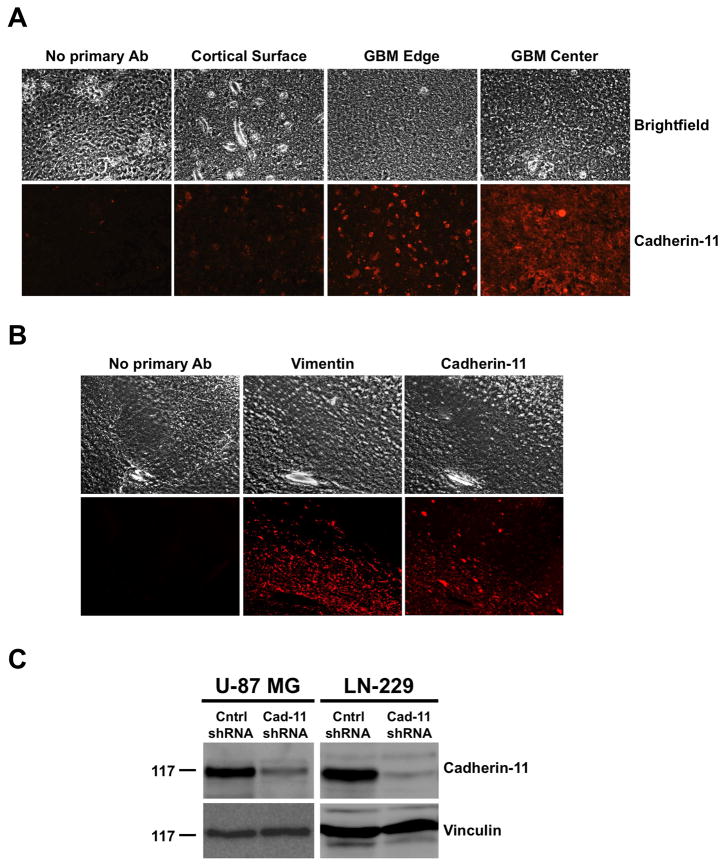
To understand the role of cadherin-11 in GBM, we searched the Oncomine™ database (40) for Cadherin-11 mRNA expression in normal brain versus glioblastoma. Three independent studies demonstrated a 1.819, 2.504 and 2.091 fold increase, respectively, in Cadherin-11 expression in glioblastoma compared to normal brain (40–43). To analyze the importance of the cadherin-11 protein in GBM, we performed immunohistochemical labeling of sections from human GBM (center and edge of tumor) and normal cortical surface samples from the same patient. Samples from 3 different patients were analyzed. Data from 3 patients showed increased expression of cadherin-11 in the center and edge of the tumor compared to the normal cortical surface (Fig. 1A) indicating that cadherin-11 may have an important function in GBM. Because human GBM tumors are proposed to arise from GSCs, cadherin-11 may be expressed in the glioma stem-like tumor initiating cells. To test this hypothesis, we performed immunolabeling for cadherin-11 expression in intracranial xenograft tumor sections that were prepared by injecting CD133+ enriched neurospheres developed from human GBM tumors into the brains of athymic nude mice. The sections were also labeled with anti-human vimentin antibody to visualize injected human tumor cells (Fig. 1B). The higher expression of cadherin-11 in the vimentin-positive human glioma stem-like cells depicts the importance of cadherin-11 in GBM (Fig. 1B). Additionally, we analyzed cadherin-11 protein expression by immunoblotting lysates from two distinct human glioma cell lines, U-87 MG and LN-229. Cadherin-11 was expressed in both glioma cell lines (Fig. 1C). To directly investigate the role of cadherin-11 in GBM, we used shRNA to reduce cadherin-11 expression. U-87 MG and LN-229 glioma cells were infected with lentivirus encoding control or cadherin-11 shRNA. 3 days post infection, cell lysates were immunoblotted for cadherin-11 (Fig. 1C). Densitometry analysis of immunoblots comparing cadherin-11 protein levels in control shRNA expressing cells to those expressing cadherin-11 shRNA showed a 77% and 93% reduction in cadherin-11 expression in cadherin-11 shRNA-expressing U-87 MG and LN-229 glioma cells, respectively (Fig. 1C).
Figure 1.
Analysis of cadherin-11 expression in human GBM tumors, human GBM stem-like cells, glioma cell lines and shRNA mediated knockdown of cadherin-11. A, Cadherin-11 is highly expressed in human GBM tumors (GBM Center and GBM Edge) compared to normal human brain (Cortical Surface). Paraffin-embedded sections of human brain tumors and cortical surface were immunohistochemically labeled with anti-human cadherin-11 antibodies and imaged using a 20x objective. B, Cadherin-11 is expressed in glioma stem-like cells. Frozen sections of intracranial xenograft nude mouse brains implanted with CW 421 human glioma stem-like cells were immunolabeled with anti-human vimentin and cadherin-11 antibodies. Representative images are shown at 200x magnification. C, Cadherin-11 is expressed in human glioma cell lines. Cell lysates were separated by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with antibodies against cadherin-11. Immunoblots were stripped and reprobed for vinculin as a loading control. Molecular weight in kDa is shown at left. Expression of cadherin-11 protein in U-87 MG and LN-229 glioma cells is shown as well as the decrease in cadherin-11 protein levels in cells infected with lentivirus encoding cadherin-11 shRNA (Cad-11) compared to control shRNA (Cntrl).
Cadherin-11 regulates glioma cell migration in vitro
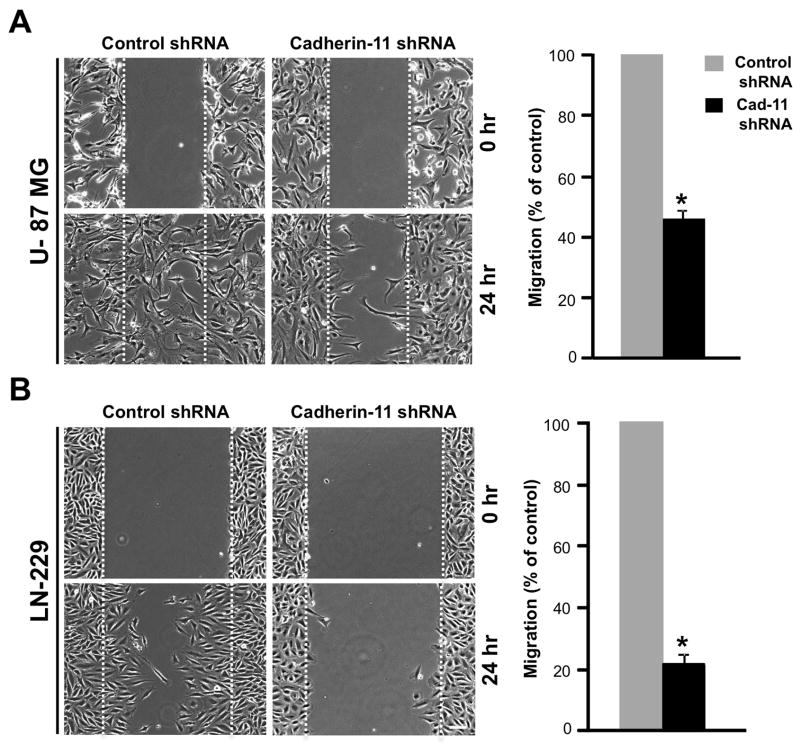
Cadherin-11 is expressed in various cancers including breast and prostate cancer, where expression of cadherin-11 is associated with increased migration and invasion (reviewed in 44). To understand the role of cadherin-11 in GBM migration, we performed scratch wound assays with U-87 MG and LN-229 glioma cells that are migratory in vitro (45). Confluent monolayers of U-87 MG and LN-229 cells expressing control or cadherin-11 shRNA were scratched with a pipette tip to form a wound. Migration of cells into the wound was measured over time by acquiring images at 0 and 24 hr post scratch (Fig. 2A and B). U-87 MG and LN-229 glioma cells expressing control shRNA migrated over a period of 24 hr to fill the wound (Fig. 2A and B). However, U-87 MG and LN-229 cells expressing cadherin-11 shRNA failed to migrate into the wound and showed a 54.26% +/− 3.32% and 78.39% +/− 2.55% (p<0.0001) decrease, respectively, in the distance migrated (Fig. 2A and B), demonstrating that cadherin-11 expression is necessary for glioma cell migration in vitro.
Figure 2.
Knockdown of cadherin-11 inhibits migration of glioma cells in scratch wound assays in vitro. Data represent mean ± SE of at least 3 independent experiments. U-87 MG (A) and LN-229 (B) cells infected with lentivirus encoding control or cadherin-11 shRNA were scratched and imaged at 0 and 24 hr. Dashed lines indicate the boundary of the edges of the wound at 0 hr. Scale bar, 100 μm. (p<0.0001, n=3 for U-87 MG and n=5 for LN-229).
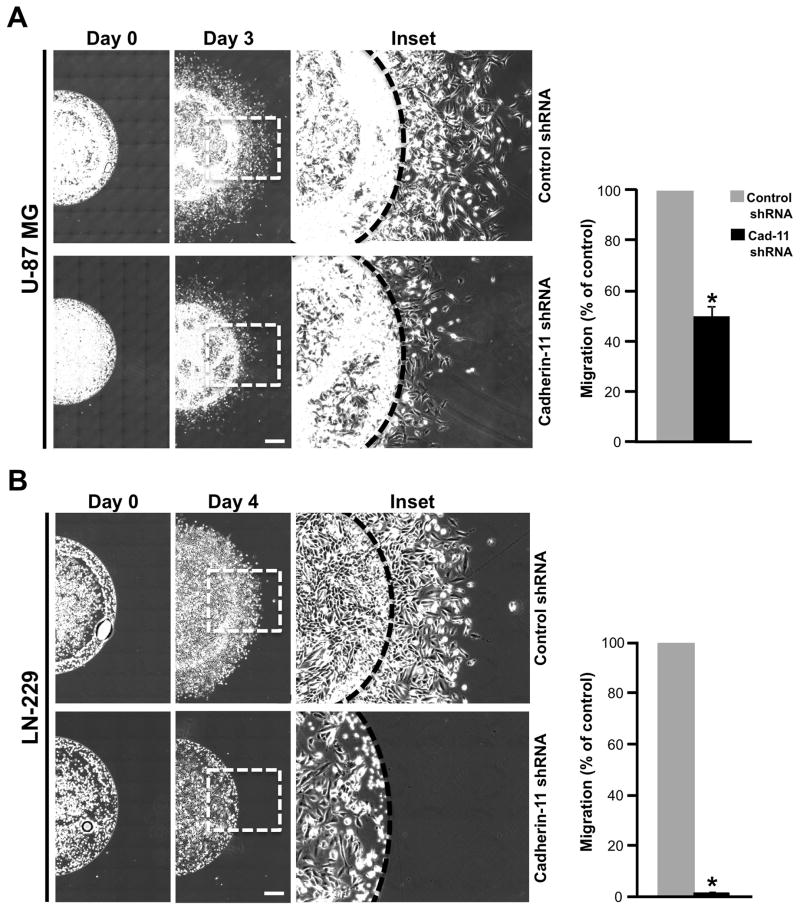
In the scratch wound assay, the cells migrate in a 2-dimensional tissue culture environment. However, in human tumors and in xenograft models of glioma in vivo, the tumor cells grow and migrate in a complex 3-dimensional environment. In an in vivo tumor mass, cells are at high density wherein cell-cell interactions exist and cells migrate away from the tumor during dispersal or invasion. We hypothesized that cadherin-11 may be an important regulator of the cell-cell interactions that occur in 3-dimensional tissues. To mimic these 3-dimensional conditions in vitro, we developed a novel spot assay to study cell migration. U-87 MG and LN-229 glioma cells infected with lentivirus encoding control or cadherin-11 shRNA for 3 days were mixed with Matrigel and spotted on a tissue culture plate followed by addition of media (Fig. 3A and B). Matrigel provides a 3-dimensional environment at the beginning of cell migration. Images were taken immediately after spotting (day 0 time point) and media was changed each day for 3 days (for U-87 MG) and 4 days (for LN-229) to supply nutrients to the migrating cells continuously. Spots were re-imaged on day 3 (for U-87 MG) and 4 (for LN-229) and the distance migrated by the cells away from the spot was quantitated. U-87 MG and LN-229 cells expressing control shRNA migrated a distance of 834.26 μm and 509.68 μm, respectively (Fig. 3A and B). In contrast, U-87 MG cells expressing cadherin-11 shRNA migrated a distance of 417.13 μm, a 50.38% +/− 4.37% (p<0.001) decrease in migration compared to controls. LN-229 cells expressing cadherin-11 shRNA failed to migrate and remained in the spot (Fig. 3A and B). At day 4, LN-229 cells expressing cadherin-11 shRNA also had an overall decrease in cell number due to reduced survival (Fig. 3B). These data provide further support that cadherin-11 expression is required for glioma cell migration in a 3-dimensional environment in vitro.
Figure 3.
Knockdown of cadherin-11 inhibits migration of glioma cells in spot assays in vitro. Data represent mean ± SE of at least 3 independent experiments. U-87 MG (A) and LN-229 (B) cells infected with lentivirus encoding control or cadherin-11 shRNA were mixed with Matrigel and spotted. Representative images depict one half of the spot at 0 and at 3 or 4 days (for U-87 MG or LN-229, respectively) post plating. Scale bar, 500 μm. (p<0.0001, n=3). *, statistically significant reduction in migration. The inset in days 3 and 4 images is enlarged and depicted. The dashed line indicates the edge of the spot at day 0.
Cadherin-11 regulates glioma cell survival in vitro
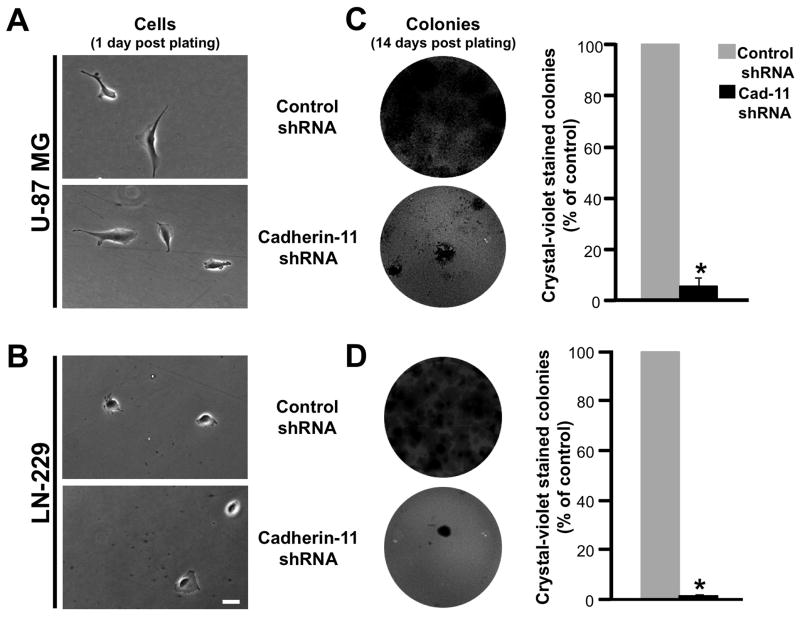
During tumor progression, cells that disperse away from the main tumor are able to survive in suboptimal environments by depending on their own autocrine/paracrine signaling to form secondary tumor masses. Since U-87 MG and LN-229 glioma cells express cadherin-11, we hypothesized that cadherin-11 may function in glioma cell survival. To directly test this possibility in vitro, we performed a growth factor-independent colony formation assay. U-87 MG and LN-229 glioma cells infected with lentivirus encoding control or cadherin-11 shRNA were plated at low density (Fig. 4A and B) and allowed to grow for 2 weeks without replenishing the medium to test their ability to survive in the absence of exogenous growth factors. 14 days post plating, colonies were stained with crystal-violet and quantitated. U-87 MG and LN-229 glioma cells expressing control shRNA formed abundant colonies (Fig. 4C and D). In contrast, following cadherin-11 knockdown, both U-87 MG and LN-229 glioma cells had a significant reduction in colony formation of 94.76% +/− 3.79% and 99.74% +/− 0.01% (p<0.0001), respectively (Fig. 4C and D). These data suggest that cadherin-11 is required for glioma cell survival in vitro.
Figure 4.
Knockdown of cadherin-11 reduces growth factor-independent survival of glioma cells in vitro. U-87 MG or LN-229 glioma cells infected with lentivirus encoding control or cadherin-11 shRNA were plated at low density and allowed to form colonies under growth factor-deprivation for 2 weeks. Representative photomicrographs show U-87 MG (A) and LN-229 (B) cells 1 day post plating at 400x magnification. C, crystal violet stained colonies from U-87 MG glioma cells expressing control or cadherin-11 shRNA are shown. Graphical representation of colony number quantification is depicted (p<0.0001, n=4). D, crystal violet stained colonies from LN-229 glioma cells expressing control or cadherin-11 shRNA are shown. Graphical representation of colony number quantification is shown (p<0.0001, n=3). Data represent mean ± SE of at least 3 independent experiments performed in triplicate. Scale bar, 50 μm. *, statistically significant reduction in colony formation.
Cadherin-11 regulates glioma cell survival in vivo
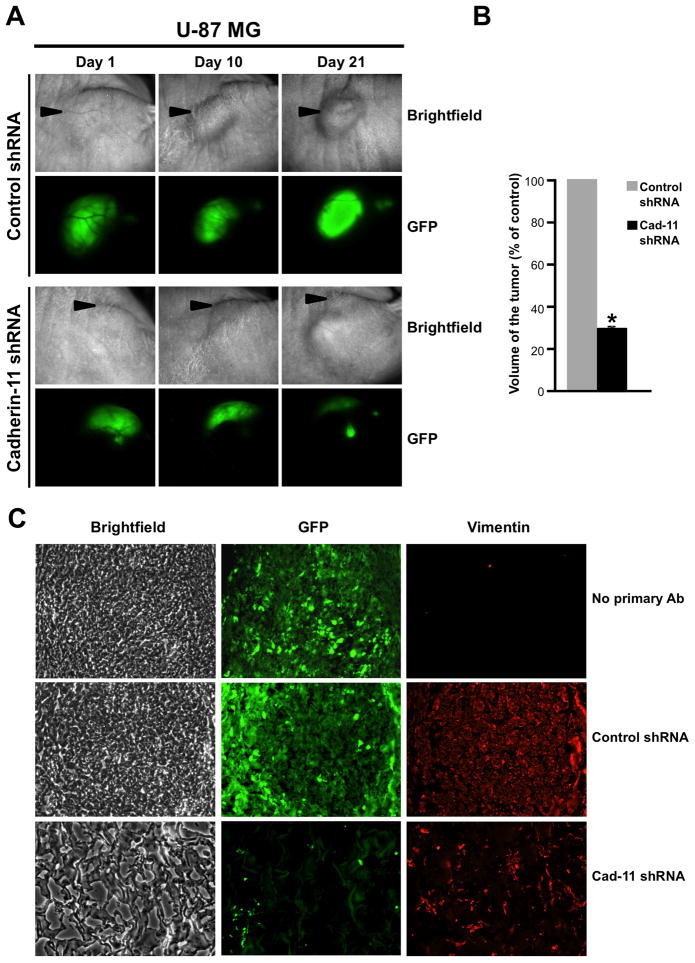
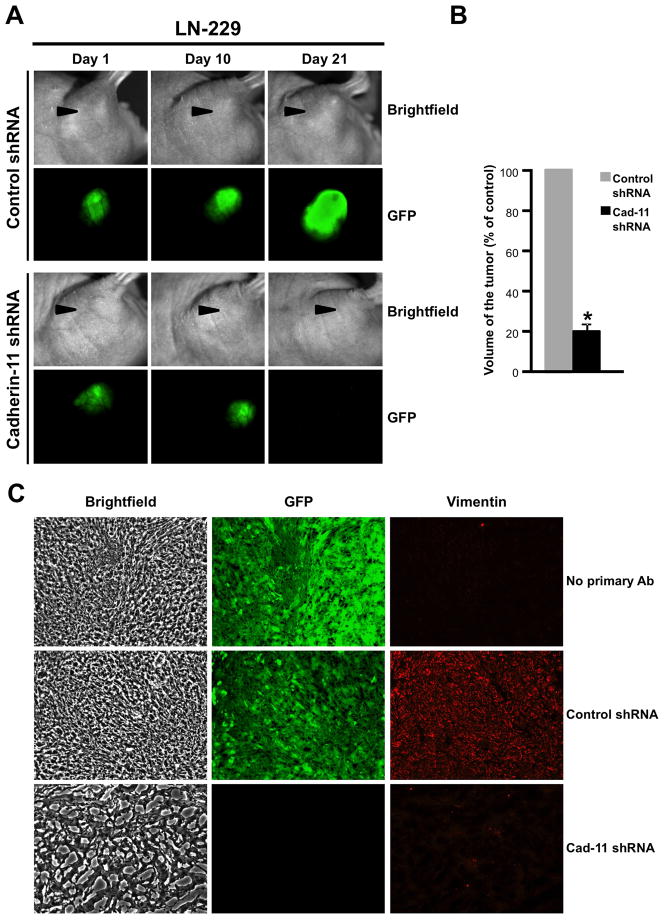
To investigate if cadherin-11 is required for glioma cell survival in an in vivo environment, U-87 MG and LN-229 glioma cells co-expressing control or cadherin-11 shRNA and GFP were mixed with Matrigel and injected subcutaneously into the right flank region of nude mice. Tumors were allowed to grow for 3 weeks during which tumor growth was assessed by imaging the tumors for GFP fluorescence in vivo at regular intervals (day 1, 5, 10, 15 and 20). Representative images from day 1, 10 and 21 are shown in Figs. 5A and 6A. U-87 MG and LN-229 glioma cells infected with lentivirus encoding control shRNA continued to grow from day 1 to day 21 as assessed by the increase in GFP fluorescence. In contrast, knockdown of cadherin-11 expression resulted in a reduction in tumor size from day 5 (data not shown) to day 21 as seen by a loss of GFP fluorescence at day 21 (Figs. 5A and 6A). On day 21, animals were sacrificed and the volume of each excised tumor was calculated (Figs. 5B and 6B). Compared to the large tumors formed by U-87 MG and LN-229 cells expressing control shRNA, those expressing cadherin-11 shRNA had a significant reduction in tumor volume (70.32 % +/− 1.23% decrease for U-87 MG, 80.86 % +/− 4.49% decrease for LN-229, p<0.0001) (Figs. 5B and 6B). To study the tumor architecture, frozen tumor sections were immunolabeled with anti-human vimentin antibodies to distinguish mouse host cells from implanted human tumor cells (Figs. 5C and 6C). Tumors expressing GFP and control shRNA showed densely packed GFP-positive tumor cells (Figs. 5C and 6C). In contrast, tumors formed by cells where cadherin-11 was knocked down had very few surviving cells as evidenced by both GFP and vimentin staining scattered in the Matrigel, which formed the bulk of the tumor (Figs. 5C and 6C).
Figure 5.
Knockdown of cadherin-11 inhibits U-87 MG glioma cell survival in a heterotopic xenograft in vivo. A, U-87 MG cells expressing control or cadherin-11 shRNA and GFP were injected into the flank of athymic nude mice and imaged for brightfield and GFP fluorescence on days 1, 10 and 21. B, volume of each excised tumor was calculated using measurements of length, width and height of the tumor and plotted. *, statistically significant reduction in tumor volume (p<0.0001, n=3). Arrowheads point to the location of the flank tumors. C, Frozen sections of excised flank tumors formed from U-87 MG cells expressing control or cadherin-11 shRNA and GFP were immunolabeled with anti-human vimentin antibodies to detect injected human tumor cells. Representative images of sections showing brightfield, GFP and vimentin are shown at 200x magnification.
Figure 6.
Knockdown of cadherin-11 inhibits LN-229 glioma cell survival in a heterotopic xenograft in vivo. A, LN-229 cells expressing control or cadherin-11 shRNA and GFP were injected into the flank of athymic nude mice and imaged for bright field and GFP fluorescence on days 1, 10 and 21. B, volume of each excised tumor was calculated using measurements of length, width and height of the tumor and plotted. *, statistically significant reduction in tumor volume (p<0.0001, n=3). Arrowheads point to the location of the flank tumors. C, Frozen sections of excised flank tumors formed from LN-229 cells expressing control or cadherin-11 shRNA and GFP were immunolabeled with anti-human vimentin antibodies to detect injected human tumor cells. Representative images of sections showing bright field, GFP and vimentin are shown at 200x magnification.
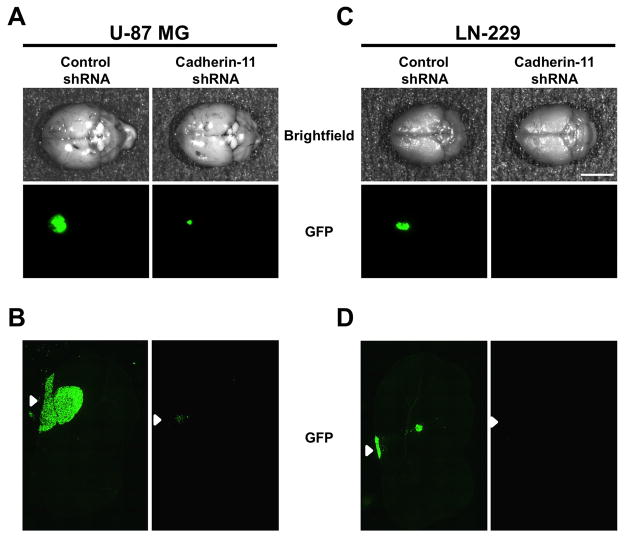
To test the influence of cadherin-11 knockdown on cell survival in an orthotopic environment, U-87 MG and LN-229 glioma cells co-infected with lentivirus encoding control or cadherin-11 shRNA and GFP were injected intracranially in the striatum in athymic nude mice. 3 weeks post injection, excised whole brains were imaged for GFP fluorescence to assess tumor growth. U-87 MG and LN-229 cells expressing control shRNA formed large tumors as assessed by the amount of GFP fluorescence at the injection site (Fig. 7A and C). In contrast, U-87 and LN-229 glioma cells where cadherin-11 expression was knocked down with shRNA did not form a tumor and showed a complete absence of GFP fluorescence in the striatum (Fig. 7A, B, C and D). However, U-87 MG cells expressing cadherin-11 shRNA had a tiny dot of fluorescence (Fig. 7A), which corresponded with a small cluster of GFP expressing cells at the site of injection (Fig. 7B). No tumors were detected in the striatum or near the corpus callosum in sections from brains injected with U-87 MG or LN-229 cells expressing cadherin-11 shRNA (Fig. 7B and D). Together, these data demonstrate that cadherin-11 is required for the survival of glioma cells in vitro as well as in heterotopic and orthotopic sites in vivo.
Figure 7.
Knockdown of cadherin-11 reduces survival of glioma cells at an orthotopic site in vivo. U-87 MG (A) and LN-229 cells (C) co-expressing control or cadherin-11 shRNA and GFP were injected intracranially into the striatum of athymic nude mice. 3 weeks following injection, animals were sacrificed and whole brains were excised and imaged. Representative images depict the dorsal surface of whole brains showing injected GFP-expressing tumor cells (n=3). Scale bar, 5 mm. Frozen sections of intracranial mouse xenografts of U-87 MG (B) and LN-229 (D) cells expressing control or cadherin-11 shRNA and GFP were imaged for GFP fluorescence. Arrowheads represent the injection site on the dorsal surface of the brain.
Discussion
Cadherins are a class of calcium-dependent cell-cell adhesion molecules that interact with catenins (α-, β- and γ-catenin). Association of an intact cadherin-catenin complex with the actin cytoskeleton is essential for cadherin-mediated cell-cell adhesion (reviewed in 15). Cell-cell adhesion, a process important for determining cell polarity and maintaining tissue architecture, is disrupted during tumorigenesis by multiple genetic and epigenetic alterations leading to increased motility and invasiveness of tumors (44). Many cadherins have been identified as potential tumor suppressors and/or oncogenic proteins depending on the type of cadherin and the type of tumor (reviewed in 44).
Cadherins are also important players in epithelial-mesenchymal transition (EMT), a process that occurs during both normal embryonic development and cancer progression (46–48). During normal tissue morphogenesis, EMT promotes plasticity of epithelial cells (47). Tumor cells activate EMT to gain characteristics of the mesenchymal phenotype including increased motility and invasiveness, stem-cell like characteristics as well as greater resistance to apoptosis, senescence and therapeutic drugs (46–49). Identification of molecular markers upregulated during EMT to promote malignancy and aggressiveness of tumors could be used to develop targeted therapies. Several global gene profiling studies have identified an aberrant mesenchymal lineage in GBM (4–9) that could contribute to the increased invasive capacity of GBM tumors. During the development of a mesenchymal phenotype through EMT in tumor progression, loss of E-cadherin expression is often associated with expression of mesenchymal cadherins like N-cadherin or cadherin-11. This phenomenon, known as cadherin switching, occurs in breast, prostate, gastric and pancreatic cancers (12, 13, 44). N-cadherin expression in tumors promotes cell migration, invasion and survival (50). Cadherin-11, a cell-surface biomarker of EMT (47), is expressed in multiple normal tissues including mesenchymal tissues of the brain during development (51). In highly malignant forms of breast and prostate cancer, cadherin-11 expression increases migration and invasion of tumor cells (24, 27, 28, 52). Additionally, in prostate cancer cells, expression of cadherin-11 increases association with osteoblasts and homing to the bone (52). In this study, we cited increased expression of Cadherin-11 mRNA (40–43) in human GBM tumors compared to normal brain and also observed an increase in cadherin-11 protein expression in human GBM tumors compared to normal brain (Fig. 1A). Cadherin-11 protein was also expressed in the two human glioma cells lines tested (Fig. 1C). Additionally, increased expression of cadherin-11 protein in human glioma stem-like cells (Fig. 1B) emphasizes the importance of cadherin-11 in human GBM. We hypothesized that cadherin-11 may play a role in either maintaining or promoting the malignant, dispersive state of GBM. A previous study by Zhou and colleagues (53) analyzed the expression of cadherin-11 in normal brains compared to astrocytoma samples from human patients by immunoblotting. In their article, Zhou et al. observed cadherin-11 expression in normal brain. An abundance of cadherin-11 in the normal brain may be due to the presence of various cell types that express cadherin-11, like neurons and vascular endothelial cells. Grade II and Grade III astrocytoma have no cadherin-11 expression whereas grade IV astrocytoma or GBM have increased cadherin-11 expression. Interestingly, comparison of Cadherin-11 mRNA expression in GBM versus other types of astrocytomas showed a 1.735 fold increase in Cadherin-11 in the data analysis from Oncomine™ (40, 54). This again suggests the importance of cadherin-11 in GBM. Interestingly, studies in other tumor types like osteosarcoma suggest reduced levels of CDH11 are important in tumor metastasis and implicate CDH11 as a prognostic marker of osteosarcoma patient survival (55).
Cadherin-11 expression is associated with increased invasiveness in breast and prostate cancer tumors (24, 27, 28). Our results demonstrate that shRNA mediated reduction of cadherin-11 inhibits glioma cell migration in vitro (Fig. 2 and 3) suggesting cadherin-11 is a pro-invasive factor in gliomas. However, one study reported cadherin-11 inhibits cell invasion in vitro in A172 glioma cells (33). The literature presents other similar discrepancies in different cell types with respect to the role of cadherin-11 in migration and invasion. For example, overexpression of cadherin-11 in the SKBR3 breast carcinoma cell line decreased their invasiveness (21). In contrast, overexpression of cadherin-11 in the BT-20 breast carcinoma cell line increased their invasive capacity (56). Interestingly, co-expression of full length cadherin-11 with a splice variant form of cadherin-11 (that has a truncated C-terminus) increased invasiveness of SKBR3 cells in vitro (21). Thus the effect of an increase or reduction in cadherin-11 levels on cancer cell migration likely depends upon the cell- and tumor-type.
The lethality of GBM is attributed to the ability of GBM cells to proliferate and disperse rapidly throughout the brain. To form secondary tumor masses, dispersed GBM cells need to depend on their own autocrine/paracrine signaling mechanisms to survive at a new site. In our studies, knockdown of cadherin-11 significantly reduced the ability of glioma cells to survive in vitro and form colonies in the absence of exogenous growth factors (Fig. 4). Importantly, reduction of cadherin-11 in glioma cells significantly reduced the formation of flank and intracranial xenograft tumors in vivo (Figs. 5, 6 and 7). Future studies are required to explore the molecular mechanism by which cadherin-11 promotes glioma cell migration and survival. In conclusion, our studies demonstrate cadherin-11 to be a pro-migratory and pro-survival factor in gliomas. Therefore, development of specific inhibitors blocking cadherin-11 function could be a potential strategy to reduce the aggressiveness and lethality of GBM tumors.
Acknowledgments
Grant support
This study was supported by the Case Center for Imaging Research, Athymic Animal Core Facility of the Comprehensive Cancer Center at Case Western Reserve University (P30-CA43703), National Institutes of Health grants to Susann M. Brady-Kalnay (R01-NS051520), National Institutes of Health grants to Susann M. Brady-Kalnay, James P. Basilion and Andrew E. Sloan (R01-NS063971). The glioma stem cell core is supported by grants from the Kimble Foundation and the Peter D. Cristal Chair to Andrew E. Sloan. This work is dedicated to Tabitha Yee-May Lou whose endowment supports brain tumor research in the Brady-Kalnay lab.
The authors thank Scott Howell, PhD, and Catherine Doller of the Visual Sciences Research Center for quantification of the data and providing histological assistance, respectively. The authors also thank Scott A. Becka for technical assistance and Sonya E. L. Craig, PhD, for critical reading of the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Louis DNOH, Wiestler OD, Cavenee WK. WHO Classification of tumours of the central nervous system. 4. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonavia R, Inda MD, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–60. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman H, Aldape K. Molecular predictors in glioblastoma: toward personalized therapy. Arch Neurol. 2008;65:877–83. doi: 10.1001/archneur.65.7.877. [DOI] [PubMed] [Google Scholar]
- 5.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–10. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–9. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–8. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 9.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tso CL, Shintaku P, Chen J, Liu Q, Liu J, Chen Z, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–19. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 12.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–63. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 13.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 14.Anastasiadis PZ. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 15.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–57. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 16.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 18.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 19.Okazaki M, Takeshita S, Kawai S, Kikuno R, Tsujimura A, Kudo A, et al. Molecular cloning and characterization of OB-cadherin, a new member of cadherin family expressed in osteoblasts. J Biol Chem. 1994;269:12092–8. [PubMed] [Google Scholar]
- 20.Hoffmann I, Balling R. Cloning and expression analysis of a novel mesodermally expressed cadherin. Dev Biol. 1995;169:337–46. doi: 10.1006/dbio.1995.1148. [DOI] [PubMed] [Google Scholar]
- 21.Feltes CM, Kudo A, Blaschuk O, Byers SW. An alternatively spliced cadherin-11 enhances human breast cancer cell invasion. Cancer Res. 2002;62:6688–97. [PubMed] [Google Scholar]
- 22.Kashima T, Kawaguchi J, Takeshita S, Kuroda M, Takanashi M, Horiuchi H, et al. Anomalous cadherin expression in osteosarcoma. Possible relationships to metastasis and morphogenesis. Am J Pathol. 1999;155:1549–55. doi: 10.1016/S0002-9440(10)65471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro SB, Turner IM, Farookhi R, Blaschuk OW, Jothy S. E-cadherin and OB-cadherin mRNA levels in normal human colon and colon carcinoma. Exp Mol Pathol. 1995;62:118–22. doi: 10.1006/exmp.1995.1013. [DOI] [PubMed] [Google Scholar]
- 24.Pishvaian MJ, Feltes CM, Thompson P, Bussemakers MJ, Schalken JA, Byers SW. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–52. [PubMed] [Google Scholar]
- 25.Shibata T, Ochiai A, Gotoh M, Machinami R, Hirohashi S. Simultaneous expression of cadherin-11 in signet-ring cell carcinoma and stromal cells of diffuse-type gastric cancer. Cancer Lett. 1996;99:147–53. doi: 10.1016/0304-3835(95)04047-1. [DOI] [PubMed] [Google Scholar]
- 26.Shimazui T, Giroldi LA, Bringuier PP, Oosterwijk E, Schalken JA. Complex cadherin expression in renal cell carcinoma. Cancer Res. 1996;56:3234–7. [PubMed] [Google Scholar]
- 27.Bussemakers MJ, Van Bokhoven A, Tomita K, Jansen CF, Schalken JA. Complex cadherin expression in human prostate cancer cells. Int J Cancer. 2000;85:446–50. [PubMed] [Google Scholar]
- 28.Tomita K, van Bokhoven A, van Leenders GJ, Ruijter ET, Jansen CF, Bussemakers MJ, et al. Cadherin switching in human prostate cancer progression. Cancer Res. 2000;60:3650–4. [PubMed] [Google Scholar]
- 29.Cavallaro U, Christofori G. Multitasking in tumor progression: signaling functions of cell adhesion molecules. Ann N Y Acad Sci. 2004;1014:58–66. doi: 10.1196/annals.1294.006. [DOI] [PubMed] [Google Scholar]
- 30.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–11. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 31.Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6:1259–67. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura D, Hiraga T, Myoui A, Yoshikawa H, Yoneda T. Cadherin-11-mediated interactions with bone marrow stromal/osteoblastic cells support selective colonization of breast cancer cells in bone. Int J Oncol. 2008;33:17–24. [PubMed] [Google Scholar]
- 33.Delic S, Lottmann N, Jetschke K, Reifenberger G, Riemenschneider MJ. Identification and functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma invasion-associated candidate genes. Neuropathol Appl Neurobiol. 2011 doi: 10.1111/j.1365-2990.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 34.Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, et al. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12:305–16. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–95. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, et al. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009;11:767–78. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 38.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, et al. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69:6960–8. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–40. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization.
- 41.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 42.Shai R, Shi T, Kremen TJ, Horvath S, Liau LM, Cloughesy TF, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–23. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 43.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–11. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–37. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 50.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–44. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simonneau L, Kitagawa M, Suzuki S, Thiery JP. Cadherin 11 expression marks the mesenchymal phenotype: towards new functions for cadherins? Cell adhesion and communication. 1995;3:115–30. doi: 10.3109/15419069509081281. [DOI] [PubMed] [Google Scholar]
- 52.Huang CF, Lira C, Chu K, Bilen MA, Lee YC, Ye X, et al. Cadherin-11 increases migration and invasion of prostate cancer cells and enhances their interaction with osteoblasts. Cancer Res. 2010;70:4580–9. doi: 10.1158/0008-5472.CAN-09-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou R, Skalli O. Identification of cadherin-11 down-regulation as a common response of astrocytoma cells to transforming growth factor-alpha. Differentiation. 2000;66:165–72. doi: 10.1046/j.1432-0436.2000.660402.x. [DOI] [PubMed] [Google Scholar]
- 54.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–89. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 55.Nakajima G, Patino-Garcia A, Bruheim S, Xi Y, San Julian M, Lecanda F, et al. CDH11 expression is associated with survival in patients with osteosarcoma. Cancer Genomics Proteomics. 2008;5:37–42. [PubMed] [Google Scholar]
- 56.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–44. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]