Abstract
Iterative highly-reducing polyketide synthases (HR-PKSs) from filamentous fungi are the most complex and enigmatic type of PKS discovered to date. Here we uncover an unusual level of programming by the hypothemycin HR-PKS, in which a single ketoreductase domain displays stereospecificity that is controlled by substrate length. Mapping of the structural domains responsible for this feature allowed the biosynthesis of an unnatural diastereomer of the natural product dehydrozearalenol.
Polyketides are secondary metabolites that display important biological activities1. Their biosynthetic construction is similar to that of fatty acid biosynthesis, but selective reduction of the keto functionalities to different extent at each cycle of chain extension results in products bearing a variety of functionalities2. Important fungal polyketides such as the cholesterol-lowering drug lovastatin are biosynthesized by Type I iterative, highly-reducing polyketide synthases (HR-PKSs)3,4, which employ reductive domains including β-ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) to tailor the polyketide backbone. The domain architecture of an HR-PKS resembles a single module of the bacterial modular PKS. In the latter, the catalytic domains are sequentially arranged in multiple versions and elongate the ketides by an assembly line process5. In contrast, fungal HR-PKS employs the same protein domains repeatedly for each stage of chain growth. The structural variations generated by HR-PKSs partially depend on the permutations of the reductive domains to transform functionalities at the β-carbon of the growing intermediates3,4. The programming rules of HR-PKS are therefore significantly more complex and far less understood compared to bacterial PKSs.
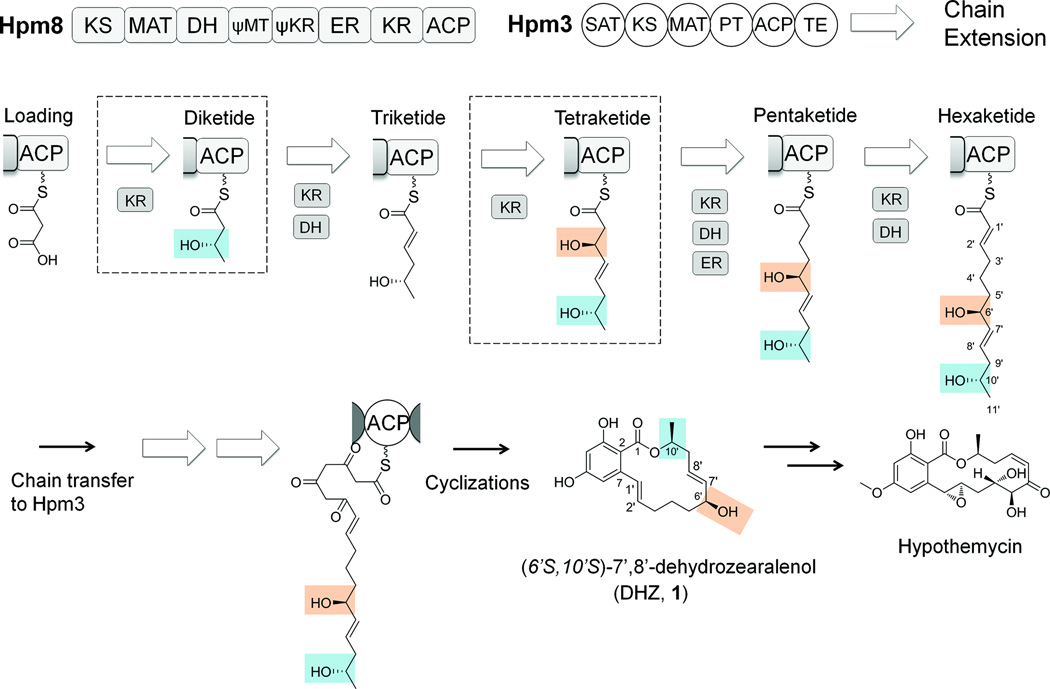
The KR domain belongs to the short chain dehydrogenase/reductase (SDR) superfamily, which reduces a β-ketone into an alcohol of l or d stereochemistry using NADPH6. A KR domain from bacterial PKSs performs the ketoreduction with stringent stereospecificity to afford a single alcohol stereoisomer. The molecular basis of this stereospecificity has been investigated in detail7–17. The geometry of the active site fixes the β-carbon relative to NADPH, facilitating a stereospecific attack by the hydride. X-ray crystal structures of bacterial KRs have suggested diagnostic residues that control KR stereospecificity by guiding the entrance of substrates into the active site groove from different sides15–17. Since only a single KR is present in the iterative HR-PKS, one would expect all the β-keto reduction to occur with the same relative stereochemistry. However, an exception was noted during our investigation of the biosynthesis of (6’S,10’S)-7’,8’-dehydrozearalenol (DHZ, 1), an intermediate to the resorcylic acid lactone (RAL) hypothemycin18,19.
The HR-PKS Hpm8 synthesizes the reduced hexaketide portion of 1 (Fig. 1) and transfers it to a non-reducing PKS (NR-PKS) Hpm3 that further extends it using three units of malonyl-CoA. The KR domain in Hpm8 catalyzes five β-ketoreduction steps involving substrates ranging in size from diketide to hexaketide. The stereochemistries of the 6’ and 10’ positions were determined by X-ray crystallography to both be S configuration18. By analogy with the nomenclature of carbohydrate chemistry20, the absolute stereochemistry of C6’ and C10’ can be further assigned as d-S-OH-C6’ and l-S-OH-C10’. The unusual opposite stereochemistry of C6’ and C10’ suggests an additional level of programming complexity by KR in fungal HR-PKSs.
Figure 1.
Biosynthesis of (6’S,10’S)-7’,8’-dehydrozearalenol (DHZ, 1) by Hpm8 and Hpm3 from Hypomyces subiculosus. In Hpm8, ψMT represents the pseudo C-methyltransferase domain and ψKR denotes the structural KR, as designated in the mammalian FAS25. The biosynthesis of the highly-reduced acyl chain of 1 by Hpm8 is displayed in detail. The two chiral centers C6’ and C10’ are generated at diketide and tetraketide stages. The stereochemistry of the stereogenic centers in 1 can be assigned as d-S-OH-C6’ and l-S-OH-C10’.
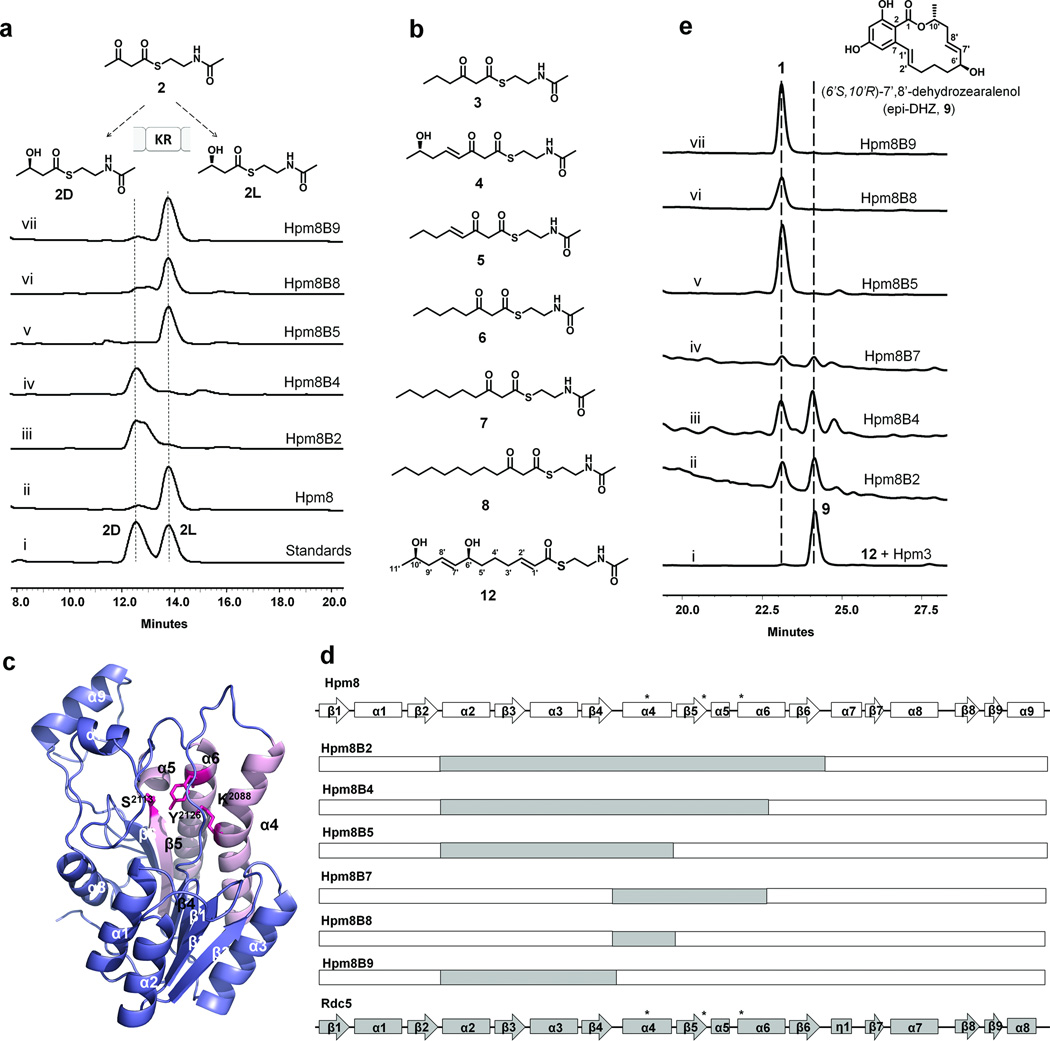
To confirm this Janus-faced stereospecificity of the Hpm8 KR domain, a series of β-keto-S-N-acetyl cysteamine (SNAC) compounds were synthesized and assayed in the presence of Hpm8 and NADPH. The active Hpm8 was expressed from Saccharomyces cerevisiae strain BJ5464-NpgA4 (Supplementary Methods). Synthetic standards of the β-hydroxyacyl-SNAC in either l or d configuration were also prepared. The Hpm8 efficiently reduced both diketide acetoacetyl-SNAC 2 and tetraketide (7S,E)-7-hydroxy-3-oxo-oct-4-enoyl-SNAC 4 into the corresponding β-hydroxyacyl-SNAC products. Hpm8 reduced 2 into 2L (l configuration) with greater than 91% stereoselectivity, whereas 4 was reduced exclusively to the d product 4D (Fig. 2a, Supplementary Results, Supplementary Fig. 1). These results are consistent with the stereochemistries of C10’ and C6’ in 1, and verify the substrate-tuned stereospecificity of the KR. Simplifying the structural complexity of 4 by using either β-ketoacyl-SNAC 5 or 6 did not alter the KR stereospecificity and yielded 5D or 6D, respectively (Fig. 2b and Supplementary Figs. 2–3). Hence substitutions distal to the β-position do not affect KR specificity, suggesting that carbon backbone length may solely dictate the product stereochemistry. Additional β-ketoacyl-SNAC substrates, including 3, 7 and 8 were assayed using Hpm8 to establish the chain-length/stereoselectivity relationship at the tri-, penta- and hexaketide stages. All substrates were quantitatively reduced to the β-hydroxyl product with d configuration (3D, 7D, or 8D, Fig. 2b, Supplementary Figs. 4–9 and Supplementary Table 1), in line with the syn stereochemistry required by the DH domain to generate a trans double bond11,13. Therefore, the Hpm8 KR domain displays strict stereospecificity to generate the d-hydroxyl product, except in the case of the diketide, which is reduced from the opposite side to the l-alcohol.
Figure 2.
Analysis and engineering of KR stereospecificity. a: The chiral HPLC traces (240 nm) of (i) the mixture of the chemical standards 2L and 2D; and the reductive products generated by (ii) Hpm8, (iii) Hpm8B2, (iv) Hpm8B4, (v) Hpm8B5, (vi) Hpm8B8 or (vii) Hpm8B9 from acetoacetyl-SNAC 2. b: The chemical structures of the synthesized β-keto SNACs for the stereochemical interrogation. c: The cartoon view of the homology model of Hpm8_cKR. The catalytic residues Ser2113, Lys2088 and Tyr2126 are shown as sticks in red. The secondary structural elements are labeled sequentially. d: Illustration of the swapped regions between Hpm8_cKR and Rdc5_cKR in different chimeric HR-PKSs. The sequences retained from Hpm8 are shown in white squares and the introduced regions from Rdc5 are shown in gray squares. e: The HPLC traces (320 nm) of (i) in vitro assay containing 10 µM Hpm3, 2 mM precursor 12 and 2 mM malonyl-CoA; the in vivo metabolites profiles from the S. cerevisiae co-transformants harboring Hpm3 and (ii) Hpm8B2, (iii) Hpm8B4, (iv) Hpm8B5, (v) Hpm8B8 or (vi) Hpm8B9. The differences in relative amounts of the d and l products generated via the in vitro assays (Fig. 2a) and in vivo biosynthesis (Fig. 2e) involving Hpm8B2 and Hpm8B4 are likely due to the selectivity for the minor reduced l-diketide isomer in the subsequent enzymatic processing towards 1 and 9.
In contrast to Hpm8, the HR-PKS Rdc5, which is involved in the biosynthesis of monocillin II en route to radicicol (Supplementary Fig. 10a), reduces the same diketide intermediate with the opposite stereochemistry to give a d-alcohol. End-to-end sequence alignment of the two KR domains revealed a highly conserved catalytic domain (cKR) with 66% identity (~190 AA), whereas the structural domains share a relatively lower sequence similarity (42% identity). By comparison to the KR subdomain of DEBS Module 1, the catalytic tetrad in Hpm8 was identified as Lys2088, Ser2113, Tyr2126 and Asn2130 (Supplementary Fig. 10b). Indeed, mutation of the first three residues abolished the activities of Hpm8 (Supplementary Fig. 11). However, alignment suggests that sequence fingerprints derived from modular KRs may not fully apply to fungal cKR (Supplementary Fig. 10 and 12). Specifically, the signature LDD motif for B-type KR (EryKR1) catalyzing “R” stereochemistry (d-OH)7,14,16 is replaced by LRD in both Hpm8 and Rdc5. With the exception of producing 2L, the retained aspartic acid residue of LRD may still be predictive of d-OH products. The signature tryptophan in A-type KR (AmpKR2) catalyzing reduction to “S” stereochemistry (l-OH)8,9,17 is substituted with either Tyr2118 in Hpm8 or Phe2143 in Rdc5. The point mutation Y2118F in Hpm8 did not lead to alteration in product stereochemistry, indicating that additional regions are involved in directing the opposite substrate orientation in Hpm8 at the diketide stage (Supplementary Fig. 11).
The homology structures of Hpm8 and Rdc5 cKR domains displayed a variant Rossmann fold21,22 with two β–α–β–α–β motifs that are connected by α3 (Fig. 2c and Supplementary Fig. 13). The β1–α1–β2–α2–β3 stretch is involved in NADPH binding, whereas the catalytic residues and the putative active site are located in the β4–α4–β5–α5/6–β6 motif, in which the second helix is fractured into α5 and α6 due to the presence of hairpin-forming proline. To map the secondary structural elements that contribute to the stereospecificity difference between the two cKRs, a series of Hpm8 mutants containing chimerical cKR sequences were constructed. Among them, Hpm8B2 covers most of the Rossmann fold and contains Rdc5 sequences from the loop between β2 and α2 to the loop between β6 and α7, while the remaining chimeras were swapped with subsets of structural elements. Solubly expressed mutants are shown in Fig. 2d.
Several Hpm8 chimeras displayed altered stereospecificity in the ketoreduction of 2 (Fig. 2a), such as Hpm8B2 that afforded 95% 2D instead of the initial major product 2L. Leaving β6 intact as in Hpm8B4 similarly produced ~95% 2D. The product stereochemistry reverted back to 2L for Hpm8B5, which does not contain the Rdc5 cKR β5–α5–α6 region in which Ser2113 and Tyr2126 are located. This result clearly indicated the importance of this structural element in dictating the orientation of the substrate 2. Swapping β5–α5–α6 alone between Hpm8 and Rdc5 was not successful as Hpm8B6 was not soluble. Two other hybrids Hpm8B8 and Hpm8B9 produced 2L as the major product, which excludes the involvement of α4 as well as the first β–α–β–α–β motif in stereoselectivity. The set of chimera megasynthases were further assayed for ketoreduction of 4, and each reduced the tetraketide stereoselectively into the d isomer 4D (Supplementary Fig. 1). Collectively, these results indicate the substrate-tuned stereoselectivity is modulated by the β5–α5–α6 motif in the catalytic site.
We next examined if the altered stereospecificity of the chimeras can produce (6’S,10’R)-7’,8’-dehydrozearalenol 9, a diastereomer of 1. This required the functions of the remaining domains of Hpm8, as well as those of the partnering Hpm3, to be tolerant to the stereochemical inversion of the diketide intermediate. Each of the Hpm8 chimeras was co-expressed with Hpm3 in BJ5464-NpgA and metabolite extract was analyzed by LCMS. Consistent with in vitro results, chimeras Hpm8B5, B8 and B9 all produced 1 as observed with the wild type (Fig. 2e). In contrast, in the presence of Hpm8B2 or Hpm8B4, a new RAL-like compound with the same UV absorption pattern and molecular weight as 1 emerged (Supplementary Fig. 14). NMR analysis suggested it is a diastereomer of 1 (Supplementary Figs. 15–18 and Supplementary Table 2).
To verify the stereochemistry of the new compound at C6’ and C10’, it was hydrogenated to zearalanol 10 (SI Scheme 8). The circular dichroism spectrum of 10 was a mirror image of that of the well characterized (6’R, 10’S) α-zearalanol 1123. Therefore, the stereochemistry of C6’ and C10’ in 10 can be deduced to be S and R respectively, thus confirming the new compound to be 9. To exclude any possible epimerization activity of Hpm3, (7S, 11R, 2E, 8E)-7,11-dihydroxy-dodeca-2,8-dienyl-SNAC 12 that incorporated the stereochemistry of the putative precursor of 9 was synthesized as a starter unit for Hpm3. Addition of 12 to Hpm3 and malonyl-CoA produced 9 as the single product (Fig. 2e).
Interestingly, the in vivo synthesis of 1 and 9 by Hpm8B7 and Hpm3 established the minimally swapped region in KR to invert diketide stereospecificity to be α4–β5–α5–α6. This further supports the role of β5–α5–α6 as a critical determinant of KR stereospecificity. Although Hpm8B2 and Hpm8B4 reduced 2 into 2D with >95% enantioselectivity, compounds 1 and 9 were produced at comparable levels in vivo. This is probably due to the intrinsic preference of both HR- and NR-PKS for the natural, S-hydroxybutyryl intermediate. The selectivity of other domains in either HR-PKS or NR-PKS towards the natural diketide isomer therefore represents a potential barrier to the rational manipulation of stereocontrol in these systems.
In conclusion, we have shown that the Hpm8 KR possesses a previously unrecognized ability to reduce the β-ketoacyl intermediates with different stereochemical outcomes based on substrate chain length. Recently, fungal KR domains were proven to unexpectedly influence the chain length of the HR-PKS products24. Discovering these additional degrees of programming complexity in the tailoring domains represents important steps towards fully deciphering the programming rules of the enigmatic IPKSs.
Supplementary Material
Acknowledgement
This work was supported by NIH 1R01GM085128 to YT, the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chair in Bioorganic and Medicinal Chemistry to JCV. We thank Dr. Chris Reeves for the hpm genes.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Contributions
H. Z., Y. T. and J.C.V. conceived the idea and designed the study. Z. G. and H. Z. performed the syntheses of all the thioester substrates in this study. H.Z. designed and performed molecular cloning. H. Z., K. Q and J. W. performed the heterologous protein expression and purification as well as in vitro and in vivo characterization of the megasynthases. All authors analyzed and discussed the results. H. Z., Y. T. and J. C. V. prepared the manuscript.
REFERENCES
- 1.Weissman KJ, Leadlay PF. Nat. Rev. Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 2.Smith S, Tsai SC. Nat. Prod. Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox RJ. Org. Biomol. Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 4.Ma SM, et al. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier JL, Burkart MD. Chem. Soc. Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- 6.Oppermann U, et al. Chem. Biol. Interact. 2003;143–144:247–253. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 7.Siskos AP, et al. Chem. Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Baerga-Ortiz A, et al. Chem. Biol. 2006;13:277–285. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 9.O'Hare HM, Baerga-Ortiz A, Popovic B, Spencer JB, Leadlay PF. Chem. Biol. 2006;13:287–296. doi: 10.1016/j.chembiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Castonguay R, He W, Chen AY, Khosla C, Cane DE. J. Am. Chem. Soc. 2007;129:13758–13769. doi: 10.1021/ja0753290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castonguay R, et al. J. Am. Chem. Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. J. Am. Chem. Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffrey P. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 14.Reid R, et al. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 15.Keatinge-Clay AT, Stroud RM. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Keatinge-Clay AT. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structure. 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H, et al. J. Am. Chem. Soc. 2010;132:4530–4531. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves CD, Hu Z, Reid R, Kealey JT. Appl. Environ. Microbiol. 2008;74:5121–5129. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celmer WD. J. Am. Chem. Soc. 1965;87:1801. doi: 10.1021/ja01086a038. -&. [DOI] [PubMed] [Google Scholar]
- 21.Rossmann MG, Argos P. Annu. Rev. Biochem. 1981;50:497–532. doi: 10.1146/annurev.bi.50.070181.002433. [DOI] [PubMed] [Google Scholar]
- 22.Filling C, et al. J. Biol. Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 23.Hagler WM, et al. Appl. Environ. Microbiol. 1979;37:849–853. doi: 10.1128/aem.37.5.849-853.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisch KM, et al. J. Am. Chem. Soc. 133:16635–16641. doi: 10.1021/ja206914q. [DOI] [PubMed] [Google Scholar]
- 25.Maier T, Leibundgut M, Ban N. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.