Abstract
The p53 tumor suppressor is embedded in a large gene network controlling diverse cellular and organismal phenotypes. Multiple signaling pathways converge onto p53 activation, mostly by relieving the inhibitory effects of its repressors, MDM2 and MDM4. In turn, signals originating from increased p53 activity diverge into distinct effector pathways to deliver a specific cellular response to the activating stimuli. Much attention has been devoted to dissecting how the various input pathways trigger p53 activation and how the activity of the p53 protein itself can be modulated by a plethora of co-factors and post-translational modifications. In this review we will focus instead on the multiple configurations of the effector pathways. We will discuss how p53-generated signals are transmitted, amplified, resisted and eventually integrated by downstream gene circuits operating at the transcriptional, post-transcriptional and post-translational level. We will also discuss how context-dependent variations in these gene circuits define the cellular response to p53 activation and how they may impact the clinical efficacy of p53-based targeted therapies.
Keywords: gene network, PUMA, p21, apoptosis, cell cycle arrest, personalized medicine, functional genomics, molecular diagnostics
Introduction
As molecular biology evolved from the study of single molecules to the study of large biological systems, the meaning of the word p53 displayed a parallel increase in sophistication. When discovered more than 30 years ago, p53 was first a protein in a gel [1–3]. Early on, p53 was characterized as an oncogene, a confounding issue derived from the study of the mutant p53 variants expressed in tumors [4–9]. Years later, the wild type version of the p53 gene was unequivocally identified as a tumor suppressor [10–12]. Eventually, the p53 protein was characterized as a DNA-binding transcription factor [13–17]. With the discovery of MDM2, a repressor of p53 [18–20], and p21 (CDKN1A), a p53 target gene and effector [21, 22], a p53 pathway was born. With the identification of numerous upstream regulators and hundreds of target genes with different functions [23–26], one pathway became many (Figure 1). Today, the notion of a p53 pathway is obsolete: p53 is a gene network. An enormous amount of knowledge has been generated while studying the p53 gene, the p53 protein, and the various p53 pathways. What can we learn from studying p53 as a gene network?
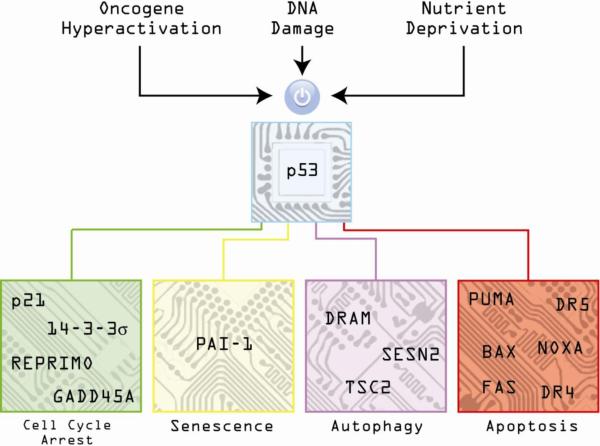
Figure 1. The p53 circuit board.
Activation of p53 by diverse stimuli such as oncogene hyperactivation, DNA damage and nutrient deprivation results in increased expression of numerous genes controlling different cellular outcomes such as cell cycle arrest, senescence, autophagy and apoptosis.
In this review, we will abandon all p53-centric views and consider p53 as mere “signal generator” within a vast gene network. We will deliberately neglect all information about the multiple events that affect the p53 molecule itself, such as dozens of post-translational modifications and hundreds of binding partners that regulate its activity. We refer those readers interested in the p53 molecule to many excellent p53-centered reviews [27–31]. Instead, in the next pages we will assume, for the sake of argument, that the p53 protein is one and the same in every context, and that its activity as a DNA binding protein and transcriptional regulator is invariant across scenarios. Having blinded ourselves to the outstanding complexity of the molecular events surrounding p53 activation, we will focus our attention on the downstream networks of genes that modulate p53-derived signals. Throughout this review we will employ an electrical engineering metaphor and it is important for us to define at this point how we will use the terms circuit, integrated circuit and circuit board.
In our metaphor, p53 is a switch turned to the `ON' position by p53 activating stimuli, thus transmitting signals to downstream circuitry. We define a circuit as the collection of factors co-regulating a given p53 target gene at the transcriptional, post-transcriptional and post-translational levels. In Section 1 we illustrate this for p21, a direct transcriptional target of p53 that mediates cell cycle arrest (Figure 2) [21, 22, 32]. An integrated circuit is defined as a group of p53 target genes that work together to drive a given cellular response. For example, the p21 and 14-3-3σ (SFN) circuits are part of an integrated circuit that mediates p53-dependent cell cycle arrest, whereas PUMA (p53-upregulated modulator of apoptosis, BBC3) and NOXA (PMA-induced protein 1, PMAIP1) are two among many circuits mediating p53-dependent apoptosis. Finally, a circuit board represents the assembly of several diverse integrated circuits, which ultimately coalesces all of the various signals into a single cellular outcome (Figure 3). In Section 2 we assemble a minimal circuit board composed of p21, 14-3-3σ, PUMA and NOXA, which includes their various gene-specific co-regulators acting at the transcriptional, post-transcriptional and post-translational levels. In Section 3 we describe how p53 circuit boards can be assembled into cell type- and stimulus-specific configurations that define context-dependent p53 responses. We discuss at this point the impact of the p53 family members p63 and p73 on these differential assemblies. Finally, in Section 4 we discuss the biomedical importance of a gene network (i.e. circuit board) approach, with an emphasis on a new generation of p53-based targeted therapies whose clinical worth is limited by the highly pleiotropic character of p53.
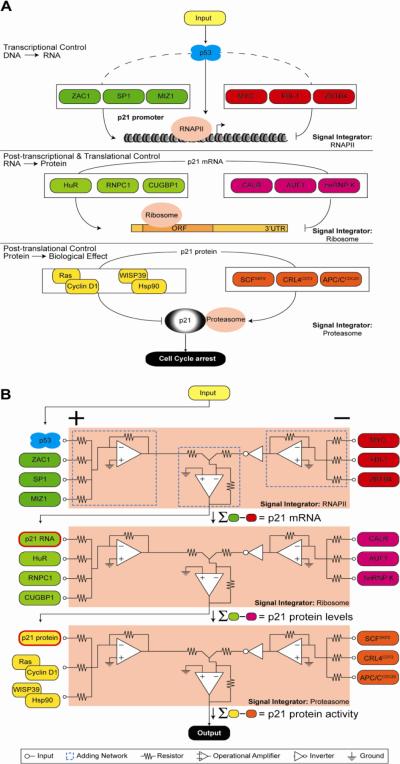
Figure 2. The p21 circuit.
The p53 target gene p21 is co-regulated by various factors acting at the transcriptional, post-transcriptional and post-translational levels, all of which can potentially affect p53/p21-dependent cell cycle arrest. In both A and B, from top to bottom, transcriptional control of p21 gene expression merges the action of positive and negative transcriptional regulators into a single output, functional mRNA. In this context, p53 is just one among many regulators feeding into RNA polymerase II (RNAPII), the signal integrator at this step, which synthesizes p21 mRNA, the input for the next level of regulation. From there, signals from factors that positively and negatively influence RNA stability and translation are combined via the ribosome signal integrator to achieve protein synthesis. Finally, the levels of active p21 protein in the cell are fine-tuned by stabilizing and destabilizing signals resolved by the proteasome. In A, the regulatory layers are displayed as cartoons of DNA, RNA and protein molecules. In B, the same processes are depicted as an electrical circuit comprising three signal integrators. At the transcriptional level, activating signals including p53 (green) are applied to the positive terminals of the RNAPII signal integrator (+) and are summed internally by an adding network (dashed blue box); inhibitory signals (red, negative terminals) are combined similarly. The difference in total activating and total inhibitory signals determines output amplitude, in this case p21 mRNA level. Integrator output can subsequently serve as input to other signal processing machinery; in this example, p21 mRNA levels become positive inputs for p21 protein production in the ribosome.
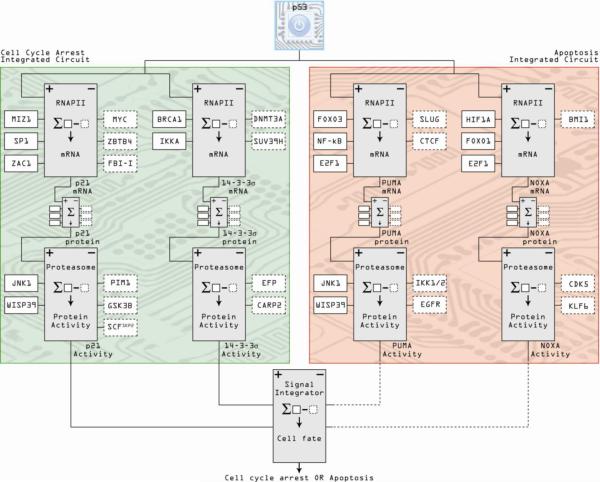
Figure 3. Integrated circuits of cell cycle arrest and apoptosis comprise a minimal p53 circuit board.
Following a similar schema to Figure 2, co-regulators of individual p53-target genes acting at the transcriptional, post-transcriptional and post-translational levels are integrated into circuits. Individual genes contributing to a particular cellular outcome following p53 activation are then combined into integrated circuits, represented here by the cell cycle arrest and apoptosis integrated circuits composed of p21/14-3-3σ and PUMA/NOXA, respectively. The two integrated circuits are then assembled into the p53 circuit board, which ultimately consolidates all positive and negative signals to define one cell fate. Positive regulators of gene activity are denoted by solid boxes, dashed boxes indicate negative regulators.
Section 1. Layered regulation of p53-derived signals: the p21 circuit
p21 is a key mediator of p53-dependent cell cycle arrest by acting as a potent inhibitor of cyclin-dependent kinases [21, 22, 32]; however, p53 is only one among dozens of regulators of p21 activity in the cell. p21 is exquisitely regulated at various stages, from transcription to protein degradation [33], and these regulatory events tune p53-dependent cell cycle arrest. Here, we will assemble a small fraction of the known regulators of p21 into a circuit (Figure 2).
1.1. Transcriptional co-regulation of p21
p53 is but one of many well documented transcriptional regulators of the p21 locus [33]. Positive and negative regulation by these other factors dictate the extent to which p53 transactivates p21. For example, MIZ1 (MYC interacting zinc-finger protein-1) [34] and SP1 (specificity protein 1) [35, 36] have been shown to regulate p21 in p53-dependent and/or -independent manners. Recently, ZAC1 (zinc-finger protein which regulates apoptosis and cell cycle arrest) was shown to synergistically transactivate p21 through a direct interaction with SP1 and through a functional interaction with p53 [37], demonstrating the hierarchical action of three p21 transactivators. In contrast, p53-dependent p21 induction is subject to myriad antagonistic factors. For example, MYC represses p21 activation in response to multiple forms of DNA damage via its interaction with MIZ1 [34, 38]. ZBTB4 (zinc-finger and BTB domain containing 4) is a member of the POZ domain-containing family of transcriptional repressors that can also inhibit p21 transcription through interaction with MIZ1 [39]. Other POZ repressors including FBI-1, ZBTB2 and ZBTB5, also regulate p21 transactivation through direct competition with p53 and/or SP1 [40–42]. These examples demonstrate the potential for regulatory complexity that exists at just one step of gene regulation. Integration of the effects from many transcription factors is required to fine-tune the amount of p21 mRNA produced, which then functions as the input for the next step in the circuit, regulation of RNA stability and translational control.
1.2. Post-transcriptional co-regulation of p21
After transcription, splicing and 3' end formation, mature mRNAs are regulated by numerous factors that influence their stability, localization and access to ribosomes for active translation. Viewed as a coordinated regulatory module, these events consolidate multiple signals into a single output: functional protein (Figure 2). Increased RNA stability plays a major role in facilitating p21 protein upregulation in response to DNA damage via the binding of HuR (ELAVL1) to an AU-rich element (ARE) in the p21 3' untranslated region (UTR) [43, 44]. RNPC1 (RBM38, RNA binding motif 38) is a direct transcriptional target of p53 which also binds to an ARE in the p21 3' UTR to stabilize it under DNA damaging conditions, thus creating a link between two adjacent levels of the circuit [45]. In fact, HuR and RNPC1 act cooperatively to bind the 3' UTR and increase the stability of the p21 mRNA [46]. In contrast, AUF1 (AU-rich binding protein 1, hnRNP D) competes with HuR to regulate p21 mRNA stability antagonistically. In an intriguing set of experiments, Lal et al. demonstrated that while AUF1 and HuR can bind to adjacent sites in the p21 3' UTR to promote nuclear export, in the absence of HuR, AUF1 shifts its binding position to promote destabilization of the mRNA [47]. In neuronal cells, hnRNP K acts in concert with HuB (ELAVL2) to block translation of p21 mRNA, an interesting finding in light of the fact that hnRNP K also positively regulates p53-dependent transcription of p21 [48, 49]. Therefore, hnRNP K is an example of a factor that can regulate expression of p53 target genes at multiple sites within the circuit. Elements in the 5' coding region of the p21 mRNA are also subject to regulation, as CUGBP1 and calreticulin (CALR) compete for binding to CG-rich elements to increase or decrease translation, respectively, in an interplay shown to affect differentiation and senescence [50, 51]. Thus, post-transcriptional regulation by competing RNA binding factors significantly influence p21 protein levels. Interestingly, p53 activates the expression of p21 both directly, at the transcription level, and indirectly, by regulating factors that increase mRNA stability. The ultimate signal integrator of post-transcriptional events is the ribosome, which synthesizes protein that can in turn be post-translationally regulated.
1.3. Post-translational co-regulation of p21
p21 is subject to a variety of post-translational modifications that alter its activity, subcellular localization and/or stability (Figure 2). The p21 protein can be phosphorylated on at least seven Ser/Thr residues with variable effects on localization and stability. For example, AKT/PKB can phosphorylate p21 on Thr145 or Ser146. This is an intriguing case because AKT phosphorylation at Thr145 was shown to increase p21 stability and block its association with PCNA, promoting cell cycle arrest, whereas the Ser146 modification activates a Cyclin D1/CDK4 complex to drive proliferation [52]. Additionally, phosphorylation of Thr145 by the oncogenic PIM1 kinase was shown to alter p21 localization, sequestering it in the cytoplasm where it is not competent to block cell cycle progression [53]. Furthermore, p21 protein stability can be altered both positively and negatively by p38α/JNK1 phosphorylation of Ser130 and GSK3β phosphorylation of Ser114, respectively [54, 55]. These examples represent only a small subset of the kinases and phosphorylation sites that regulate p21, but they surely serve to demonstrate how a single type of post-translational modification can modulate p21 activity well after p53-dependent transactivation.
The final layer of regulation in this circuit comes at the level of proteasomal degradation. Ubiquitin-mediated degradation of p21 occurs at different phases of the cell cycle, directed by different E3 ligases including SCFSKP2, APC/CCdc20 and CRL4CDT2 [56–58]. It has also been reported that p21 can be degraded by the proteasome independent of its ubiquitination status [59, 60]. p21 can be stabilized without interceding post-translational modifications through the formation of complexes with Cyclin D1, mediated by RAS overexpression, or by binding to WISP39/HSP90, both of which prevent the association of p21 with the proteasome [61, 62]. This last layer of regulation can amplify or dampen the signal initiated by active p53 to determine the level and activity of p21 in the cell, as well as its final effect on the cell cycle.
Section 2. Assembling circuits into integrated circuits and circuit boards: the minimal case of p21 and 14-3-3σ versus PUMA and NOXA
As illustrated for p21, the regulation of individual p53 target genes is a complex process analogous to a circuit. Determination of the ultimate biological outcome following p53 activation owes to the convergence of these regulatory processes on numerous target genes. The groups of genes that work together to affect a given phenotypic outcome are akin to an integrated circuit where numerous circuits operate in parallel. In turn, integrated circuits assemble into circuit boards governing the overall cellular response to a specific stimulus. In the next few paragraphs we will assemble a minimal circuit board composed of the manifold gene-specific transcriptional, post-transcriptional and post-translational regulators affecting four p53 target genes: p21, 14-3-3σ, PUMA and NOXA (Figure 3).
2.1. Gene-specific transcriptional regulation of p21, 14-3-3σ, PUMA and NOXA
Cell cycle arrest in response to p53 activation is mediated by two main proteins: p21, which arrests cells mainly in the G1 phase of the cell cycle, and 14-3-3σ, which arrests cells at the G2/M transition [63]. 14-3-3σ is a direct transcriptional target of p53 that binds to the mitotic phosphatase CDC25 and sequesters it in the cytoplasm, thus preventing it from activating CDK1-cyclin B complexes [63, 64]. p21 and 14-3-3σ work coordinately to enforce cell cycle arrest in response to various DNA damaging agents and protect from apoptosis [65, 66]. Relatively little is known about additional transcriptional coregulators of 14-3-3σ; however, BRCA1 has been shown to increase 14-3-3σ mRNA levels in a p53-dependent fashion [67]. In mouse ES cells, BRCA1 is required for full expression of 14-3-3σ, a finding which translated to several p53 wild-type human cancer cell lines where overexpression of BRCA1 resulted in upregulation of 14-3-3σ [67]. Negative regulation of 14-3-3σ transcription by promoter DNA hypermethylation occurs in many normal tissues and is widespread in human cancers, but the mechanisms by which it is established remain largely elusive [68–70]. Of note, the IKKα kinase shields 14-3-3σ from silencing in keratinocytes by binding to the promoter and blocking the action of the histone- and DNA-methyl-transferases SUV39H1 and DNMT3A, respectively [71].
The apoptotic module of the p53 response involves numerous factors, including the BCL2-homology domain 3 (BH3)-only proteins PUMA and NOXA, which are both p53 target genes that inhibit pro-survival BCL2 family members such as BCL2, BCL2L1 (BCL-xL) and MCL1, thus activating the intrinsic apoptotic pathway [72–75]. PUMA is also known to directly activate BAX, the pore-forming protein required for permeabilization of the outer mitochondrial membrane [76]. Each of these genes is subject to additional regulation at the transcriptional level. PUMA, for example, can be directly activated by the FOXO3 (Forkhead box O3a) transcription factor and may be repressed by MYC [77, 78]. FOXO3-dependent induction of PUMA synergizes with transactivation-independent p53 activity to induce apoptosis [78, 79]. PUMA is also induced by the transcription factor IRF1 as well as by NF-κB in response to TNF-α (tumor necrosis factor-α) [80, 81]. In some cell types, the p53 target SLUG (SNAI2) directly represses PUMA transcription in response to ionizing radiation [82]. Finally, we have demonstrated that PUMA is subject to a novel mode of gene-specific transcriptional repression whereby the insulator protein CTCF (CCCTC-binding factor) establishes a repressive chromatin boundary within the coding region of the gene [83].
A large cohort of factors similarly regulates NOXA transcription. NOXA is transactivated under hypoxic conditions by HIF1A (Hypoxia inducible factor-1 alpha) independently of p53 [84]. Proteasome inhibition and cytotoxic stimuli both result in NOXA upregulation mediated by MYC and FOXO1 (Forkhead box O1), respectively [85, 86]. Conversely, transcriptional repression of NOXA has been demonstrated in several instances, including silencing by the polycomb group protein BMI1 and by glucocorticoids in acute lymphoblastic leukemia (ALL) cells [87, 88]. E2F1 is a particularly interesting component in the apoptotic integrated circuit as it can bind to and activate the promoters of PUMA, NOXA and several other BH3-only genes [89]. The integration of these complex and diverse regulatory signals generates a set of mature mRNAs that serve as input for the post-transcriptional and post-translational stages of the circuit board.
2.2. Gene-specific regulation of p21, 14-3-3σ, PUMA and NOXA at the post-transcriptional and post-translational levels
In addition to p21, the cell cycle arrest integrated circuit is modulated by post-transcriptional regulators of 14-3-3σ (Figure 3). 14-3-3σ protein levels are regulated by several E3-ubiquitin ligases. EFP (estrogen-responsive finger protein, TRIM25), a protein commonly overexpressed in breast cancers, specifically targets 14-3-3σ for proteolysis and, consistent with this function, knockdown of EFP reduces tumor growth in vivo [90]. CARP2 (caspase 8/10 associated RING protein 2) has also been implicated in proteasomal degradation of 14-3-3σ, although the exact mechanism of CARP2 action remains to be elucidated [91].
The apopoptic integrated circuit includes post-transcriptional regulators of PUMA and NOXA. A number of microRNAs, including miR-125b, -221/222 and -296, have been shown to inhibit the production of PUMA protein in various cell types. miR-125b promotes tumor growth in vivo by targeting several pro-apoptotic mRNAs, including the 3' UTR of the PUMA transcript [92]. miR-221/222 have been shown to down-regulate PUMA expression in both glioblastoma and epithelial cancers [93, 94]. The IKK1/IKK2/NEMO kinase complex was found to trigger phosphorylation of PUMA protein at Ser10 to promote its proteasomal degradation, a mechanism that can be initiated by inputs that compete with p53 activation such as cytokine signaling [95, 96]. PUMA activity is also regulated at the level of subcellular localization. In glioblastoma cells, EGFR (epidermal growth factor receptor) binds directly to PUMA and sequesters it in the cytoplasm, preventing its activity at the mitochondria [97]. Regulation of NOXA protein translation is less well understood, although cytoplasmic sequestration has been reported as a mechanism to reduce its activity at the mitochondrial membrane [98]. In hematopoietic cells, NOXA is constitutively expressed and phosphorylated on Ser13 by CDK5, resulting in its cytoplasmic localization and inhibition. Glucose deprivation removes this mark, activating the apoptotic potential of NOXA [98]. NOXA protein is also subject to proteasomal degradation. In particular, the Kruppel-like tumor suppressor, KLF6-SV1 (splice variant 1), has been shown to promote NOXA turnover using a mechanism that involves the E3-ubiquitin ligase activity of MDM2, thereby promoting cancer progression and chemoresistance in vivo [99].
Clearly, the examples highlighted so far are a minimal representation of the p53 circuit board. If more p53 target genes were included in the analysis, the complexity would become staggering. Instead, we will leave the circuit board in this minimal form and explore how its differential assembly in different contexts modulates the cellular response to p53 activation.
Section 3. Context-dependent configurations of the p53 circuit board
Context is the hallmark of biological processes. The biological response observed upon activation of any node within a gene network is affected by the contextual connectivity of said node. The ON-OFF switch of a food blender operates in basically the same way as the ON-OFF switch of a bedside lamp. The outcome of flipping the switch to the ON position is not defined by the switch itself, but rather by the downstream circuitry and machinery. Such context-dependent outcomes are readily apparent in the p53 network and can be reasonably resolved into two major types, cell type-specific and stimulus-specific. The same p53 activating stimulus triggers starkly different responses across cell types. A single cell type undergoes very different p53-dependent outcomes in response to distinct p53-activating stimuli. In this section, examples of context-specific assemblies of the p53 circuit board will be discussed, with careful attention paid to how p53-dependent and p53-independent events are integrated to produce specific outcomes.
3.1. Cell type-specific configurations of the p53 circuit board
The two hundred or so differentiated cell types in the adult human body are genetically identical, yet the fraction of their genomes that is expressed is clearly different. Although p53 is ubiquitously expressed across tissues, its surrounding network is massively different from one cell type to another. As epigenetic mechanisms silence much of the genome in a cell type-specific fashion, the availability of p53 target genes and their co-regulators varies immensely, and so does the cellular response to p53 activation.
A clear example of this pleiotropy is provided by elegant mouse studies where the wild type p53 gene was replaced by p53ERTAM, a 4-hydroxy-tamoxifen (4-OHT)-dependent variant of p53 [100, 101]. Tissues and cells derived from these knock-in mice can be toggled between p53-deficient and p53-proficient states by systemic administration and withdrawal of 4-OHT. In the presence of MDM2, addition of 4-OHT does not suffice to trigger a p53 response, which requires additional activating stimuli such as DNA damage or oncogene hyperactivation [101]. However, in p53ERTAM Mdm2−/− mice, 4-OHT elicits full p53 activation [100]. In this scenario, activation of p53ERTAM triggered efficient apoptosis in all classically radiosensitive tissues such as bone marrow, thymus, spleen white pulp and small and large intestine, which led to their rapid atrophy [100]. In contrast, classically radio-insensitive tissues like lung, heart, brain, liver and kidney remained phenotypically normal. In the case of testis, although apoptosis was not observed, atrophy was still obvious, which was seemingly caused by a profound proliferation arrest. Importantly, the authors confirmed that p21 and Puma mRNAs were effectively induced in all tissues regardless of outcome. Thus, although the switch was flipped to the ON position in every tissue and specific circuits within the circuit board were available and activated in every cell type tested (i.e. p21 and PUMA), the final output was markedly different in each cell type, which could only be explained by as yet undefined differences in the rest of the circuit board.
Although p21 is a key mediator of p53-dependent cell cycle arrest, its loss does not completely abrogate the ability of p53 to halt proliferation, which demonstrates the existence of redundant and cooperative pathways [102, 103]. 14-3-3σ, GADD45A and REPRIMO (RPRM) are additional p53 target genes which have been proposed to collaborate with p21, mainly to deliver G2 arrest in specific cell types [63, 66, 104, 105]. Another mediator of proliferation arrest is the p53-inducible microRNA miR-34a, which is a direct transcriptional target of p53. miR-34a over-expression induces arrest of various cell types in the presence or absence of p21, whereas reducing miR-34a activity compromises the arrest response [106–109]. Is the cell cycle arrest integrated circuit assembled in a cell type-specific fashion? Indeed. 14-3-3σ was first identified as an epithelial cell antigen exclusively expressed in epithelia, and it is silenced via DNA methylation in many normal tissues [68]. In the context of cancer, 14-3-3σ, REPRIMO and miR-34a are common targets of aberrant silencing via DNA methylation, such that their availability varies across cancer cell types [69, 70, 110–117]. Furthermore, enhanced turnover of the p21 mRNA and impaired processing of the primary transcript for miR-34a prevent their accumulation upon p53 activation in some cell types [118].
Cell type-specific configurations of the apoptotic circuit are also obvious. p53 transactivates genes in various apoptotic pathways, including components of the intrinsic (mitochondrial) pathway such as PUMA, NOXA, BID, BAX and APAF1 [72, 73, 75, 119–121], as well as members of the extrinsic (death receptor) pathway such as FAS, DR4 and DR5/Killer [122–124]. The specific contribution of these target genes to p53-dependent apoptosis varies greatly across tissues. For example, in the thymus and developing central nervous system of mice, apoptosis induced by ionizing radiation requires p53 and PUMA, but not BAX [125]. In mice where either PUMA or NOXA were disrupted, both factors contributed to DNA damage-induced apoptosis in fibroblasts, but only loss of PUMA protected lymphocytes from cell death [126]. Careful analysis of double knockout Puma−/−, Dr5−/− mice demonstrated an unexpected interdependency of the intrinsic and extrinsic pathways for the execution of p53-dependent apoptosis in some, but not all tissues [127]. In this comparative study, apoptosis in vivo following sub-lethal whole-body IR was almost exclusively p53-dependent in the bone marrow, spleen, thymus and GI tract. Both Dr5 and Puma contributed significantly to cell death in the spleen and thymus, yet Puma was the main contributor to cell death in the GI-tract and bone marrow.
Expectedly, pro-apoptotic factors tend to be lost or inactivated during tumor development, including various p53 targets, which then leads to multiple possible assemblies of the p53 apoptotic integrated circuit across cancer cell types. For example, PUMA is often silenced in lymphomas independently of p53 mutations [128]. In small cell lung carcinomas, various combinations of p53 target genes in the extrinsic pathway are often silenced via DNA methylation, including FAS, DR4 and caspase 8 [129]. A recent genomics study of somatic copy number alterations in 680 tumors representing 17 major cancer types revealed multiple possible assemblies of the apoptotic circuit as defined by deletions of PUMA and the BH3-only protein BOK and copy number gain for MCL1 and BCL-xL [130].
Taken together, these observations reveal a great diversity in the assembly of the p53 circuit board across both normal and cancerous cell types.
3.2. Stimulus-specific configurations of the p53 circuit board
In addition to the cell type-specific configurations of the p53 network, a given cell type can also adopt alternative p53-dependent cell fates in a stimulus-specific manner. There are disappointingly few publications wherein distinct stimuli that elicit different p53-dependent responses have been compared in a single cell type in an attempt to understand how alternative cell fates are established.
p21 has provided an excellent paradigm to understand stimulus-specific regulation within the p53 network. Not all p53-activating agents cause p21 upregulation. p53 activation by Nutlin-3, 5-fluorouracil (5-FU), doxorubicin, daunorubicin and ionizing radiation (IR) results in effective accumulation of the p21 protein in diverse cancer cell types, whereas p53 activation by Ultraviolet Light C (UVC) or hydroxyurea (HU) does not [131–135]. Interestingly, all these agents produce equivalent accumulation of p53, effective binding of p53 to the enhancers in the p21 promoter and p53-dependent recruitment of specific p53 cofactors such as histone acetyl-transferases and subunits of the core Mediator complex [131–134]. However, in the case of UVC, after a transient wave of early transcription, the transcriptional apparatus at the p21 promoter is partially disassembled, as defined by loss of: i) the CDK8-module of the Mediator complex, ii) the general transcription factors TFIIF and TFIIB, iii) transcription elongation factors and iv) elongating RNA polymerase II (RNAPII) [132, 133]. Interestingly, the TAF1 protein, a subunit of the TFIID general transcription factor known to downregulate p21 expression, is strongly recruited to the p21 promoter after UVC [132–134]. In the case of HU, p21 inactivation occurs not at early transcriptional events, but rather at late elongation steps [131, 135]. During the S-phase checkpoint triggered by HU, p53 activation leads to efficient recruitment and assembly of the transcriptional apparatus and RNAPII escape from the p21 promoter, but RNAPII fails to complete elongation throughout the p21 coding region. Interestingly, this block in late elongation is relieved by genetic or pharmacological ablation of the stress-induced kinase CHK1 [131]. Thus, parallel signals created by UVC and HU can impact the transcriptional machinery acting at the p21 promoter and intragenic region, respectively, to block the activating signal generated by p53 at the enhancers. Importantly, these effects are gene-specific, as other p53 target genes do not display such stimulus-specific regulation [131–135].
The p53 apoptotic circuit can also be assembled in a stimulus-specific fashion. In response to DNA damage, mouse embryonic fibroblasts (MEFs) undergo p53-dependent G1 arrest. However, when MEFs are transformed with the adenoviral E1A oncoprotein, the same stimulus leads to p53-dependent apoptosis. Using this cellular system and a subtractive cloning strategy, the Jacks group identified PERP as an “apoptosis-specific gene” that was expressed in a p53-dependent manner at much higher levels in the apoptotic setting [136]. PERP is a membrane protein whose overexpression is sufficient to cause cell death in various cell types [137, 138]. Strikingly, several other p53 targets in the arrest and apoptosis integrated circuits were equally activated in each scenario, including p21, BAX and DR5. Curiously, the p53 target gene IGFBP3 was expressed only in arresting cells. In order to understand how the proliferative signals generated by E1A resulted in differential expression of PERP, the authors showed that E2F1 overexpression activates PERP, but only in the presence of p53. Thus, oncogenic transformation creates an extra signal that enables the activation of an additional apoptotic p53 target.
3.3. Cell type- and stimulus-specific action of the p53 family members p63 and p73
The p53 family of transcription factors includes p63 and p73 [139–141], which share with p53 an N-terminal transactivation domain (TA), a highly conserved DNA binding domain (DBD), and an oligomerization domain (OD) (reviewed in [142], Figure 4). However, p63 and p73 diverge from p53 at their C-termini, which contain a sterile active motif (SAM) domain and a transcription inhibition domain (TID) [143, 144]. p63 and p73 are subject to alternative promoter usage and alternate splicing, which creates a myriad number of isoforms varying in their N-termini and C-termini, respectively. Of note, several p53 isoforms have also been identified, whose functions remain enigmatic. We direct the reader to an excellent review by Marcel et al. on the current state of knowledge related to the p53 isoforms [145].
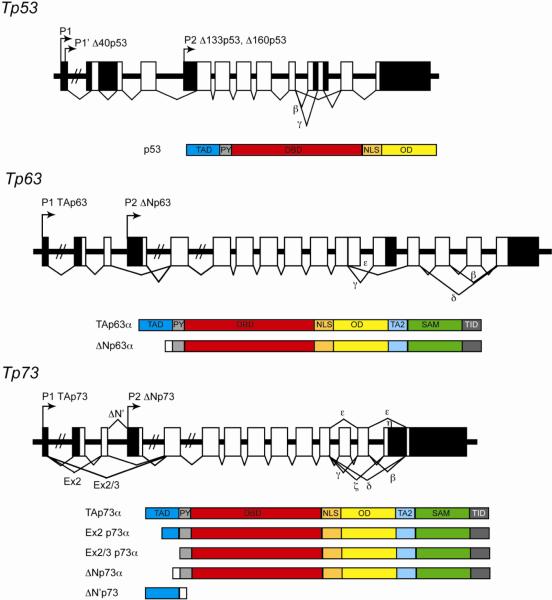
Figure 4. The p53 family of transcription factors.
Schematic of the gene architecture and well characterized isoforms of p53 and the closely related factors p63 and p73. Arrows represent alternative promoters, boxes represent exons (black segments are untranslated regions). TAD: transactivaiton domain. TA2: second transactivation domain. PY: proline rich domain. DBD: DNA binding domain. NLS: nuclear localization signal. OD: oligomerization domain. SAM: sterile-alpha motif. TID: trans-inhibitory domain.
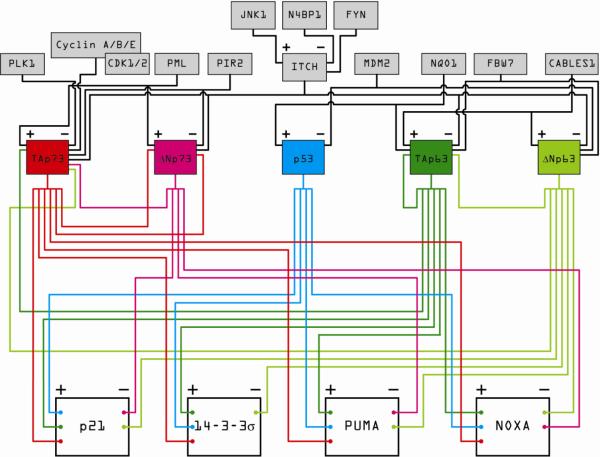
As the DNA binding domains of both p63 and p73 exhibit high identity with the p53 DBD [140], the consensus binding sites for all three factors are virtually indistinguishable from one another [146–148]. However, the fact that the various isoforms contain different transcription activation and repression domains creates a unique opportunity to positively or negatively regulate gene expression in a combinatorial fashion. In general, the TAp63 and TAp73 isoforms are positive transcriptional regulators within the p53 network, and the ΔNp63 and ΔNp73 isoforms have been described mostly as negative regulators. Unlike p53, the p63 and p73 isoforms are not ubiquitously expressed and show instead exquisite tissue-specific patterns of expression, which then contributes to cell type-specific assemblies of the p53 circuit board.
3.3.1. Cousins in different places
The TA and ΔN isoforms of p63 clearly affect the p53 circuit board in a cell type-specific manner. TAp63 has been shown to transactivate the cell cycle arrest genes p21 and 14-3-3σ [149–151] as well as the pro-apoptotic genes PUMA and NOXA [149, 150, 152] (Figure 5). Conversely, ΔNp63α has been reported as a transcriptional repressor of p21, 14-3-3σ, PUMA and NOXA [152–157]. TAp63 is highly expressed in germ cells of the ovary and testis [158], and is essential for DNA damage-induced oocyte death [159], thereby protecting the germline genome in a p53-independent manner. Furthermore, TAp63 is essential for RAS-induced senescence of fibroblasts, and therefore may serve as a p53-independent tumor suppressor in mesenchymal tissues [160]. In contrast, ΔNp63 is a potent pro-proliferative factor that is highly expressed in the basal cells of all stratified epithelia, including skin, cervix, vaginal epithelium, urothelium and prostate [140]. The role of ΔNp63 in epithelial stem cell maintenance and proliferation is well established, due to a number of insightful studies performed using p63 transgenic mice. Mice lacking all p63 isoforms (p63−/−) exhibit profound developmental defects, lacking all stratified epithelial tissues, hair follicles, teeth, mammary, lachrymal and salivary glands and limbs [139, 161]. Genetic complementation of p63−/− mice with either ΔNp63 or TAp63 demonstrate that there are unique roles for the p63 isoforms in regulating epithelial development; ΔNp63 is required for epidermal stem cell proliferation, whereas TAp63 may contribute to the differentiation of suprabasal keratinocytes and formation of the mature pluristratified epithelium [162]. Thus, while TAp63 feeds activating signals into the p53 circuit board in germ cells, mesenchymal tissue and suprabasal keratinocytes, triggering differentiation, cell cycle arrest, senescence and/or apoptosis depending on the context, ΔNp63 attenuates signaling within the circuit board to allow the continued proliferation of stem cells in the epithelium.
Figure 5. The impact of p63 and p73 isoforms on the p53 circuit board.
This wiring diagram illustrates the ability of various p53 family members to co-regulate, both positively and negatively, canonical p53 target genes involved in cell cycle arrest and apoptosis. Different isoforms of p63 and p73 are expressed in diverse cell types, thus providing cell type-specific regulatory capacity. Furthermore, p63 and p73 isoforms are positively and negatively regulated by diverse factors that do not affect p53 directly (gray boxes on top), thus providing additional opportunities for stimulus-specific regulation within the circuit board.
Somewhat less is known about the cell type-specific action of the p73 isoforms. Like p53 and TAp63, TAp73 can increase the transcription of p21, 14-3-3σ, PUMA and NOXA [152, 156, 160, 163–165] thereby activating the p53 circuit board in response to DNA damage and cell stress. Conversely, ΔNp73 has been shown to repress the transcription of p21, PUMA, and NOXA [166–168], and may directly bind to and inactivate p53, TAp63 and/or TAp73 [169], thereby serving to dampen signaling in the circuit board. Initial characterization of the p73−/− mouse determined that the loss of all p73 isoforms precipitated profound neurological defects [170]. Furthermore, two independently generated ΔNp73 knockout mouse strains show signs of neurodegeneration and brain atrophy [171, 172], indicating that ΔNp73 plays a crucial role in regulating neuronal survival. Indeed, TAp73 and ΔNp73 have been shown to antagonize one another in neural tissue. TAp73 stimulates neuronal apoptosis through the p53-independent activation of PUMA expression, a phenomenon which may be suppressed by the exogenous expression of ΔNp73 [152]. ΔNp73 can also inhibit the pro-apoptotic effects of NGF (nerve growth factor) withdrawal, thereby mediating neuronal survival through p53-dependent and –independent mechanisms [173]. As ΔNp73 is the predominant isoform expressed in sympathetic neurons [173] and in numerous parts of the brain [172], this demonstrates a cell type-specific role for ΔNp73 in promoting neuronal survival. In comparison, studies using the TAp73−/− mouse demonstrate its role as a bona fide tumor suppressor and in maintaining genomic stability in a broad range of tissues [174]. Thus, tissue-specific expression of the TA and ΔN isoforms of p73 allow for the exquisite regulation of their functions in the p53 network in a cell type-specific manner.
3.3.2. Cousins with different lifestyles
p63 and p73 isoforms are regulated by signaling pathways which do not affect p53 directly, thus creating additional regulatory diversity for the stimulus-specific assembly of the p53 circuit board. Unlike for p53, MDM2 does not appear to function as an E3 ubiquitin ligase for p63 or p73. Instead, the E3 ubiquitin ligase ITCH has been shown to target the TA and ΔN isoforms of both p63 and p73 for proteasomal degradation [175, 176]. ITCH itself may be positively modulated by phosphorylation of Ser199, Thr222 and Ser232 by JNK1 [177], negatively modulated by phosphorylation of Tyr371 by the Src kinase FYN [178] or by association with the competitive inhibitor N4BP1 (Nedd4-binding partner 1) [179], thereby creating unique opportunities for stimulus-specific regulation of p63 and p73 levels in the cell. Other E3 ubiquitin ligases targeting p63 and p73 for degradation are SCFFbw7 [180] and PIR2 [181], respectively, which add other entry points for regulation at the protein turnover level. On the other hand, CABLES1 (CDK 5 and Abl enzyme substrate 1) has been shown to protect TAp63 and ΔNp63 from proteasomal degradation and CABLES1 expression is required for maximal stabilization of TAp63 and subsequent induction of apoptosis in female germ cells following DNA damage [182]. Similarly, NQO1 (NAD(P)H quinone oxidoreductase) and the PML (promyelocytic leukemia) protein have been shown to protect TAp63 and TA/ΔNp73 from proteasomal degradation, respectively [183, 184]. Finally, phosphorylation of TAp73 by PLK1 (polo-like kinase 1) [185] or various CDKs [186] may inhibit p73's transactivation abilities. These examples provide mechanisms by which specific factors outside of the p53 network may modulate the expression, stability, and activation of the p53 family members in a cell-type and/or stimulus-specific manner, thereby providing additional avenues for input into the regulation of the p53 circuit board.
Section 4. Clinical relevance of understanding p53 as a circuit board
4.1. p53-based targeted therapies: promise and obstacles
As the paradigm for cancer therapy shifts from blunt cocktails of genotoxic agents, antimetabolites and mitotic poisons to biologically targeted therapies, the development of non-genotoxic p53-based therapies is gaining momentum. The validity of p53 as a therapeutic target has been elegantly proven in animal models where specific activation of p53 in tumors leads to their demise via apoptosis or senescence [187, 188]. Virtually every tumor expresses p53. In about half of these tumors p53 has been inactivated by a single point mutation that disrupts its tumor suppression function and, in some cases, confers upon it oncogenic properties (reviewed in [189]). Since mutant p53 can not transactivate MDM2, these tumors express large amounts of the former. In the other half of tumors wild type p53 is expressed at low levels and kept in check by hyperactivation of its repressors MDM2 and/or MDM4. Therefore, two p53-based targeted therapies can be envisioned: i) small molecules that convert (even if partially) mutant p53 back to the wild type form, and ii) small molecule inhibitors of MDM2 and/or MDM4. Approximately 11 million patients worldwide would benefit from each type of therapy [190]. It is hard to envision a more widely useful type of targeted therapy - perhaps only inhibitors of the RAS oncogene would match this potential. Currently, both classes of p53-based therapies are being tested in the clinic.
The first molecules targeting p53 activity with any degree of specificity were described in the late 1990's and early 2000's. CP-31398 was discovered by screening small molecules for the ability to restore conformational stability to p53 using antibodies specific for the active and denatured forms of p53. CP-31398 was shown to rescue the ability of mutant p53 to transactivate a p53 reporter and inhibit tumor growth in vivo [191]. A subsequent screen for compounds to reactivate mutant p53 identified PRIMA-1 (p53 reactivation and induction of massive apoptosis) [192]. This small molecule was found to restore p53 activity in cells with mutant p53 and suppress tumor growth in a mouse xenograft model. The first small molecules inhibitors of the MDM2-p53 interaction, Nutlins, were described in 2004 [193]. Nutlin-3 mimics three hydrophobic residues on p53 required for MDM2 binding, thus working as a competitive inhibitor of the interaction [193]. Additional drugs in this group include another MDM2 inhibitor, MI-219, and RITA, which binds instead to p53 to disrupt the p53-MDM2 interaction [194, 195]. Nutlin-3 was shown to bind MDM2 with nanomolar efficiency, activate p53 and suppress tumor growth, all without inducing the cytotoxic side effects associated with traditional chemotherapeutics [193]. Unfortunately, it quickly became clear that in a majority of cancer cell lines, the effects of Nutlin-3 treatment were cytostatic rather than apoptotic [196], limiting its therapeutic efficacy.
If we agree that p53 is a mere signal generator upstream of a vast gene circuit controlling starkly diverse cellular responses, it is then obvious to anticipate that p53-based targeted therapeutics will induce tumor regression in a small fraction of patients. It is fair to assume that these successful cases will be due to rapid commitment to apoptosis or establishment of senescence within the tumors. Cell cycle arrest and autophagy are deemed the least preferred outcomes from a therapeutic perspective, as they are reversible and would lead only to a temporary stall in tumor growth for as long as the therapy is administered. Furthermore, it should be noted that the effects of Nutlin-3 are rapidly reversible. Within half an hour of washing Nutlin-3 from cell cultures, p53 levels drop drastically to low basal levels, most likely due to the fact that MDM2 accumulated during the treatment [197]. In contrast, genotoxic p53 activating agents produce long lasting signaling, which may explain their potency in the clinic. Some unavoidable questions then arise: What molecular mechanisms define cell type-specific responses to Nutlin-3? How does the p53 circuit board react to genotoxic versus non-genotoxic p53 activating agents? Can drugs like Nutlin-3 be used in the clinic, alone or in combination, to elicit potent cell death in tumors but without the systemic long lasting side effects of genotoxic agents?
4.2. The impact of cell type- and stimulus-specific assemblies of the p53 circuit board on the efficacy of p53-based therapies
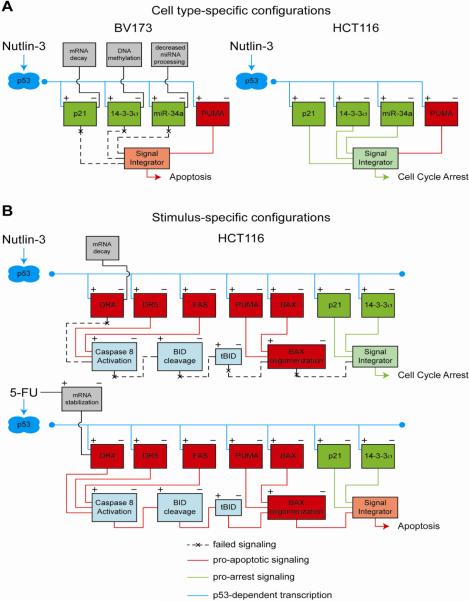
While studying the mechanisms of cell fate choice in response to Nutlin-3, we uncovered combinatorial assemblies of the cell cycle arrest integrated circuit across cancer cell types, which impacted the decision to undergo arrest or commit to apoptosis (Figure 6A) [118]. Upon Nutlin-3-treatment, some cancer cell lines undergo reversible cell cycle arrest without apoptosis (e.g. HCT116, colorectal cancer; A549, lung cancer), others arrest at first and then commit to apoptosis (e.g. SJSA, osteosarcoma; LNCaP, prostate cancer), and a few select types undergo apoptosis without signs of arrest (e.g. BV173, chronic myelogenous leukemia). Although p53 and several apoptotic targets were equally activated in all cell lines, we observed a clear correlation between the expression of p21, 14-3-3σ and miR-34a and the outcome adopted. HCT116 and A549 cells expressed high levels of the three arrest genes, SJSA and LNCaP cells expressed high levels of p21, mid-levels of miR-34a and no 14-3-3σ, and BV173 cells expressed none of these three p53 targets. Mechanistic studies showed that the observed differences were due to a combination of cell type-specific degradation of p21 mRNA, 14-3-3σ promoter DNA methylation and impaired processing of the primary miR-34a transcript [118]. Importantly, we showed that attenuation of miR-34a function in HCT116 p21−/−, 14-3-3σ−/− cells led to a significant increase in apoptosis upon Nutlin-3 treatment relative to HCT116 cells expressing all three genes, suggesting that the concerted action of p21, 14-3-3σ and miR-34a protects cells from p53-dependent apoptosis, and that their expression level may determine, at least partially, the choice of p53 response.
Figure 6. Context-dependent configurations of the p53 circuit board define the efficacy of p53 based therapies.
A. An example of cell type-specific p53 responses is provided by non-genotoxic p53 activation by Nutlin-3 across cancer cell lines. In BV173 cells (left), the cell cycle arrest integrated circuit is impaired by p21 mRNA decay, 14-3-3σ promoter methylation and impaired processing of miR-34a. In contrast, these three cell cycle arrest genes are effectively activated in HCT116 cells (right), where they function coordinately to establish a cell cycle arrest response, even though potent apoptotic genes such as PUMA have also been induced. B. Stimulus-specific assembly of the p53 circuit in response to p53 activation by Nutlin-3 versus 5-FU in HCT116 cells. Both Nutlin-3 and 5-FU strongly activate genes involved in both cell cycle arrest and apoptosis; however, only 5-FU treatment results in p53-independent stabilization of DR4 mRNA and concomitant upregulation of DR4 protein levels, which is required for caspase 8 activation and proteolytic activation of BID into tBID. Activation of the DR4:tBID axis by 5-FU drives the apoptotic response by promoting oligomerization of poised BAX at the mitochondria.
In a comparative study of Nutlin-3 versus genotoxic agents, we identified stimulus-specific assemblies of the apoptotic circuit that define the p53 response (Figure 6B). Interestingly, we found that the same cell lines that adopt a cell cycle arrest response upon Nutlin-3 treatment can effectively undergo p53-dependent apoptosis when treated with the antimetabolite 5-FU. Using this paradigm, we investigated the contribution of ~20 components of the p53 network to cell fate choice [198]. Strikingly, we found that arresting cells displayed effective transactivation of PUMA concurrent with translocation of BAX to the mitochondria. However, these cells failed to release cytochrome C into the cytosol and activate caspases, which was explained by the failure of BAX to oligomerize at the mitochondria. On the other hand, cells undergoing p53-dependent apoptosis accumulated p21, 14-3-3σ and other genes involved in cell cycle arrest, but they failed to arrest and showed instead p53-dependent activation of caspases. Genetic dissection of different branches of the apoptotic integrated circuit revealed that the key stimulus-specific events were: i) p53-dependent activation of caspase 8, ii) caspase 8-dependent activation of the BH3-only protein BID, and iii) BID-dependent activation and oligomerization of poised BAX. Interestingly, we found that the death receptor DR4 was strongly induced only in 5-FU-treated cells via a combination of p53-dependent transactivation and p53-independent mRNA stabilization. In fact, DR4 was required for activation of caspase 8, BID and BAX. Thus, parallel signals generated by 5-FU complemented p53 action to tip the balance toward apoptosis.
4.3. Personalized p53 medicines: the urgent need for combinatorial therapies, functional genomics and molecular diagnostics
Given the stochastic nature of the mutations driving cancer progression, it is safe to assume that even across a unique tumor type, the p53 circuit board will adopt a large number of possible configurations, with the consequent variability in the efficacy of p53-based targeted therapies. One way of circumventing these shortcomings is to combine Nutlin-3 with other agents that would tip the balance toward a rapid apoptotic response. Many recent efforts have been devoted to such combinatorial strategies, using both traditional and targeted therapeutics. For example, Nutlin-3 promotes apoptosis in concert with ionizing radiation in otherwise radioresistant lung cancer cells [199]. Similarly, Nutlin-3 exhibits synergy with genotoxic drugs in diverse cancers ranging from chronic lymphocytic leukemia to hepatocellular carcinoma [200, 201]. These potential treatments represent progress, but remain a rather blunt approach that merely combines DNA damage with high levels of p53 induction. Of note, Nutlin-3-induced cell cycle arrest protects from the killling effects of mitotic poisons such as paclitaxel and taxol, an observation that led to the hypothesis that Nutlin-3 could be used for cyclotherapy, protecting healthy, dividing cells from the harmful effects of certain drugs meant to target cancer cells [202, 203]. Nutlin-3 is also more effective when combined with more specific drugs and small molecules. For example, several groups have reported that CDK inhibitors potentiate the apoptotic effects of Nutlin treatment [204, 205] and similar results have that been reported for MAPK inhibition in AML cells [206]. In a more directed approach, Nutlin-3 was combined with ABT-737, a small molecule sequesters pro-survival BCL2 family members, in studies that demonstrated striking synergy between the drugs in AML and breast cancer cells [207, 208]. A recent study showed that inhibition of Hsp90 by 17AAG strongly activates p53-mediated apoptosis in response to Nutlin-3 treatment, both in vitro and in vivo [209]. Multiple studies have also shown that Nutlin-3 increases susceptibility to TRAIL via upregulation of death receptors [210, 211]. These examples represent only a fraction of the drug combinations tested with Nutlin, but all carry the same challenges as single drug strategies: which combinatorial therapies will be effective on any given tumor? Certainly, tumors where TRAIL receptors have been silenced via DNA methylation will not respond to a [Nutlin-3 + TRAIL] combination [129]. Thus, there is an increasing need for both a large menu of possible combinatorial strategies and the diagnostic tools to predict which strategy will be most effective for a given tumor.
With the advent of functional genomics it is now possible to interrogate the entire genome for pathways displaying synthetic lethality with targeted therapeutics. In fact, RNAi based screens for sensitizers to several targeted therapies approved for clinical use have identified additional drug targets that increase the efficacy of the drugs [212, 213]. These screens could be adapted to identify novel combinatorial strategies that improve the efficacy of p53-based targeted therapies. These “synthetic lethal with p53 activation pathways” (SLPAPs) may behave as such in some, but not all cancer types. We can envision the existence of “core” and “facultative” SLPAPs, depending on their universality. However, core SLPAPs may also function as such in non-cancerous tissues, which would limit their applicability due to side effects of the combinatorial therapy. Upon identification of these pathways, the challenge will move then to defining which patients will benefit from which combinatorial therapies, highlighting the need for molecular diagnostics. Obviously, Nutlin-3 and other MDM2 inhibitors have no effects in p53 mutant cells [202]. Thus, p53 mutational status should be the first biomarker to be analyzed before deciding on a therapeutic strategy. However, great variability is to be expected across wild type p53 tumors, as some may undergo cell cycle arrest and others apoptosis, and no biomarker is available yet to distinguish these subclasses. It is reasonable to predict that tumors with impaired arrest integrated circuits (as defined by loss of p21, 14-3-3σ and/or miR-34a) may respond better to these drugs. However, loss of just one of these arrest factors does little to change the response to Nutlin-3 [118, 214]. Thus, intensive gene expression profiling of tumors displaying variable responses to the drug seem necessary to identify a more powerful gene signature. Recently, synthetic lethal screens have been proposed to accelerate biomarker discovery [215]. The hypothesis is that many synthetic lethal genes, which protect “resistant” cell lines from the drug, will be expressed at lower levels in the “sensitive” cells. Thus, cross-referencing gene profiling and synthetic lethality datasets will provide the gene signature with highest prognostic value. However, whereas gene profiling can be easily performed in tumor samples, RNAi screens are technically amenable only to established cell cultures, and it is unclear if synthetic lethality observed in vitro can be extrapolated to the clinical setting. Technical difficulties aside, the potential of these approaches is undeniable, and they set clear research goals for the field.
Final Remarks
We ask the reader to picture the following scenario, not too far into the future. During an annual check up, the reader is found to carry a non-resectable tumor. Pathologists report that is a tumor type for which no single agent therapy has proven useful. The pathology report indicates that the tumor is wild type for p53, MDM2 positive, MDM4 negative. The oncologist orders two assays: i) a genome wide shRNA screen for synthetic lethality with an approved MDM2 inhibitor, to be performed on a growing explant derived from a fresh tumor biopsy; and ii) a global RNA and protein profile of gene expression for the tumor tissues and a few select normal tissues. Two weeks later the results arrive as a list of possible synthetic lethal drug combinations and digital blueprints of the p53 circuit boards in the tumor and normal tissues. A week later treatment begins with sequential combinations of an MDM2 inhibitor with other targeted therapies predicted to be synthetic lethal only in tumor cells. All drugs are administered at low doses and for short periods of time. Within days, the tumor regresses and the reader is considered cancer free. Science fiction or rational optimism?
“The rung of a ladder was never meant to rest upon, but only to hold a man's foot long enough to enable him to put the other somewhat higher.” ~Thomas H. Huxley.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci U S A. 1979;76:2420–2424. doi: 10.1073/pnas.76.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- [3].Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- [4].Eliyahu D, Raz A, Gruss P, Givol D, Oren M. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature. 1984;312:646–649. doi: 10.1038/312646a0. [DOI] [PubMed] [Google Scholar]
- [5].Jenkins JR, Rudge K, Currie GA. Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature. 1984;312:651–654. doi: 10.1038/312651a0. [DOI] [PubMed] [Google Scholar]
- [6].Lane DP. Cell immortalization and transformation by the p53 gene. Nature. 1984;312:596–597. doi: 10.1038/312596a0. [DOI] [PubMed] [Google Scholar]
- [7].Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- [8].Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, Hamilton S, Vogelstein B. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- [9].Hinds PW, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- [10].Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- [11].Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- [12].Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- [13].Fields S, Jang SK. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- [14].Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- [16].Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- [17].Zambetti GP, Bargonetti J, Walker K, Prives C, Levine AJ. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- [18].Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- [19].Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res. 1993;53:2231–2234. [PubMed] [Google Scholar]
- [20].Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- [21].el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- [22].Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- [23].Tokino T, Thiagalingam S, el-Deiry WS, Waldman T, Kinzler KW, Vogelstein B. p53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- [24].Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc Natl Acad Sci U S A. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- [26].Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- [27].Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- [28].Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- [29].Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- [30].Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- [31].Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- [33].Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- [35].Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem. 2001;276:29116–29125. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]
- [36].Nakano K, Mizuno T, Sowa Y, Orita T, Yoshino T, Okuyama Y, Fujita T, Ohtani-Fujita N, Matsukawa Y, Tokino T, Yamagishi H, Oka T, Nomura H, Sakai T. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J Biol Chem. 1997;272:22199–22206. doi: 10.1074/jbc.272.35.22199. [DOI] [PubMed] [Google Scholar]
- [37].Liu PY, Hsieh TY, Liu ST, Chang YL, Lin WS, Wang WM, Huang SM. Zac1, an Sp1-like protein, regulates human p21(WAF1/Cip1) gene expression in HeLa cells. Exp Cell Res. 2011;317:2925–2937. doi: 10.1016/j.yexcr.2011.09.018. [DOI] [PubMed] [Google Scholar]
- [38].Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- [39].Weber A, Marquardt J, Elzi D, Forster N, Starke S, Glaum A, Yamada D, Defossez PA, Delrow J, Eisenman RN, Christiansen H, Eilers M. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J. 2008;27:1563–1574. doi: 10.1038/emboj.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Koh DI, Choi WI, Jeon BN, Lee CE, Yun CO, Hur MW. A novel POK family transcription factor, ZBTB5, represses transcription of p21CIP1 gene. J Biol Chem. 2009;284:19856–19866. doi: 10.1074/jbc.M109.025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jeon BN, Choi WI, Yu MY, Yoon AR, Kim MH, Yun CO, Hur MW. ZBTB2, a novel master regulator of the p53 pathway. J Biol Chem. 2009;284:17935–17946. doi: 10.1074/jbc.M809559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Choi WI, Jeon BN, Yun CO, Kim PH, Kim SE, Choi KY, Kim SH, Hur MW. Protooncogene FBI-1 represses transcription of p21CIP1 by inhibition of transcription activation by p53 and Sp1. J Biol Chem. 2009;284:12633–12644. doi: 10.1074/jbc.M809794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gorospe M, Wang X, Holbrook NJ. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38:2256–2267. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- [49].Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- [50].Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li Y, Dowbenko D, Lasky LA. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- [53].Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim Biophys Acta. 2002;1593:45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- [54].Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- [55].Lee JY, Yu SJ, Park YG, Kim J, Sohn J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol Cell Biol. 2007;27:3187–3198. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- [57].Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cayrol C, Ducommun B. Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene. 1998;17:2437–2444. doi: 10.1038/sj.onc.1202189. [DOI] [PubMed] [Google Scholar]
- [60].Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday MJ. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Coleman ML, Marshall CJ, Olson MF. Ras promotes p21(Waf1/Cip1) protein stability via a cyclin D1-imposed block in proteasome-mediated degradation. EMBO J. 2003;22:2036–2046. doi: 10.1093/emboj/cdg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell. 2005;17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- [63].Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- [64].Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- [65].Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- [66].Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- [67].Aprelikova O, Pace AJ, Fang B, Koller BH, Liu ET. BRCA1 is a selective co-activator of 14-3-3 sigma gene transcription in mouse embryonic stem cells. J Biol Chem. 2001;276:25647–25650. doi: 10.1074/jbc.C100265200. [DOI] [PubMed] [Google Scholar]
- [68].Bhatia K, Siraj AK, Hussain A, Bu R, Gutierrez MI. The tumor suppressor gene 14-3-3 sigma is commonly methylated in normal and malignant lymphoid cells. Cancer Epidemiol Biomarkers Prev. 2003;12:165–169. [PubMed] [Google Scholar]
- [69].Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, Sukumar S. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lodygin D, Hermeking H. Epigenetic silencing of 14-3-3sigma in cancer. Semin Cancer Biol. 2006;16:214–224. doi: 10.1016/j.semcancer.2006.03.008. [DOI] [PubMed] [Google Scholar]
- [71].Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y. IKKalpha shields 14-3-3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol Cell. 2007;27:214–227. doi: 10.1016/j.molcel.2007.05.042. [DOI] [PubMed] [Google Scholar]
- [72].Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- [73].Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- [74].Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- [75].Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- [76].Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, Villunger A, Mak TW. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Amente S, Zhang J, Lubrano Lavadera M, Lania L, Avvedimento EV, Majello B. Myc and PI3K/AKT signaling cooperatively repress FOXO3a-dependent PUMA and GADD45a gene expression. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].You H, Yamamoto K, Mak TW. Regulation of transactivation-independent proapoptotic activity of p53 by FOXO3a. Proc Natl Acad Sci U S A. 2006;103:9051–9056. doi: 10.1073/pnas.0600889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang P, Qiu W, Dudgeon C, Liu H, Huang C, Zambetti GP, Yu J, Zhang L. PUMA is directly activated by NF-kappaB and contributes to TNF-alpha-induced apoptosis. Cell Death Differ. 2009;16:1192–1202. doi: 10.1038/cdd.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gao J, Senthil M, Ren B, Yan J, Xing Q, Yu J, Zhang L, Yim JH. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 2010;17:699–709. doi: 10.1038/cdd.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wu W-S, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- [83].Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J Exp Med. 2004;199:113–124. doi: 10.1084/jem.20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nikiforov MA, Riblett M, Tang WH, Gratchouck V, Zhuang D, Fernandez Y, Verhaegen M, Varambally S, Chinnaiyan AM, Jakubowiak AJ, Soengas MS. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci U S A. 2007;104:19488–19493. doi: 10.1073/pnas.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Valis K, Prochazka L, Boura E, Chladova J, Obsil T, Rohlena J, Truksa J, Dong LF, Ralph SJ, Neuzil J. Hippo/Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Res. 2011;71:946–954. doi: 10.1158/0008-5472.CAN-10-2203. [DOI] [PubMed] [Google Scholar]
- [87].Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, Hasegawa A, Motohashi S, Iwama A, Nakayama T. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008;205:1109–1120. doi: 10.1084/jem.20072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ploner C, Rainer J, Lobenwein S, Geley S, Kofler R. Repression of the BH3-only molecule PMAIP1/Noxa impairs glucocorticoid sensitivity of acute lymphoblastic leukemia cells. Apoptosis. 2009;14:821–828. doi: 10.1007/s10495-009-0355-5. [DOI] [PubMed] [Google Scholar]
- [89].Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–8634. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- [90].Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- [91].Yang W, Dicker DT, Chen J, El-Deiry WS. CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2. Cell Cycle. 2008;7:670–682. doi: 10.4161/cc.7.5.5701. [DOI] [PubMed] [Google Scholar]
- [92].Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, White RW. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2011;71:538–549. doi: 10.1002/pros.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37:1621–1626. doi: 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- [95].Fricker M, O'Prey J, Tolkovsky AM, Ryan KM. Phosphorylation of Puma modulates its apoptotic function by regulating protein stability. Cell Death Dis. 2010;1:e59. doi: 10.1038/cddis.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sandow JJ, Jabbour AM, Condina MR, Daunt CP, Stomski FC, Green BD, Riffkin CD, Hoffmann P, Guthridge MA, Silke J, Lopez AF, Ekert PG. Cytokine receptor signaling activates an IKK-dependent phosphorylation of PUMA to prevent cell death. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhu H, Cao X, Ali-Osman F, Keir S, Lo HW. EGFR and EGFRvIII interact with PUMA to inhibit mitochondrial translocalization of PUMA and PUMA-mediated apoptosis independent of EGFR kinase activity. Cancer Lett. 2010;294:101–110. doi: 10.1016/j.canlet.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lowman XH, McDonnell MA, Kosloske A, Odumade OA, Jenness C, Karim CB, Jemmerson R, Kelekar A. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell. 2010;40:823–833. doi: 10.1016/j.molcel.2010.11.035. [DOI] [PubMed] [Google Scholar]
- [99].Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G, Martignetti JA. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res. 2009;69:4733–4741. doi: 10.1158/0008-5472.CAN-08-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- [101].Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- [102].Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- [103].Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- [104].Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- [105].Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, Taniguchi T. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J Biol Chem. 2000;275:22627–22630. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- [106].He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007 doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential Regulation of microRNAs by p53 Revealed by Massively Parallel Sequencing: miR-34a is a p53 Target That Induces Apoptosis and G(1)-arrest. Cell Cycle. 2007;6 doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- [108].Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Henrique R, Jeronimo C, Hoque MO, Carvalho AL, Oliveira J, Teixeira MR, Lopes C, Sidransky D. Frequent 14-3-3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol. 2005;24:264–269. doi: 10.1089/dna.2005.24.264. [DOI] [PubMed] [Google Scholar]
- [111].Kunze E, Wendt M, Schlott T. Promoter hypermethylation of the 14-3-3 sigma, SYK and CAGE-1 genes is related to the various phenotypes of urinary bladder carcinomas and associated with progression of transitional cell carcinomas. Int J Mol Med. 2006;18:547–557. [PubMed] [Google Scholar]
- [112].Hamilton JP, Sato F, Jin Z, Greenwald BD, Ito T, Mori Y, Paun BC, Kan T, Cheng Y, Wang S, Yang J, Abraham JM, Meltzer SJ. Reprimo methylation is a potential biomarker of Barrett's-Associated esophageal neoplastic progression. Clin Cancer Res. 2006;12:6637–6642. doi: 10.1158/1078-0432.CCR-06-1781. [DOI] [PubMed] [Google Scholar]
- [113].Sato N, Fukushima N, Matsubayashi H, Iacobuzio-Donahue CA, Yeo CJ, Goggins M. Aberrant methylation of Reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma. Cancer. 2006;107:251–257. doi: 10.1002/cncr.21977. [DOI] [PubMed] [Google Scholar]
- [114].Wong TS, Kwong DL, Sham JS, Wei WI, Yuen AP. Methylation status of Reprimo in head and neck carcinomas. Int J Cancer. 2005;117:697. doi: 10.1002/ijc.21208. [DOI] [PubMed] [Google Scholar]
- [115].Suzuki M, Shigematsu H, Takahashi T, Shivapurkar N, Sathyanarayana UG, Iizasa T, Fujisawa T, Gazdar AF. Aberrant methylation of Reprimo in lung cancer. Lung Cancer. 2005;47:309–314. doi: 10.1016/j.lungcan.2004.08.006. [DOI] [PubMed] [Google Scholar]
- [116].Takahashi T, Suzuki M, Shigematsu H, Shivapurkar N, Echebiri C, Nomura M, Stastny V, Augustus M, Wu CW, Wistuba, Meltzer SJ, Gazdar AF. Aberrant methylation of Reprimo in human malignancies. Int J Cancer. 2005;115:503–510. doi: 10.1002/ijc.20910. [DOI] [PubMed] [Google Scholar]
- [117].Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- [118].Paris R, Henry RE, Stephens SJ, McBryde M, Espinosa JM. Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle. 2008;7:2427–2433. doi: 10.4161/cc.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- [120].Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- [121].Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- [122].Muller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M, Friedman SL, Galle PR, Stremmel W, Oren M, Krammer PH. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Liu X, Yue P, Khuri FR, Sun SY. p53 upregulates death receptor 4 expression through an intronic p53 binding site. Cancer Res. 2004;64:5078–5083. doi: 10.1158/0008-5472.CAN-04-1195. [DOI] [PubMed] [Google Scholar]
- [124].Wu GS, Burns TF, McDonald ER, 3rd, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, el-Deiry WS. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- [125].Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- [126].Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- [127].Kuribayashi K, Finnberg N, Jeffers JR, Zambetti GP, El-Deiry WS. The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo. Cell Cycle. 2011;10:2380–2389. doi: 10.4161/cc.10.14.16588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, Sample JT, Cleveland JL, Zambetti GP. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hopkins-Donaldson S, Ziegler A, Kurtz S, Bigosch C, Kandioler D, Ludwig C, Zangemeister-Wittke U, Stahel R. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- [130].Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-Specific Transcriptional Regulation Within the p53 Network. Cell Cycle. 2007;6 doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 Is a Stimulus-Specific Positive Coregulator of p53 Target Genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- [135].Mattia M, Gottifredi V, McKinney K, Prives C. p53-Dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol. 2007;27:1309–1320. doi: 10.1128/MCB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- [137].Reczek EE, Flores ER, Tsay AS, Attardi LD, Jacks T. Multiple response elements and differential p53 binding control Perp expression during apoptosis. Mol Cancer Res. 2003;1:1048–1057. [PubMed] [Google Scholar]
- [138].Ihrie RA, Reczek E, Horner JS, Khachatrian L, Sage J, Jacks T, Attardi LD. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- [139].Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- [140].Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- [141].Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- [142].Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- [143].Thanos CD, Bowie JU. p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 1999;8:1708–1710. doi: 10.1110/ps.8.8.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ghioni P, Bolognese F, Duijf PH, Van Bokhoven H, Mantovani R, Guerrini L. Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol Cell Biol. 2002;22:8659–8668. doi: 10.1128/MCB.22.24.8659-8668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, Olivier M, Hall J, Mollereau B, Hainaut P, Bourdon JC. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–1824. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006;580:4544–4550. doi: 10.1016/j.febslet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- [147].Perez CA, Ott J, Mays DJ, Pietenpol JA. p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene. 2007;26:7363–7370. doi: 10.1038/sj.onc.1210561. [DOI] [PubMed] [Google Scholar]
- [148].Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, Bolat E, Jurgelenaite R, Huynen MA, Hoischen A, Veltman JA, Brunner HG, Roscioli T, Oates E, Wilson M, Manzanares M, Gomez-Skarmeta JL, Stunnenberg HG, Lohrum M, van Bokhoven H, Zhou H. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- [150].Trink B, Osada M, Ratovitski E, Sidransky D. p63 transcriptional regulation of epithelial integrity and cancer. Cell Cycle. 2007;6:240–245. doi: 10.4161/cc.6.3.3803. [DOI] [PubMed] [Google Scholar]
- [151].Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, Puisieux A, Hainaut P, Caron de Fromentel C. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–281. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- [152].Mundt HM, Stremmel W, Melino G, Krammer PH, Schilling T, Muller M. Dominant negative (DeltaN) p63alpha induces drug resistance in hepatocellular carcinoma by interference with apoptosis signaling pathways. Biochem Biophys Res Commun. 2010;396:335–341. doi: 10.1016/j.bbrc.2010.04.093. [DOI] [PubMed] [Google Scholar]
- [153].Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Lu H, Yang X, Duggal P, Allen CT, Yan B, Cohen J, Nottingham L, Romano RA, Sinha S, King KE, Weinberg WC, Chen Z, Van Waes C. TNF-alpha Promotes c-REL/{Delta}Np63alpha Interaction and TAp73 Dissociation from Key Genes That Mediate Growth Arrest and Apoptosis in Head and Neck Cancer. Cancer Res. 2011;71:6867–6877. doi: 10.1158/0008-5472.CAN-11-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- [157].Ramsey MR, He L, Forster N, Ory B, Ellisen LW. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 2011;71:4373–4379. doi: 10.1158/0008-5472.CAN-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Kurita T, Cunha GR, Robboy SJ, Mills AA, Medina RT. Differential expression of p63 isoforms in female reproductive organs. Mech Dev. 2005;122:1043–1055. doi: 10.1016/j.mod.2005.04.008. [DOI] [PubMed] [Google Scholar]
- [159].Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- [160].Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- [162].Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, Knight RA, Melino G. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- [163].Yamamura Y, Lee WL, Goh MX, Ito Y. Role of TAp73alpha in induction of apoptosis by transforming growth factor-beta in gastric cancer cells. FEBS Lett. 2008;582:2663–2667. doi: 10.1016/j.febslet.2008.06.046. [DOI] [PubMed] [Google Scholar]
- [164].Shi Y, Takenobu H, Kurata K, Yamaguchi Y, Yanagisawa R, Ohira M, Koike K, Nakagawara A, Jiang LL, Kamijo T. HDM2 impairs Noxa transcription and affects apoptotic cell death in a p53/p73-dependent manner in neuroblastoma. Eur J Cancer. 2010;46:2324–2334. doi: 10.1016/j.ejca.2010.05.026. [DOI] [PubMed] [Google Scholar]
- [165].Sang M, Li Y, Ozaki T, Ono S, Ando K, Yamamoto H, Koda T, Geng C, Nakagawara A. p73-dependent induction of 14-3-3sigma increases the chemo-sensitivity of drug-resistant human breast cancers. Biochem Biophys Res Commun. 2006;347:327–333. doi: 10.1016/j.bbrc.2006.06.079. [DOI] [PubMed] [Google Scholar]
- [166].Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- [167].Fricker M, Papadia S, Hardingham GE, Tolkovsky AM. Implication of TAp73 in the p53-independent pathway of Puma induction and Puma-dependent apoptosis in primary cortical neurons. J Neurochem. 2010;114:772–783. doi: 10.1111/j.1471-4159.2010.06804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kartasheva NN, Lenz-Bauer C, Hartmann O, Schafer H, Eilers M, Dobbelstein M. DeltaNp73 can modulate the expression of various genes in a p53-independent fashion. Oncogene. 2003;22:8246–8254. doi: 10.1038/sj.onc.1207138. [DOI] [PubMed] [Google Scholar]
- [169].Chan WM, Siu WY, Lau A, Poon RY. How many mutant p53 molecules are needed to inactivate a tetramer? Mol Cell Biol. 2004;24:3536–3551. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- [171].Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci U S A. 2009;106:16871–16876. doi: 10.1073/pnas.0903191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Lee AF, Ho DK, Zanassi P, Walsh GS, Kaplan DR, Miller FD. Evidence that DeltaNp73 promotes neuronal survival by p53-dependent and p53-independent mechanisms. J Neurosci. 2004;24:9174–9184. doi: 10.1523/JNEUROSCI.1588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G. The ubiquitin-protein ligase Itch regulates p73 stability. Embo J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Yang C, Zhou W, Jeon MS, Demydenko D, Harada Y, Zhou H, Liu YC. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol Cell. 2006;21:135–141. doi: 10.1016/j.molcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- [179].Oberst A, Malatesta M, Aqeilan RI, Rossi M, Salomoni P, Murillas R, Sharma P, Kuehn MR, Oren M, Croce CM, Bernassola F, Melino G. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci U S A. 2007;104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Galli F, Rossi M, D'Alessandra Y, De Simone M, Lopardo T, Haupt Y, Alsheich-Bartok O, Anzi S, Shaulian E, Calabro V, La Mantia G, Guerrini L. MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J Cell Sci. 2010;123:2423–2433. doi: 10.1242/jcs.061010. [DOI] [PubMed] [Google Scholar]
- [181].Sayan BS, Yang AL, Conforti F, Tucci P, Piro MC, Browne GJ, Agostini M, Bernardini S, Knight RA, Mak TW, Melino G. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci U S A. 2010;107:12877–12882. doi: 10.1073/pnas.0911828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Wang N, Guo L, Rueda BR, Tilly JL. Cables1 protects p63 from proteasomal degradation to ensure deletion of cells after genotoxic stress. EMBO Rep. 2010;11:633–639. doi: 10.1038/embor.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Hershkovitz Rokah O, Shpilberg O, Granot G. NAD(P)H quinone oxidoreductase protects TAp63gamma from proteasomal degradation and regulates TAp63gamma-dependent growth arrest. PLoS One. 2010;5:e11401. doi: 10.1371/journal.pone.0011401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Soond SM, Barry SP, Melino G, Knight RA, Latchman DS, Stephanou A. p73-mediated transcriptional activity is negatively regulated by polo-like kinase 1. Cell Cycle. 2008;7:1214–1223. doi: 10.4161/cc.7.9.5777. [DOI] [PubMed] [Google Scholar]
- [186].Gaiddon C, Lokshin M, Gross I, Levasseur D, Taya Y, Loeffler JP, Prives C. Cyclin-dependent kinases phosphorylate p73 at threonine 86 in a cell cycle-dependent manner and negatively regulate p73. J Biol Chem. 2003;278:27421–27431. doi: 10.1074/jbc.M300251200. [DOI] [PubMed] [Google Scholar]
- [187].Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- [189].Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- [191].Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- [192].Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- [193].Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- [194].Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- [196].Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Gomes NP, Espinosa JM. Disparate chromatin landscapes and kinetics of inactivation impact differential regulation of p53 target genes. Cell Cycle. 2010;9:3428–3437. doi: 10.4161/cc.9.17.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Henry RE, Andrysik Z, Paris R, Galbraith MD, Espinosa JM. A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX. EMBO J. 2012 doi: 10.1038/emboj.2011.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Cao C, Shinohara ET, Subhawong TK, Geng L, Woon Kim K, Albert JM, Hallahan DE, Lu B. Radiosensitization of lung cancer by nutlin, an inhibitor of murine double minute 2. Mol Cancer Ther. 2006;5:411–417. doi: 10.1158/1535-7163.MCT-05-0356. [DOI] [PubMed] [Google Scholar]
- [200].Coll-Mulet L, Iglesias-Serret D, Santidrian AF, Cosialls AM, de Frias M, Castano E, Campas C, Barragan M, de Sevilla AF, Domingo A, Vassilev LT, Pons G, Gil J. MDM2 antagonists activate p53 and synergize with genotoxic drugs in B-cell chronic lymphocytic leukemia cells. Blood. 2006;107:4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- [201].Zheng T, Wang J, Song X, Meng X, Pan S, Jiang H, Liu L. Nutlin-3 cooperates with doxorubicin to induce apoptosis of human hepatocellular carcinoma cells through p53 or p73 signaling pathways. J Cancer Res Clin Oncol. 2010;136:1597–1604. doi: 10.1007/s00432-010-0817-8. [DOI] [PubMed] [Google Scholar]
- [202].Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65:1918–1924. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- [203].Tokalov SV, Abolmaali ND. Protection of p53 wild type cells from taxol by nutlin-3 in the combined lung cancer treatment. BMC Cancer. 2010;10:57. doi: 10.1186/1471-2407-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Ribas J, Boix J, Meijer L. (R)-roscovitine (CYC202, Seliciclib) sensitizes SH-SY5Y neuroblastoma cells to nutlin-3-induced apoptosis. Exp Cell Res. 2006;312:2394–2400. doi: 10.1016/j.yexcr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- [205].Cheok CF, Dey A, Lane DP. Cyclin-dependent kinase inhibitors sensitize tumor cells to nutlin-induced apoptosis: a potent drug combination. Mol Cancer Res. 2007;5:1133–1145. doi: 10.1158/1541-7786.MCR-07-0161. [DOI] [PubMed] [Google Scholar]
- [206].Kojima K, Konopleva M, Samudio IJ, Ruvolo V, Andreeff M. Mitogen-activated protein kinase kinase inhibition enhances nuclear proapoptotic function of p53 in acute myelogenous leukemia cells. Cancer Res. 2007;67:3210–3219. doi: 10.1158/0008-5472.CAN-06-2712. [DOI] [PubMed] [Google Scholar]
- [207].Kojima K, Konopleva M, Samudio IJ, Schober WD, Bornmann WG, Andreeff M. Concomitant inhibition of MDM2 and Bcl-2 protein function synergistically induce mitochondrial apoptosis in AML. Cell Cycle. 2006;5:2778–2786. doi: 10.4161/cc.5.23.3520. [DOI] [PubMed] [Google Scholar]
- [208].Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–1982. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Vaseva AV, Yallowitz AR, Marchenko ND, Xu S, Moll UM. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with Nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis. 2011;2:e156. doi: 10.1038/cddis.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Hori T, Kondo T, Kanamori M, Tabuchi Y, Ogawa R, Zhao QL, Ahmed K, Yasuda T, Seki S, Suzuki K, Kimura T. Nutlin-3 enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through up-regulation of death receptor 5 (DR5) in human sarcoma HOS cells and human colon cancer HCT116 cells. Cancer Lett. 2010;287:98–108. doi: 10.1016/j.canlet.2009.06.002. [DOI] [PubMed] [Google Scholar]
- [211].Tseng HY, Jiang CC, Croft A, Tay KH, Thorne RF, Yang F, Liu H, Hersey P, Zhang XD. Contrasting effects of nutlin-3 on TRAIL- and docetaxel-induced apoptosis due to upregulation of TRAIL-R2 and Mcl-1 in human melanoma cells. Mol Cancer Ther. 2010;9:3363–3374. doi: 10.1158/1535-7163.MCT-10-0646. [DOI] [PubMed] [Google Scholar]
- [212].Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- [213].Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O'Hare T, Zaberezhnyy V, Williams RT, Druker BJ, Perrotti D, Degregori J. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010;18:74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Xia M, Knezevic D, Vassilev LT. p21 does not protect cancer cells from apoptosis induced by nongenotoxic p53 activation. Oncogene. 2011;30:346–355. doi: 10.1038/onc.2010.413. [DOI] [PubMed] [Google Scholar]
- [215].Bernards R. It's diagnostics, stupid. Cell. 2010;141:13–17. doi: 10.1016/j.cell.2010.03.018. [DOI] [PubMed] [Google Scholar]