Abstract
Background and aims
Molecular analyses of biliary brushings using microarray and qPCR have the potential to provide valuable information on the biology of biliary diseases. Microarray analysis of biliary strictures has rarely been applied to endoscopic biliary brushings.
Methods
Biliary brushings were obtained from patients with benign and malignant biliary disease at the time of ERCP. Microarray analysis of mRNA isolated using brushings from ten patients was validated for a selection of genes by qPCR using the same source mRNA and a second fresh set of nine biliary brushings as well as surgical resection tissue. Cultured cholangiocytes were used to assess the impact of bile or x-ray contrast solution on RNA quality.
Results
RNA was of variable quantity (100–1500 ng) and poor quality (Agilent RNA Integrity Number (RIN) <5, estimated to be fragments 100 to 600 base pairs long). Reliable qPCR results required primer pairs designed to produce amplicons <130bp. Differential gene expression by microarray analysis identified 1,140 up-regulated genes and 1,001 down-regulated genes between benign and malignant biliary strictures. The trends in a selection of 45 upregulated genes, including various HOX genes, collagens, PVT1, MUC4, MUC5AC and LEF1, were validated by qPCR using RNA from biliary strictures with a moderate to strong correlation coefficient between microarray and qPCR (r=0.41 to r=0.57). Immunohistochemistry of surgical resection tissues (n=23) showed elevated CD9, SERPINA3 and PNMA2 protein expression in cancer samples.
Conclusions
RNA isolated from biliary brushings, is suitable for molecular analysis of biliary diseases using qPCR and microarray.
Keywords: biliary brush cytology, cholangiocarcinoma, ERCP, gene expression
INTRODUCTION
Clinical samples of bile and biliary brush cytology taken at the time of endoscopic retrograde cholangiopancreatography (ERCP) have the potential to provide valuable information on the biology of benign and malignant biliary diseases and may be useful in developing much needed prognostic and predictive biomarkers of treatment benefit in biliary tract cancer. Until recently, the quantities and quality of RNA isolated from ERCP samples have been inadequate for further laboratory investigation. Advances in RNA isolation and gene expression techniques now allow small quantities of degraded RNA, such as that extracted from formalin fixed paraffin embedded (FFPE) tissues, to be successfully applied to downstream applications such as quantitative real time PCR (qPCR) and microarray analysis [1–5]. Expression data using RNA from paired fresh frozen and FFPE tissue samples are comparable, as long as appropriate methodologies, such as use of primer pairs amplifying short nucleic acid sequences, are employed [4–5].
These advances allow further investigation of archived pathology samples as well as potentially allowing more thorough evaluation of degraded RNA from other sources such as bile or biliary brushings. Successful application of whole genome RNA expression profiling has been applied to similar clinical samples of bronchial brushings in patients with suspected lung cancer and urinary sediment in patients with bladder cancer [6–7]. One of these studies using degraded RNA isolated from urine showed that results of gene expression profiling by microarray were similar to those obtained from freshly frozen primary tumour [7].
Gene expression profiling of biliary tissue using fresh frozen surgical resection material has been reported [8–10]. However, there are few published data on the quantity, quality, primary source of RNA in bile or biliary brushings, or its suitability for further investigation such as RNA expression analysis using microarray, most likely due to the small quantities and highly degraded nature of RNA isolated from such samples [11–13].
Potential causes of RNA degradation in biliary samples include a direct effect of bile or the x-ray contrast agents used during ERCP procedures during which clinical samples are collected, both of which have been shown to be cytotoxic to biliary epithelial cells in culture [14–15]. A potential confounding factor in biliary samples is leukocyte RNA contamination as a result of biliary infection, particularly in those with biliary obstruction. Previous work has shown that RNA from bile and/or biliary brushings can be useful for qPCR when measuring small numbers of highly expressed genes [12–13].
Here we demonstrate that samples of biliary brushings provide useful RNA for more extensive evaluation of gene expression in biliary strictures using microarray and qPCR validation. The techniques described may allow more thorough investigation of cholangiocarcinoma and other biliary diseases using similar clinical materials in the future.
MATERIALS AND METHODS
Clinical samples and cell lines
Informed consent was obtained from each patient included in the study. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institution’s human research committee (National Research Ethics Service reference 06/Q0152/106). Samples of bile and biliary brushings (Table 1) were taken at the time of clinically indicated diagnostic or therapeutic ERCP by aspiration of bile from a major bile duct using standard biliary catheters. Biliary brushings were collected from macroscopically normal bile ducts and/or benign or malignant biliary strictures using a wire guided, sheathed endobiliary brush (Combocath Microinvasive, Boston Scientific, Notick, MA, USA). Clinical samples were immediately snap frozen in liquid nitrogen and stored at −80°C until further use. Bile samples for expression analysis were used in 0.5 ml aliquots of whole unfiltered bile.
Table 1.
Summary of clinical samples of biliary brushings used for microarray and qPCR analysis
| Biliary brush samples used for microarray and initial qPCR validation
| |||
|---|---|---|---|
| Diagnosis | Age | Gender | |
| Benign | Papillary Stenosis | 74 | M |
| SOD | 55 | F | |
| CBD Stricture (CP) | 48 | F | |
| CBD Stricture (CP) | 58 | M | |
| Mean 59 | |||
|
| |||
| Malignant | CC (CBD) | 50 | M |
| CC (Hilar) | 64 | M | |
| CC (CBD) | 79 | F | |
| CC (Hilar) | 59 | M | |
| CC (Hilar) | 79 | F | |
| Mean 66 | |||
|
| |||
|
Second independent set of biliary brushings used for qPCR validation
| |||
| Benign | PSC | 35 | M |
| CBD stricture and stones | 65 | M | |
| IAC | 69 | M | |
| SOD | 38 | F | |
| Mean 53 | |||
|
| |||
| Malignant | CC (CBD) | 79 | M |
| CC (CBD) | 63 | M | |
| CC (Hilar) | 60 | F | |
| CC (CBD) | 59 | M | |
| CC (CBD) | 71 | F | |
| Mean 67 | |||
|
| |||
|
Surgical samples used for qPCR validation
| |||
| Diagnosis | Age | Gender | |
|
| |||
| Benign | Benign ischaemic CBD stricture | 66 | M |
| Cystic duct at cholecystectomy | 39 | F | |
| Benign ischaemic CBD stricture | 64 | M | |
| Cystic duct at cholecystectomy | 38 | F | |
| Left hepatic duct (stones) | 53 | F | |
| Benign CBD stricture (IAC) | 55 | M | |
| Mean 53 | |||
|
| |||
| Malignant | CC (CBD) | 60 | M |
| CC (Hilar) | 65 | M | |
| CC (Hilar) | 59 | F | |
| CC (CBD) | 69 | M | |
| CC (Hilar) | 71 | M | |
| CC (CBD) | 64 | F | |
| CC (Hilar) | 76 | M | |
| CC (Hilar) | 72 | M | |
| CC (Hilar) | 67 | M | |
| Mean 67 | |||
SOD; sphincter of Oddi dysfunction: CP; chronic pancreatitis: PSC; primary sclerosing cholangitis: IAC; IgG4 associated cholangitis
A human extra-hepatic cholangiocarcinoma cell line (TFK-1) (DSMZ, Germany) cultured as per published methods, was used as a control [16]. Briefly, cells were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 incubator at 37°C. Fresh bile for cell spiking experiments was collected from free draining percutaneous biliary drains in patients with cholangiocarcinoma, or gallbladder puncture in patients having undergone planned cholecystectomy, and was filtered using sterile 0.45μm filters in order to remove cells in the donor bile, and separated into 0.5 ml aliquots ready for cell spiking. For spiking experiments, TFK-1 cells were cultured until confluence, harvested by trypsinisation and re-cultured in bile or x-ray contrast solution as described below.
RNA isolation and purification
RNA was isolated using the well established guanidium isothiocyanate plus acid phenol method to maximise the yield and quality of the RNA from complex tissues, and then further cleaned up and concentrated using silica matrix columns. Biliary brushes were agitated in 1ml of TRI Reagent® (Ambion Inc, Austin, TX, USA) until most of the visible tissue material had been disrupted from the brush and total RNA isolated as per the manufacturer’s instructions. Bile appears to impair the efficiency of RNA extraction using the TRI Reagent®, as a result of the volumes of bile required and possibly by chemical interaction interrupting the phase separation. We found that the optimum practical volumes for RNA isolation are 0.5ml of bile added to 1ml of TRI Reagent®. Bile also results in extensive DNA contamination requiring a DNase digestion step that was performed using 4U of TurboDNase (Ambion Inc, Austin, TX, USA) for 30 minutes at 37°C. Total RNA was further purified using the RNEasy MinElute clean up kit (Qiagen Ltd, Crawley, UK) and re-dissolved in 12μl RNase free water as per the manufacturer’s instructions.
Estimation of total RNA quantity and quality was first performed using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc, Wilmington, USA). An Agilent 2100 Bioanalyzer was used to further assess RNA quantity and quality. The Agilent RNA integrity number (RIN) and electropherogram plots were used to accurately document RNA quality and calculate RNA fragment size.
In addition to the brushings and bile, plastic biliary stents removed at the time of ERCP were also used to isolate cellular material in the early stages of testing the methodology (data not shown).
Culture of biliary epithelial cells in bile and x-ray contrast agent
In order to test whether the RNA degradation occurs in vivo as a result of cytotoxicity from bile or x-ray contrast agents, or ex vivo during the RNA isolation process, a series of control and cell spiking experiments were performed. A concentrated cell suspension containing 250,000 TFK-1 cells was spiked into 0.5ml aliquots of filtered bile or Iohexol x-ray contrast solution (Omnipaque®, GE Healthhcare Inc, Princeton, NJ, USA) at 25% and 50% concentrations diluted in RPMI culture medium. Further aliquots of cells were transferred into an Eppendorf tube and immediately lysed in TRI reagent to assess the RNA integrity from cells at baseline. The spiked aliquots were incubated at 37°C for 15 minutes, 30 minutes, 1, 3, 6, and 24 hours. After the appropriate incubation time, cells were lysed with TRI reagent and stored at −80°C until RNA isolation. RNA quality was assessed by Nanodrop spectrophotometer, Agilent Bioanalyzer and gel separation on a 1% Agarose gel
Quantitative real time PCR
Real time quantitative PCR was used to assess the suitability of RNA for gene expression analysis and to assess the impact of fragment size and primary source of RNA (ie biliary epithelium or leukocyte). Purified total RNA was reverse transcribed using the iScript Select cDNA Synthesis Kit (Bio-Rad) as per the manufacturer’s protocol. In view of the fragmented nature of the RNA, cDNA synthesis reactions were primed using random hexamers in 20μl volumes using 200ng total RNA. PCR reactions were performed in 25μl volumes using the qPCR Master Mix Plus dNTP Kit (Eurogentec) with 2μl of cDNA (equivalent to 20ng RNA) sample template per reaction. PCR reactions were performed in duplicate using the SYBR Green detection method and an Applied Biosystems 7500 Real Time PCR system (Applied Biosystems Inc, Austin, TX, USA). Thermal cycling conditions were set at 50°C for 2 minutes, 95°C for 10 minutes followed by 40 repeats of 95°C for 15 seconds and 60°C for 1 minute. Relative quantification values for gene expression were calculated using the comparative ΔΔCt method normalizing to GAPDH (NM_002046, 87bp amplicon length) or 18S ribosomal RNA (NR_003286.2, 98bp amplicon length) [17]. When using clinical samples of bile and biliary brushings, expression of CK19 (NM 002276.3) and CD45 (NM 080922.1), markers for biliary epithelial cells and leukocytes respectively, were measured in order to ascertain the primary origin of the RNA. Expression of MUC4 (AF058803), and CD9 (NM_001769.2) were also measured in biliary brushings and bile. Primer pairs used are listed in Supplementary Table 1.
Microarray analysis
RNA isolated from biliary brushings of 4 patients with normal or benign biliary disease and 6 patients with malignant biliary strictures (cholangiocarcinoma) were used for the microarray analysis (Table 1). RNA amplification and cDNA synthesis was performed with a starting quantity of 100ng total purified RNA using the WT-Ovation FFPE RNA Amplification V2 kit (Nugen) that is designed for processing of degraded RNA. cDNA labelling was performed using the FL-Ovation cDNA Biotin module v2 kit (Nugen). Samples were hybridized to Affymetrix Human Genome U133 plus 2.0 GeneChip arrays and data analysed using LIMMA software to assess differential gene expression in cancer compared to benign disease. Internal quality control of microarray data included mas 5.0 normalisation plots, mismatch to perfect match comparisons, MvA plots, RNA degradation plots and analysis using the SimpleAffy software (Patterson Institute for Cancer Research, UK). Data were corrected for multiple hypothesis testing using the Benjamini-Hochberg (FDR) method. A fold change of 2 with p value of <0.05 (modified T-test) was then used for identification of genes with significant alteration in gene expression.
Validation of microarray data using qPCR
A selection of the up- and down-regulated genes identified by microarray were assessed using qPCR. Custom TaqMan Array qPCR cards (Applied Biosystems Inc, Austin, TX, USA) formatted to 48 genes per fill reservoir were used in order to test multiple genes using the limited quantity of RNA isolated from the biliary brushings. cDNA was synthesized from a template of 150ng total RNA using the High Capacity RNA to cDNA Synthesis Kit (Applied Biosystems Inc, Austin, TX, USA). qPCR reactions were performed as per the manufacturers instructions using TaqMan Universal PCR Master Mix (2X) (Applied Biosystems Inc, Austin, TX, USA) and 37.5ng cDNA used for each fill reservoir. Thermal cycling was set at 95°C for 10 minutes followed by 40 repeats of 95°C for 15 seconds and 60°C for 1 minute using a 7900HT Real Time PCR system (Applied Biosystems Inc, Austin, TX, USA). 45 genes were tested in duplicate using the same source RNA used for the microarray analysis (n=9) as well as a second validation using RNA isolated from a fresh sample set (n=9). The qPCR cards also included 3 reference genes (GAPDH, 18S and ACTB) for normalisation. Relative quantification of gene expression was calculated using the ΔΔCt method after pooling cancer versus benign samples with expression of genes in the benign set normalised to 1 using 18S ribosomal RNA and/or GAPDH as the calibrator [17].
Use of surgical resection material for analysis of gene expression using qPCR
Following written, informed patient consent, small sections of surgical resection tissues were snap frozen in liquid nitrogen. Sections of large bile duct were collected from patients with benign biliary disease (n=6) and a portion of the tumour bulk (n=9) removed from patients with cholangiocarcinoma. Following physical homogenisation of 2–3mm3 of tissue, RNA isolation and purification was completed as described above. qPCR was performed using custom designed TaqMan Array cards (Applied Biosystems) as described above.
Immunohistochemistry
Immunohistochemistry using an independent set of biliary tissues was performed in order to test whether changes in gene expression identified by microarray and qPCR were translated to the protein level. Formalin fixed paraffin embedded (FFPE) tissue sections from patients with benign (n=11, benign bile duct and gall bladder resections) and malignant (n=12, cholangiocarcinoma and gallbladder cancer) biliary disease were stained using haematoxylin and eosin (H&E) and serial sections stained for proteins of interest using immunohistochemistry (IHC). Antibodies used (as per manufacturers protocols) included; anti-CD9 (NCL-CD9, Novocastra Laboratories, Burlington, Canada), anti-POU5F1 (NCL-L-oct3/4, Leica Microsystems, Milton Keynes, UK), anti-SERPINA3 (H0000012-M02, Abnova, Jhongli City, Taiwan), anti-PNMA2 (HPA001936, Atlas Antibodies, Stockholm, Sweden) and anti-HOXA10 (SC-17159, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Intensity of staining was reported by a histopathologist using a widely used semi quantitative scoring system (0; no staining, +; weak, ++; moderate and +++; strongly positive staining) [18]. The number of biliary epithelial cells stained positive were also scored (0; <5%, +; 5%–20%, ++; 20%–50% and +++; >50% stained positive [12]. Comparison of results was performed for both number of cells positively stained and intensity of staining.
Statistical analysis
Data are presented as a median with range for RNA quantification. Microarray data were corrected for multiple hypothesis testing using the Benjamini-Hochberg (FDR) method prior to listing genes with a fold change of +/− 2 and p value of <0.05 (modified T-test). qPCR results are presented as the mean with 95% confidence intervals (CI) obtained by applying the general formula for the propagation of errors to the initial standard deviations of the replicates measured for each sample. Analysis of differential expression by IHC was assessed using the Wilcoxon test
RESULTS
Suitability of RNA recovered from bile and biliary brushings for microarray and qPCR analysis
The quantities of purified total RNA isolated from the clinical samples were as follows. Unfiltered bile (0.5ml aliquots, n=26); median total RNA 148 ng (range 0 – 535 ng), 260/280 ratio 2.17 (1.34 – 3.25). Biliary brushings (n=51); median RNA 759 ng, range 44 – 2640 ng, 260/280 ratio 2.04 (1.2 to 2.74).
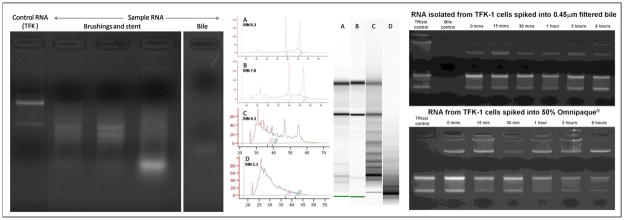
In the clinical samples assessed by gel electrophoresis, faint smears of RNA were seen, suggesting the RNA was highly degraded (Figure 1, A). In agreement with this hypothesis, Agilent Bioanalyzer plots of purified RNA from bile and biliary brushings demonstrated highly degraded RNA with low RIN scores and short fragments of RNA estimated to be primarily 100 to 600 nt long (Figure 1, B). Median RIN scores were 2.4 (range 1 to 3.9) for bile and 2.4 (range 1 to 5.8) for biliary brushings.
Fig 1. RNA quality.
Assessment of total RNA quality shown by representative: (A) agarose gel separation showing smears of degraded RNA in clinical samples (left image); (B) Agilent bioanalyzer plots (centre) of RNA isolated from i) control TFK cells [intact], ii) TFK cells incubated in bile (6 hours) [minimal degradation], iii) biliary brushings, [partly degraded], and iv) bile [highly degraded]; (C) clear bands of intact ribosomal RNA by agarose gel separation (right) after incubation of TFK-1 cells in bile or Omnipaque for up to 6 hours.
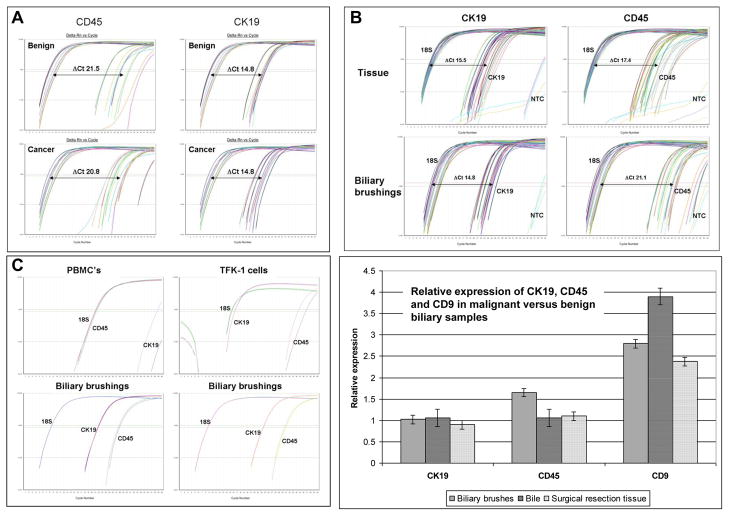
When assessing other genes of interest (CK19, MUC4, CD9 and CD45) in clinical samples, primers amplifying regions ≤100bp and the ΔΔCt method can be used to assess relative gene expression [17] (Figure 2 and Supplementary Figure). In biliary brush samples (n=14), the relative expression of epithelial RNA (mean ΔCt 14.8) was consistently higher than that for leukocyte RNA (mean ΔCt 21.1) suggesting that the RNA isolated from bile or biliary brushings is primarily of epithelial origin (Figure 2). Relative expression of CK19 was similar between benign and malignant samples (1.02, CI 0.98–1.06) suggesting no significant difference in epithelial RNA in the different patient groups. Relative expression of CD45 was slightly higher in the cancer group (1.65, CI 1.48–1.83) suggesting a greater number of leukocytes in these samples. However, as demonstrated above, epithelial RNA is far more abundant and so the small increase in leukocyte RNA is not likely significant for overall RNA expression analysis.
Fig 2. quantitative real time PCR.
A) qPCR demonstrating similar relative expression of epithelial and leukocyte reference genes in benign and malignant ERCP biliary brushings. B) Comparison of CK19 and CD45 expression in biliary resection tissue and ERCP biliary brushings (note the higher CD45 expression in biliary tissues). Box C shows higher expression of epithelial than leukocyte RNA in individual samples of biliary brushings. Note that in the PBMC and BTC cell line controls, CK45 and CK19 respectively are expressed at high levels near to that of GAPDH. Box D shows the similar relative expression by qPCR of leukocyte (CD45) and epithelial (CK19) markers in cancer versus benign controls (normalised to 1) as well as upregulated CD9 in samples of bile, biliary brushings and tumour tissue from patients with biliary tract cancer.
Similar qPCR results were obtained using highly degraded RNA isolated from bile (n=13). Gene expression was higher for CK19 (Mean ΔCt 18.2) than for CD45 (Mean ΔCt 24.7), again showing a much higher contribution from epithelial than leukocyte RNA (Figure 2). Relative expression of CD45 (1.06, CI 0.66–1.69) and CK19 (1.06, CI 0.89–1.26) was not different between benign and malignant samples suggesting no difference in leukocyte or epithelial RNA in the different patient groups.
Purified RNA isolated from freshly cultured TFK-1 cells resulted in high quality intact RNA as assessed by both agarose gel electrophoresis and Agilent Bioanalyzer (RIN scores >8) demonstrating that the methodologies for RNA isolation were not responsible for the RNA degradation found in the clinical samples (Figure 1, A & B). The quality of RNA extracted from TFK-1 cells cultured for 1 to 24 hours in bile was excellent (RIN scores 7.8 to 9.0) (Figure 1, B & C). Similar findings were found in TFK-1 cultured in 25% and 50% Omnipaque® x-ray contrast agent (RIN scores 7.8 to 8.9) (Figure 1, C). These data suggest that short term exposure to bile or x-ray contrast agents are not responsible for the RNA degradation found in clinical samples and that this degradation occurs in vivo.
Microarray analysis using degraded RNA from biliary brushings
One sample (hilar CC) failed the microarray quality control outlined in methods and was excluded prior to data analysis. Therefore, despite the degraded nature of the RNA and low RIN scores, 9 of the 10 original samples were considered suitable for data analysis. Using a fold change cut-off of +/− 2 and p<0.05 after correction for multiple hypothesis testing, we identified 1,140 up-regulated genes, 1,001 down-regulated genes and 34,057 genes with no significant difference between benign and malignant biliary strictures (data submitted to the Gene Expression Omnibus (GEO)).
Validation of gene expression profiling in biliary brushings using qPCR
In order to validate the results, a selection of genes of interest were further assessed using qPCR using the same source RNA as per the methods above (Table 2). On the assumption that a relative gene expression of >2 represents upregulated expression, overall 79% (34/43) of the validated genes were upregulated by both microarray and qPCR analysis. The fold change varied but the trend in gene expression was considered similar with a moderate Pearson correlation coefficient of r=0.41. Using the second, fresh validation set of patients with benign and malignant biliary disease (n=9), 36 out of 43 (83%) upregulated genes showed upregulated gene expression using both platforms and the trend in overall level of expression was strong with a Pearson correlation coefficient of r=0.57.
Table 2.
Summary of clinical resection tissues used for immunohistochemistry
| Samples used for immunohistochemistry
| |||
|---|---|---|---|
| Diagnosis | Age | Gender | |
| Benign | CBD (Choledochal cyst) | 74 | F |
| GB (Chronic cholecystitis) | 63 | F | |
| GB (Chronic cholecystitis) | 44 | M | |
| GB (Chronic cholecystitis) | 30 | F | |
| GB (Chronic cholecystitis) | 39 | F | |
| GB (Chronic cholecystitis) | 43 | F | |
| Cystic duct (Chronic cholecystitis) | 40 | F | |
| GB(Chronic cholecystitis) | 52 | F | |
| GB (Chronic cholecystitis) | 57 | M | |
| CBD (Chronic pancreatitis) | 50 | M | |
| GB (Chronic cholecystitis) | 67 | M | |
| Mean 51 | |||
|
| |||
| BTC | Tumour mass (CC) | 55 | M |
| Tumour mass (GBCa) | 63 | F | |
| Tumour mass (CC) | 65 | M | |
| Tumour mass (CC) | 59 | M | |
| Tumour mass (CC) | 75 | F | |
| Tumour mass (CC) | 53 | M | |
| Tumour mass (CC) | 69 | M | |
| Tumour mass (CC) | 49 | M | |
| Tumour mass (CC) | 55 | M | |
| Tumour mass (GBCa) | 78 | M | |
| Tumour mass (CC) | 61 | M | |
| Tumour mass (CC) | 56 | F | |
| Mean 62 | |||
CC; cholangiocarcinoma: GB; gallbladder
Validation of a further set of biliary brush samples by individual gene qPCR using the SYBR Green method also confirmed upregulation of MUC4 (fold change 21.4 [95% CI 19.1 to 24.1], n=9) and CD9 (fold change 2.8 [95% CI 2.6 to 3.0], n=11).
Comparison of gene expression between biliary brush RNA and biliary surgical resection tissues
RNA quantity and quality assessed by Nanodrop spectrophotometer was higher in the surgical resection tissues than that of biliary brushings with median total quantity of 6675ng (range 1140 to 14820) and 260/280 ratio of 2.06 (range 1.75 to 2.17).
Using SYBR Green qPCR, expression of CK19 relative to 18S was similar in cancer (ΔCt 15.23) and benign (ΔCt 15.77) groups demonstrating comparable relative quantities of epithelial cells in malignant compared to benign samples. When comparing relative expression of leukocyte RNA using CD45, there were similar contributions of leukocyte RNA in both benign and malignant samples (ΔCt 17.4 and 17.9 respectively). However, when comparing the relative contribution of leukocyte RNA in brushes and tissues, there was a much higher contribution of leukocyte RNA in surgical resection material (ΔCt 17.4) compared to ERCP biliary brushings (ΔCt 21.1) suggesting that biliary brushings are a purer source of epithelial cells with less stromal contamination than surgical resection material analysed without laser capture microdissection
Analysis of upregulated genes (cut-off of ≥2) by TaqMan array qPCR in the tissue samples had a lower concordance with biliary brush mRNA expression with 7/43 (16%) upregulated in both groups and the trend in level of expression was weak (Pearson correlation coefficient r=0.13). Genes that were consistently elevated between microarray and qPCR of biliary brush samples with similar elevated expression in the tissue samples were MUC4, COL17A1, COL1A1, HOXA10, ITGB8, LIF, and PVT1. Other genes were not considered significantly upregulated in the tissue samples and others appeared downregulated in tissue despite upregulation being shown in the biliary brush samples (CEACAM1, PRKCB1, RAB27A, SERPINA3, and TM4SF18).
Assessment of tissue protein expression using immunohistochemistry
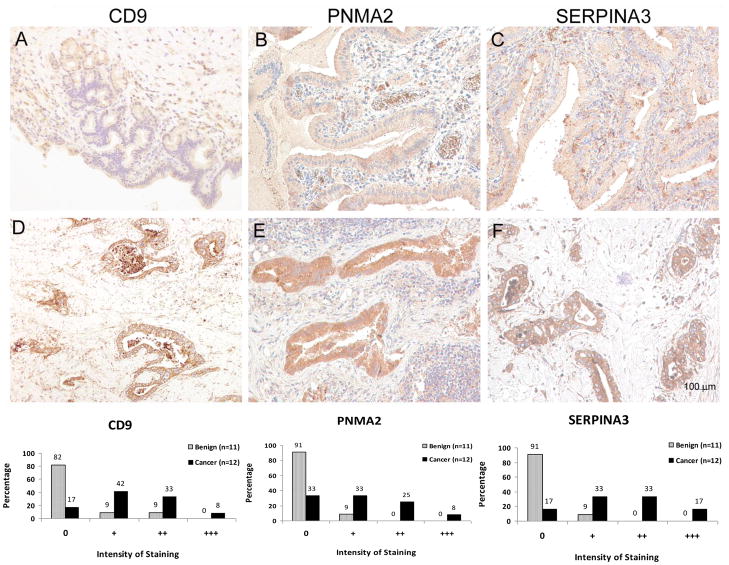
The elevated gene expression of CD9, SERPINA3 and PNMA2 was confirmed at the protein levels by immunohistochemistry (n=23) (Figure 3). There was significantly higher levels of both intensity of staining (CD9 p=0.008, SERPINA3 p=0.001, PNMA2 p=0.01) and proportion of cells stained (CD9 p=0.009, SERPINA3 p=0.001, PNMA2 p=0.01) in biliary tract cancer (Wilcoxon test). Using a cut-off of 0 or + being negative and ++ or +++ positive, staining for CD9, SERPINA3 and PNMA2 had relatively low sensitivity (42%, 50% and 33% respectively) but high specificity (83%, 100% and 100% respectively) for the presence of biliary tract cancer. These results support a role for larger prospective studies of the measurement of these and other identified proteins by immunohistochemistry and/or ELISA using clinical samples such as bile.
Fig 3. Assessment of protein expression using immunohistochemistry.
Immunohistochemistry of benign (A, B and C) and malignant (D, E and F) biliary tissues. In malignant biliary epithelium there was significantly increased protein expression of CD9 (A and D), PNMA2 (B and E) and SERPINA3 (C and F). The graphs demonstrate a moderate proportion of malignant tissues staining positive for the respective proteins but a very high percentage (90% to 100%) of benign tissues having very weak staining or negative staining.
However, POU5F1 (oct3/4) protein expression was negative in both benign and malignant groups. HOXA10 and COL17A1 staining were negative in all samples including positive controls and thus the antibodies were considered unsuitable for further analysis.
DISCUSSION
In this study, RNA isolated from clinical samples of bile and biliary brushings is shown to be highly degraded. However, application of methodology developed for similarly degraded RNA extracted from FFPE tissues allows gene expression profiling that has, as yet, been rarely applied to endoscopically obtained biliary samples. Although there are reports of qPCR for individual highly expressed genes in biliary brushings, we believe that this is the first comprehensive study of the utility of ERCP biliary brushings for whole genome expression analysis. The in vitro experiments demonstrate that the well established and validated methodology described for RNA isolation is suitable and can provide high quality purified total RNA. In contrast to published data in biliary epithelial cell lines that suggest a cytotoxic effect of both bile and x-ray contrast agents [14–15], our data suggest that the RNA degradation does not occur as a result of short term cell exposure to these potentially noxious agents. These fluids appear to have little effect on the quality of RNA isolated from BTC cells cultured for up to 24 hours. One explanation may be that the cancer cell line used differs from normal biliary epithelial cells in their ability to survive in such environments. These data also suggest that the RNA degradation occurs in vivo and as such is not amenable to methodological variations to prevent this. Maximisation of data acquisition is therefore dependent on the use of appropriate cDNA synthesis, qPCR methods and data analysis after isolation of the RNA.
Our results demonstrate that methodology used to assess gene expression in bile or biliary brushings should be similar to that developed for use on degraded RNA from FFPE tissues. Methods used for analysis of good quality intact RNA may not be suitable for use in these clinical samples. In particular, cDNA synthesis kits using primarily oligo-dT primers and PCR primer sets producing amplicon lengths greater than approximately 130bp should be avoided.
In addition to snap frozen biliary brush samples, we isolated RNA from some biliary samples collected at room temperature using RNA later® solution (Ambion Inc, Austin, TX, USA) and found similar results in levels of RNA degradation and gene expression by qPCR in biliary brush samples (data not shown). Other RNA preserving reagents may therefore provide similar quality data and allow a simpler methodology for collection of clinical samples of biliary brushings.
As we have shown previously in cell lines, the concordance in gene expression patterns in the same clinical samples using two different platforms (ie microarray and qPCR), provides strong evidence for the reliability of the gene expression profiling obtained using the microarray method [19]. This suggests that microarray analysis should be suitable for whole genome expression profiling of other benign and malignant biliary diseases such as primary sclerosing cholangitis and autoimmune pancreatitis/IgG4 associated cholangitis. These techniques may be useful in identifying pre-malignant gene signatures in patients with PSC and surveillance for cholangiocarcinomas in patients with dominant biliary strictures.
A potential source of significant ‘noise’ and false positive results in biliary samples is the presence of leukocytes which are commonly found in the bile of patients with biliary disease and biliary obstruction. Relative gene expression of epithelial cell markers (CK19) and leukocyte markers (CD45) using the ΔCt method suggest that the primary source of RNA in our samples was epithelial in origin and leukocyte RNA contributes a relatively small proportion of the overall RNA. In addition, leukocyte specific genes, such as those used for cell sorting (CD45, leukocyte specific protein-1 (LSP1), leukosialin (CD43), CD18 (MHM23), cathepsin G, leukocyte alkaline phosphatise (LAP), CD11 and CD166), were not differentially expressed by microarray analysis in either group providing strong evidence against a significant leukocyte confounding effect. We are therefore confident that we have representative RNA expression data from our samples based primarily on epithelial cell origin.
With regards to the surgical resection tissues, the patterns of relative expression of epithelial and leukocyte RNA suggest that biliary brushings may be a purer source of epithelial RNA than complex, homogenised whole fragments of biliary tissues. The high stromal component of BTC tissue is well recognised and studies comparing microdissected epithelium from macrodissected tissue including tumour stroma demonstrate significant variance in results [20]. Also, cells at the surface of the tumour bulk behave differently from those in the centre. This phenomenon of tumour heterogeneity is well recognised and reported in other cancers such as pancreatic cancer [21]. Possibly for these and other reasons, including a higher contribution of leukocyte RNA in resected cancer tissues, when compared with gene expression in ERCP biliary brushings, elevated mRNA expression in surgical resection material had a concordance of only 16%. In order to reduce this difference in surgical material, methods such as laser capture microdissection to isolate more purified biliary epithelial cells are required. However, a selection of genes with consistently elevated mRNA expression in all sample sets have been identified and deserve further investigation. These include genes with high plausibility for a role in cancer biology such as PVT1, HOXA10, LEF1, and POU5F1 (oct 3/4). We are further investigating the role of these genes in the biology of BTC as well as investigating the role of some other protein markers such as COL17A1 as biomarkers.
In summary, our findings suggest that RNA isolated from bile and biliary brushings is suitable for gene expression profiling. The clinical importance of this study is primarily related to the fact that biliary samples obtained at ERCP are far more readily available than relatively rare surgical resection specimens and thus, provide a potential route for development of new biomarkers for the diagnosis of malignant biliary strictures, as well as to further investigate the biology of other biliary diseases such as primary sclerosing cholangitis.
Supplementary Material
qPCR amplification plots and mean ΔCt values (+/− 1 standard deviation) for representative samples using four different GAPDH primer pairs with increasing size of PCR amplicon length (87bp, 130bp, 220bp and 405bp). Note that using primer pairs producing larger PCR product lengths, there is a fall in measured gene expression of the reference gene GAPDH in the more degraded RNA samples. Thus experiments using RNA from clinical samples of bile or ERCP biliary brushings require design of primer pairs that generate PCR products <130nt in length.
Supplementary Table 1. Primer pairs used for SYBR Green qPCR experiments.
Table 3.
qPCR validation of upregulated genes identified by microarray analysis (normalised to 18S ribosomal RNA)
| Gene | Assay ID | Mean fold change (cancer versus benign) using RNA isolated from biliary brushings | Mean fold change using RNA isolated from biliary Tissue | ||
|---|---|---|---|---|---|
| Microarray (n=9) | qPCR validation (n=9) | Second qPCR validation set (n=9) | qPCR Tissue (n=15) | ||
| 18S | Hs99999901_s1 | 1.0 | 1.0 | 1.0 | 1.0 |
| MUC4 | Hs00366414_m1 | 3.2 | 29.4 | 15.5 | 4.1 |
| MUC5AC | Hs01365616_m1 | 3.6 | 7.4 | 5.1 | 1.2 |
| CD9 | Hs00233521_m1 | 19 | 2.2 | 4.0 | 1.5 |
| NOTCH3 | Hs00166432_m1 | 6 | 8.0 | 4.7 | 1.7 |
| ASPHD1 | Hs00736180_m1 | 10 | 1.7 | 3.6 | 1.0 |
| ATP6V0A2 | Hs00429389_m1 | 10 | 1.1 | 3.1 | 1.0 |
| CEACAM1 | Hs00236077_m1 | 3.5 | 2.1 | 3.1 | 0.6 |
| CELSR1 | Hs00183906_m1 | 5.2 | 7.2 | 7.3 | 1.6 |
| CFDP1 | Hs01041483_m1 | 5.3 | 1.1 | 1.7 | 0.5 |
| COL17A1 | Hs00990073_m1 | 16 | 10.9 | 4.9 | 15.6 |
| COL1A1 | Hs00164004_m1 | 7.2 | 21.6 | 3.4 | 2.5 |
| COL6A3 | Hs00915120_m1 | 8.8 | 26.7 | 2.3 | 1.6 |
| CSPG4 | Hs00426981_m1 | 8.5 | 19.5 | 1.6 | 1.3 |
| HOXA10 | Hs00172012_m1 | 24 | 67.0 | 35.8 | 6.2 |
| HOXB6 | Hs00255831_s1 | 20 | 5.0 | 10.2 | 1.5 |
| ITGB8 | Hs01110394_m1 | 2.7 | 4.0 | 2.6 | 2.0 |
| ITIH5 | Hs00228960_m1 | 20 | 12.4 | 1.5 | 0.3 |
| KRAS | Hs00364282_m1 | −2.5 | 1.1 | 1.5 | 0.8 |
| LAMC2 | Hs01043711_m1 | 2.5 | 2.8 | 3.3 | 1.4 |
| LEF1 | Hs00212390_m1 | 15 | 7.1 | 4.9 | 1.6 |
| LIF | Hs00171455_m1 | 4.7 | 7.5 | 2.0 | 3.1 |
| LUM | Hs00158940_m1 | 11 | 3.7 | 1.1 | 0.6 |
| MAML2 | Hs00418423_m1 | 3.5 | 3.9 | 1.4 | 0.8 |
| MAML3 | Hs00298519_s1 | −8.5 | −1.3 | 3.7 | N/A |
| MAPK1 | Hs01046830_m1 | 2.35 | 1.6 | 2.2 | 0.8 |
| MCM4 | Hs00381539_m1 | 2.2 | 1.5 | 2.5 | 1 |
| MMP2 | Hs01548727_m1 | 2.6 | 11.0 | 2.3 | 0.9 |
| MXRA5 | Hs00377849_m1 | 24 | 10.4 | 3.4 | 1.5 |
| MYC | Hs99999003_m1 | 2.7 | 6.0 | 11.5 | 0.9 |
| NCSTN | Hs00299716_m1 | 3.4 | 1.0 | 1.9 | 0.8 |
| PEAR1 | Hs01378394_m1 | 10.3 | 6.6 | 2.7 | 0.8 |
| PNMA2 | Hs00246721_s1 | 39 | 3.9 | 112.3 | 0.9 |
| POU5F1 | Hs00999632_g1 | 3.3 | 2.5 | 5.5 | 1.4 |
| PRKCB1 | Hs00176998_m1 | 8.3 | 6.8 | 3.1 | 0.5 |
| PTK2 | Hs00178587_m1 | 3 | 1.9 | 1.8 | 0.8 |
| PVT1 | Hs00413039_m1 | 11 | 12.1 | 12.6 | 5.2 |
| RAB27A | Hs00608302_m1 | 2.5 | 2.7 | 2.7 | 0.6 |
| RAD51 | Hs00153418_m1 | 4.3 | 3.0 | 4.2 | 1.5 |
| SERPINA3 | Hs00153674_m1 | 37.8 | 46.7 | 4.4 | 0.1 |
| SPARC | Hs00234160_m1 | 7.1 | 41.4 | 3.1 | 1.1 |
| STAT1 | Hs01014005_m1 | 2.5 | 1.9 | 2.8 | 0.8 |
| TGFBI | Hs00932734_m1 | 12.3 | 3.3 | 2.5 | 1.2 |
| TM4SF18 | Hs00298933_m1 | 10.9 | 0.9 | 5.3 | 0.6 |
| TRIB2 | Hs00222224_m1 | 4.3 | 3.4 | 3.1 | 1.0 |
| VEGFA | Hs99999070_m1 | 2.5 | 2.6 | 2.9 | 1.0 |
Acknowledgments
Financial support
This work was supported by NIH grant PO1CA84203, MRC grant G0801588, the British Liver Trust (with thanks to the Brian Mercer Trust) and liver research funds from the Centre for Hepatology, and undertaken at UCLH/UCL which receive a proportion of funding from the Department of Health’s National Institute for Health Research (NIHR) Biomedical Research Centres funding scheme.
We gratefully acknowledge the help of Scientific Services and The Bloomsbury Centre for Bioinformatics for processing and analysis of the microarrays and Graham Clark in the CR-UK equipment park at Lincoln’s Inn Fields.
Abbreviations used in this paper
- BTC
biliary tract cancer
- CC
cholangiocarcinoma
- ERCP
endoscopic retrograde cholangiopancreatography
- FFPE
formalin fixed paraffin embedded
- qPCR
quantitative real time polymerase chain reaction
- PSC
primary sclerosing cholangitis
Footnotes
Conflicts of interest
None of the authors have a conflict of interest to declare.
CONFLICT OF INTEREST DISCLOSURE
All authors have none to declare.
PERMISSIONS
None required
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael H. Chapman, Email: mhchapman@doctors.org.uk.
Robert Tidswell, Email: rxt560@bham.ac.uk.
James S. Dooley, Email: jdooley@medsch.ucl.ac.uk.
Neomal S. Sandanayake, Email: nsan6752@uni.sydney.edu.au.
Virginie Cerec, Email: v.cerec@ucl.ac.uk.
Maesha Deheragoda, Email: maeshadeheragoda@yahoo.co.uk.
Alvin J.X. Lee, Email: alvin.lee@cancer.org.uk.
Charles Swanton, Email: charles.swanton@cancer.org.uk.
Fausto Andreola, Email: f.andreola@ucl.ac.uk.
Stephen P. Pereira, Email: stephen.pereira@ucl.ac.uk.
References
- 1.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 2.Coudry RA, Meireles SI, Stoyanova R, Cooper HS, Carpino A, Wang X, et al. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9:70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007;87:383–391. doi: 10.1038/labinvest.3700529. [DOI] [PubMed] [Google Scholar]
- 5.Linton KM, Hey Y, Saunders E, Jeziorska M, Denton J, Wilson CL, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer. 2008;98:1403–1414. doi: 10.1038/sj.bjc.6604316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 7.Mengual L, Burset M, Ars E, Ribal MJ, Lozano JJ, Minana B, et al. Partially degraded RNA from bladder washing is a suitable sample for studying gene expression profiles in bladder cancer. Eur Urol. 2006;50:1347–1355. doi: 10.1016/j.eururo.2006.05.039. discussion 1355–1346. [DOI] [PubMed] [Google Scholar]
- 8.Hansel DE, Rahman A, Hidalgo M, Thuluvath PJ, Lillemoe KD, Shulick R, et al. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am J Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obama K, Ura K, Li M, Katagiri T, Tsunoda T, Nomura A, et al. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology. 2005;41:1339–1348. doi: 10.1002/hep.20718. [DOI] [PubMed] [Google Scholar]
- 10.Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, et al. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025–1038. doi: 10.1002/hep.21330. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann G, Nattermann J, Nischalke HD, Gorschluter M, Kuntzen T, Ahlenstiel G, et al. Detection of human aspartyl (asparaginyl) beta-hydroxylase and homeobox B7 mRNA in brush cytology specimens from patients with bile duct cancer. Endoscopy. 2006;38:604–609. doi: 10.1055/s-2006-925065. [DOI] [PubMed] [Google Scholar]
- 12.Matull WR, Andreola F, Loh A, Adiguzel Z, Deheragoda M, Qureshi U, et al. MUC4 and MUC5AC are highly specific tumour-associated mucins in biliary tract cancer. Br J Cancer. 2008;98:1675–1681. doi: 10.1038/sj.bjc.6604364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito H, Satoh K, Hamada S, Hirota M, Kanno A, Ishida K, et al. The evaluation of MSX2 mRNA expression level in biliary brush cytological specimens. Anticancer Research. 2011;31:1011–1017. [PubMed] [Google Scholar]
- 14.Benedetti A, Alvaro D, Bassotti C, Gigliozzi A, Ferretti G, La Rosa T, et al. Cytotoxicity of bile salts against biliary epithelium: a study in isolated bile ductule fragments and isolated perfused rat liver. Hepatology. 1997;26:9–21. doi: 10.1002/hep.510260102. [DOI] [PubMed] [Google Scholar]
- 15.Ju YM, Kim MH, Lee SK, Seo DW, Min YI, Kim JY. Comparative cytotoxicity of low-osmolar nonionic and high-osmolar ionic contrast media to dog gallbladder epithelial cells. Gastrointest Endosc. 2002;55:382–386. doi: 10.1067/mge.2002.121595. [DOI] [PubMed] [Google Scholar]
- 16.Saijyo S, Kudo T, Suzuki M, Katayose Y, Shinoda M, Muto T, et al. Establishment of a new extrahepatic bile duct carcinoma cell line, TFK-1. Tohoku J Exp Med. 1995;177:61–71. doi: 10.1620/tjem.177.61. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Adams EJ, Green JA, Clark AH, Youngson JH. Comparison of different scoring systems for immunohistochemical staining. J Clin Pathol. 1999;52:75–77. doi: 10.1136/jcp.52.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee AJ, East P, Pepper S, Nicke B, Szallasi Z, Eklund AC, et al. Concordance of exon array and real-time PCR assessment of gene expression following cancer cell cytotoxic drug exposure. Cell Cycle. 2008;7:3947–3948. doi: 10.4161/cc.7.24.7217. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama Y, Sugiyama K, Hirai Y, Akiyama F, Hasumi K. Microdissection is essential for gene expression profiling of clinically resected cancer tissues. Am J Clin Pathol. 2002;117:109–116. doi: 10.1309/G1C8-39MF-99UF-GT2K. [DOI] [PubMed] [Google Scholar]
- 21.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR amplification plots and mean ΔCt values (+/− 1 standard deviation) for representative samples using four different GAPDH primer pairs with increasing size of PCR amplicon length (87bp, 130bp, 220bp and 405bp). Note that using primer pairs producing larger PCR product lengths, there is a fall in measured gene expression of the reference gene GAPDH in the more degraded RNA samples. Thus experiments using RNA from clinical samples of bile or ERCP biliary brushings require design of primer pairs that generate PCR products <130nt in length.
Supplementary Table 1. Primer pairs used for SYBR Green qPCR experiments.