Abstract
Age-related cognitive decline is likely promoted by accumulated brain injury due to chronic conditions of aging, including neurodegenerative and vascular disease. Since common neuronal mechanisms may mediate the adaptation to diverse cerebral insults, we hypothesized that susceptibility for age-related cognitive decline may be due in part to a shared genetic network. We have therefore performed a genome-wide association study using a quantitative measure of global cognitive decline slope, based on repeated measures of 17 cognitive tests in 749 subjects from the Religious Orders Study. Top results were evaluated in three independent replication cohorts, consisting of 2,279 additional subjects with repeated cognitive testing. As expected, we find that the Alzheimer’s disease (AD) susceptibility locus, APOE, is strongly associated with rate of cognitive decline (PDISC=5.6×10−9; PJOINT=3.7×10−27). We additionally discover a variant, rs10808746, which shows consistent effects in the replication cohorts and modestly improved evidence of association in the joint analysis (PDISC=6.7×10−5; PREP=9.4×10−3; PJOINT=2.3×10−5). This variant influences the expression of two adjacent genes, PDE7A and MTFR1, which are potential regulators of inflammation and oxidative injury, respectively. Using aggregate measures of genetic risk, we find that known susceptibility loci for cardiovascular disease, type II diabetes, and inflammatory diseases are not significantly associated with cognitive decline in our cohort. Our results suggest that intermediate phenotypes, when coupled with larger sample sizes, may be a useful tool to dissect susceptibility loci for age-related cognitive decline and uncover shared molecular pathways with a role in neuronal injury.
INTRODUCTION
Decline in cognitive performance occurs with advancing age and is associated with a variety of common, age-related chronic medical conditions. Alzheimer’s disease (AD) is the most prevalent cause of dementia (Reitz et al., 2011a); however, many other common adult illnesses, including type II diabetes (Croxson and Jagger, 1995; Grodstein et al., 2001; Reijmer et al., 2010), cerebrovascular disease (Desmond et al., 2000; Pendlebury and Rothwell, 2009) as well as other cardiovascular risk factors (Desmond et al., 1993; Warsch and Wright, 2010), and inflammatory disorders (Lucin and Wyss-Coray, 2009) have been implicated in age-related cognitive decline. Based on autopsy series from community-based cohorts, most individuals with dementia have multiple contributory pathologies at the time of death (Neuropathology Group, 2001; Sonnen et al., 2007; Troncoso et al., 2008). It is likely that diverse forms of brain injury interact to accelerate cognitive decline. For example, it has been suggested that vascular-related brain injury may promote the development of AD pathology or, less directly, the clinical manifestation of AD-related cognitive decline (Launer et al., 2008; Schneider and Bennett, 2010; Warsch and Wright, 2010). Results from a variety of experimental paradigms for the study of neuronal injury and repair indicate that overlapping cellular and molecular mechanisms likely mediate the response to a diversity of central nervous system insults (Bishop et al., 2010; Cho et al., 2010; Lucin and Wyss-Coray, 2009; Martinez-Vicente and Cuervo, 2007; Ross and Poirier, 2004). Besides the local reactions to brain lesions, the ability of neuronal networks to adapt to and compensate for an accumulated burden of injury, sometimes referred to as cognitive reserve (Stern, 2009), likely has a substantial impact on the trajectory of cognitive decline. Studies of elder twins suggest substantial heritability in cognitive performance in late life (McClearn et al., 1997; Swan et al., 1990), and we hypothesize that a core genetic network might therefore impact susceptibility for rate of age-related cognitive decline.
Recently, genome-wide association studies have proven a successful strategy for discovering susceptibility genes for complex human traits, including neurologic disorders, such as AD (Bertram and Tanzi, 2009). Besides the apolipoprotein E locus (APOE), these studies have identified common variants in ABCA7, BIN1, CD2AP, CD33, CLU, CR1, EPHA1, MS4A4/MS4A6E, and PICALM as associated with AD susceptibility (Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Naj et al., 2011; Seshadri et al., 2010). While elucidating the functional impact of disease-associated genetic variants remains an active area of investigation, there is evidence that these genes may have important roles beyond AD pathogenesis in affecting other disorders potentially relevant to cognitive decline. For example, in addition to the well-known effect of the APOE locus in promoting AD risk, this locus has also been associated with dyslipidemia, cardiovascular disease, and increased cerebral infarcts (Eichner et al., 2002; Kim et al., 2003; McCarron et al., 1999). Similarly, polymorphisms in the CR1 gene, encoding a complement receptor, have previously been associated with susceptibility for infectious disease, particularly malaria (Cockburn et al., 2004; Rowe et al., 1997). We have shown that polymorphisms in both APOE (Wilson et al., 2002a, 2002b) and CR1 (Chibnik et al., 2011) have a measurable impact on age-related cognitive decline, including in subjects without dementia, and further, that these associations are mediated in part by an effect on promoting amyloid plaque pathology (Bennett et al., 2005a; Chibnik et al., 2011).
The Religious Orders Study (ROS) is following more than 1,100 older Catholic nuns, priests and brothers who have completed up to 16 years of annual cognitive testing. Here, we have leveraged available genotyping data for 749 subjects of European ancestry with longitudinal cognitive data to conduct a genome scan for loci associated with the rate of age-related cognitive decline. We report efforts to replicate the best results using data from two complementary, community-based studies, the Rush Memory and Aging Project (MAP) and Chicago Health and Aging Project (CHAP), as well as a predominantly clinic-derived subject sample from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), and offer evidence in support of replication for one variant. Finally, we explore whether known genetic susceptibility factors associated with other illnesses that are known to influence the risk of dementia, such as AD, cardiovascular disease and type II diabetes, also affect age-related cognitive decline.
METHODS
Subjects
Subjects are participants from four longitudinal studies, which are each described below. The number of study subjects with genotyping data, included in the genetic analyses, are described in the Genotyping Methods sub-section, and also summarized in Table 1.
Table 1.
Demographic and clinical characteristics of study cohorts.
| ROS | MAP | CHAP3 | ADNI | |
|---|---|---|---|---|
| N | 749 | 825 | 737 | 717 |
| Age at Enrollment | 75.3 (±7.2) | 80.8 (±6.6) | 72.0 (±5.5) | 75.3 (±6.9) |
| Education (years) | 18.2 (±3.4) | 14.8 (±2.9) | 14.6 (±3.2) | 15.6 (±3.0) |
| Male | 254 (34.0%) | 222 (26.9%) | 291 (39.5%) | 422 (58.9%) |
| Cognitive Decline Slope | −0.007 (±0.1) | 0.012 (±0.1) | 0.001 (±0.04) | −0.001 (±0.04) |
| Cognitively Normal1 | 444 (59.4%) | 460 (55.8%) | 265 (68.7%) | 211 (29.4%) |
| Mild Cognitive Impairment1 | 151 (20.2%) | 200 (24.2%) | 71 (18.4%) | 205 (28.6%) |
| Dementia1,2 | 152 (20.4%) | 165 (20.0%) | 46 (11.9%) | 301 (42.0%) |
Cognitive status at time of last evaluation.
19.7% of ROS, 19% MAP, and 11% CHAP met NINCDS criteria for possible or probable AD.
Diagnoses of dementia and mild cognitive impairment only available for a subset (n=386) of CHAP.
The Religious Order Study (ROS), started in 1994, enrolled Catholic priests, nuns and brothers, aged 53 or older from about 40 groups in 12 states. Since January 1994, 1,132 participants completed their baseline evaluation, of whom 1,001 are non-Hispanic white, and the follow-up rate of survivors of exceeds 90%. Participants were free of known dementia at enrollment and agreed to annual clinical evaluations (Bennett et al., 2006a). More detailed description of the ROS can be found in prior publications (Bennett et al., 2006a).
The Rush Memory and Aging Project (MAP), started in 1997, enrolls older men and women from assisted living facilities in the Chicago area with no evidence on dementia at baseline. Since October 1997, 1285 participants completed their baseline evaluation, of whom 1118 were non-Hispanic white. The follow-up rate of survivors of exceeds 90%. Similar to ROS, participants agreed to annual clinical evaluations. More detailed descriptions can be found in previous studies (Bennett et al., 2006a, 2005b).
The Chicago Health and Aging Project (CHAP) is a biracial (63% African American, 37% non-Hispanic European American) longitudinal population study of all participating residents 65-years-of-age-and-older of four adjacent neighborhoods of the south side of Chicago that examines risk factors for cognitive decline and Alzheimer’s Disease (AD). Participation was 78.6% of all community residents at baseline and 80% to 85% retention of survivors at follow-up. The study began in 1993 and enrolls successive age cohorts of community residents as they attain the age of 65 years. To date, 10,712 subjects have contributed data. The subjects included in these analyses are a stratified random sample of European American subjects. More details may be found in prior publications (Bienias et al., 2003; Evans et al., 2003).
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a 5-year public-private partnership for the study of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co- investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The cohort includes 800 adults, ages 55 to 90, to participate in the research – approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). For up-to-date information, see www.adni-info.org.
Clinical and cognitive evaluation
Supplementary Table 1 summarizes the specific longitudinal cognitive testing data that was available within our discovery and replication cohorts. The ROS and MAP studies annually administer 21 cognitive tests, of which 17 tests in common were incorporated into summary measures of 5 domains of cognitive function--episodic memory (7 tests), visuospatial ability (2 tests), perceptual speed (2 tests), semantic memory (3 tests) and working memory (3 tests)--as previously described (Bennett et al., 2002, 2005b; Wilson et al., 2002c, 2005). The tests from each area of cognition were converted to z-scores, using the mean and SD from the baseline evaluation of all participants, and averaged to yield summary measures of each area of cognitive function as previously described (Bennett et al., 2002, 2005b; Wilson et al., 2002c, 2005). The global cognition summary measure used in our primary analyses was computed by averaging the 5 summary scores for each cognitive subdomain. Summary measures have the advantage of minimizing floor and ceiling effects, and other sources of random variability. A valid summary score required that at least half of the component scores be present. The global cognition summary measures in the CHAP and ADNI cohorts were constructed using identical procedures as ROS and MAP on the available longitudinal testing data (Supplementary Table 1). In CHAP, cognitive testing included a subset of 3 instruments from ROS and MAP. In ADNI, 8 tests overlapped with ROS and MAP, and we additionally incorporated available longitudinal data from 11 other tests.
The clinical diagnoses of dementia and AD were made following the recommendations of the joint working group of the National Institute of Neurologic and Communicative Disorders and Stroke and the AD and Related Disorders Association (McKhann et al., 1984), as previously described in detail (Bennett et al., 2006b). Mild cognitive impairment (MCI) referred to those individuals rated as cognitively impaired by the neuropsychologist but not demented by the examining physician, as previously described (Bennett et al., 2002).
Statistical modeling of cognitive decline
Mixed effects models were used to characterize individual paths of change in the cognitive summary measures, including terms for age, sex, and years of education as fixed effects (Laird and Ware, 1982; Wilson et al., 2000, 2002c). In this approach, each individual’s path is assumed to follow the mean path of the group except for random effects that cause the initial level of function (i.e. intercept) to be higher or lower and the rate of change (i.e. slope) to be faster or slower. These random effects are assumed to follow a bivariate normal distribution. The random and fixed effects were then used to estimate individual trajectories of cognitive decline. Residual, individual cognitive decline slope terms were extracted from the mixed models, after adjustment for the effects of age, sex, and education. Person-specific, adjusted residual slopes were then used as a quantitative outcome phenotype for the genetic association analyses. These estimates equate to the difference between an individual’s slope and the predicted slope of an individual of the same age, sex and education level. This method, including adjustment for age, sex and education was used for all four cohorts.
Genotyping
DNA from ROS and MAP subjects was extracted from whole blood, lymphocytes or frozen post-mortem brain tissue and genotyped on the Affymetrix Genechip 6.0 platform at either the Broad Institute’s Center for Genotyping (n=1,204) or the Translational Genomics Research Institute (n=674). These two sets of data underwent the same quality control (QC) analysis in parallel, and genotypes were pooled. Only self-declared non-Hispanic Caucasians were genotyped to minimize population heterogeneity. The PLINK toolkit (http://pngu.mgh.harvard.edu/~purcell/plink/) (Purcell et al., 2007) was used to implement our QC pipeline. We applied standard quality control measures for subjects (genotype success rate >95%, genotype-derived gender concordant with reported gender, excess inter/intra-heterozygosity) and for single nucleotide polymorphisms (SNPs) (HWE p > 0.001; MAF > 0.01, genotype call rate > 0.95; misshap test > 1×10−9). Subsequently, EIGENSTRAT (Price et al., 2006) was used to identify and remove population outliers using default parameters. At the conclusion of the QC pipeline, data on 672,266 SNPs was available for 1,709 total ROS and MAP subjects. 749 ROS subjects and 825 MAP subjects with longitudinal cognitive data and high-quality genotyping data were available for the discovery and replication analyses, respectively (Table 1).
For the replication analysis, the top 50 independent SNPs (P<10−4, minor allele frequency > 10%) based on the ROS discovery stage analysis were extracted from the quality-controlled MAP genome-wide dataset (above). The generation and quality control procedures for the ADNI genotyping dataset was previously described (Biffi et al., 2010). 717 ADNI subjects with longitudinal cognitive testing data and genotypes were available for our analyses. CHAP subject DNA was extracted from whole blood and genotyping of top SNPs from the discovery stage genome-wide scan using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry on a MassARRAY platform (Sequenom). After excluding subjects for failed genotyping exceeding the 10% threshold, 414 individuals remained for subsequent analysis (genotyping rate in these subjects was >99%). All SNP allele frequencies satisfied Hardy-Weinberg equilibrium (p > 0.001). For evaluation of AD susceptibility alleles and for the development of the aggregate genetic risk scores, SNP genotypes in ROS and MAP were imputed using MACH software (version 1.0.16a) (Scott et al., 2007) and HapMap release 22 CEU (build 36) (Frazer et al., 2007) as a reference.
Genome-wide association analysis
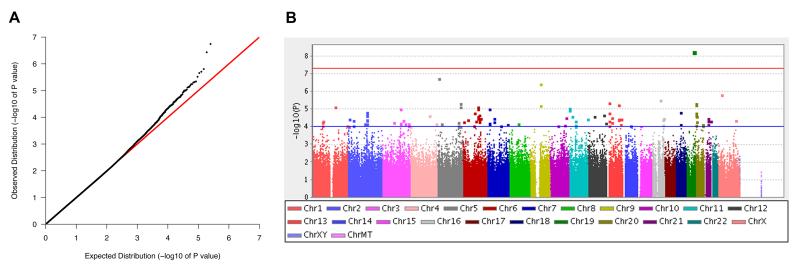
The genome-wide association analysis in the ROS discovery cohort, as well as the targeted association analysis of the top 50 SNPs in the MAP, CHAP, and ADNI replication cohorts, were performed using linear regression implemented in PLINK software (Purcell et al., 2007). As described above, the outcome phenotype was the residual cognitive decline slope extracted from the mixed effects models, after adjustment for age, gender and education. The association analysis in the ROS discovery cohort was additionally adjusted for the first three ancestry principal components calculated using EIGENSTRAT (Price et al., 2006). In order to perform a joint replication and study-wide meta-analysis, the cohort-specific PLINK association output was subsequently analyzed using METAL software (Willer et al., 2010). The default METAL parameters were used, in which meta-analysis is based on p-values and direction of effect, and weighted by sample size in each cohort. The Manhattan plot (Figure 2B) was generated using Haploview software (Barrett et al., 2005), and the association plot (Figure 3B) was generated using the SNAP web-tool (Johnson et al., 2008).
Figure 2.
A genome-wide association scan for age-related cognitive decline. Using the residual cognitive decline slope as an outcome trait, associations were evaluated for 672,266 SNPs in the discovery cohort consisting of 749 ROS subjects. (A) Quantile-quantile plot. (B) Manhattan plot. Thresholds for suggestive (P<10−4, blue) and genome-wide (P<5×10−8, red) significance are indicated.
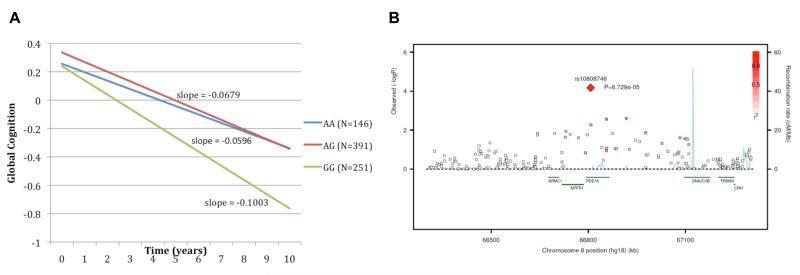
Figure 3.
Association of rs10808746 at the PDE7A/MTFR1 locus with rate of cognitive decline. (A) Mean linear trajectories of cognitive decline within the ROS discovery cohort for each of the rs10808746 genotype classes, demonstrating evidence that the rs10808746G allele is associated with increased rate of cognitive decline. (B) Plot showing rs10808746 association peak over the PDE7A, MTFR1, and ARMC1 genes.
Aggregate Genetic Risk Scores
We developed four disease specific cumulative genetic risk scores (GRSs) based on published genome wide significant (p<10−8) SNPs for the following disease categories: Alzheimer’s Disease; Cardiovascular Disease (including Myocardial Infarction, LDL Cholesterol, HDL Cholesterol, Triglycerides, Hypertension, and Stroke); Inflammatory Disease (including Celiac Disease, Crohn’s Disease, Irritable Bowel Disease, Multiple Sclerosis, Psoriasis, Rheumatoid Arthritis, Systemic Lupus Erythematosus, Type 1 Diabetes, and Ulcerative Colitis); and Type 2 Diabetes. Genome wide significant SNPs for each disease category were identified through a literature review, primarily using the Catalogue of Published Genome-Wide Association Studies available online at http://www.genome.gov/26525384. For the cardiovascular disease and inflammatory disease categories, which have multiple component outcomes, SNPs that had reference alleles which were both protective for one disease and a risk factor for a second disease were removed. To limit the amount of correlation between SNPs, we further refined this list by identifying all pairs of SNPs with a linkage disequilibrium r2≥0.5 and keeping only the SNP in those pairs with the lowest p-value. Once we identified a final SNP set, the four GRSs were created by summing up the number of category specific risk alleles for each individual. To assure that our scores were comprehensive, we used imputed allelic dosages for SNPs that were not genotyped within our population. We then examined the associations between the resulting genetic risk scores and global cognitive decline using linear regression models adjusted for age, sex, and education. The final list of SNPs and risk alleles included in each score, as well as relevant references, can be found in Supplementary Table 7.
Gene expression QTL analysis
Gene expression levels were quantified using mRNA derived from peripheral blood mononuclear cells (PBMCs) of 228 subjects of European ancestry with Relapsing Remitting (RR) Multiple Sclerosis (MS) using an Affymetrix Human Genome U133 Plus 2.0. These data were collected between July 2002 and October 2007, as part of the Comprehensive Longitudinal Investigation of MS at the Brigham and Women’s Hospital. The data is available on the Gene Expression Omnibus website (GSE16214) (De Jager et al., 2009). DNA from each individual was genotyped on the Affymetrix GeneChip 6.0 platform as a part of a case-control multiple sclerosis meta-analysis (De Jager et al., 2009). Using a Spearman Rank Correlation, we tested for association using an additive model for allelic dosage as an independent variable and residuals of expression as the dependent variable. Significance w as established by comparing the association p-values to the empiric distribution of p-values generated by permuting expression phenotypes 10,000 times independently for each gene. Similar methods were used to evaluate expression within the publically available lymphoblastic cell line expression dataset from 60 CEU individuals in the HapMap project (Stranger et al., 2007).
RESULTS
Characteristics of the discovery cohort
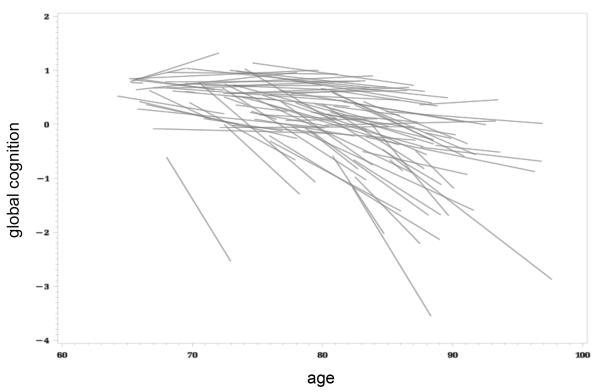
Following quality control, genome-wide genotype data (672,266 SNPs) were available on 749 non-Hispanic, white subjects from the ROS with longitudinal cognitive testing. Detailed cohort characteristics are presented in Table 1. The mean age at enrollment was 75 years, and subjects were followed for 9 years, on average (range 1-15 years of follow-up). Cognitive decline trajectories were quantified based on annual performance of 17 distinct neuropsychological tests sampling 5 cognitive domains (episodic memory, perceptual speed, semantic memory, visuospatial ability, and working memory) (Supplementary Table 1). As previously described (Wilson et al., 2002c), a subject’s performance on each test was standardized and an average, aggregate measure of global cognitive performance was computed. At recruitment, all subjects were without known dementia. At their last evaluation, 59% of subjects retained normal cognition, 20% had mild cognitive impairment and 20% had a diagnosis of dementia. We used linear mixed effects modeling, including all available longitudinal cognitive testing, to obtain a residual cognitive decline slope for each individual, adjusting for age at enrollment, gender, and years of education, (Figure 1). This slope parameter describes the person-specific rate of global cognitive change as a quantitative, continuous outcome and is the primary outcome measure that we used in our association study.
Figure 1.
Variability in the rate of age-related cognitive decline. Linear cognitive trajectories are shown for 100 random subjects from the ROS cohort, based on mixed effect modeling of repeated measures of the global cognition summary score, incorporating 17 distinct cognitive tests. Trajectories are adjusted for the effects baseline age, gender, and education. The residual cognitive decline slope was used as an outcome for the genome-wide association analysis. The distribution of the cognitive decline trait for the entire ROS discovery cohort is shown in Supplementary Figure 1.
Genome-wide association scan for age-related cognitive decline
We implemented our genome-wide association scan using the residual cognitive decline slope for each individual as an outcome trait (Figure 2). The genomic inflation factor was 1.009, indicating no significant inflation of our test statistics. A selection of top results from the scan, based on our replication analysis, are shown in Table 2, and complete results (P<10−4) are provided in the Supplementary Material (Table S8 and S9). As expected from prior studies (Feskens et al., 1994; Haan et al., 1999; Henderson et al., 1995; Hyman et al., 1996; Jonker et al., 1998; McQueen et al., 2007; Wilson et al., 2002a, 2002b), the strongest associations with the rate of cognitive decline were found for markers at the APOE locus. No other locus association surpassed the genome-wide significance threshold (P<5×10−8). However, numerous polymorphisms demonstrate suggestive evidence of association (P<10−4) with rate of cognitive decline, including several that fall within or adjacent to candidate genes previously implicated in cognition (CTNND2, rs2973488, P=1.8×10−7) (Israely et al., 2004; Medina et al., 2000) or AD susceptibility (SORCS1, rs12219216, P=8.0×10−5) (Reitz et al., 2011b). As we have previously reported (Chibnik et al., 2011), we find nominal evidence of association at the CR1 locus (rs6656401, p=0.048), but do not detect evidence of association for other known AD susceptibility variants with the rate of cognitive decline in our discovery cohort (Table S2).
Table 2.
Top results of genome-wide scan for rate of cognitive decline.
| Discovery (ROS) | Replication | Joint | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | C | A1/2 | MAF | Beta (95% CI) | P | PMAP | PCHAP | PADNI | PREP | P | Direction | Gene(s) |
| rs4420638 | 19 | C/T | 0.19 | −0.039 (−0.051,−0.026) | 5.62E-09 | 9.38E-09 | 5.06E-06 | 5.31E-08 | 9.12E-20 | 3.74E-27 | ++++ | TOMM40,APOE |
| rs10808746 | 8 | T/C | 0.43 | 0.021 (0.011,0.031) | 6.73E-05 | 0.091 | 0.274 | 0.089 | 9.44E-03 | 2.30E-05 | ++++ | PDE7A,MTFR1 |
| rs10769565 | 11 | G/A | 0.34 | −0.023 (−0.034,−0.013) | 9.09E-06 | 0.455 | 0.159 | N/A | 0.131 | 1.63E-04 | +++? | OR56A4,OR56A1 |
| rs9387454 | 6 | A/G | 0.36 | −0.023 (−0.033,−0.012) | 2.12E-05 | 0.870 | 0.014 | 0.710 | 0.199 | 1.26E-03 | −−−+ | |
| rs11939527 | 4 | T/C | 0.25 | −0.023 (−0.034,−0.011) | 9.55E-05 | 0.876 | 0.196 | 0.154 | 0.149 | 1.41E-03 | −+−− | EVC |
| rs376535 | 2 | C/T | 0.10 | −0.034 (−0.050,−0.018) | 4.38E-05 | 0.113 | 0.031 | 0.097 | 0.230 | 2.10E-03 | +−++ | SLC8A1 |
| rs2571577 | 6 | C/T | 0.20 | −0.027 (−0.039,−0.014) | 3.42E-05 | 0.688 | 0.039 | 0.715 | 0.255 | 2.32E-03 | +−++ | LAMA2 |
| rs13015892 | 2 | A/G | 0.17 | −0.031 (−0.044,−0.017) | 1.50E-05 | 0.790 | 0.589 | 0.162 | 0.347 | 3.04E-03 | −+−− | |
| rs9602785 | 13 | C/T | 0.15 | −0.033 (−0.047,−0.019) | 5.71E-06 | 0.478 | 0.770 | 0.843 | 0.482 | 4.17E-03 | ++++ | SLITRK6 |
| rs11701130 | 21 | A/C | 0.26 | 0.024 (0.013,0.036) | 5.06E-05 | 0.141 | 0.101 | 0.116 | 0.349 | 4.67E-03 | +−++ | PCBP3 |
| rs4760608 | 12 | C/A | 0.21 | −0.027 (−0.040,−0.015) | 2.68E-05 | 0.674 | 0.100 | 0.779 | 0.398 | 4.77E-03 | +−++ | COL2A1 |
| rs2973488 | 5 | T/A | 0.11 | −0.044 (−0.061,−0.028) | 1.81E-07 | 0.823 | 0.688 | 0.524 | 0.792 | 5.06E-03 | ++−+ | CTNND2 |
| rs7606972 | 2 | G/T | 0.36 | 0.022 (0.012,0.033) | 2.08E-05 | 0.541 | 0.803 | 0.811 | 0.520 | 7.57E-03 | − − − − | |
| rs9875783 | 3 | A/C | 0.25 | −0.024 (−0.036,−0.012) | 5.78E-05 | 0.894 | 0.803 | 0.075 | 0.437 | 7.62E-03 | −++− | |
| rs524398 | 2 | A/G | 0.42 | −0.022 (−0.032,−0.012) | 2.45E-05 | 0.155 | 0.971 | 0.736 | 0.519 | 7.84E-03 | −−++ | THSD7B |
| rs610315 | 10 | A/T | 0.26 | −0.024 (−0.036,−0.013) | 3.00E-05 | 0.634 | 0.090 | 0.918 | 0.535 | 8.94E-03 | −+−+ | AFAP1L2 |
| rs6919160 | 6 | A/T | 0.31 | −0.024 (−0.035,−0.013) | 1.72E-05 | 0.562 | 0.354 | 0.521 | 0.589 | 9.14E-03 | −+−− | |
| rs347311 | 1 | C/G | 0.31 | −0.025 (−0.036,−0.014) | 7.62E-06 | 0.446 | 0.094 | 0.791 | 0.729 | 0.012 | −+−+ | NOS1AP |
| rs4129955 | 4 | T/C | 0.47 | −0.020 (−0.030,−0.010) | 6.69E-05 | 0.195 | 0.589 | 0.123 | 0.693 | 0.020 | −+−− | |
| rs3212701 | 19 | A/G | 0.25 | 0.052 (0.028,0.075) | 2.69E-05 | 0.761 | 0.661 | 0.397 | 0.456 | 0.021 | +−++ | JAK3 |
| rs1595781 | 3 | G/A | 0.31 | −0.023 (−0.034,−0.012) | 6.70E-05 | 0.665 | 0.617 | 0.551 | 0.717 | 0.022 | +−++ | |
| rs276420 | 13 | A/C | 0.45 | −0.021 (−0.031,−0.011) | 3.25E-05 | 0.226 | 0.947 | 0.266 | 0.887 | 0.028 | −−−+ | |
| rs12660119 | 6 | G/A | 0.11 | −0.038 (−0.054,−0.021) | 7.28E-06 | 0.912 | 0.792 | 0.827 | 0.970 | 0.029 | +−+− | WASF1 |
| rs1041676 | 1 | T/C | 0.32 | −0.023 (−0.034,−0.012) | 4.73E-05 | 0.567 | 0.998 | 0.384 | 0.882 | 0.032 | −++− | NEGR1 |
| rs2086366 | 15 | T/C | 0.37 | 0.020 (0.010,0.030) | 4.77E-05 | 0.992 | 0.392 | 0.827 | 0.706 | 0.039 | +−+− | |
| rs12219216 | 10 | A/G | 0.41 | 0.020 (0.010,0.031) | 8.01E-05 | 0.777 | 0.359 | 0.415 | 0.915 | 0.040 | ++−+ | SORCS1 |
| rs1534378 | 7 | T/C | 0.11 | −0.030 (−0.044,−0.015) | 5.51E-05 | 0.848 | 0.729 | 0.654 | 0.714 | 0.042 | −+−− | |
| rs12362733 | 11 | C/T | 0.19 | 0.027 (0.015,0.040) | 2.72E-05 | 0.267 | 0.168 | 0.643 | 0.891 | 0.049 | −+−+ | LUZP2 |
Summary results are shown for SNPs with the best evidence of association in the study-wide meta-analysis (PJOINT < 0.05). The top 50 independent SNPs from the ROS discovery cohort (MAF > 10% and LD-pruned based on r2>0.5) were evaluated for replication (complete results presented in Supplementary Table 8). C=Chromosome, A1/2=minor allele/major allele, MAF=minor allele frequency in ROS cohort. Beta (95% confidence interval) and P-values are shown for association with rate of cognitive decline in the discovery stage. P-values are also shown for associations in the MAP, CHAP, and ADNI replication cohorts. Meta-analytic P-values are shown for the replication cohorts (PREP) and for the study-wide analysis including all cohorts (PJOINT). Direction of effect (+ = protective, − = risk) is indicated for all 4 cohorts.
Replication of variants associated with rate of cognitive decline
To replicate the results of our genome scan, the top 50 independent SNPs (P<10−4, minor allele frequency > 10%) from our discovery stage were evaluated in three additional cohorts of older individuals with longitudinal measures of cognitive performance (Table 2 and Table S8). MAP, CHAP, and ADNI are described in the Methods and the cohort characteristics are summarized in Table 1. A summary of all cognitive testing data used from each study is provided in Supplementary Table 1. As in ROS, available cognitive data for MAP, CHAP, and ADNI subjects were incorporated into aggregate global cognition scores, and mixed effect modeling of longitudinal data was used to compute residual, adjusted global cognitive decline slopes for all subjects. The SNPs selected for replication were evaluated in each of the three replication cohorts, and meta-analysis was implemented to compute summary association statistics (PREP) across the replication cohorts, consisting of 2,279 subjects. We also determined a study-wide association meta-analysis statistic (PJOINT), consisting of 3,028 total subjects, including the results from the discovery cohort and those of the three replication cohorts. The results of the replication and joint analyses are shown in Table 2. We hypothesized that discovered variants that truly impact rate of cognitive decline would show robust and consistent effects across these sample collections, despite modest differences in cohort make-up, such as mean age, cognitive testing procedures, and years of follow-up.
As expected, a SNP at the APOE locus, rs4420638, showed significant replication for association with rate of cognitive decline across each independent sample evaluated and in the pooled replication cohort (PMAP=9.4×10−9, PCHAP=5.1×10−6, PADNI=5.3×10−8, PREP=9.1×10−20), and the overall association was strongly enhanced in the joint analysis (PROS=5.6×10−9, PJOINT=3.7×10−27). Although no other variant met a threshold of genome-wide significance at the conclusion of the replication study, a chromosome 8 SNP, rs10808746, showed consistent direction of effect on the rate of cognitive decline in each cohort (the major allele rs10808746G is associated with increased risk of cognitive decline) and suggestive evidence of replication in the combined replication cohort (PREP=0.009). Further supporting replication, in the study-wide joint analysis, rs10808746 shows modest evidence of enhanced association once all available data are considered (PROS=6.7×10−5, PJOINT=2.3×10−5).
Figure 3A shows the mean trajectory of cognitive decline within the discovery cohort for the three genotype classes of rs10808746. These data are supportive of the major allele rs10808746G being associated with more rapid cognitive decline. To better understand the impact of this variant on cognition, we evaluated associations with cognitive decline based on measures for each of the 5 domains of the ROS global cognitive score (Table S3). This polymorphism was associated with the rate of cognitive decline in episodic memory (P=3×10−4), perceptual speed (P=0.048), semantic memory (P=0.015), and working memory (P=0.0046), but not visuospatial processing (P=0.13), suggesting that, while it appears to have a predominant effect on decline in episodic memory, its role may not be limited to a single, functionally distinct anatomic region or circuit.
Analysis of rs10808746 effect on local gene expression
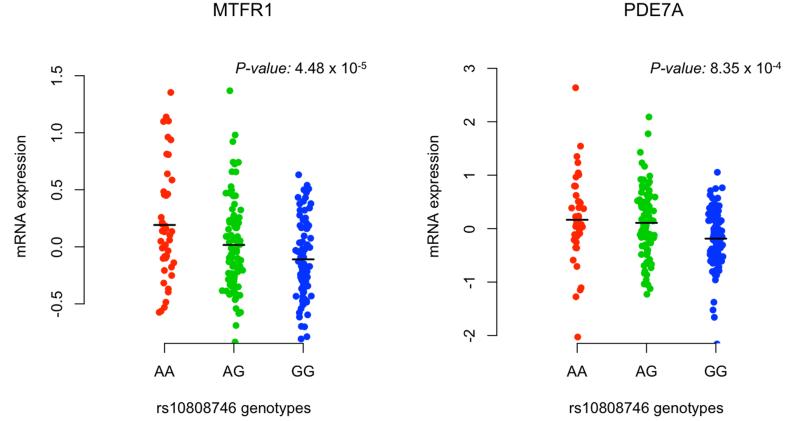
The rs10808746 SNP falls within an intron of Phosphodiesterase 7A (PDE7A) and is near two flanking genes, mitochondrial fission regulator 1 (MTFR1) and armadillo repeat-containing protein 1 (ARMC1), that fall within a linkage disequilibrium block identified by the association peak (Figure 3B). In order to begin to characterize the effect of the discovered variant, we attempted to evaluate its impact on local gene expression in available datasets. In an analysis of gene expression data from peripheral blood mononuclear cells (PBMCs) of 228 individuals with demyelinating disease (De Jager et al., 2009), representing a set of subjects with an activated immune system, we found evidence for association between the discovered variant and expression of both PDE7A (P=8.4×10−4) and MTFR1 (P=4.5×10−5), but not ARMC1 (P=0.21) (Figure 4 and Supplementary Table 4). The significance of the rs10808746 association with PDE7A and MTFR1 expression in PBMCs was robust to gene-based permutation testing. Published data from a different cell type, HapMap lymphoblastic cell lines (Stranger et al., 2007), also revealed associations with the expression of PDE7A (P=5×10−3) and MTFR1 (P=0.041), but not ARMC1 (P=0.17). However, the direction of effect for the association between this SNP and gene expression was not consistent between these two sets of data generated from different cell populations: the risk allele was associated with decreased expression of both genes in the PBMCs from MS patients, but a more modest increase in both PDE7A and MTFR1 expression in the HapMap cell lines, which are B cells transformed by Epstein-Barr virus. Based on available databases of gene expression data, such as BioGPS (http://biogps.gnf.org), both PDE7A and MTFR1 are widely expressed in diverse tissue types, including the central nervous system (Rhead et al., 2010; Su et al., 2002; Wu et al., 2009); however, a publically available brain expression dataset (Myers et al., 2007) with genome-wide genotyping did not include probes for either MTFR1 or PDE7A. We were therefore unable to readily confirm our findings in this tissue context.
Figure 4.
Association of rs10808746 with MTFR1 and PDE7A gene expression. The relation of rs10808746 genotype was evaluated with locus transcript levels in an available gene expression dataset from 228 subjects with demyelinating disease. The rs10808746G allele is associated with decreased expression of MTFR1 (P=4.5×10−5) and PDE7A (P=8.4×10−4), but not ARMC1 (P=0.21).
Aggregate genetic risk profiles for prediction of cognitive decline
In addition to a per SNP genome-wide analysis, we pursued a complementary approach taking into account the aggregate effect of multiple validated susceptibility alleles for other diseases. Specifically, we developed 4 separate genetic risk score models incorporating known susceptibility variants for medical conditions known or hypothesized to promote cognitive decline, including AD, cardiovascular disease, type II diabetes, and inflammatory disease. In our ROS discovery cohort, a diagnosis of AD or of type II diabetes mellitus was significantly associated with cognitive decline (Table S5), consistent with prior studies (Arvanitakis et al., 2004). Table S7 summarizes the SNPs included in each aggregate genetic risk score, along with the relevant references. For both the AD and cardiovascular risk models, we excluded APOE from consideration, since this locus’ strong effect on cognitive decline would overwhelm the contribution of loci with more modest effect sizes. As shown in Table 3, none of the aggregate risk models returned evidence of association in our ROS discovery cohort, and only the AD model demonstrated nominal association to cognitive decline in a more powerful joint analysis that included both the ROS and MAP cohorts. Our modest sample size limits our power in these analyses, but validating our strategy, we do see the expected correlation of the aggregate estimate of genetic risk for type II diabetes with a diagnosis of diabetes in our subjects (Table S6).
Table 3.
Associations between aggregate genetic risk scores and rate of cognitive decline.
| ROS | MAP | ROS + MAP | ||||
|---|---|---|---|---|---|---|
| GRS | β (95% CI) | p | β (95% CI) | p | β (95% CI) | p |
| AD | −0.001 (−0.005,0.003) | 0.50 | −0.004 (−0.008,0.000) | 0.06 | −0.003 (−0.005,0.000) | 0.07 |
| CVD | −0.0004 (−0.002,0.001) | 0.61 | 0.001 (−0.001,0.002) | 0.49 | 0.0001 (−0.001,0.001) | 0.87 |
| I | 0.0003 (−0.0006,0.001) | 0.49 | 0.0001 (−0.0008,0.001) | 0.82 | 0.0002 (−0.0004,0.001) | 0.52 |
| T2D | −0.001 (−0.003,0.001) | 0.28 | −0.0001 (−0.002,0.002) | 0.96 | −0.001 (−0.002,0.001) | 0.48 |
β for association of aggregate genetic risk score (GRS) and rate of cognitive decline. Separate GRS were developed based on published SNP associations with Alzheimer’s disease (AD), cardiovascular disease (CVD), inflammatory disease (I), and type 2 diabetes (T2D)
Power for discovery and replication of genetic variants associated with cognitive decline
The genetic architecture of cognitive decline is currently not known. Given our results and the size of our discovery and replication sample sets, it is likely that cognitive decline, like most other complex traits, is influenced by many variants of modest effect, in addition to the APOE locus which has a strong effect on this trait. Using the PDE7A/MTFR1 and CR1 loci as putative susceptibility loci with which to calibrate the design of future studies exploring cognitive decline, we performed power calculations to assess the sample size that would be necessary for a robust discovery study. For CR1, we observe an effect size (Beta) of −0.0130 and a minor allele frequency (MAF) of 0.20 in the ROS cohort; thus, we would need 7,241 subjects to observe a genome-wide significant effect on the rate of cognitive decline (P<5×10−8) with 80% power, and 8,288 subjects for 90% power. For PDE7A/MTFR1, the effect size in the ROS cohort (Beta=0.0210) is probably inflated given that this is the discovery cohort for this variant. However, given this magnitude of an effect and a MAF of 0.43, we would need 1,794 subjects to have 80% power to observe a genome-wide significant effect (P<5×10−8), and 2,053 subjects for 90% power. However, in the MAP cohort, the PDE7A/MTFR1 has an effect size of 0.008, yielding an alternative estimate of 12,175 subjects needed for 80% power and 13,937 subjects for 90% power. The latter estimate is likely to more accurately reflect the true effect size of our variant since it comes from a replication cohort. The requirement for a larger sample size is consistent with what has been observed for other human traits (Park et al., 2010) and likely explains, at least in part, why we have not yet discovered susceptibility variants for cognitive decline reaching the genome-wide significance level. These estimates will guide the design of our future efforts: it appears that sample sizes between 7,241 and 12,175 subjects will be needed to have reasonable power to assemble convincing evidence of association between genetic variation and the rate of cognitive decline.
DISCUSSION
We report the results of a genome-wide scan in 749 elder subjects to identify loci associated with the rate of cognitive decline, using a mixed effects model that incorporates repeated cognitive measures. Consistent with numerous prior studies (Feskens et al., 1994; Haan et al., 1999; Henderson et al., 1995; Hyman et al., 1996; Jonker et al., 1998; Wilson et al., 2002a, 2002b), we found robust evidence for association between the APOE locus and the rate of age-related cognitive decline. Besides APOE, in a replication analysis including 2,279 additional subjects, we identified one other locus (PDE7A/MTFR1) with consistent direction of effect on cognitive decline in all cohorts and improved evidence of association in a joint analysis. While additional replication efforts will be needed to validate the role of this locus in cognitive decline, our replication and extension analyses strongly suggest that this locus has an effect on the rate of cognitive decline in aging individuals.
To begin to understand the function of this variant, we explored its possible role in influencing the expression of nearby genes using available gene expression datasets. Although further work is needed to better understand the effect of this variant in different cell types (particularly in the brain for which data were not available), two of the three genes found in the block of LD surrounding the associated variant – PDE7A and MTFR1 – appear to be influenced by rs10808746, which is found in an intron of PDE7A. MTFR1 has been shown to induce mitochondrial fission in a variety of cell types (Tonachini et al., 2004), and in Mtfr1null mice, testicular cells show increased oxidative injury and reduced expression of free radical scavengers, such as Glutathione Peroxidase 3 (Monticone et al., 2007). Thus, MTFR1 is a reasonable candidate gene for further validation in the context of cognitive decline since it could be involved in modulating the response of CNS cells to oxidative stress, implicated in many different models of neuronal injury including AD, Amyotrophic Lateral Sclerosis, and Parkinson’s disease (Cho et al., 2010). PDE7A is another strong candidate: it is expressed in both neuronal and non-neuronal CNS cells and regulates intracellular signaling by affecting the concentration of cyclic dinucleotides (Miró et al., 2001; Pérez-Torres et al., 2003). Data on its possible function in CNS cells is limited, but one report suggests that inhibiting this molecule may contribute to enhancing cell death in response to an apoptotic signal in lymphocytes (Dong et al., 2010). There is also mixed evidence that it may have a role in mediating the expression of pro-inflammatory cytokines during T cell activation (Kadoshima-Yamaoka et al., 2009; Yang et al., 2003). Thus, PDE7A is also a reasonable candidate for mediating the effect of the rs10808746 variant, and it is further possible that the variant influences the expression and function of both genes, possibly by affecting shared pathways involved in neuronal degeneration.
Based on our power calculations, it is clear that substantially larger sample sizes including 10,000-15,000 subjects will be needed for a definitive investigation of the genetic architecture of the rate of cognitive decline. This estimate is in line with power calculations for other complex traits (Park et al., 2010). The distribution of results from the ROS cohort suggest that it is unlikely that loci with a strong effect on cognitive decline exist outside of APOE. The existence of loci with more modest effects on cognitive decline is consistent with the suggestive evidence presented in support of CR1, a known AD susceptibility locus, and possibly PDE7A/MTFR1. We attempted to enhance our ability to test detect the role of genetic variation in cognitive decline by assessing aggregate measures of genetic risk for conditions that are well-known (AD, cerebrovascular disease, and type II diabetes) or hypothesized (inflammatory diseases) to influence this trait. Our aggregate measure of AD risk is nominally associated with cognitive decline, suggesting that this may be a fruitful strategy to deploy in future studies that leverage larger sample sizes and additional susceptibility alleles that will emerge in the near future.
Our study was based on the hypothesis that common genetic pathways mediate the response and adaptation to diverse forms of brain pathology, and that variation in such loci might therefore impact age-related cognitive decline. This hypothesis is supported by observations from a variety of experimental paradigms of neuronal injury that several core cellular pathways, including protein misfolding/aggregation (Ross and Poirier, 2004), autophagy (Martinez-Vicente and Cuervo, 2007), mitochondrial dynamics (Cho et al., 2010), and inflammation (Lucin and Wyss-Coray, 2009), play important roles in multiple neurodegenerative diseases. In prospective, community-based autopsy series of dementia, most brains are found to have multiple pathologies (Neuropathology Group, 2001; Schneider et al., 2007; Sonnen et al., 2007), which likely interact to produce the clinical manifestations of age-related cognitive decline (Wilson et al., 2010). Our strategy of using cognitive trajectories as an outcome trait for a genome-wide scan holds great promise for the identification of core genetic mechanisms mediating cognitive reserve and adaptation to injury. However, our current study provides evidence that the genetic architecture of this trait may be similar to that of other complex human traits, involving common variants of modest effects. We provide estimates of the substantially larger sample size required to achieve effective statistical power for robust gene discovery efforts, highlighting the need to combine additional cohorts with those reported here.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sara Pulit for help with assembling a comprehensive list of SNPs associated with inflammatory diseases. The authors are also grateful to the participants in the Religious Orders Study, the Chicago Health & Aging Project, the Memory and Aging Project, and the AD Neuroimaging Initiative. This work is supported by the National Institutes of Health [R01 AG30146, P30 AG10161, R01 AG17917, R01 AG15819, K08 AG034290, P30 AG10161 and R01 AG11101], and the Illinois Department of Public Health. JMS is additionally supported by the Clinical Investigator Training Program: Beth Israel Deaconess Medical Center and Harvard/MIT Health Sciences and Technology, in collaboration with Pfizer Inc. and Merck & Co. AJM is supported by NIA grant R01 AG034504 and the Johnnie B Byrd Institute. MJH, JC and EMR are supported by grants from Kronos Life Sciences Laboratories, the National Institute on Aging (Arizona Alzheimer’s Disease Center P30 AG19610, R01 AG023193, Mayo Clinic Alzheimer’s Disease Center P50 AG16574 and Intramural Research Program), the National Alzheimer’s Coordinating Center (U01 AG016976), and the state of Arizona. Additional funding was obtained from the NIH Neuroscience Blueprint (U24NS051872), the ENDGAME Consortium (U01HL084744), a National Institute on Aging grant to Carl Cotman (University of California, Irvine, P50 AG23173), National Institute on Aging (K01AG024079) and the state of Arizona. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A full listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wpcontent/uploads/how_to_apply/ADNI_Authorship_List.pdf
DISCLOSURE The authors have no conflicts of interest to report related to this work.
REFERENCES
- Arvanitakis Z, Wilson RS, Bienias JL, Evans DA, Bennett DA. Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Archives of Neurology. 2004;61(5):661–6. doi: 10.1001/archneur.61.5.661. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bennett D, Schneider J, Arvanitakis Z, Kelly J, Aggarwal N, Shah R, Wilson R. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006a;66(12):1837–44. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- Bennett D, Schneider J, Wilson R, Bienias J, Berry-Kravis E, Arnold S. Amyloid mediates the association of apolipoprotein E e4 allele to cognitive function in older people. J Neurol Neurosurg Psychiatry. 2005a;76(9):1194–9. doi: 10.1136/jnnp.2004.054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Wilson R, Schneider J, Evans D, Beckett L, Aggarwal N, Barnes L, Fox J, Bach J. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59(2):198–205. doi: 10.1212/wnl.59.2.198. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Aggarwal NT, Arvanitakis Z, Shah RC, Kelly JF, Fox JH, Cochran EJ, Arends D, Treinkman AD, Wilson RS. Decision rules guiding the clinical diagnosis of Alzheimer’s disease in two community-based cohort studies compared to standard practice in a clinic-based cohort study. Neuroepidemiology. 2006b;27(3):169–76. doi: 10.1159/000096129. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Buchman AS, de Leon C. Mendes, Bienias JL, Wilson RS. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005b;25(4):163–75. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer’s disease. Human Molecular Genetics. 2009;18(R2):R137–45. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienias J, Beckett L, Bennett D, Wilson R, Evans D. Design of the Chicago Health and Aging Project (CHAP) J Alzheimers Dis. 2003;5(5):349–55. doi: 10.3233/jad-2003-5501. [DOI] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67(6):677–85. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464(7288):529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, Hardy JA, Huentelman MJ, Corneveaux JJ, Reiman EM, Evans DA, Bennett DA, De Jager PL. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011 doi: 10.1002/ana.22277. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D-H, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67(20):3435–47. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Mackinnon MJ, O’Donnell A, Allen SJ, Moulds JM, Baisor M, Bockarie M, Reeder JC, Rowe JA. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2004;101(1):272–7. doi: 10.1073/pnas.0305306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxson SC, Jagger C. Diabetes and cognitive impairment: a community-based study of elderly subjects. Age Ageing. 1995;24(5):421–4. doi: 10.1093/ageing/24.5.421. [DOI] [PubMed] [Google Scholar]
- De Jager P, Jia X, Wang J, de Bakker P, Ottoboni L, Aggarwal N, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S, Consortium IMG, Baranzini S, McCauley J, Pericak-Vance M, Haines J, Gibson R, Naeglin Y, Uitdehaag B, Matthews P, Kappos L, Polman C, McArdle W, Strachan D, Evans D, Cross A, Daly M, Compston A, Sawcer S, Weiner H, Hauser S, Hafler D, Oksenberg J. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009 doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond DW, Moroney JT, Paik MC, Sano M, Mohr JP, Aboumatar S, Tseng CL, Chan S, Williams JB, Remien RH, Hauser WA, Stern Y. Frequency and clinical determinants of dementia after ischemic stroke. Neurology. 2000;54(5):1124–31. doi: 10.1212/wnl.54.5.1124. [DOI] [PubMed] [Google Scholar]
- Desmond DW, Tatemichi TK, Paik M, Stern Y. Risk factors for cerebrovascular disease as correlates of cognitive function in a stroke-free cohort. Archives of Neurology. 1993;50(2):162–6. doi: 10.1001/archneur.1993.00540020040015. [DOI] [PubMed] [Google Scholar]
- Dong H, Zitt C, Auriga C, Hatzelmann A, Epstein PM. Inhibition of PDE3, PDE4 and PDE7 potentiates glucocorticoid-induced apoptosis and overcomes glucocorticoid resistance in CEM T leukemic cells. Biochem Pharmacol. 2010;79(3):321–9. doi: 10.1016/j.bcp.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol. 2002;155(6):487–95. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- Evans D, Bennett D, Wilson R, Bienias J, Morris M, Scherr P, Hebert L, Aggarwal N, Beckett L, Joglekar R, Berry-Kravis E, Schneider J. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003;60(2):185–9. doi: 10.1001/archneur.60.2.185. [DOI] [PubMed] [Google Scholar]
- Feskens EJ, Havekes LM, Kalmijn S, de Knijff P, Launer LJ, Kromhout D. Apolipoprotein e4 allele and cognitive decline in elderly men. BMJ. 1994;309(6963):1202–6. doi: 10.1136/bmj.309.6963.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodstein F, Chen J, Wilson RS, Manson JE, Study NH. Type 2 diabetes and cognitive function in community-dwelling elderly women. Diabetes Care. 2001;24(6):1060–5. doi: 10.2337/diacare.24.6.1060. [DOI] [PubMed] [Google Scholar]
- Haan MN, Shemanski L, Jagust WJ, Manolio TA, Kuller L. The role of APOE epsilon4 in modulating effects of other risk factors for cognitive decline in elderly persons. JAMA. 1999;282(1):40–6. doi: 10.1001/jama.282.1.40. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Easteal S, Jorm AF, Mackinnon AJ, Korten AE, Christensen H, Croft L, Jacomb PA. Apolipoprotein E allele epsilon 4, dementia, and cognitive decline in a population sample. Lancet. 1995;346(8987):1387–90. doi: 10.1016/s0140-6736(95)92405-1. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ERLC, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Rüther E, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Gallacher J, Hüll M, Rujescu D, Giegling I, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MMB, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues J-F, Tzourio C, Alpérovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snædal J, Björnsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossù P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Gomez-Isla T, Briggs M, Chung H, Nichols S, Kohout F, Wallace R. Apolipoprotein E and cognitive change in an elderly population. Ann Neurol. 1996;40(1):55–66. doi: 10.1002/ana.410400111. [DOI] [PubMed] [Google Scholar]
- Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14(18):1657–63. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker C, Schmand B, Lindeboom J, Havekes LM, Launer LJ. Association between apolipoprotein E epsilon4 and the rate of cognitive decline in community-dwelling elderly individuals with and without dementia. Archives of Neurology. 1998;55(8):1065–9. doi: 10.1001/archneur.55.8.1065. [DOI] [PubMed] [Google Scholar]
- Kadoshima-Yamaoka K, Murakawa M, Goto M, Tanaka Y, Inoue H, Murafuji H, Nagahira A, Hayashi Y, Nagahira K, Miura K, Nakatsuka T, Chamoto K, Fukuda Y, Nishimura T. ASB16165, a novel inhibitor for phosphodiesterase 7A (PDE7A), suppresses IL-12-induced IFN-gamma production by mouse activated T lymphocytes. Immunol Lett. 2009;122(2):193–7. doi: 10.1016/j.imlet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Kim J-S, Han S-R, Chung S-W, Kim B-S, Lee K-S, Kim Y-I, Yang D-W, Kim K-S, Kim J-W. The apolipoprotein E epsilon4 haplotype is an important predictor for recurrence in ischemic cerebrovascular disease. J Neurol Sci. 2003;206(1):31–7. doi: 10.1016/s0022-510x(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38(4):963–74. [PubMed] [Google Scholar]
- Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, Investigators E.A.s.D.I., de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues J-F, Tzourio C, Gut I, van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Launer LJ, Petrovitch H, Ross GW, Markesbery W, White LR. AD brain pathology: vascular origins? Results from the HAAS autopsy study. Neurobiol Aging. 2008;29(10):1587–90. doi: 10.1016/j.neurobiolaging.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64(1):110–22. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet neurology. 2007;6(4):352–61. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- McCarron MO, Delong D, Alberts MJ. APOE genotype as a risk factor for ischemic cerebrovascular disease: a meta-analysis. Neurology. 1999;53(6):1308–11. doi: 10.1212/wnl.53.6.1308. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA, Plomin R. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997;276(5318):1560–3. doi: 10.1126/science.276.5318.1560. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McQueen MB, Bertram L, Lange C, Becker KD, Albert MS, Tanzi RE, Blacker D. Exploring candidate gene associations with neuropsychological performance. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(8):987–91. doi: 10.1002/ajmg.b.30500. [DOI] [PubMed] [Google Scholar]
- Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63(2):157–64. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- Miró X, Pérez-Torres S, Palacios JM, Puigdomènech P, Mengod G. Differential distribution of cAMP-specific phosphodiesterase 7A mRNA in rat brain and peripheral organs. Synapse. 2001;40(3):201–14. doi: 10.1002/syn.1043. [DOI] [PubMed] [Google Scholar]
- Monticone M, Tonachini L, Tavella S, Degan P, Biticchi R, Palombi F, Puglisi R, Boitani C, Cancedda R, Castagnola P. Impaired expression of genes coding for reactive oxygen species scavenging enzymes in testes of Mtfr1/Chppr-deficient mice. Reproduction. 2007;134(3):483–92. doi: 10.1530/REP-07-0199. [DOI] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, Jager PLD, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, George-Hyslop PS, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, Dekosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, Mccormick WC, Mccurry SM, Mcdavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Deerlin VMV, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuropathology Group of the Medical Research Council Cognitive Function and Ageing Study Pathological correlates of late-onset dementia in a multicentre, community-based population in England and Wales. Neuropathology Group of the Medical Research Council Cognitive Function and Ageing Study (MRC CFAS) Lancet. 2001;357(9251):169–75. doi: 10.1016/s0140-6736(00)03589-3. [DOI] [PubMed] [Google Scholar]
- Park J-H, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–5. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendlebury ST, Rothwell PM. Prevalence, incidence, and factors associated with pre-stroke and post-stroke dementia: a systematic review and meta-analysis. Lancet neurology. 2009;8(11):1006–18. doi: 10.1016/S1474-4422(09)70236-4. [DOI] [PubMed] [Google Scholar]
- Pérez-Torres S, Cortés R, Tolnay M, Probst A, Palacios JM, Mengod G. Alterations on phosphodiesterase type 7 and 8 isozyme mRNA expression in Alzheimer’s disease brains examined by in situ hybridization. Experimental Neurology. 2003;182(2):322–34. doi: 10.1016/s0014-4886(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmer YD, van den Berg E, Ruis C, Kappelle LJ, Biessels GJ. Cognitive dysfunction in patients with type 2 diabetes. Diabetes Metab Res Rev. 2010;26(7):507–19. doi: 10.1002/dmrr.1112. [DOI] [PubMed] [Google Scholar]
- Reitz C, Brayne C, Mayeux R. Epidemiology of Alzheimer disease. Nature Publishing Group. 2011a;7(3):137–52. doi: 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tokuhiro S, Clark LN, Conrad C, Vonsattel J-P, Hazrati L-N, Palotás A, Lantigua R, Medrano M, Z Jiménez-Velázquez I, Vardarajan B, Simkin I, Haines JL, Pericak-Vance MA, Farrer LA, Lee JH, Rogaeva E, George-Hyslop PS, Mayeux R. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer’s disease risk. Ann Neurol. 2011b;69(1):47–64. doi: 10.1002/ana.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38(Database issue):D613–9. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(7):S10–S7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388(6639):292–5. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- Schneider J, Arvanitakis Z, Bang W, Bennett D. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69(24):2197–204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- Schneider JA, Bennett DA. Where Vascular Meets Neurodegenerative Disease. Stroke. 2010;41(10, Supple 1):S144–S6. doi: 10.1161/STROKEAHA.110.598326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EMC, Ramirez-Lorca R, Debette S, Longstreth WT, Janssens ACJW, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MMB, Consortium C, Consortium G, Consortium E. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen JA, Larson EB, Crane PK, Haneuse S, Li G, Schellenberg GD, Craft S, Leverenz JB, Montine TJ. Pathological correlates of dementia in a longitudinal, population-based sample of aging. Ann Neurol. 2007;62(4):406–13. doi: 10.1002/ana.21208. [DOI] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve. Neuropsychologia. 2009;47(10):2015–28. doi: 10.1016/j.neuropsychologia.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavaré S, Deloukas P, Dermitzakis ET. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2002;99(7):4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Reed T, Harshfield GA, Fabsitz RR, Eslinger PJ. Heritability of cognitive performance in aging twins. The National Heart, Lung, and Blood Institute Twin Study. Archives of Neurology. 1990;47(3):259–62. doi: 10.1001/archneur.1990.00530030025010. [DOI] [PubMed] [Google Scholar]
- Tonachini L, Montcone M, Puri C, Tacchetti C, Pinton P, Rizzuto R, Cancedda R, Tavella S, Castagnola P. Chondrocyte protein with a poly-proline region (CHPPR) is a novel mitochondrial protein and promotes mitochondrial fission. J Cellular Physiology. 2004;201(3):470–82. doi: 10.1002/jcp.20126. [DOI] [PubMed] [Google Scholar]
- Troncoso JC, Zonderman AB, Resnick SM, Crain B, Pletnikova O, O’Brien RJ. Effect of infarcts on dementia in the Baltimore longitudinal study of aging. Ann Neurol. 2008;64(2):168–76. doi: 10.1002/ana.21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsch JRL, Wright CB. The aging mind: vascular health in normal cognitive aging. J Am Geriatr Soc. 2010;58(Suppl 2):S319–24. doi: 10.1111/j.1532-5415.2010.02983.x. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Bienias J, Berry-Kravis E, Evans D, Bennett D. The apolipoprotein E epsilon 2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002a;73(6):672–7. doi: 10.1136/jnnp.73.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Schneider J, Barnes L, Beckett L, Aggarwal N, Cochran E, Berry-Kravis E, Bach J, Fox J, Evans D, Bennett D. The apolipoprotein E epsilon 4 allele and decline in different cognitive systems during a 6-year period. Arch Neurol. 2002b;59(7):1154–60. doi: 10.1001/archneur.59.7.1154. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Barnes LL, Krueger KR, Hoganson G, Bienias JL, Bennett DA. Early and late life cognitive activity and cognitive systems in old age. J Int Neuropsychol Soc. 2005;11(4):400–7. [PubMed] [Google Scholar]
- Wilson RS, Beckett LA, Barnes LL, Schneider JA, Bach J, Evans DA, Bennett DA. Individual differences in rates of change in cognitive abilities of older persons. Psychol Aging. 2002c;17(2):179–93. [PubMed] [Google Scholar]
- Wilson RS, Gilley DW, Bennett DA, Beckett LA, Evans DA. Person-specific paths of cognitive decline in Alzheimer’s disease and their relation to age. Psychol Aging. 2000;15(1):18–28. doi: 10.1037//0882-7974.15.1.18. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Leurgans SE, Boyle PA, Schneider JA, Bennett DA. Neurodegenerative basis of age-related cognitive decline. Neurology. 2010;75(12):1070–8. doi: 10.1212/WNL.0b013e3181f39adc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, McIntyre KW, Townsend RM, Shen HH, Pitts WJ, Dodd JH, Nadler SG, McKinnon M, Watson AJ. Phosphodiesterase 7A-deficient mice have functional T cells. J Immunol. 2003;171(12):6414–20. doi: 10.4049/jimmunol.171.12.6414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.