Abstract
Cell fusion in vitro has been used to study cancer, gene mapping and regulation, and the production of antibodies via hybridomas. However, in-vivo heterosynkaryon formation by cell-cell fusion has received less attention. This investigation describes the spontaneous fusion of a human glioblastoma with normal hamster cells after xenogeneic transplantation, resulting in malignant cells that express both human and hamster genes and gene products, and retention of glioblastoma traits with an enhanced ability to metastasize. Three of 7 human genes found showed translation of their proteins during serial propagation in vivo or in vitro for years; namely, CD74, CXCR4, and PLAGL2, each implicated with malignancy or glioblastoma. This supports the thesis that genetic hybridization of cancer and normal cells can transmit malignancy and also, as first described herein, regulatory genes involved in the tumor’s organotypic morphology. Evidence also is increasing that even cell-free human cancer DNA can induce malignancy and transfer genetic information to normal cells. Hence, we posit that the transfer of genetic information between tumor and stromal cells, whether by cell-cell fusion or other mechanisms, is implicated in the progression of malignancy, and may further define the crosstalk between cancer cells and their stromal neighbors.
Keywords: cell fusion, cancer stroma, metastasis, gene transfer, glioblastoma
Introduction
It is 4 decades since the first reports of in-vivo fusion of human tumor and normal animal host cells based on heterosynkaryon formation and biochemical evidence of both species,1–7 suggesting that cell fusion is a mechanism for the horizontal transmission of malignancy and tumor progression.1,6,7 This was proposed already at the beginning of the 20th century by various German pathologists.1,8 Subsequently, numerous studies confirmed in-vivo intra- and cross-species fusion of malignant and normal cells, leading to increased malignancy in the progeny,9–14 and that fusions with hematopoietic or myeloid cells occur experimentally and possibly clinically.11,14–16 The role of cell fusion in biology, particularly organogenesis and tissue regeneration, viral transfer, and cancer has been the subject of recent reviews.15,17–22 However, despite reports of the induction of malignancy in rodent hosts given human tumor cells, there is a paucity of evidence that these are stable hybrid tumors retaining human genes, or even expressing their products during serial in-vivo propagation. Usually, most human chromosomes are lost with subcultivation of hybrid cells, either in vitro or in vivo.23 With current techniques for disclosing DNA and its protein products in paraffin sections, we determined if human genes were retained and were functional over long-term passage of a spontaneous human-hamster hybrid tumor. This cross-species model provides a means of distinguishing the genetic contribution of a cancer from its mesenchymal stroma (defined broadly as fibroblasts, endothelial cells, macrophages, etc.) by having the latter from another species. In contrast, homotypic cell-cell fusions have relied on genetic or sex chromosome differences between donor and recipient in the absence of synkaryon formation.16,21,24 We now report that the hybrid tumor that became a serially propagated, metastatic, stable cell line in hamsters retained at least 7 human genes (of 12 tested), and continued to express, surprisingly, at least 3 human gene products, each implicated with oncogenesis or with the glioblastoma morphological phenotype. This indicates that horizontal gene transfer by cell-cell interaction can occur between mammalian cells of the same or a different species with profound implications in biology and pathology. We believe this is the first report of the serial propagation of heterosynkaryons expressing dual-species genes, including human malignant and organotypic genes in a predominately hamster host cell.
Materials and Methods
Transplantation studies
As described previously,6 an aliquot of a glioblastoma multiforme (GBM) from the brain of a 44-year-old female was injected as a cell suspension into the cheek pouches of 9 adult male Syrian golden hamsters not receiving any immunosuppressive conditioning. One resulting cheek pouch tumor was further passaged serially to successive hamster generations both in cheek pouches and intraperitoneally during the next year. A portion of this tumor was cloned and propagated in-vitro over 31 months and then regrafted in hamster cheek pouches to confirm stable morphology and metastasizability. Cell cultures were derived from cheek pouch tumors that were trypsinized and the resulting diluted cells grown in flasks containing Eagle’s Minimum Essential Medium with Hanks’ salts and fetal calf serum (Gibco, Grand Island, NY), supplemented with antibiotics and mycostatin, as described.25 Animal studies were conducted with approval of the Institution’s Animal Care and Use Committee.
Antibodies
Humanized monoclonal antibodies hLL1 (milatuzumab; anti-CD74) and hA20 (veltuzumab; anti-CD20, used as a non-binding control) were provided by Immunomedics, Inc. (Morris Plains, NJ). Polyclonal goat anti-GFAP (C-19), polyclonal goat anti-PLAGL2 (C-16) and mouse anti-fusin (CXCR4; clone 12G5) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A considerable number of other anti-human protein antibodies evaluated for their reactivity with GB-749 were found to be negative, and are listed in Supplementary Information (Supplementary Table S1) online.
Fluorescence in-situ hybridization (FISH)
Tissue sections were deparaffinized and processed by FISH for hamster X chromosome and human paracentromeric probes, as described in the Supplementary Information online.
Preparation of DNA from human lymphoma cells
Human and hamster genomic DNA were purified from Raji and CHO cells, respectively, (ATCC, Manassas, VA) as positive controls. DNA was extracted from 106 cells using DNeasy Tissue Kit (Qiagen, Germantown, MD), according to manufacturer’s instructions.
Preparation of DNA from paraffin-embedded tissues
Paraffin-embedded tissues of human and hamster normal tissues and tumors, or GB-749 transplants, were cut in 4–5-μ sections. Following the dissolution of paraffin in xylene, DNA was extracted from each section using QIAamp DNA FFPE Tissue Kit (Qiagen, Germantown, MD), according to manufacturer’s instructions.
PCR
The primers for CD74 gene were designed for the largest exon (exon 2) using Vector NTI (Invitrogen, Carlsbad, CA); CXCR4 and PLAGL2 gene primers were selected from UniSTS database of NCBI. The primers and their sources for other genes are provided in Table S2 in the Supplementary Information online. All primers were custom-made by Eurofins MWG Operon (Huntsville, AL).
Each PCR sample contained 1 µL of DNA, 2.5 µL of 10 X PCR buffer, 2.5 µL of the respective primer pairs (20 µM each), and 5 units of AmpliTaq DNA polymerase. The PCR was repeated for 50 cycles, each consisting of denaturation at 94 °C for 30 sec, annealing at 58 °C for 30 sec, and polymerization at 72°C for 30 sec. The amplified fragments were analyzed on 2% agarose gel. The 10X PCR buffer and the AmpliTaq DNA polymerase were purchased from Applied Biosystems (Foster city, CA).
Immunohistochemistry (IHC)
Paraffin-embedded specimens were cut to 4-μ sections on superfrost plus adhesive slides (Thermo Scientific, Waltham, MA), and deparaffinized by routine methods. Primary antibodies, along with the appropriate species non-binding controls, were then used at concentrations ranging from 1–10 µg/mL. An appropriate species-specific ABC Vectastain kit (Vector Laboratories, Burlingame, CA) was then used as per the manufacturer’s instructions for labeling tissues. For interpretation, a positive reaction was considered to be staining >5% of the appropriate tissue/cells. Murine irrelevant myeloma antibody (P3-X63-Ag8.653) from ATCC served as a negative control.
Results
Transplantation history, morphology and karyology
After transplantation of the primary human GBM (Fig. 1A), the resulting cheek pouch tumor initially showed a more uniform anaplastic character with abundant cytoplasm in the first transplant generation (Fig. 1B), but had resemblance morphologically to the original GBM in later cheek pouch grafts and distant metastases, with a pseudopalisading, lobulated pattern and/or sheets of cells, even after 32 transplant generations over 2 years (Fig. 1C). The very first generation animal grafted expired after 4 weeks, with metastases to its lungs (Fig. 1D), liver, and other major organs. Over the course of 3 years, tumor aliquots were transplanted to other hamsters, both in the cheek pouch and intraperitoneally as an ascites cell population, were grown in cell culture for up to 31 months and reestablished in vitro and in vivo, becoming a continuous cell line (designated GB-749). In all transplant settings, from the very first graft, GB-749 was widely metastatic and lethal. The grafting of human tumor cells to hamsters resulting in highly malignant serial transplants has been reproduced by us 14 times with other human tumors of diverse histopathology, of which 3 that were evaluated, GW-127, GW-478, and GB-749, had evidence of undergoing in vivo cell fusion based on karyological, isoenzyme, or immunological analyses.1–6
Figure 1.
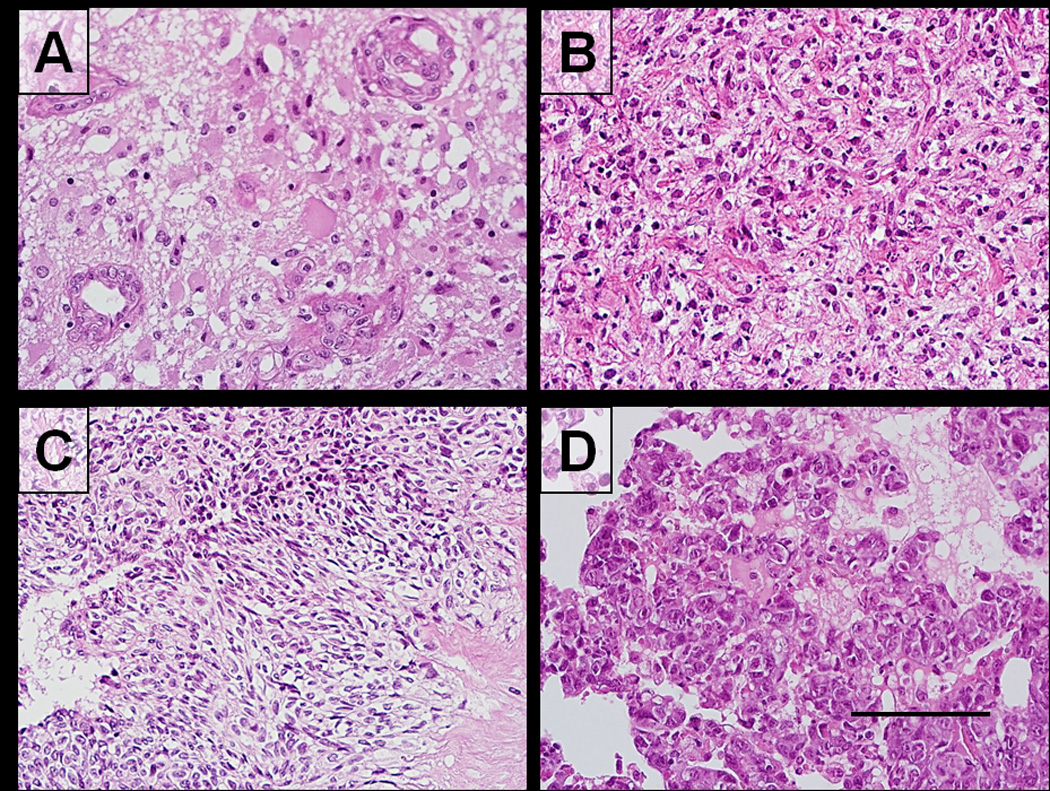
Microscopic morphology of primary human glioblastoma and growth as a xenograft (GB-749) in the hamster cheek pouch; representative hematoxylin-eosin staining of tumors. A: The primary clinical specimen exhibits a moderately cellular neoplasm with vascular hyperplasia and little necrosis. B: In the first generation hamster transplant, a more uniform anaplastic nature with more abundant cytoplasm is observed. C: Later xenograft generations show greater similarity to the patient’s primary tumor, with a pseudopalisading, lobulated pattern and/or sheets of cells consistent with a glioblastoma morphology. D: The highly aggressive nature of GB-749 is reflected by numerous spontaneous metastases in the lungs and other hamster organs, which are necrotic, replacing most of the normal parenchyma. Scale bar in D corresponds to 100 µm (A,B,C,D).
Cells from early GB-749 transplants were cultivated in vitro and subjected to cytogenetic analysis by the trypsin-Giemsa banding method, including also the cancer patient’s peripheral blood cells for comparison, and contained both hamster and human chromosomes, with human chromosomes 2, 3, 7, 9, 12, 18, and 21 being identical to those of the patient’s cells.6 Up to 11 human chromosomes were identified in various frequencies in over 20 karyotypes evaluated, with the highest number being observed for human chromosomes 2, 3, 11, 18, and 21. However, the majority were hamster chromosomes.6
With evidence of both human and hamster chromosomes in the same cells of the transplants, thus confirming heterosynkaryons, we asked if human genes survived continuous propagation of these highly malignant hybrid tumors by examining both human and hamster genomic DNA by fluorescence in-situ hybridization (FISH), human genes by polymerase chain reaction (PCR), as well as expression of human gene products by immunohistochemistry (IHC). Both cheek pouch transplants and their metastases during long-term passage were tested.
FISH
The third generation GB-749 transplant (after 4 weeks of growth in hamsters) and control GW-39 human colonic carcinoma grown in the hamster cheek pouch or nude mice for over 40 years and proven to be a completely human cell line26 were hybridized with human pancentromeric and hamster X-chromosome probes. Figs. 2A–2C demonstrate that signals for the hamster and human probes were detected within the same cells, thus again proving human-hamster heterosynkaryons. The control GW-39 human colon cancer transplant (Figs. 2D–2F), in contrast, showed that the green signal of the hamster probe was observed only in the surrounding hamster tissue, while the red signal for the human pancentromeric probe was specific to the human malignant signet-ring carcinoma cells typical for GW-39.26 The overlay (Fig. 2F) emphasizes that the human and hamster probes hybridize mutually exclusively with either hamster cheek pouch or human tumor xenograft cells, indicating that in this case, no chromosome transfer occurred and that the GW-39 xenograft remained a truly human tumor during its 40-year transplant history. These FISH results confirm the chromosome analysis that suggested that GB-749 was a spontaneous human-hamster hybrid transplant.6
Figure 2.
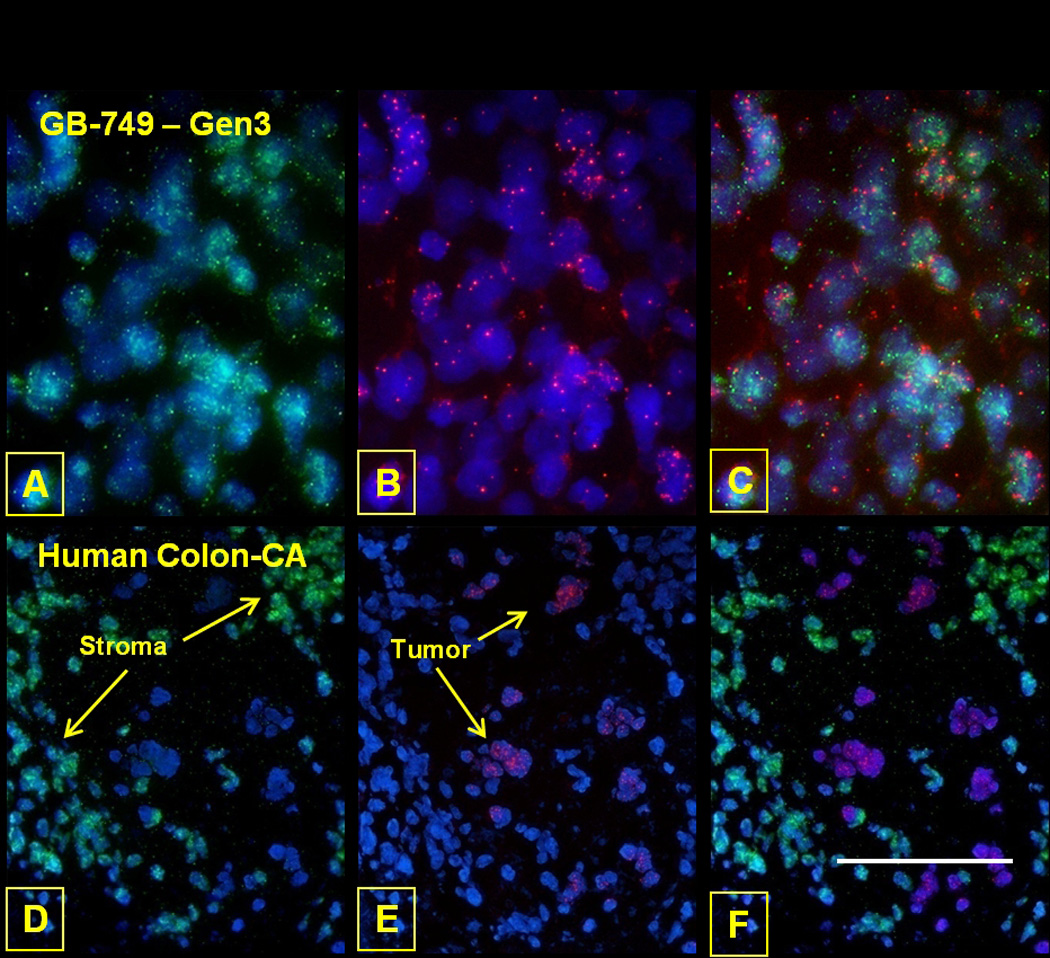
FISH of GB-749 tumor cells was performed with the hamster X-chromosome (green) and human pancentromeric (red) probes to evaluate the co-existence of species-specific genomic material within these cells. A human colon carcinoma (GW-39) grown in the hamster cheek pouch served as a positive control. Within the GB-749 tumor cells, the hamster (green) and human (red) probes are identified in most cells and co-localized to the same cells (Photoshop composite). On the other hand, human and hamster chromosomes show distinct, cell-specific localization within the colon carcinoma xenograft, with hamster stromal tissue and human colon tumor cells identified. DAPI background staining was performed on all tissues to identify the nuclei. Scale bar in F corresponds to 50 µm (A,B,C) and 75 µm (D,E,F).
PCR for human genes
Since human chromosomes were present in these heterosynkaryons that showed a highly malignant phenotype during serial propagation, we determined if certain human genes were retained and were possibly also functional after long-term passage in a foreign host. The first genes of interest were 12 implicated with oncogenesis and particularly glioma formation. The primers of 12 genes of interest were designed for the largest exon (Supplementary Information Table S2), and then used for PCR of human genomic DNA purified from paraffin sections of various GB-749 transplant generations, as well as control human and hamster normal tissues and tumors. In Fig. 3, the agarose gels show the retention of human genomic DNA for CD74, CXCR4, PLAGL2 in the GB-749 transplants, but not in the control hamster tissue. Normal human tissue and Raji Burkitt lymphoma cells served as positive controls, confirming the preservation of these human genes for over a year of GB-749’s propagation in vivo. Table 1 lists human genomic DNA for 4 additional genes, glial fibrillary acidic protein (GFAP), vimentin (VIM), p53 (TP53), and epidermal growth factor receptor (EGFR), which also were retained in early and late GB-749 transplants in the hamster. The human chromosomal assignments are provided for all 7 genes, and indicate the retention of genomic DNA from at least 6 human chromosomes. These correspond to many of the human chromosomes identified previously by Giemsa-banding in these transplants, such as chromosomes 2, 7, 9, and 10.6 It appears that two genes on chromosome 17 (GFAP and TP53) were retained. However, other human genes tested, such as, TNC (tenascin C), MIF (macrophage migration-inhibitory factor), CXCL12, and CDKN2A (p16), were not detectable by the method used (Table 1). Whether the 7 human genes propagated in GB-749 transplants are part of intact human chromosomes or fragments, or translocated onto hamster chromosomes as ‘jumping genes,’ is not known, but intriguing to speculate.
Figure 3.
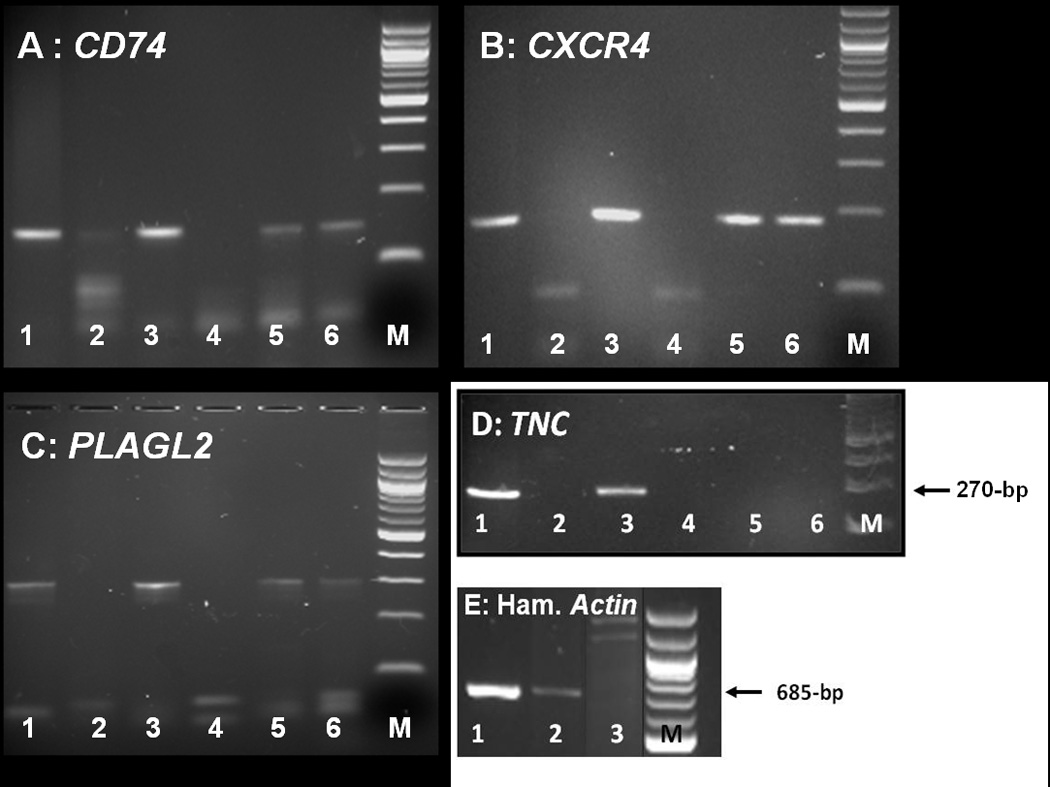
PCR evaluation of GB-749 for the presence of CD74, CXCR4, PLAGL2 and TNC genes. DNA was extracted from formalin-fixed, paraffin-embedded specimens to be used as templates for PCR amplification with primers, as described in Supplementary Table S2. The individual panels demonstrate the presence or absence of each gene within human spleen (lane 1), hamster spleen (lane 2), Raji human lymphoma cells (lane 3), primers alone (lane 4), GB-749 generation-4 (lane 5), and GB-749 generation-29 (lane 6). Size markers are shown in lane M. A band at the correct size (145 bp) was observed for both GB-749 specimens and positive controls (Raji and human spleen) when evaluated for CD74. The presence of CXCR4 and PLAGL2 are shown within their respective panels with a band at the correct size (190 bp for CXCR4 and 304 bp for PLAGL2) detected for both generations of GB-749 and positive controls, whereas TNC (tenascin C) was not detected in either generation of GB-749 xenotransplants. Hamster spleen (lane 2) was negative for all human genes tested; however, panel E demonstrates the presence of hamster DNA (β-actin) in Chinese hamster ovary (CHO) cell line (lane 1) and hamster spleen (lane 2), but not in Raji human lymphoma cells (lane 3).
Table 1.
PCR and immunohistochemical detection of human genes and their respective proteins in GB-749 xenograft and control tissues.
| Gene | Chromosome Locus |
PCR Results | IHC Results |
|||||
|---|---|---|---|---|---|---|---|---|
| Human Spleen |
Hamster Spleen |
Raji- NHL |
Primers Alone |
GB-749 Gen-4 |
GB-749 Gen-29 |
GB-749 | ||
| CD74 | 5q32 | + | − | + | − | + | + | + |
| CXCR4 | 2q22.1 | + | − | + | − | + | + | + |
| PLAGL2 | 20q11.21 | + | − | + | − | + | + | + |
| GFAP | 17q21.31 | + | − | + | − | + | + | − |
| VIM | 10p13 | + | − | + | − | + | + | − |
| EGFR | 7p11.2 | + | − | + | − | + | + | − |
| EGFRvIII | 7p11.2 | − | − | + | − | − | − | − |
| TP53 | 17p13 | + | − | + | − | + | + | − |
| TNC | 9q33.1 | + | − | + | − | − | − | − |
| MIF | 22q11.23 | + | − | + | − | − | − | − |
| CXCL12 | 10q11.21 | + | − | + | − | − | − | − |
| CDKN2A | 9p21.3 | + | − | + | − | − | − | − |
Protein expression by IHC
IHC staining of paraffin sections of early and late cheek pouch and ascites tumor generations of GB-749 showed that human CD74, CXCR4, and PLAGL2 proteins were expressed in serial transplants of GB-749, but not in control normal or malignant hamster tissues (Fig. 4 and Table 2), nor when using an irrelevant murine myeloma antibody. However, proteins related to many human genes, 4 of which were present in GB-749 (GFAP, VIM, EGFR, and TP53), were not detected, at least at the level of sensitivity provided by the immunohistochemical method employed (Tables 1 and 2). We also found that 7 of 8 other patient GBM specimens tested had elevated expression of CD74 (Table 2).
Figure 4.
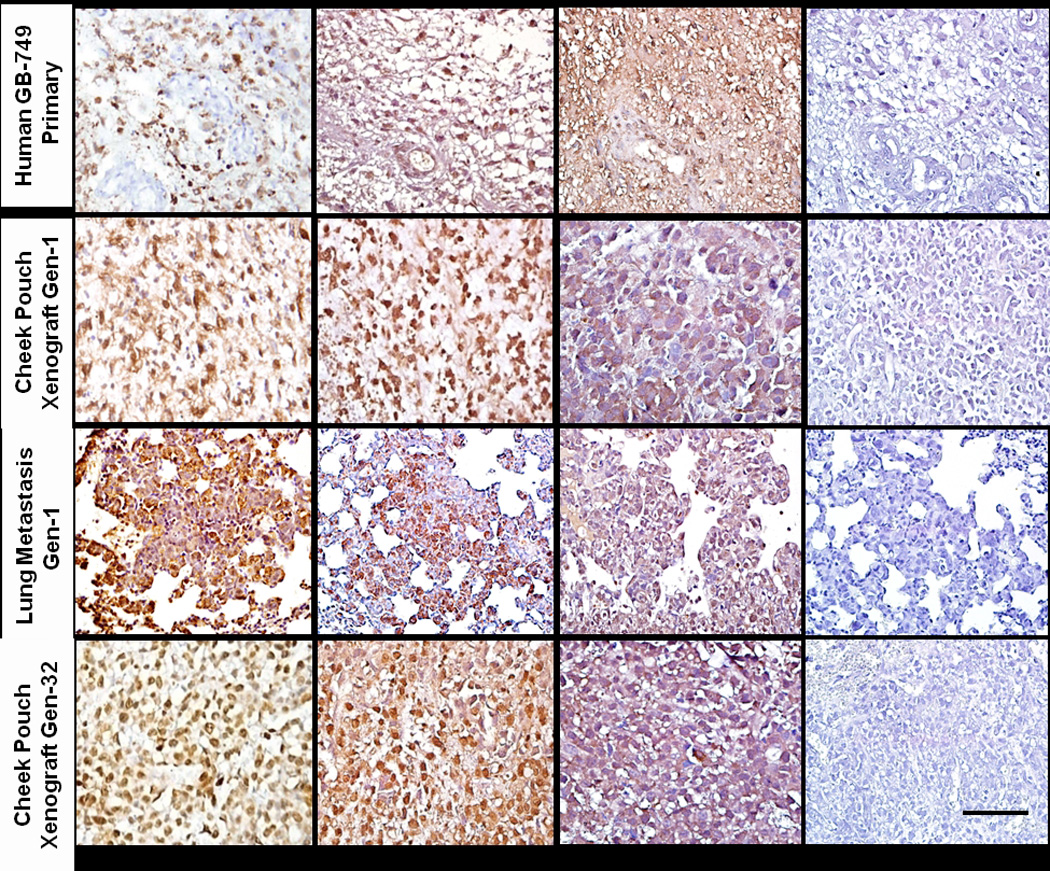
Staining of human CD74, CXCR4, PLAGL2 and GFAP (latter not shown) human proteins is observed within the tumor cells of the primary GBM. GB-749 xenotransplants, whether growing in the hamster cheek pouch or metastatic sites, show evidence of human CD74, CXCR4 and PLAGL2 expression. Staining is within the tumor cytoplasm and nucleus, similar in generations 1 through 32. Irrelevant, isotype-matched, control antibody gave negative results with all tissues evaluated. Scale bar in bottom right corresponds to 100 µm for all photomicrographs.
Table 2.
Immunohistochemical detection of biomarkers in GB-749 xenografts and control tissues.
| Specimen | CD74 | CXCR4 | PLAGL2 |
|---|---|---|---|
| GB-749 Primary | + | + | + |
| Hamster Cheek-Pouch GB-749 Xenograft Tumors | |||
| Gen-1 | + | + | NDa |
| Gen-2 | + | + | + |
| Gen-3 | + | + | + |
| Gen-4 | + | + | + |
| Gen-6 | + | + | + |
| Gen-7 | + | + | + |
| Gen-31 | + | + | +b |
| Gen-32 | + | + | + |
| Gen-45 | + | + | + |
| GB-749 Gen-1 Metastases | |||
| Lung | + | + | ND |
| Liver | + | + | ND |
| Normal Hamster Tissues | |||
| Cheek Pouch | − | − | − |
| Lung | − | − | − |
| Liver | − | − | − |
| Spleen | − | − | − |
| Kidney | − | − | − |
| Control Grafted Tumors | |||
| Hamster Melanoma | − | − | − |
| Human Colon Cancer (GW-39) | + | − | − |
| Human Lymphoma (Raji) | + | + | + |
| Other Primary GBM Tumors | + (7/8) | + (6/8) | + (5/7) |
ND - Not Done (insufficient tissue);
Result is for Gen-29.
Discussion
In addition to the elucidation of cancer oncogenes, tumor-suppressing genes, and microRNA genes, recent attention has focused on the interaction of the tumor parenchyma with its stroma, including fibroblasts, endothelial cells, macrophages, and diverse inflammatory cells.27–30 This stromal component was originally thought to be supportive and even reactive (‘desmoplasia’) to the growth of cancer cells, but recent evidence describes an interplay between these two components, and that this interaction can affect tumor progression, either by promotion or inhibition.27–30 Cancer-associated fibroblasts (CAF) that are oncogenic to epithelial cells have been described.27 However, little is known how altered stromal cells in a cancer gain and maintain their cancer-enhancing phenotype, and what genetic or epigenetic mechanisms are involved in this crosstalk between tumor and stromal components, including those derived from bone marrow.30
Our findings now demonstrate the genetic transfer of malignancy and certain gene functions between a human cancer and the stroma of its xenogeneic host, suggesting that heterosynkaryon formation, presumably by cell-cell fusion, is the mechanism by which a genetic transfer occurs between the tumor and its new stroma, facilitating the progression of malignancy to the aggressive state of metastasis. In addition, both malignancy and the organotypic features of the original human glioblastoma were stable in the hybrid tumor transplants despite loss of most human chromosomes. However, which stromal cell (e.g., macrophage or fibroblast) of the hamster was the partner in this fusion is not known. Although this is not an autochtonous model of cancer within its original host, cell-cell fusion or, more generally, DNA transfer, may be an important mechanism by which a tumor’s parenchyma and stroma orchestrate cancer development and progression. These results also suggest that typically non-metastatic glioblastoma does have the potential to acquire this property in the different setting of a xenogeneic host, similar to our experience with other human cancer types,1–5 and other recent evidence that hybrid tumors can access new traits from their parental cells.24 This report of GBM fusion with normal hamster cells is not unique, since we have observed the generation of highly malignant serial transplants in hamsters receiving human tumors of diverse histopathology 14 times, proving in-vivo cell fusion by karyological, enzymatic, and immunological methods,1–6 but without assessing the stable maintenance and expression of human genes.
A corollary investigation of interspecies hybridization similar to these in-vivo studies with human transplants was undertaken by the in-vitro fusion of a murine melanoma with normal hamster cheek pouch cells, resulting in hybrid tumor cells that were more hamster than murine in terms of antigenic and isoenzyme profiles, but with retention of murine melanoma marker chromosomes among a majority of hamster chromosomes.25 Interestingly, the hybrid cells were more malignant in the hamster than the original melanoma cells were in mice, whereas the murine melanoma could not grow in hamsters nor could the hybrid tumor cells proliferate in genetically compatible mice.
Despite evidence of artificially fused tumor/normal cells being cultivated in vitro and sporadic evidence of spontaneous fusion in vivo,9–14 little attention has been given to the in-vivo stability of such hybrid tumors, particularly the continuous retention of genes from the original tumor and an analysis of which genes are active in translating their respective proteins. Herein, we show, for the first time, that a human GBM grafted to hamster cheek pouches produced a hybrid human/hamster tumor with segregation of mostly human chromosomes happening in the first transplant generation (within a week), and with evidence widespread metastasis, but with the retention over more than 3 years of at least 7 genes from 6 human chromosomes, of which 3 continued to translate their respective proteins.
It is intriguing to consider why CD74, CXCR4, and PLAGL2 gene products were all translated and expressed continuously in GB-749 transplants, whereas those of other human genes retained, such as GFAP, VIM, TP53, and EGFR, were not. Do the 3 functional genes play a role in the hybrid tumor’s growth behavior and pattern? CD74 is the membrane form of invariant chain (Ii) of HLA class II, whose function is as a chaperone molecule controlling antigenic peptide loading and transport to the plasma membrane. It is also important for the regulation of B-cell survival by macrophage migration-inhibitory factor (MIF), and is expressed in a number of solid tumors, even being a prognostic marker in pancreatic and other cancers.31 In this study, we found its expression in 7 of 8 primary GBM specimens in addition to GB-749. Interestingly, it has been shown that CD74 interacts with CXC chemokine receptor, CXCR4 (also called fusin), the second chemokine receptor that mediates MIF-specific signaling,32 and which is also expressed in GB-749. CXCR4 is a G-protein receptor for stromal-derived-factor-1 (SDF-1 or CXCL12). Together with CXCL12, CXCR4 has been implicated with the proliferation, motility, homing, and metastasis of cancer cells, especially glioma cell invasion,33 where it is associated with regions of necrosis and angiogenesis.34 It was also characterized as a GBM oncogene.35 Further, both CXCL12 and CXCR4 are expressed in pseudopalisading cells localized around necrosis, a hallmark of glioblastoma and GB-749.
The third human protein expressed by GB-749 is coded by the Pleiomorphic Adenoma Like 2 (PLAGL2) gene, originally associated with pleiomorphic adenomas, and implicated in the development of malignancies, such as glioblastoma, where it also has been identified as an oncogene.36 Thus, all 3 functional genes found in GB-749 transplants have either been implicated in cancer survival, invasion and metastases, or have a particular role in the biology or genesis of glial tumors, including oncogenic properties (CXCR4 and PLAGL2).
It is not clear, however, how such horizontally-transmitted human DNA transcribes specific functions of the original human tumor, such as its organotypic morphology, in the progeny hybrid tumor cells. In another model, the transfection of human small-cell carcinoma DNA into murine host cells in vitro resulted in fibrosarcomas with a neuroepithelial phenotype, confirming that such organoid regulatory gene sequences can be transcribed and expressed.37 In the current studies, it appears that the typical GBM phenotype was genetically transferred and translated, because glioblastoma-like pseudopalisading, necrosis, and a lobulated growth pattern with sheets of cells, including vascular hyperplasia, were retained in long-term transplants. This observation needs to be examined in other instances of highly metastatic tumors arising rapidly after human tumor transplantation, in order to assess if this method of cell fusion could allow assigning organoid properties to specific gene loci. Indeed, cell fusion has also indicated organ-specific metastasis gene signatures.38
Related to our observations is the grafting of normal human hematopoietic stem cells to fetal pigs, resulting in hybrid cells containing porcine and human chromosomal DNA that express both species’ proteins.17 It was also shown that the hybrid cells contained porcine endogenous retroviral DNA that permitted viral transmission to uninfected human cells in vitro. This evidence of spontaneous fusion in vivo between normal cells and subsequent retroviral transmission has been discussed as a mechanism for the potential transfer of viral or other infectious organisms between species by cell-cell contact.17 Indeed, the role of tumor viruses in cell fusion and malignancy (with the identification of prevalent fusogenic viruses), particularly involving macrophages, has been suggested by others.19,39
Our results with GB-749 and other human tumor-hybrid transplants support the notion that genomic change in the transplanted tumor by means of interaction with host stroma is implicated in malignant progression, whereas non-hybrid tumor xenografts rarely show evidence of spontaneous metastasis. From a clinical perspective, it is important to ascertain (i) whether and how frequently such events occur in human cancers, (ii) when they may occur in a tumor’s development, (iii) if fusions with stromal cells (fibroblasts, endothelial cells, macrophages, etc.) are restricted to the primary tumor or can also involve metastases, and (iv) its role in evoking chemoresistance,19,22 and escape from host immunity.1
These tumor-host genetic transfer results support an emerging view that genomic alterations in the stroma can be induced by the tumor and vice versa, affecting the genetic composition and behavior of the neoplasm. One explanation is that stromal components of epithelial tumors are implicated in the tumor’s phenotype and contribute genetically to the tumor’s progression. Supportive evidence is based on alterations of the genomic DNA of stromal cells during the establishment of human tumor cell lines in nude mice,40 and also by the genetic analysis of stromal cells within human breast cancer xenografts.41 Conversely, as mentioned, there is an expanding literature disclosing oncogenesis in non-malignant epithelial cell lines induced by CAFs,17 and also CAFs being important constituents of a tumor that metastasizes by actually traveling with the metastases.42
Indeed, evidence that stromal cells within a tumor can be truly malignant was reported already in 1981, when fibroblasts from human epithelial tumor transplants in nude mice developed invasive and metastatic murine fibrosarcomas that killed either the nude mice or genetically-compatible, immunocompetent mice of the same genetic background.43 Also, a human ovarian cancer transplant to nude mice showed both epithelial and sarcomatous malignant cell populations, being of human and murine origin, respectively.44 Only the murine sarcoma cells, presumably induced by the human carcinoma, were metastatic and lethal in both nude and immunocompetent mice of the same genetic background. In fact, the development of a stromal malignancy within an epithelial tumor is not a new finding, having been observed repeatedly.45 Already in 1905, Ehrlich and Apolant speculated that ‘chemical substances’ produced by rodent carcinoma cells could transform adjacent connective tissue, since they observed a sarcomatous transformation in vivo.46 This collateral induction of malignancy is consistent with the experiences of human histopathology, where mixed tumor types and lineage transformation are not infrequent. In this regard, we propose that dedifferentiation, which is associated with aneuploidy and enhanced malignancy, can result from a genetic interaction between tumor and its stromal cells, increasing ploidy and having variable effects on organoid or tissue-specific morphology.
A critical question is whether cell-cell fusion or yet other mechanisms of human gene transmission are essential in the horizontal transfer of malignancy and organotypic features. Recent studies indicate that DNA, as well as epigenetic factors, can be released from human tumors and transform normal cells, such as via membrane-derived vesicles, or exosomes,47 or DNA in cancer patient plasma.52 The latter authors showed that embryonic murine fibroblasts were transformed after incubation with plasma from cancer patients but not from normal individuals. They also reported that mutated KRAS sequences could be detected in both the murine tumors induced as well as in distant organs of the mice transplanted with murine cells transformed by human patient plasma.48 This is consistent with the finding of human proteins expressed in various porcine tissues after normal human bone marrow cells fused in vivo with fetal pig cells.17
In conclusion, we describe, for the first time, the transfer of at least 7 human genes to normal xenogeneic host cells via heterosynkaryon formation, and their retention in vivo during long-term serial passage. Of these, at least 3, CD74, CXCR4, and PLAGL2, all implicated with oncogenesis and tumor progression, were translated. In particular, PLAGL2 and CXCR4 have been reported as oncogenes for glioblastoma,35,36 and CXCR4 has been associated with the glioblastoma organoid phenotype, which is also expressed by the serially-propagated hybrid tumor, GB-749. Our results confirm that genetic exchanges can occur between tumors and the stroma of their microenvironment, either by a mechanism of cell-cell fusion or, as cited from the literature, other methods of DNA transfer, thus providing the tumor with new attributes for survival, yet can retain regulatory proteins that control a tumor’s morphological phenotype and possibly even gain new traits. These events are consistent with emerging concepts that cancers and their disseminated cells are influenced by local and distant environmental factors derived from both normal and malignant cells.27–30 Indeed, the morphological diversity observed occasionally in different parts of the same cancer, as well as between primary and metastatic tumors and even between different metastases of the same tumor, is consistent with the genetic disparity observed between primary tumors and their metastases.49 This stimulates speculation that the mechanism of this diversity between different lineages within a tumor and between different tumors within the same patient is the horizontal transmission of genes and gene products between cells, whether by cell-cell fusion or by incorporation of circulating genetic or epigenetic factors derived from tumor and/or stromal cells. This could also explain the induction of the cancer-enhancing phenotype described for stromal cells within a cancer,27–30 and how cancers become metastatic.1,6,15 Evidence in a murine model that intestinal inflammation and epithelial proliferation are both involved in promoting cell fusion50 further supports the hypothesis that this mechanism also may be implicated in early oncogenic events, as proposed many years ago.1
Novelty.
We report the long-term propagation in hamsters of a human glioblastoma which had spontaneously fused with hamster stromal cells in vivo. The hybrid tumor retained functional human genes associated with malignancy and the original human tumor’s morphologic phenotype during serial transplantation and cell culture. Three of these genes were translated to their respective proteins. We believe that this is the first report of functional human genes surviving long-term xenogeneic passage of a human tumor, yet being highly metastatic in the animal host.
Significance.
This paper discusses these findings in relation to the expanding literature on in-vivo cell fusion and horizontal gene transfer involving oncogenesis, as well as, more generally, the interaction between cancer parenchymal and stromal cells in the progression of malignancy.
Supplementary Material
Acknowledgements
We are grateful to the late Dr. Gary Stone and to Dr. Polina Perelman from the National Cancer Institute, Frederick Campus, for the generous gift of the golden hamster X-chromosome probe and expert advice. We appreciate crucial advice and discussions with Dr. Hesed Padilla-Nash, Genetics Branch, National Cancer Institute, regarding interspecies FISH hybridization, and the preparation of FISH figures by Buddy Chen. Supported in part by the intramural research program of the National Cancer Institute, NIH (T.R.), and by the American Cancer Society (Grant IN-581), the Damon Runyon Memorial Fund for Cancer Research and USPHS grant CA12374 from the National Institutes of Health (D.M.G.) for establishing the GB-749 tumor.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Goldenberg DM. Űber die Progression der Malignität: Eine Hypothese [On the progression of malignancy: A hypothesis] Klin Wochenschr. 1968;46:898–899. doi: 10.1007/BF01746251. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg DM. Stathmokinetic effect of colcemid on a presumptive human-hamster hybrid tumor, GW-478. Exp Mol Pathol. 1971;14:134–137. doi: 10.1016/0014-4800(71)90059-1. [DOI] [PubMed] [Google Scholar]
- 3.Goldenberg DM, Götz H. On the ‘human’ nature of highly malignant heterotransplantable tumors of human origin. Eur J Cancer. 1968;4:547–548. doi: 10.1016/0014-2964(68)90011-x. [DOI] [PubMed] [Google Scholar]
- 4.Götz H, Goldenberg DM. Antigenic characterization of a heterotransplanted human tumor, GW-127. Experientia. 1968;24:957–958. doi: 10.1007/BF02138681. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg DM, Bhan RD, Pavia RA. In vivo human-hamster somatic cell fusion by glucose-6-phosphate dehydrogenase and lactate dehydrogenase profiles. Cancer Res. 1971;31:1148–1152. [PubMed] [Google Scholar]
- 6.Goldenberg DM, Pavia RA, Tsao MC. In vivo hybridisation of human tumour and normal hamster cells. Nature. 1974;250:649–651. doi: 10.1038/250649a0. [DOI] [PubMed] [Google Scholar]
- 7.Goldenberg DM, Pavia RA. Horizontal transmission of malignant conditions rediscovered. N Engl J Med. 1981;305:283–284. doi: 10.1056/nejm198107303050513. [DOI] [PubMed] [Google Scholar]
- 8.Aichel O. Über Zellverschmelzung mit qualitative abnormer Chromosomenverteilung als Ursache der Geschwulstbildung [On cell fusion with qualitative abnormal chromosome distribution as the cause of tumor formation] In: Roux W, editor. Vorträge und Aufsätze über Entwicklungsmechanik der Organismen. Vol. 13. Leizig: Wilhelm Engelmann; 1911. [Google Scholar]
- 9.Chakraborty A, Sodi S, Rachkovsky M, Kolesnikova N, Platt JT, Bolognia JL, Pawelek JM. A spontaneous murine melanoma lung metastasis comprised of host x tumor hybrids. Cancer Res. 2000;60:2512–2519. [PubMed] [Google Scholar]
- 10.DeBaetselier P, Roos E, Brys L, Remels L, Gobert M, Dekegel D, Segal S, Feldman M. Nonmetastatic tumor cells acquire metastatic properties following somatic hybridization with normal cells. Cancer Metastasis Rev. 1984;3:5–24. doi: 10.1007/BF00047690. [DOI] [PubMed] [Google Scholar]
- 11.Larizza L, Schirrmacher V. Somatic cell fusion as a source of genetic rearrangement leading to metastatic variants. Cancer Metastasis Rev. 1984;3:193–222. doi: 10.1007/BF00048385. [DOI] [PubMed] [Google Scholar]
- 12.Lagarde AE, Kerbel RS. Somatic cell hybridization in vivo and in vitro in relation to the metastatic phenotype. Biochim Biophys Acta. 1985;823:81–110. doi: 10.1016/0304-419x(85)90008-3. [DOI] [PubMed] [Google Scholar]
- 13.Miller FR, Mohamed AN, McEachern D. Production of more aggressive tumor cell variant by spontaneous fusion of two mouse tumor subpopulations. Cancer Res. 1989;49:4316–4321. [PubMed] [Google Scholar]
- 14.Jacobsen BM, Harrell JC, Jedlicka P, Borges, Varella-Garcia M, Horwitz KB. Sponteneous fusion with and transformation of mouse stroma by malignant human breast cancer epithelium. Cancer Res. 2006;66:8274–8279. doi: 10.1158/0008-5472.CAN-06-1456. [DOI] [PubMed] [Google Scholar]
- 15.Pawelek JM, Chakrabarty AK. Fusion of tumor cells with bone marrow-derived cells: a unifying explanation of metastasis. Nat Rev Cancer. 2008;8:377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 16.Yilmaz Y, Lazova R, Qumsiyeh M, Cooper D, Pawelek J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005;35:1021–1024. doi: 10.1038/sj.bmt.1704939. [DOI] [PubMed] [Google Scholar]
- 17.Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005;6:567–575. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- 18.Vignery A. Macrophage fusion: are somatic and cancer cells possible partners? Trends Cell Biol. 2005;15:188–193. doi: 10.1016/j.tcb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Duelli D, Lazebnik Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer. 2007;7:968–976. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- 20.Larsson L-I, Bjerregaard B, Talts JF. Cell fusions in mammals. Histochem Cell Biol. 2008;129:551–561. doi: 10.1007/s00418-008-0411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar T, Nagler C, Schwitalla S, Reith G, Niggemann B, Zänker KS. Recurrence cancer stem cells – Made by cell fusion? Med Hypotheses. 2009;73:542–547. doi: 10.1016/j.mehy.2009.05.044. [DOI] [PubMed] [Google Scholar]
- 23.Harris H. The analysis of malignancy by cell fusion: The position in 1988. Cancer Res. 1988;48:3302–3306. [PubMed] [Google Scholar]
- 24.Powell AE, Anderson EC, Davies PS, Silk AD, Pelz C, Impey S, Wong MH. Fusion between intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldenberg DM, Pavia RA. Oncogenesis by interspecific interaction of malignant murine and non-malignant hamster cells in vitro. Int J Cancer. 1975;15:282–300. doi: 10.1002/ijc.2910150214. [DOI] [PubMed] [Google Scholar]
- 26.Goldenberg DM, Pavia RA, Smith L. Karyology of a human colonic carcinoma (GW-39) serially xenografted in hamsters and nude mice. Cancer Lett. 1977;2:195–200. doi: 10.1016/s0304-3835(77)80021-9. [DOI] [PubMed] [Google Scholar]
- 27.Tlsty TD. Stromal cells can contribute oncogenic signals. Cancer Biol. 2001;11:97–104. doi: 10.1006/scbi.2000.0361. [DOI] [PubMed] [Google Scholar]
- 28.Mueller MM, Fusenig NE. Friends or foe – biopolar effects of the tumor stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 29.Proia DA, Kuperwasser C. Stroma. tumor agonist or antagonist. Cell Cycle. 2005;4:1022–1025. doi: 10.4161/cc.4.8.1903. [DOI] [PubMed] [Google Scholar]
- 30.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J, Govindan S, Goldenberg DM. CD74: A new candidate target for the immunotherapy of B-cell neoplasms. Clin Cancer Res. 2000;23(Suppl):5556s–5563s. doi: 10.1158/1078-0432.CCR-07-1167. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. A functional heteromeric MIF receptor formed by CD74 and CXCR4 . FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehtesham M, Winston JA, Kabos P, Thompson RC. CXCR4 expression mediates glioma cell invasiveness. Oncogene. 2006;25:2801–2806. doi: 10.1038/sj.onc.1209302. [DOI] [PubMed] [Google Scholar]
- 34.Rempel SA, Dudas S, Ge S, Gutiérrez JA. Identification and localization of the cytokine SDF1 and its receptor, CXC chemokine receptor 4, to regions of necrosis and angiogenesis in human glioblastoma. Clin Cancer Res. 2000;6:102–111. [PubMed] [Google Scholar]
- 35.Sehgal A, Ricks S, Boynton AL, Warrick J, Murphy GP. Molecular characterization of CXCR-4: a potential brain tumor-associated gene. J Surg Oncol. 1998;69:239–248. doi: 10.1002/(sici)1096-9098(199812)69:4<239::aid-jso9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 36.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, Hu J, Vinjamoori A, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta V, Rajaraman S, Gadson P, Costanzi JJ. Primary transfection as a mechanism for transformation of host cells by human tumor cells implanted in nude mice. Cancer Res. 1987;47:5194–5201. [PubMed] [Google Scholar]
- 38.Lu X, Kang Y. Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc Natl Acad Sci USA. 2009;106:9385–9390. doi: 10.1073/pnas.0900108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Current Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 40.Pelham RJ, Rodgers L, Hall I, Lucito R, Nguyen KC, Navin N, Hicks J, Mu D, Powers S, Wigler M, Botstein D. Identification of alterations in DNA copy number in host stromal cells during tumor progression. Proc Natl Acad Sci USA. 2006;103:19848–19853. doi: 10.1073/pnas.0609635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monifer F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60:2562–2566. [PubMed] [Google Scholar]
- 42.Duda DG, Duyverman AM, Kohno M, Snuderl M, Steller EJ, Fukumura D, Jain RK. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci USA. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldenberg DM, Pavia RA. Malignant potential of murine stromal cells after transplantation of human tumors into nude mice. Science. 1981;212:65–67. doi: 10.1126/science.7209521. [DOI] [PubMed] [Google Scholar]
- 44.Goldenberg DM, Pavia RA. In vivo horizontal oncogenesis by a human tumor in nude mice. Proc Natl Acad Sci USA. 1982;79:2389–2392. doi: 10.1073/pnas.79.7.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beattie GM, Knowles AF, Jensen FC, Baird SM, Kaplan NO. Induction of sarcomas in athymic mice. Proc Natl Acad Sci USA. 1982;79(9):3033–3036. doi: 10.1073/pnas.79.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrlich P, Apolant H. Beobachtungen über maligne Mäusetumoren [Observations on malignant murine tumors] Berl Klin Wochenschr. 1905;42:871–874. [Google Scholar]
- 47.Holmgren L, Bergsmedh A, Spetz AL. Horizontal transfer of DNA by uptake of apoptotic bodies. Vox Sang. 2002;83(Suppl 1):S305–S306. doi: 10.1111/j.1423-0410.2002.tb05323.x. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Olmo DC, Domínguez C, García-Arranz M, Anker P, Stroun M, García-Verdugo JM, Garcia-Olmo D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70:560–567. doi: 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]
- 49.Stoecklin NH, Klein CA. Genetic disparity between primary tumors, disseminated tumor cells, and manifest metastasis. Int J Cancer. 2010;126:589–598. doi: 10.1002/ijc.24916. [DOI] [PubMed] [Google Scholar]
- 50.Davies PS, Powell AE, Swain JR, Wong MH. Inflammation and proliferation act together to mediate intestinal cell fusion. PLoS ONE. 2009;4:e6530. doi: 10.1371/journal.pone.0006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


