Abstract
HSV type 1 (HSV-1) expresses its genes sequentially as immediate early (α), early (β), leaky late (γ1), and true late (γ2), where viral DNA synthesis is an absolute prerequisite only for γ2 gene expression. The γ1 protein glycoprotein B (gB) contains a strongly immunodominant CD8+ T cell epitope (gB498–505) that is recognized by 50% of both the CD8+ effector T cells in acutely infected trigeminal ganglia (TG) and the CD8+ memory T cells in latently infected TG. Of 376 predicted HSV-1 CD8+ T cell epitopes in C57BL/6 mice, 19 (gB498–505 and 18 subdominant epitopes) stimulated CD8+ T cells in the spleens and TG of HSV-1 acutely infected mice. These 19 epitopes identified virtually all CD8+ T cells in the infected TG that represent all or the vast majority of the HSV-specific CD8+ TCR repertoire. Only 11 of ∼84 HSV-1 proteins are recognized by CD8+ T cells, and most (∼80%) are expressed before viral DNA synthesis. Neither the immunodominance of gB498–505 nor the dominance hierarchy of the subdominant epitopes is due solely to MHC or TCR affinity. We conclude that the vast majority of CD8+ T cells in HSV-1 acutely infected TG are HSV specific, that HSV-1 β and γ1 proteins that are expressed before viral DNA synthesis are favored targets of CD8+ T cells, and that dominance within the TCR repertoire is likely due to the frequency or expansion and survival characteristics of CD8+ T cell precursors.
Herpes simplex virus infects a high percentage of the world population and establishes a latent infection in which the viral genome is retained in sensory neurons, but no virions are produced. Periodic reactivation of the virus from this latent state results in lesions that can affect the mucosal surfaces of the mouth and lips, genital tract, and cornea of the eye, and less frequently the skin and brain. HSV-2 can be lethal to newborns who acquire it from the birth canal; corneal HSV-1 infections are a leading infectious cause of blindness; and brain HSV-1 infections account for approximately one quarter of cases of viral encephalitis that can be fatal. HSV-1 vaccines that have made their way to clinical trials have been primarily designed for Ab production and have been largely ineffective (1, 2). Evidence suggests a significant role for CD8+ T cells in controlling HSV infections in both mice and humans (3, 4). However, only limited information is available about the HSV epitopes that are recognized by human CD8+ T cells or the types of viral proteins they target (5–7).
In mice, viruses typically induce a CD8+ T cell response that targets a very small fraction of the viral proteins and the potential epitopes they contain. Those viral epitopes that are targeted typically fall into a dominance hierarchy consisting of one or a few dominant epitopes and several other subdominant epitopes. The CD8+ T cell response to HSV-1 follows this pattern, where it has been estimated that 70% of the HSV-specific CD8+ TCR repertoire recognizes a single immunodominant epitope on HSV glycoprotein B (gB498–505) (8). One subdominant CD8+ T cell epitope was identified on ribonucleotide reductase 1 (RR1822–829), which is also known as infected cell protein (ICP)6 (9). During lytic infections, HSV expresses its genes sequentially as immediate early (α) genes, early (β) genes, leaky late (γ1) genes, and true late (γ2) genes (10–12). The α and β genes are fully expressed before viral DNA synthesis, the γ1 genes are expressed at low levels before and at much higher levels after viral DNA synthesis, whereas γ2 gene expression is absolutely dependent on prior initiation of viral DNA replication (12). Thus, both gB (a γ1 protein) and RR1 (a β protein) are expressed early in the viral life cycle before viral DNA replication.
We and others have demonstrated that CD8+ T cells infiltrate the HSV-1–infected trigeminal ganglion of C57BL/6 mice, reaching maximal numbers 8 d after corneal infection coincident with the establishment of viral latency. These cells closely associate with infected neurons and remain in direct apposition to, and in some cases forming an immunological synapse with, neurons during a lifelong latent infection (13). The composition of both the CD8+ T cell effector population in the trigeminal ganglion at 8 d postinfection (dpi) and the contracted memory population during latency is highly consistent with 50% showing specificity for the immunodominant gB498–505 epitope, ∼5% specific for the known subdominant RR1822–829 epitope, and the remainder of unknown specificity. Our previous findings strongly suggested that the CD8+ T cell of unknown specificity in the trigeminal ganglion is HSV-1 specific (14). The current study confirms those findings and defines the entire HSV-specific CD8+ T cell repertoire in C57BL/6 mice.
Materials and Methods
Mice and virus
Wild-type HSV-1 strain RE was grown in Vero cells, and intact virions were isolated on Optiprep gradients according to the manufacturer's instructions (Accurate Chemical and Scientific, Westbury, NY). Six- to eight-week-old female wild-type C57BL/6 mice were anesthetized by i.p. injection of 2.0 mg ketamine hydrochloride and 0.04 mg xylazine (Phoenix Scientific, San Marcos, CA) in 0.2 ml HBSS (Bio Whittaker, Walkersville, MD). The abraded central corneas of anesthetized mice were infected by topical application of 3 μl RPMI 1640 (Bio Whittaker) containing 1 × 105 PFU HSV-1. All animal experiments were conducted in accordance with guidelines established by the University of Pittsburgh Institutional Animal Care and Use Committee.
Epitope prediction
The entire HSV-1 proteome (GI 9629378) was scanned for peptide sequences predicted to have a high-affinity binding capacity for the MHC class I molecules H-2 Kb or H-2 Db. According to the reported peptide length preference of these MHC molecules, peptides comprising 8 and 9 amino acids were analyzed for H-2 Kb and H-2 Db binding, respectively. For both H-2 alleles, the ANN and SMM binding prediction methods available from the Immune Epitope Database (http://www.iedb.org) were used along with matrices derived from combinatorial peptide libraries reported by Udaka et al. (15). Each method assigns a score to a peptide based on its sequence that predicts its binding affinity for the respective MHC molecule. For each prediction method, the peptides were ranked corresponding to their predicted binding capacity. To construct a consensus from the prediction methods, the median rank of each peptide for all applicable prediction methods was taken. Finally, for each allele, the 188 peptides with the highest median ranks, corresponding with the top 0.5% scoring peptides, were selected for screening.
Peptides used in the HSV screening studies were synthesized as crude material by Mimotopes (Clayton, VIC, Australia). Peptides used as radiolabeled ligands for binding assays, were synthesized by A and A Labs (San Diego, CA), and purified to >95% homogeneity by reverse-phase HPLC. Purity of these peptides was determined using analytical reverse-phase HPLC and amino acid analysis, sequencing, and/or mass spectrometry. Peptides were radiolabeled with the chloramine T method, as described elsewhere (16).
MHC peptide binding assays
MHC purification, and quantitative assays to measure the binding affinity of peptides to purified H-2 Kb and H-2 Db molecules were performed as previously described (16, 17). Briefly, 1–10 nM of radiolabeled peptide was co-incubated at room temperature with 1 μM to 1 nM of purified MHC in the presence of 1–3 μM human β2-microglobulin (Scripps Laboratories, San Diego, CA) and a mixture of protease inhibitors. After a 2-d incubation, binding of the radiolabeled peptide to the corresponding MHC class I molecule was determined by capturing MHC–peptide complexes on Greiner Lumitrac 600 microplates (Greiner Bio-one, Longwood, FL) coated with either the Y3 (anti–H-2 Kb) or 28-14-8s (anti–H-2 Db, Ld, and Dq) Ab and measuring bound cpm using the TopCount microscintillation counter (Packard Instrument Co.).
For competition assays, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Peptides were typically tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent assays. Under the conditions used, where (label) < (MHC) and IC50 ≥ (MHC), the measured IC50 values are reasonable approximations of the true Kd values (18, 19).
Reagents
PE-conjugated H-2Kb tetramers complexed with the gB498–505, RR1982–989, RR1822–829, or ICP8876–883, peptides and PE-conjugated H-2Db tetramers complexed with the ICP8168–176, RR1372–380, or ribonucleotide reductase 2 (RR2279–287) peptide were provided by the National Institute of Allergy and Infectious Diseases Tetramer Core Facility (Emory University Vaccine Center, Atlanta, GA). Rat anti-mouse Pacific blue-conjugated anti-CD8α (clone 53-6.7), allophycocyanin-conjugated anti–IFN-γ (XMG1.2), PerCP-conjugated anti-CD45 (30-F11), PE-Cy7–conjugated anti–TNF-α (MP6-XT22), allophycocyanin-conjugated anti-granzyme B (GB11), and BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit were purchased from BD Pharmingen (San Diego, CA). The appropriate isotype control Abs were purchased from BD Pharmingen (San Diego, CA). All flow cytometry samples were collected on a FACSAria cytometer and analyzed by FACSDiva software (BD Biosciences).
Tissue preparation
At 8 dpi, anesthetized mice were injected with 0.3 ml heparin 1000 U/ml and euthanized by exsanguination. Trigeminal ganglia (TG) were harvested and digested in 100 μl per gangion of DMEM (Bio Whittaker) containing 10% FCS and 400 U/ml collagenase type I (Sigma-Aldrich) for 1 h at 37°C. TG were dispersed into single-cell suspensions by trituration through a p-200 pipette tip. Spleens were dispersed mechanically and treated with RBC lysis buffer prior to use. Tissue harvest and preparation were performed under sterile conditions.
Intracellular cytokine staining
B6WT3 fibroblast targets were pulsed with peptide at a concentration of 0.8 μg/ml for 45 min at 37°C/5% CO2. Dispersed TG or spleen cells were stimulated with peptide-pulsed fibroblasts in the presence of Golgi-Plug (BG Biosciences) for 6 h at 37°C/5% CO2. After stimulation, cells were stained for surface expression of CD8α, followed by intracellular staining for IFN-γ and TNF-α after permeabilization and fixation via Cytofix/Cytoperm (BD Biosciences).
Phenotypic analysis of T cells
Dispersed TG or spleens were stained with anti-CD45, CD8α, and tetramer for 1 h at room temperature. After incubation, cells were permeabilized and fixed with Cytofix/Cytoperm (BD Biosciences) and stained for intracellular granzyme B.
Tetramer release assay
Tetramer release assay was performed as described (20). Single-cell TG suspensions were stained with gB498–505 or RR1982–989 for 1 h at 37°C, and the cells were washed and then incubated with anti–H-2Db/Kb Ab to avoid tetramer rebinding (28-8-6; BD Pharmingen) at 37°C for the designated times. Cells were then stained with anti-CD8 and anti-CD45 and analyzed via flow cytometry to observe loss of tetramer over time.
Results
HSV-specific CD8+ TCR repertoire
Potential HSV-1 CD8+ T cell epitopes in C57BL/6 mice we identified using a suite of MHC binding algorithms, as described in Materials and Methods. In total, 376 peptides representing the top 0.5% scoring Kb and Db predicted binders were selected for analysis. Our previous findings suggested that acutely infected TG represent a source of highly enriched HSV-specific CD8+ T cells that could provide sensitive, though possibly incomplete detection of HSV-1 CD8+ T cell epitopes (14). To determine how faithfully the TCR repertoire in TG reflects that generated in the lymphoid organs, we additionally analyzed the spleen because it exhibits a higher frequency of HSV-specific CD8 T cells than is observed in the lymph nodes after primary HSV-1 corneal infection. Accordingly, the 376 peptides that were predicted to represent CD8+ T cell epitopes were used to stimulate CD8+ T cells in dispersed TG and spleens obtained from mice at 8 dpi. Responding T cells were quantified in a flow cytometry-based 6-h intracellular IFN-γ detection assay. This screening identified 19 individual peptides that were recognized by CD8+ T cells from infected TG (Fig. 1). These included two previously identified epitopes; an immunodominant epitope on gB (gB498–505) and a subdominant epitope on RR1 (RR1822–829). In addition, 17 previously undefined subdominant epitopes were identified. The MHC binding affinity of the 19 peptides for their respective predicted MHC was tested in in vitro binding assays, as described in Materials and Methods. As shown in Table I, 15 (79%) of the peptides bound their putative restricting molecule with an affinity of 500 nM, or better, including 11 that bound with affinities <100 nM. Notably, both the immunodominant gB498–505 and subdominant RR1822–829 epitopes were shown to bind H-2 Kb with very high affinities, with IC50 of 0.8 and 2.1 nM, respectively.
Figure 1.
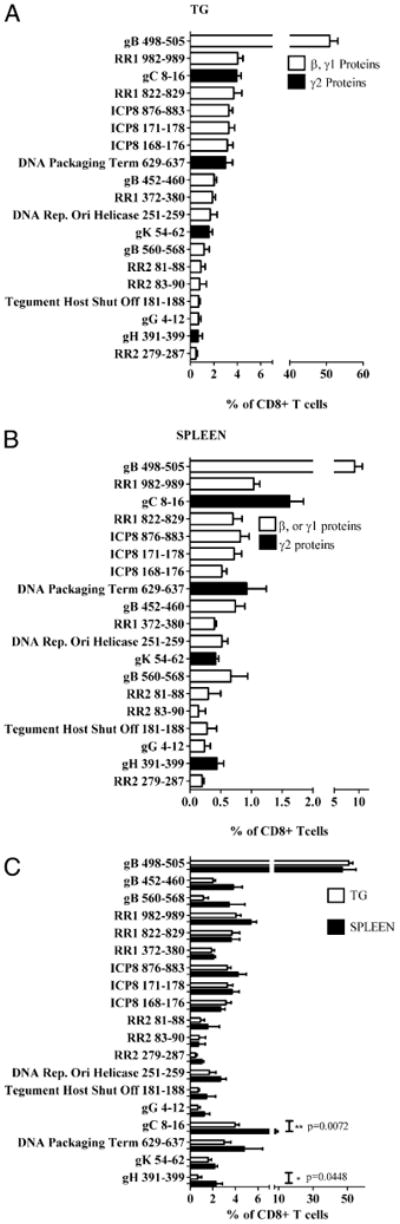
One dominant and 18 subdominant epitopes define the HSV-specific CD8+ TCR repertoire in the trigeminal ganglion and spleen of C57BL/6 (B6) mice. TG (A, C) and spleens (B, C) from 8 dpi B6 mice were dispersed into single-cell suspensions. Tissues were then incubated for 6 h with peptideloaded B6WT3 fibroblasts in the presence of Golgi-Plug, followed by intracellular staining for IFN-γ. A, Bars represent the mean percentage of TG-resident CD8+ T cells that produce IFN-γ ± SEM (n = 5). B, Bars represent the mean percentage of CD8+ T cells in the spleen that produce IFN-γ ± SEM(n = 5). C, Comparison of HSV-specific CD8+ T cells in the TG and spleen that respond to HSV-1 epitopes. All data are from two independent experiments.
Table I. Epitopes recognized by HSV-specific CD8+ T cells in spleens and TG.
| Sequence | Pro. | Type | Pos. | MHC (H-2) | Binding Capacity (IC50, nM) |
Spleen | TG Percentage of CD8b | TG Percentage of Spleenc | |
|---|---|---|---|---|---|---|---|---|---|
| CD8 (%) | HSV-CD8+ (%)a | ||||||||
| SSIEFARL | gB | γ1 | 498 | Kb | 0.78 | 9.14 | 46.07 | 57.9 | 125.81 |
| YQPLLSNTL | gB | γ1 | 452 | Db | 5153 | 0.74 | 3.73 | 2.30 | 61.62 |
| SARMLGDVM | gB | γ1 | 560 | Db | 9206 | 0.67 | 3.38 | 1.32 | 39.05 |
| Total gB | 10.55 | 53.18 | 61.58 | 115.80 | |||||
| GAMRAVVPI | gG | γ1 | 4 | Db | 499 | 0.24 | 1.21 | 0.78 | 64.46 |
| Total γ1 | 10.79 | 54.39 | 62.36 | 114.65 | |||||
| FAPLFTNL | RR1 | β | 982 | Kb | <0.5 | 1.04 | 5.24 | 4.60 | 87.79 |
| QTFDFGRL | RR1 | β | 822 | Kb | 2.1 | 0.7 | 3.53 | 4.19 | 118.70 |
| FGLLNYALV | RR1 | β | 372 | Db | 1.0 | 0.4 | 2.02 | 2.14 | 105.94 |
| Total RR1 | 2.14 | 10.79 | 10.93 | 101.30 | |||||
| GAINFINL | ICP8 | β | 876 | Kb | 6.2 | 0.82 | 4.13 | 3.74 | 90.56 |
| INNTFLHL | ICP8 | β | 171 | Kb | 2.0 | 0.72 | 3.63 | 3.74 | 103.03 |
| AVCINNTFL | ICP8 | β | 168 | Db | 450 | 0.52 | 2.62 | 3.60 | 137.41 |
| Total ICP8 | 2.06 | 10.38 | 11.08 | 106.74 | |||||
| SFYRFLFA | RR2 | β | 81 | Kb | 64 | 0.3 | 1.51 | 1.03 | 68.21 |
| YRFLFAFL | RR2 | β | 83 | Kb | 5.4 | 0.14 | 0.71 | 0.89 | 125.35 |
| AAIENYVRF | RR2 | β | 279 | Db | 38 | 0.2 | 1.01 | 0.52 | 51.49 |
| Total RR2 | 0.64 | 3.23 | 2.44 | 75.54 | |||||
| FLPRLGTEL | UL9 | β | 251 | Db | 3049 | 0.52 | 2.62 | 1.91 | 72.90 |
| LGYAYINS | UL41 | β | 181 | Kb | 10 | 0.28 | 1.41 | 0.80 | 56.74 |
| Total β | 5.64 | 28.43 | 27.16 | 95.53 | |||||
| LAVVLWSLL | gC | γ2 | 8 | Db | 2781 | 1.62 | 8.17 | 4.51 | 55.20 |
| YSVENVGLL | UL28 | γ2 | 629 | Db | 1.9 | 0.93 | 4.69 | 3.42 | 72.92 |
| FAFVNAAHA | gH | γ2 | 391 | Db | 4.9 | 0.44 | 2.22 | 0.75 | 33.78 |
| WMKMNQTLL | gK | γ2 | 54 | Db | 131 | 0.42 | 2.12 | 1.80 | 84.91 |
| Total γ2 | 3.41 | 17.20 | 10.18 | 59.20 | |||||
| Total subdominant | 10.70 | 53.95 | 36.90 | 68.40 | |||||
| Total | 19.84 | 100.02 | 99.7 | ||||||
Mathematical algorithms identified 376 HSV-1 peptides with predicted capacity to bind to H-2Kb or H-2Db of which 19 proved to be CD8+ T cell epitopes. Also depicted are the percentages of CD8+ T cells that are stimulated by these epitopes.
The % of CD8+ T cells stimulated by the epitope divided by the total % of CD8+ T cells stimulated with all HSV epitopes (19.84%).
The % of CD8+ T cells stimulated by the epitope divided by the total % of CD8+ T cells stimulated with all HSV epitopes (87.77%).
% TG divided by % spleen.
Pos., starting amino acid position; Pro., protein; UL9, DNA replication origin-binding helicase; UL28, DNA replication origin-binding helicase; UL41, tegument host shutoff protein.
Two of the subdominant epitopes on HSV-1 ICP8 (ICP8168–176 and ICP8171–178) overlapped by 6 amino acids but were predicted to be separate epitopes restricted by H2-Db and H-2Kb, respectively (Table I). This was confirmed by the additive response to the pooled peptides and the 50% reduction in the response to the pooled peptides in the presence of anti–H-2Kb Ab (Supplemental Fig. 1).
HSV-specific CD8+ T cells selectively target a highly restricted array of HSV-1 proteins
The 19 HSV-1 epitopes are derived from only 11 of ∼84 HSV-1 proteins (Table I). Of the four kinetic classes of HSV-1 proteins, no a gene products are targeted, 11 of 19 defined epitopes are derived from β gene products, 4 of 19 epitopes are derived from γ1 gene products, and the remaining 4 epitopes are from γ2 gene products (Table I). Thus, nearly 80% of the epitopes are derived from viral proteins that are produced early in the viral life cycle prior to viral DNA synthesis.
Relative frequency of epitope-specific CD8+ T cells in the spleen and TG
The frequency of TG or splenic CD8+ T cells responsive to these epitopes was then determined in five separate cell preparations (Fig. 1A, 1B). In aggregate, 87.77 ± 9.22% of TG CD8+ T cells and 19.84 ± 4.13% of splenic CD8+ T cells responded to defined HSV-1 epitopes (Fig. 1), and a nearly identical frequency, 17.3 ± 2.0%, of splenic CD8+ T cells responded to HSV-1–infected fibroblasts. Together these findings suggest that these 19 epitopes define the entire HSV-1–specific TCR repertoire. Based on the assumption that 19.84% of splenic CD8+ T cells and that 87.77% of TG CD8+ T cells are HSV-1 specific, we calculated the relative frequency of CD8+ T cells specific for each epitope within the HSV-specific CD8+ T cell populations in the spleen and TG (Fig. 1C, Table I).
Approximately 50% of the HSV-specific CD8+ T cells in the TG and spleen recognize the immunodominant gB498–505 epitope (Fig. 1C, Table I). Thus, CD8+ T cells specific for subdominant epitopes represented about half of the overall HSV-specific CD8+ T cell population in the spleen and TG. Although some variation was observed in the frequency of splenic and TG CD8+ T cells responsive to individual epitopes, the overall frequencies of CD8+ T cells specific for β and γ1 gene products (produced before viral DNA synthesis) were nearly identical, representing 83 and 90% of HSV-specific CD8+ T cells in the TG and spleen, respectively. CD8+ T cells specific for γ2 gene products (produced only after viral DNA synthesis) represented only 17 and 10% of HSV-specific CD8+ T cells in the spleen and TG, respectively. Although HSV-specific CD8+ T cells recognize 19 epitopes from 11 viral proteins, nearly three quarters of the HSV-specific CD8+ T cells in the TG and spleen target epitopes on the following three HSV-1 proteins: gB (53.18% spleen/61.58% TG), RR1 (10.79% spleen/10.93% TG), and ICP8 (10.38% spleen/11.08% TG). Overall, our data demonstrate that the entire HSV-1 CD8+ T cell repertoire generated in the spleen is represented and not significantly modified in the acutely infected TG.
CD8+ effector T cells specific for subdominant epitopes express granzyme B and are multifunctional
Optimal control of many viruses has been associated with CD8+ T cells that express granzyme B and produce both IFN-γ and TNF-α. Consistent with previous reports (14, 21), nearly all of the CD8+ effector T cells in the TG at 8 dpi that are specific for the immunodominant gB498–505 epitope express granzyme B (Fig. 2A, 2B). We observed only a slight (though statistically significant, p ≤ 0.025) reduction in the frequency of granzyme B+ cells among effector CD8+ T cells specific for subdominant epitopes compared with that among gB-specific CD8+ T cells (Fig. 2A, 2B). Again, this is consistent with a previous observation of a small reduction in granzyme B expression by CD8+ T cells in the TG that are not specific for the immunodominant gB498–505 epitope (14).
Figure 2.
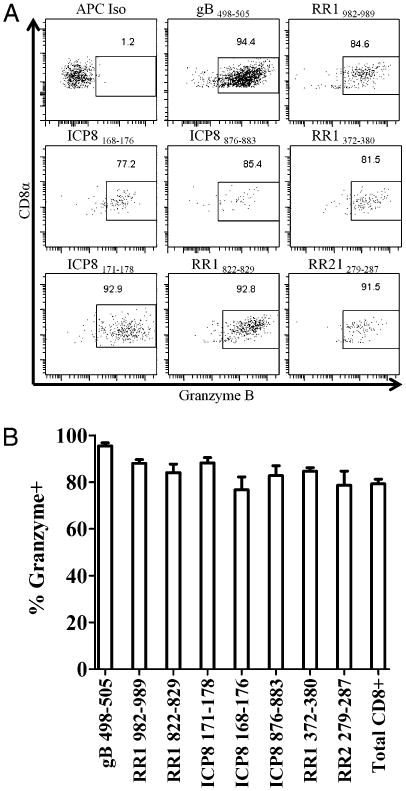
CD8+ T cells specific for subdominant epitopes from 8 dpi TG express granzyme B. Dispersed TG were stained for 1 h at room temperature with anti-CD45, CD8α, and tetramers loaded with sub-dominant epitopes followed by intracellular staining for granzyme B expression. A, Representative dot plots of granzyme B expression of select HSV-specific CD8+ T cells. B, Bars represent percentage of peptide-specific CD8+ T cells that express granzyme B ± SEM (n = 4) from two independent experiments. Granzyme B expression is lower in non-gB-specific CD8+ T cells compared with gB-specific CD8+ T cells. The p value for all specificities is <0.025.
To compare the level of functionality of the CD8+ T cells specific for immunodominant and subdominant epitopes, CD8+ T cells obtained from the TG at 8 dpi were stimulated for 6 h with targets pulsed with each of the 19 defined epitopes and stained for intracellular IFN-γ and TNF-α. The frequency of multifunctional CD8+ T cells that produced both IFN-γ and TNF-α (as opposed to IFN-γ only) is illustrated in Fig. 3A. The frequency of multifunctional CD8+ T cells reactive to subdominant epitopes varied widely. The CD8+ T cells specific for some subdominant epitopes had a high frequency of multifunctional cells (≥60%), which is similar to the frequency of multifunctional cells among CD8+ T cells specific for the immunodominant gB498–505 epitope (Fig. 3A). The frequency of multifunctional cells among CD8+ T cells specific for other subdominant epitopes ranged as low as 21.85%. We observed a significant correlation (p = 0.0337) between the frequency of CD8+ T cells specific for a given subdominant epitope in the TG and the frequency of multifunctional cells within that population (Fig. 3B).
Figure 3.
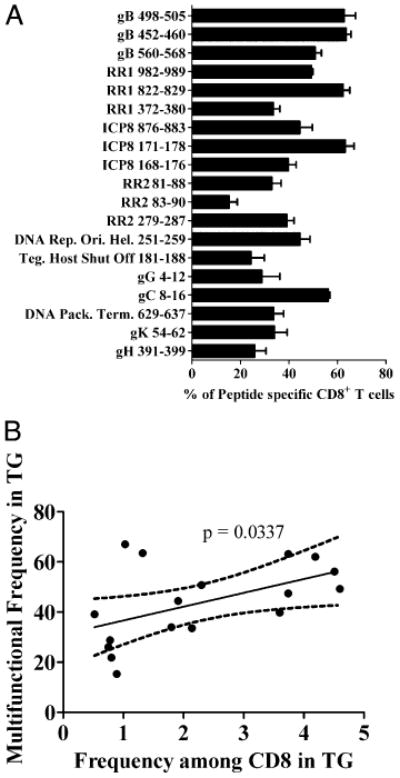
Non-gB-specific CD8+ T cells are multifunctional; multifunctionality correlates with immunodominance. TG from 8 dpi B6 mice were dispersed into single-cell suspensions. Tissues were then incubated for 6 h with peptide-loaded B6WT3 fibroblasts in the presence of Golgi-Plug, followed by intracellular staining for IFN-γ and TNF-α. A, Bars represent the mean percentage of peptide-specific CD8+ T cells that secrete IFN-γ and TNF-α when stimulated in vitro ± SEM (n = 4) from at least two independent experiments. B, Data points represent percentage abundance of individual epitopes in the TG at 8 dpi versus the percentage of epitope-specific cells that are multifunctional. Linear regression shows a significant correlation (p = 0.0337) between the prevalence in the TG and multifunctionality.
Immunodominance of HSV-1 CD8+ T cell epitopes is not determined solely by TCR affinity
Factors influencing dominance hierarchy among T cell epitopes include MHC binding affinity of the epitope, TCR binding affinity, efficiency of peptide processing and loading on MHC, and the frequency of CD8+ T cell precursors specific for a specific epitope. We ruled out MHC binding affinity as the crucial factor determining immunodominance based on the fact that several peptides representing subdominant epitopes had MHC binding affinities that were as high or higher than that of the immunodominant gB498–505 epitope (Table I). To determine if TCR binding affinity is a crucial factor determining immunodominance, we used a tetramer release assay to compare the TCR affinity of the immunodominant gB498–505 epitope with that of one of the subdominant epitopes (RR1982–989). The two epitopes exhibited comparable TCR binding affinity as indicated by the fact that the slopes of the tetramer release curves were not significantly different (Fig. 4).
Figure 4.
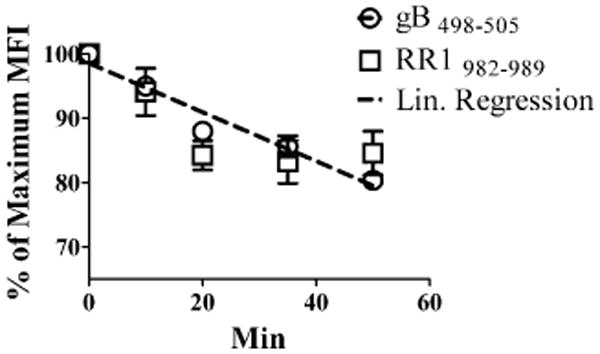
Immunodominance is not solely determined by TCR affinity for MHC–peptide complexes. Dispersed TG were stained with gB498–505 or RR1982–989 tetramer for 1 h at 37°C. Anti–H-2Kb/H-2Db blocking Ab was added, and TG were incubated for the designated times at 37°C to observe tetramer dissociation. Data represent the mean fluorescence intensity (MFI) of CD8+ tetramer+ cells as a percentage ± SEM (n = 3) of the maximum MFI observed at time zero. Linear regression represents the slopes of both gB498–505 and RR1982–989. Data are representative of three independent experiments.
Discussion
After infection at mucosal sites, HSV-1 is transported to the nuclei of sensory neurons where it establishes a latent infection. We and others have demonstrated that only activated CD8+ T cells infiltrate the TG of mice during the acute phase of a primary corneal infection and remain closely associated with neuronal cell bodies as the virus enters a life-long latent state (13). Half of the CD8+ T cells in the acutely infected TG are specific for a strongly immunodominant epitope on HSV gB, and we recently provided circumstantial evidence in support of the concept that the remaining cells in the TG are also HSV specific (14). This study was undertaken to identify the epitopes that are recognized by the 50% of CD8+ T cells in the TG that are not specific for the immunodominant epitope. Based on our previous findings, we anticipated that CD8+ T cells in the acutely infected TG would represent a highly enriched source of HSV-specific CD8+ T cells that could provide sensitive detection of HSV-1 CD8+ T cell epitopes (14).
Candidate H-2Kb and H-2 Db epitopes were identified using a consensus prediction approach, as previously described (22). An analysis of HSV-1 open reading frames identified 376 peptide sequences with the potential to serve as CD8+ T cell epitopes. Screening of these 376 candidate peptides identified 19 that were capable of stimulating CD8+ T cells obtained from HSV-1 acutely infected TG. These 19 epitopes appear to identify the entire HSV-specific CD8+ T cell repertoire in C57BL/6 mice based on the following observations. First, the 19 peptides stimulated nearly all of the CD8+ T cells in the TG. Second, there was complete concordance in the epitopes that stimulated TG and splenic CD8+ T cells. Third, in aggregate the 19 peptides stimulated 19% of the CD8+ T cells in the spleens of acutely infected mice, similar to the 17% that were stimulated by virus-infected targets.
Infiltration of CD8+ T cells into the TG begins at 6 dpi, and the population reaches maximum size by 8 dpi (23). The expansion of the CD8+ T cell population from 6 to 8 dpi reflects both infiltration and proliferation within the TG. The frequency of HSV-specific CD8+ T cells reactive to each of the 19 epitopes is quite similar in the TG and the spleen of infected mice, suggesting uniform expansion of all CD8+ T cell specificities in the TG. The only possible exception is found in CD8+ T cells reactive to epitopes on γ2 (true late) gene products, which were somewhat underrepresented in the TG (Table I).
The HSV-1 genome encodes at least 84 open reading frames (24). Thus, the CD8+ T cell response is highly selective, targeting only 19 epitopes on 11 viral proteins. The selective targeting of a minute portion of thousands of available peptide sequences is commonly seen in viral infections (22, 25–28). The mechanism of such selective targeting is unknown, but certain patterns emerge. For instance, viral immediate early genes are infrequent targets of CD8+ T cells (29), consistent with our observation that no HSV-1 immediate early proteins are targeted. Previous studies revealed that 87% of vaccinia virus epitopes and a majority of CD8+ T cell epitopes in human lesions are on proteins that are produced early in the viral life cycle (30, 31). This is consistent with our observation that ∼80% of the HSV-1 CD8+ T cell epitopes are on proteins that are produced before viral DNA synthesis, and nearly 80% of HSV-specific CD8+ T cells target these epitopes. The kinetic class of proteins that is most frequently targeted is the Early proteins that contain 58 and 65% of CD8+ T cell epitopes in HSV-1 and vaccinia virus, respectively. This is not a simple reflection of the frequency of this class of genes in the viral genome because Early genes represent only 14.2% (32) and 35% of HSV-1 and vaccinia virus genes, respectively. Although the mechanism responsible for selective targeting of early genes is not known, the advantages to the host are obvious. Viral DNA can be infectious, and the HSV-1 genome copy number in latently infected sensory ganglia is directly correlated with reactivation frequency (33).
Recent studies have established that initial expansion of HSV-specific CD8+ T cells in the draining lymph nodes of mice is induced by lymph node resident CD8α+ dendritic cells (DCs) (34– 37). Presumably these CD8α+ DCs cross-present viral Ags acquired from migratory DC from the site of infection (38). Both mature and immature DCs express HSV receptors nectin-1 and nectin-2 and herpesvirus entry mediator and are susceptible to HSV infection (39–41). However, only immature DCs are susceptible to productive infection. Moreover, productive infection of immature DCs leads to apoptosis that is dependent on both early and late HSV gene products (42). Infection of mature DCs results in an abortive infection in which HSV-1 α, β, and γ1 genes are expressed, but no infectious virions are produced (43). HSV-1 infection of mature DC results in downregulation of CD83 expression and impaired T cell stimulatory capacity. These findings are consistent with a model in which DCs that are matured at sites of infection before being infected with HSV are primarily responsible for transporting viral Ags to the lymph nodes for cross-presentation by CD8α+ DCs. Because these DCs permit only abortive HSV infections, the viral proteins they produce appear to be primarily those produced prior to viral DNA synthesis. Such a model would explain the fact that 80% of HSV-1 CD8+ T cell epitopes are derived from viral proteins that are produced before HSV-1 DNA synthesis. At later stages of infection, DCs that infiltrate sites of the lesion might begin to phagocytose apoptotic or necrotic parenchymal cells containing γ2 gene products, accounting for the small but significant response to several of these proteins.
Several factors can potentially contribute to the dominance hierarchy of viral epitopes. These include the efficiency of peptide generation by proteasomes (44), viral gene expression (29), the peptide binding affinity for MHC (27), the epitope affinity for TCR, the frequency of epitope-specific CD8+ T cell precursors (27), and the ability of epitope-specific CD8+ T cell precursors to expand and survive. The strong immunodominance of the gB498–505 epitope cannot be attributed solely to the peptide affinity for MHC as several of the subdominant epitopes have similar or higher affinities. Moreover, a tetramer release assay demonstrated a similar TCR affinity for the immunodominant and subdominant RR1982–989 epitopes. The RR1 protein is also expressed at high levels in infected cells (9). These findings suggested that the determining factor in the dominance of the gB498–505 over the RR1982–989 epitope is related either to the efficiency of peptide generation or to the characteristics of the epitope-specific CD8+ T cell precursors (frequency, proliferative capacity, or survival characteristics).
Targeting 50% of the HSV-specific CD8+ T cell repertoire to a single immunodominant epitope would seem to place the host at significant risk should that peptide be mutated. However, a recent study showed that a recombinant virus in which the gB498–505 epitope was mutated failed to expand gB498–505-specific CD8+ T cells, but the magnitude of the HSV-specific CD8+ T cell response and viral clearance were not altered (45). In this study, we show that CD8+ T cells specific for most of the subdominant epitopes exhibit functional characteristics similar to those reactive to the dominant epitope. Therefore, it is likely that elevation of any of the subdominant epitopes in the dominance hierarchy would have little impact on CD8+ T cell protection.
Our studies have important implications for the development of HSV vaccines. Murine studies show that immunization with CD8+ T cell epitopes generates effector and memory CD8+ T cells and reduces HSV lethality and viral load in the CNS (46, 47). Although the exact epitopes recognized by human CD8+ T cells will likely be different (48, 49), our findings and the study from Laing et al. (48) suggest that generating CD8+ T cells targeting β and γ1 proteins before infection might prove to be a good strategy for vaccine development. Moreover, although epitopes from HSV-1 γ2 proteins are recognized by only 20% of the HSV-specific CD8+ T cell repertoire in mice, our previous findings suggest that IFN-γ can block HSV-1 reactivation from latency even at a point after γ2 gene expression (50). Thus, targeting these gene products might also be useful in controlling both lytic infections and reactivation from latency.
Supplementary Material
Acknowledgments
We thank Dawn Maker and Jessica Spehar for technical assistance, Nancy Zurowski for flow cytometry acquisition, and the National Institute of Allergy and Infectious Diseases Tetramer Core Facility (Emory University Vaccine Center, Atlanta, GA) for supplying tetramers.
This work was supported by National Institutes of Health Grants P30-EY08098 (to R.L.H.), R01-EY005945 (to R.L.H.), and T32-EY017271 (to A.J.S.), an unrestricted grant from Research to Prevent Blindness (New York, NY), the Eye and Ear Foundation of Pittsburgh, and by National Institutes of Health-National Institute of Allergy and Infectious Diseases Contract HHSN272200700048C (to A.S.).
Abbreviations used in this article
- DC
dendritic cell
- dpi
days postinfection
- gB
glycoprotein B
- ICP
infected cell protein
- RR1
ribonucleotide reductase 1
- RR2
ribonucleotide reductase 2
- TG
trigeminal ganglia
Footnotes
Disclosures: The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Cohen J. Immunology. Painful failure of promising genital herpes vaccine. Science. 2010;330:304. doi: 10.1126/science.330.6002.304. [DOI] [PubMed] [Google Scholar]
- 2.de Bruyn G, Vargas-Cortez M, Warren T, Tyring SK, Fife KH, Lalezari J, Brady RC, Shahmanesh M, Kinghorn G, Beutner KR, et al. A randomized controlled trial of a replication defective (gH deletion) herpes simplex virus vaccine for the treatment of recurrent genital herpes among immuno-competent subjects. Vaccine. 2006;24:914–920. doi: 10.1016/j.vaccine.2005.08.088. [DOI] [PubMed] [Google Scholar]
- 3.Theil D, Derfuss T, Paripovic I, Herberger S, Meinl E, Schueler O, Strupp M, Arbusow V, Brandt T. Latent herpesvirus infection in human trigeminal ganglia causes chronic immune response. Am J Pathol. 2003;163:2179–2184. doi: 10.1016/S0002-9440(10)63575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verjans GMGM, Hintzen RQ, van Dun JM, Poot A, Milikan JC, Laman JD, Langerak AW, Kinchington PR, Osterhaus ADME. Selective retention of herpes simplex virus-specific T cells in latently infected human trigeminal ganglia. Proc Natl Acad Sci USA. 2007;104:3496–3501. doi: 10.1073/pnas.0610847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong L, Li P, Oenema T, McClurkan CL, Koelle DM. Public TCR use by herpes simplex virus-2-specific human CD8 CTLs. J Immunol. 2010;184:3063–3071. doi: 10.4049/jimmunol.0903622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koelle DM, Chen HB, Gavin MA, Wald A, Kwok WW, Corey L. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J Immunol. 2001;166:4049–4058. doi: 10.4049/jimmunol.166.6.4049. [DOI] [PubMed] [Google Scholar]
- 7.Chentoufi AA, Zhang X, Lamberth K, Dasgupta G, Bettahi I, Nguyen A, Wu M, Zhu X, Mohebbi A, Buus S, et al. HLA-A*0201-restricted CD8+ cytotoxic T lymphocyte epitopes identified from herpes simplex virus glyco-protein D. J Immunol. 2008;180:426–437. doi: 10.4049/jimmunol.180.1.426. [DOI] [PubMed] [Google Scholar]
- 8.Wallace ME, Keating R, Heath WR, Carbone FR. The cytotoxic T-cell response to herpes simplex virus type 1 infection of C57BL/6 mice is almost entirely directed against a single immunodominant determinant. J Virol. 1999;73:7619–7626. doi: 10.1128/jvi.73.9.7619-7626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvucci LA, Bonneau RH, Tevethia SS. Polymorphism within the herpes simplex virus (HSV) ribonucleotide reductase large subunit (ICP6) confers type specificity for recognition by HSV type 1-specific cytotoxic T lymphocytes. J Virol. 1995;69:1122–1131. doi: 10.1128/jvi.69.2.1122-1131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith IL, Hardwicke MA, Sandri-Goldin RM. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 11.Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson PA, MacLean C, Marsden HS, Dalziel RG, Everett RD. The product of gene US11 of herpes simplex virus type 1 is expressed as a true late gene. J Gen Virol. 1986;67:871–883. doi: 10.1099/0022-1317-67-5-871. [DOI] [PubMed] [Google Scholar]
- 13.Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity. 2003;18:593–603. doi: 10.1016/s1074-7613(03)00112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheridan BS, Cherpes TL, Urban J, Kalinski P, Hendricks RL. Reevaluating the CD8 T-cell response to herpes simplex virus type 1: involvement of CD8 T cells reactive to subdominant epitopes. J Virol. 2009;83:2237– 2245. doi: 10.1128/JVI.01699-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udaka K, Wiesmüuller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- 16.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1803s31. Chapter 18: Unit 18.3. [DOI] [PubMed] [Google Scholar]
- 17.Vitiello A, Yuan L, Chesnut RW, Sidney J, Southwood S, Farness P, Jackson MR, Peterson PA, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 18.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 19.Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 20.La Gruta NL, Doherty PC, Turner SJ. A correlation between function and selected measures of T cell avidity in influenza virus-specific CD8+ T cell responses. Eur J Immunol. 2006;36:2951–2959. doi: 10.1002/eji.200636390. [DOI] [PubMed] [Google Scholar]
- 21.Frank GM, Lepisto AJ, Freeman ML, Sheridan BS, Cherpes TL, Hendricks RL. Early CD4(+) T cell help prevents partial CD8(+) T cell exhaustion and promotes maintenance of Herpes Simplex Virus 1 latency. J Immunol. 2010;184:277–286. doi: 10.4049/jimmunol.0902373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotturi MF, Peters B, Buendia-Laysa F, Jr, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang A, Nikolich-Zugich J. Development and migration of protective CD8+ T cells into the nervous system following ocular herpes simplex virus-1 infection. J Immunol. 2005;174:2919–2925. doi: 10.4049/jimmunol.174.5.2919. [DOI] [PubMed] [Google Scholar]
- 24.Rajcáni J, Andrea V, Ingeborg R. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes. 2004;28:293–310. doi: 10.1023/b:viru.0000025777.62826.92. [DOI] [PubMed] [Google Scholar]
- 25.Freeman ML, Lanzer KG, Cookenham T, Peters B, Sidney J, Wu TT, Sun R, Woodland DL, Sette A, Blackman MA. Two kinetic patterns of epitope-specific CD8 T-cell responses following murine gamma-herpesvirus 68 infection. J Virol. 2010;84:2881–2892. doi: 10.1128/JVI.02229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moutaftsi M, Salek-Ardakani S, Croft M, Peters B, Sidney J, Grey H, Sette A. Correlates of protection efficacy induced by vaccinia virus-specific CD8+ T-cell epitopes in the murine intranasal challenge model. Eur J Immunol. 2009;39:717–722. doi: 10.1002/eji.200838815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak MA, May RM, Phillips RE, Rowland-Jones S, Lalloo DG, McAdam S, Klenerman P, Köppe B, Sigmund K, Bangham CRM, McMichael AJ. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature. 1995;375:606–611. doi: 10.1038/375606a0. [DOI] [PubMed] [Google Scholar]
- 29.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Moutaftsi M, Tscharke DC, Vaughan K, Koelle DM, Stern L, Calvo-Calle M, Ennis F, Terajima M, Sutter G, Crotty S, et al. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010;5:221–239. doi: 10.2217/fmb.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikloska Z, Kesson AM, Penfold MET, Cunningham AL. Herpes simplex virus protein targets for CD4 and CD8 lymphocyte cytotoxicity in cultured epidermal keratinocytes treated with interferon-γ. J Infect Dis. 1996;173:7–17. doi: 10.1093/infdis/173.1.7. [DOI] [PubMed] [Google Scholar]
- 32.Knipe DM, Howley PM, Griffin DE, editors. Fields' Virology. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 33.Hoshino Y, Pesnicak L, Cohen JI, Straus SE. Rates of reactivation of latent herpes simplex virus from mouse trigeminal ganglia ex vivo correlate directly with viral load and inversely with number of infiltrating CD8+ T cells. J Virol. 2007;81:8157–8164. doi: 10.1128/JVI.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith CM, Belz GT, Wilson NS, Villadangos JA, Shortman K, Carbone FR, Heath WR. Cutting edge: conventional CD8 alpha+ dendritic cells are preferentially involved in CTL priming after footpad infection with herpes simplex virus-1. J Immunol. 2003;170:4437–4440. doi: 10.4049/jimmunol.170.9.4437. [DOI] [PubMed] [Google Scholar]
- 35.Allan RS, Smith CM, Belz GT, van Lint AL, Wakim LM, Heath WR, Carbone FR. Epidermal viral immunity induced by CD8alpha+ dendritic cells but not by Langerhans cells. Science. 2003;301:1925–1928. doi: 10.1126/science.1087576. [DOI] [PubMed] [Google Scholar]
- 36.Allan RS, Waithman J, Bedoui S, Jones CM, Villadangos JA, Zhan Y, Lew AM, Shortman K, Heath WR, Carbone FR. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity. 2006;25:153–162. doi: 10.1016/j.immuni.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Jirmo AC, Nagel CH, Bohnen C, Sodeik B, Behrens GMN. Contribution of direct and cross-presentation to CTL immunity against herpes simplex virus 1. J Immunol. 2009;182:283–292. doi: 10.4049/jimmunol.182.1.283. [DOI] [PubMed] [Google Scholar]
- 38.Sprecher E, Becker Y. Langerhans cell density and activity in mouse skin and lymph nodes affect herpes simplex type 1 (HSV-1) pathogenicity. Arch Virol. 1989;107:191–205. doi: 10.1007/BF01317916. [DOI] [PubMed] [Google Scholar]
- 39.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 41.Chiu YG, Bowers WJ, Lim ST, Ryan DA, Federoff HJ. Effects of herpes simplex virus amplicon transduction on murine dendritic cells. Hum Gene Ther. 2009;20:442–452. doi: 10.1089/hum.2008.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikloska Z, Bosnjak L, Cunningham AL. Immature monocyte-derived dendritic cells are productively infected with herpes simplex virus type 1. J Virol. 2001;75:5958–5964. doi: 10.1128/JVI.75.13.5958-5964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruse M, Rosorius O, Krätzer F, Stelz G, Kuhnt C, Schuler G, Hauber J, Steinkasserer A. Mature dendritic cells infected with herpes simplex virus type 1 exhibit inhibited T-cell stimulatory capacity. J Virol. 2000;74:7127–7136. doi: 10.1128/jvi.74.15.7127-7136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stock AT, Jones CM, Heath WR, Carbone FR. CTL response compensation for the loss of an immunodominant class I-restricted HSV-1 determinant. Immunol Cell Biol. 2006;84:543–550. doi: 10.1111/j.1440-1711.2006.01469.x. [DOI] [PubMed] [Google Scholar]
- 46.Blaney JE, Jr, Nobusawa E, Brehm MA, Bonneau RH, Mylin LM, Fu TM, Kawaoka Y, Tevethia SS. Immunization with a single major histocompatibility complex class I-restricted cytotoxic T-lymphocyte recognition epitope of herpes simplex virus type 2 confers protective immunity. J Virol. 1998;72:9567–9574. doi: 10.1128/jvi.72.12.9567-9574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonneau RH, Salvucci LA, Johnson DC, Tevethia SS. Epitope specificity of H-2Kb-restricted, HSV-1-, and HSV-2-cross-reactive cytotoxic T lymphocyte clones. Virology. 1993;195:62–70. doi: 10.1006/viro.1993.1346. [DOI] [PubMed] [Google Scholar]
- 48.Laing KJ, Magaret AS, Mueller DE, Zhao L, Johnston C, De Rosa SC, Koelle DM, Wald A, Corey L. Diversity in CD8(+) T cell function and epitope breadth among persons with genital herpes. J Clin Immunol. 2010;30:703–722. doi: 10.1007/s10875-010-9441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wagener F, Elliott M, Grabstein K, Posavad C, Corey L. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J Virol. 2006;80:5509–5515. doi: 10.1128/JVI.02659-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decman V, Kinchington PR, Harvey SAK, Hendricks RL. Gamma interferon can block herpes simplex virus type 1 reactivation from latency, even in the presence of late gene expression. J Virol. 2005;79:10339–10347. doi: 10.1128/JVI.79.16.10339-10347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


