Abstract
Owing to their potential for differentiation into multiple cell types, multipotent stem cells extracted from many adult tissues are an attractive stem cell resource for the replacement of damaged tissues in regenerative medicine. The requirements for cellular differentiation of an adult stem cell are a loss of proliferation potential and a gain of cell-type identity. These processes could be restricted by epigenetic modifications that prevent the risks of lineage-unrelated gene expression or the undifferentiated features of stem cells in adult somatic cells. In this review, we focus on the role of DNA methylation in controlling the transcriptional activity of genes important for self-renewal, the dynamism of CpG methylation of tissue-specific genes during several differentiation programs, and whether the multilineage potential of adult stem cells could be imposed early in the original precursor stem cells through CpG methylation. Additionally, we draw attention to the role of DNA methylation in adult stem cell differentiation by reviewing the reports on spontaneous differentiation after treatment with demethylating agents and by considering the evidence provided by reprogramming of somatic cells into undifferentiated cells (that is, somatic nuclear transfer or generation of induced pluripotent cells). It is clear from the evidence that DNA methylation is necessary for controlling stem cell proliferation and differentiation, but their exact contribution in each lineage program is still unclear. As a consequence, in a clinical setting, caution should be exerted before employing adult stem cells or their derivatives in regenerative medicine and appropriate tests should be applied to ensure the integrity of the genome and epigenome.
Introduction
Multipotent stem cells extracted from many adult tissues are an attractive stem cell resource for the replacement of damaged tissues in regenerative medicine and have been identified in many organs and tissues, including bone marrow, peripheral blood, fat, skeletal muscle, brain, skin, cornea, heart, gut, liver, ovarian epithelium, and testis. Multipotent stem cells are all defined as undifferentiated cells, are able to self-renew with a high proliferative rate, and have the potential to differentiate into specialized cells with specific functions [1]. Unlike pluripotent embryonic stem (ES) cells, multipotent stem cells are usually restricted to a particular lineage (mesodermal, endodermal, or ectodermal) but have the potential to differentiate into distinct somatic cell types with appropriate stimulation (Figure 1). Two main advantages for their use in clinical applications are that they avoid some ethical issues associated with pluripotent ES cells, resulting in a more timely approval for research and therapeutic use, and that adult stem cells and tissues derived from them are currently believed to be less likely to initiate rejection after transplantation.
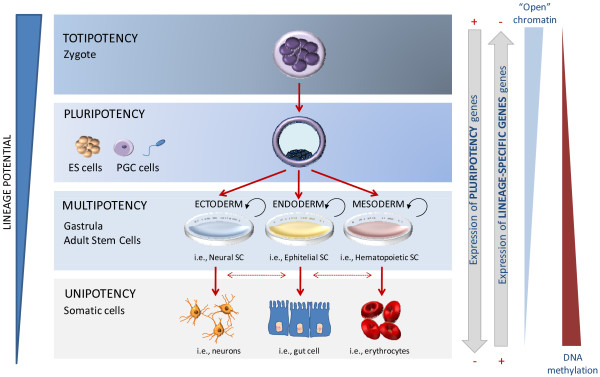
Figure 1.
Lineage restriction of human developmental potency. Totipotent cells at the morula stage have the ability to self-renew and differentiate into all of the cell types of an organism, including extraembryonic tissues. Pluripotent cells - for example, in vitro embryonic stem (ES) cells established at the blastocyst stage and primordial germ cells (PGCs) from the embryo - lose the capacity to form extraembryonic tissues like placenta. Restriction of differentiation is imposed during normal development, going from multipotent stem cells (SCs), which can give rise to cells from multiple but not all lineages, to the well-defined characteristics of a somatic differentiated cell (unipotent). Specific chromatin patterns and epigenetic marks can be observed during human development since they are responsible for controlling transcriptional activation and repression of tissue-specific and pluripotency-related genes, respectively. Global increases of heterochromatin marks and DNA methylation occur during differentiation.
Although human adult stem cells represent a promising tool for applying new clinical concepts in support of cellular therapy, many aspects remain to be explored in order to guarantee appropriate quality assurance and control of these cells, such as avoiding inappropriate gene expression in transplanted cells or the undesirable traits of tumorigenesis. Gene expression potential in stem cell renewal and differentiation could be regulated by epigenetic processes that confer a specific chromatin conformation of the genome, of which DNA methylation is the best characterized (Figure 1) [2]. DNA methylation, the addition of a methyl group to the carbon 5 of the cytosine into CpG contexts, is known to be an essential process in development and cellular differentiation [3]. It is involved in gene regulation of housekeeping and tissue-type genes, silencing of one allele of imprinted genes, and compensation of the extra copy of the X chromosome in females. It acts as a defense mechanism, preventing genomic instability due to transposon movements or insertion of endoparasitic sequences in the genome [4]. It must be pointed out that DNA methylation does not work alone in controlling chromatin conformation since histone modifications and non-coding RNA regulation also collaborate in its control. So, we must consider the existence of an 'epigenetic code' in which several epigenetic factors act in a gradual and progressive manner for controlling chromatin structure.
Currently, much attention is being paid to the effects of CpG methylation on stemness and differentiation. The first piece of evidence came from the observation that important genes for the maintenance of ES cells, such as Oct4 and Nanog genes, are usually hypomethylated when activated but became hypermethylated during differentiation [5,6]. Knowledge of the genome-wide contribution of CpG methylation to stem cell maintenance and differentiation has increased in recent years, mainly because of the development of technical approaches for assessing epigenetic factors. High-throughput strategies demonstrate that human ES cells have a unique CpG methylation signature that, in combination with histone modifications, drives stem cell differentiation through the restriction of the developmental potential of progenitor cells [7,8]. In comparison with the broad-ranging information obtained from ES cells, the role of CpG methylation in regulating differentiation of adult stem cells has been less extensively examined. In this review, we consider the reported evidence of how the developmental potential of adult stem cells could be restricted by the gain of DNA methylation of self-renewal genes (preventing the undifferentiated features of stem cells in adult somatic cells) and the DNA methylation-dependent control of tissue-specific genes (abolishing the risks of lineage-unrelated gene expression). The opportunities that this presents for manipulating the epigenome by means of pharmacological treatments and its consequences for stem cell differentiation and reprogramming will be analyzed.
DNA methylation, global chromatin context, and stemness
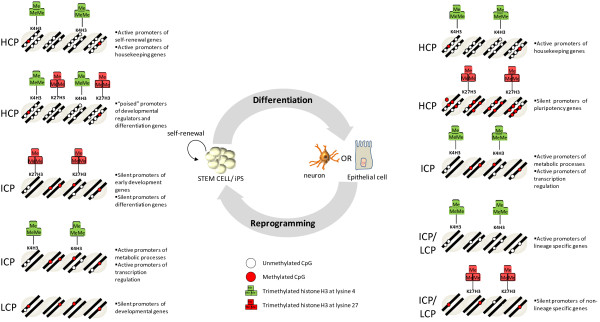
It is important to point out that the relationship between promoter DNA methylation and promoter activity depends on the CpG content of the promoters: high CpG promoters (HCPs), intermediate CpG promoters, or low CpG promoters (LCPs). In ES cells and multipotent progenitor cells, HCP promoters are characterized by low DNA methylation levels, whereas LCP promoters are enriched in DNA methylation [6,8,9] (Figure 2). Furthermore, specific histone modifications (that is, H3K4me3 and H3K27me3) in HCPs appear to be more decisive for expression of the corresponding genes and suggest a degree of protection from DNA methylation [10] (Figure 2). Conversely, methylated LCP promoters are depleted of bivalent histone marks and are mostly repressed in ES cells [6,8,9] (Figure 2). It is suggested that silencing of pluripotency-related genes occurs by means of CpG promoter hypermethylation, whereas gain of differentiation features is defined by gene regulation of Polycomb targets [8].
Figure 2.
Chromatin states during differentiation of adult stem cells and reprogramming of somatic cells. The figure shows the epigenetic alterations of genes associated with pluripotency, cell maintenance, and lineage specification during stem cell differentiation. CpG methylation is grouped into three types of promoters classified with respect to their CpG content: high CpG promoter (HCP), intermediate CpG promoter (ICP), and low CpG promoter (LCP). Bivalent domains of histone marks (trimethylated histone H3 at lysine 4 and trimethylated histone H3 at lysine 27) are represented. Reprogramming of somatic cells toward pluripotent cells - that is, induced pluripotent cells (iPS) - implies a turnover of the epigenetic landscape of adult differentiated cells, including silencing of tissue-specific genes and activation of pluripotency-related genes.
Specific epigenetic features at a global level also underpin the pluripotency of ES cells. Recent studies have demonstrated that ES cell chromatin is in a highly dynamic state with global DNA hypomethylation and a general abundance of transcriptionally active chromatin marks such as H3K4me3 and acetylation of histone H4, which is reflected in the relatively decondensed chromatin of ES cells [2,11]. This global lack of DNA methylation in stem cells could be associated with the ability of such cells to activate a wide range of cell type-specific genes during the differentiation programs [2]. It must not be forgotten that DNA methylation and histone modifications do not work alone and that the epigenetic inactivation of differentiation-specific genes in stem cells (that is, Hox and Pax family of genes) is usually repressed by alternative chromatin remodeling factors, such as Polycomb proteins [11,12]. Consequently, further study of the interplay of all of the chromatin regulators is essential for understanding the dynamism of transcriptional control during stem cell renewal and differentiation.
DNA methylation-dependent regulation of genes associated with self-renewal of stem cells
It has been widely reported that maintenance of the pluripotency state is conferred by a set of development-associated transcription factors - such as OCT4, NANOG, and SOX2 - that occupy promoters of active genes associated with self-renewal [13,14]. Expression of the aforementioned transcription regulators is usually controlled by CpG promoter methylation, and differentiation of ES cells is accomplished by partial or full methylation of pluripotency-associated genes, resulting in their downregulation [6,15-17]. The opposite association has been found in the reprogramming of induced pluripotent stem (iPS) cells from differentiated cells, in which unmethylated active promoters of ES cell-specific genes were described [18] (Figure 2). Despite the considerable information about silencing of pluripotency ES genes during differentiation, very little is known about the epigenetic control of genes associated with self-renewal and maintenance of multipotent adult stem cells. In adipose-derived stem cells (ASCs) and mesenchymal stem cells from bone marrow (BM-MSCs), OCT4 is silenced by promoter hypermethylation, whereas Nanog and Sox2 are unmethylated despite the repressed state of the genes [19]. The same patterns of methylation were found in differentiated fibroblasts and keratinocytes [19]. It seems that, whereas Oct4 regulation is strongly influenced by CpG promoter hypermethylation, the control of Nanog and Sox2 expression could be due to other repressive mechanisms such as histone modification patterns [19]. Enrichment of H3K27me3 and H3K9me3 and reduction of H3K79me3 have been described in the Nanog and Sox2 promoters of ASCs and differentiated cells but not in pluripotent cells [20]. These results demonstrate that the transcriptional repression mechanisms could vary depending on the gene and the state of cellular differentiation (that is, multipotency versus differentiation) [19] and could constitute a mechanism for preventing aberrant reactivation of pluripotency and minimizing the risk of de-differentiation [21]. In line with this hypothesis, ES cells with genetic mutations of DNA methyltransferase result in rapid apoptosis-mediated cell death [22,23].
The promoter methylation status of additional stem cell-determining genes for self-renewal (not exclusively markers of pluripotency) has also been investigated [24]. Silencing of the mesodermal transcription factor Brachyury gene during differentiation from BM-MSCs to mesodermal lineages involves hypermethylation of its promoter but not changes in promoter hypermethylation of genes such as LIN28, NESTIN, or ZFP42. This could be associated with changes of expression during differentiation of BM-MSCs [24]. Currently, we have a limited understanding of how multipotency is established and maintained in adult stem cells, and it would be very interesting to study the CpG promoter methylation status of transcription factors that confer multipotency on adult stem cells beyond the traditional role of pluripotency genes such as Oct4, Nanog, and Sox2.
Specific CpG methylation during differentiation of multipotent stem cells
Cellular differentiation is determined by a loss of proliferation potential and a gain of cell-type identity. This reduction of developmental potential could be restricted by epigenetic modifications that prevent the risks of lineage-unrelated gene expression or undifferentiated features of stem cells in adult cells [3]. However, the role of specific promoter methylation in controlling gene differentiation remains a matter of controversy. On one hand, there are some clues in favor of the hypothesis that cell type-specific patterns of DNA methylation influence cell type-specific gene expression and, by extension, cellular differentiation. For example, promoter methylation of SERPINB5 is inversely correlated with the unique expression of SERPINB5 in epithelial cells [25], and the rSPHK1 and hSLC6A8 promoter hypermethylation associated with gene silencing in specific tissues allows expression in unmethylated brain tissue only [26,27]. On the other hand, genome-wide analysis of CpG methylation changes during the conversion of human pluripotent/multipotent stem cells into differentiated somatic cells reveals small changes in DNA methylation at promoter regions [8,9,28-30]. For example, lineage commitment of neural progenitor cells into terminally differentiated neurons occurs with a very moderate number of promoter DNA hypermethylated genes as cells differentiate [8]. Further work is needed to test whether these weak associations between gene repression and CpG hypermethylation during differentiation are due to limitations of the analytical techniques or to the existence of additional methylation-independent regulatory mechanisms.
Does CpG methylation of multipotent stem cells restrict lineage specification?
One of the main features of adult stem cells is their multipotency (that is, their ability to differentiate into a number of cell types), but, in contrast to pluripotent cells, they are restricted to those of a closely related family of cells. For example, BM-MSCs primarily form mesodermal-specific cell types such as chondrocytes, myocytes, adipocytes, or osteoblasts [1]. However, we should remember that, given the information collected in recent years, this could be a very general statement, and there is some evidence to suggest that lineage restriction could be more permissive. For instance, BM-MSCs could be differentiated into cells of all three germ layers and generate tissues such as osteocytes (mesoderm), hepatocytes (endoderm), or neurons (ectoderm) [31-33]. Multipotent cells isolated from different tissues have common in vitro phenotypic and functional characteristics (for example, MSCs share fibroblast-like morphology, plastic adherence, proliferation ability, and clonogenicity) but differ in the expression of specific lineage markers (for example, ASCs and BM-MSCs differ in the expression of the surface markers CD90, CD105, CD106, and adhesion molecules [34,35] and in their differentiation potential). Since gene expression in adult stem cells is regulated by epigenetic processes, a question arises: is the differentiation potential in adult stem cells predicted by DNA methylation of specific lineage promoters? There is some evidence in favor of a differentiation restriction imposed by promoter hypermethylation in progenitor stem cell states, whereas promoter hypomethylation does not have any predictive value with respect to differentiation potential [35,36]. Characterization of DNA methylation profiles of all human RefSeq promoters in mesenchymal adult stem cells from various origins, including adipose, hematopoietic, and neural progenitors and muscle tissue, shows that the majority of the lineage-specific genes are hypomethylated even if the progenitor is not able to differentiate into this specific lineage [10]. There are some examples of epigenetic silencing associated with restriction to differentiation: endothelial markers such as CD31 and CD144 are strongly methylated in ASCs that show very limited capacity for endothelial differentiation [36] or osteogenic and adipogenic restriction of C2C12 myoblast cell line differentiation [37]. Furthermore, the restriction for differentiation in specific programs imposed by means of DNA methylation is established early in development, in the progenitor state, and persists after differentiation, as most of the hypermethylated promoters in undifferentiated cells remain hypermethylated in somatic cells [10,37]. This is in agreement with the low level of de novo methylation described after differentiation of adult stem cells [8,9]. Results lead to the conclusion that the differentiation restriction associated with promoter hypermethylation clearly differs between pluripotent and multipotent cells: lineage-specific promoters are mostly hypermethylated in ES cells [6] in contrast to the low-percentage hypermethylation found in MSCs [35].
Treatment with demethylating agents results in spontaneous differentiation
The involvement of DNA methylation in controlling the differentiation potential of stem cells has been supported by several reports of spontaneous differentiation after treatment with demethylating agents (Table 1). For example, the use of 5-aza-2'-deoxycytidine (5-ADC) promotes differentiation of ASCs into cardiac myogenic cells [38]. Pretreatment with 5-ADC also drives the osteogenic differentiation of BM-MSCs by enhancing the expression of osteogenic genes (such as Dlx5) associated with demethylation of its CpG shore [39,40]. However, we must remember that DNA methylation is just one component of the epigenetic machinery and that removing DNA methylation is often insufficient to reactivate gene expression (Table 1). Treatments with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) enhance chondrogenic differentiation of BM-MSCs accomplished by increased expression of Sox9 [41]. Similarly, neural induction was achieved when multipotent stem cells were exposed to TSA simultaneously with 5-ADC [42]. Furthermore, the effects of treatments with HDAC inhibitors are known to alter DNA methylation levels [41,42]. Additional evidence for the role of epigenetic control in differentiation comes from the functional consequences of defects in enzymes of the epigenetic machinery. For example, recovery of the expression of a defect in a histone modifier (NSD1) suppresses cell growth and increases the differentiation of neuroblastoma cell lines [43]. Recovery of epigenetic patterns, by treatment with epigenetic drugs or by genetic models, highlights the potential of epigenetic modifiers, possibly in combination with other factors, to enhance the ability of multipotent stem cells to form functional differentiated cells and has significant therapeutic implications. Some consistent lines of evidence support this therapeutic application since epigenetic drugs, among them demethylating agents, have shown significant antitumor activity and the US Food and Drug Administration has approved the use of some of them to treat patients with cancer [4]. Indeed, new HDAC inhibitors (that is, romidepsin, belinostat, or givinostat) that are currently being tested in clinical trials for anticancer therapy [44] must also be considered as candidate molecules for assessing stem cell differentiation. Further understanding of the epigenetic regulation of tissue-specific genes along with the development of additional specific epigenetic drugs may hold the key to our ability to reset the epigenome successfully during stem cell differentiation.
Table 1.
Effects on differentiation potential of multipotent/pluripotent stem cells after treatment with epigenetic drugs
| Stem cell classification | Epigenetic drug | Differentiation after treatment | Reference |
|---|---|---|---|
| Multipotent stem cells | |||
| Adipose-derived stem cells | 5-aza-2'-deoxycytidine | Cardiomyocytes | [38] |
| Adipose-derived stem cells | 5-aza-2'-deoxycytidine; trichostatin A | Cardiomyocytes | [57] |
| Bone marrow mesenchymal stem cells | 5-aza-2'-deoxycytidine | Osteocytes | [39,40] |
| Bone marrow mesenchymal stem cells | 5-aza-2'-deoxycytidine | Cardiomyocytes | [45,58] |
| Bone marrow mesenchymal stem cells | 5-aza-2'-deoxycytidine; trichostatin A | Osteocytes; chondrocytes | [41] |
| Bone marrow mesenchymal stem cells | 5-aza-2'-deoxycytidine; trichostatin A | Neural-like cells | [42] |
| Bone marrow mesenchymal stem cells | Sodium butyrate | Osteocytes | [59] |
| Cardiac progenitor stem cells | 5-aza-2'-deoxycytidine | Cardiomyocytes | [60] |
| Neural progenitor stem cells | Trichostatin A | Neuronal cells | [61] |
| Neural progenitor stem cells | Valproic acid | Neuronal cells | [62] |
| Umbilical cord mesenchymal stem cells | 5-aza-2'-deoxycytidine | Cardiomyocytes | [63] |
| Pluripotent stem cells | |||
| Embryonic stem cells | 5-aza-2'-deoxycytidine | Cardiomyocytes | [64,65] |
| Embryonic stem cells | 5-aza-2'-deoxycytidine | Endothelial cells | [66] |
| Embryonic stem cells | Trichostatin A | Cardiomyocytes | [67,68] |
Concluding remarks
It is clear that cell differentiation of multipotent stem cells is a result of a complex and dynamic network of transcriptional regulators, among them epigenetic factors that play a central role through controlling the expression/repression of tissue-specific genes and multipotency-related genes. However, it is not currently possible to manipulate cell differentiation even if we consider all of the genetic and epigenetic knowledge available for a specific lineage commitment. For example, epigenetic treatments may have a pleiotropic effect on the differentiation of stem cells, depending on multiple factors, mainly the origin of the precursor cell and environment conditions (presence of growth factors, transcriptional regulators, and so on) [39,41,45], suggesting that global epigenetic modifications, though necessary, are not sufficient to transdifferentiate by themselves [46]. These findings underline the necessity of evaluating in more detail the importance of epigenetic chromatin remodeling for establishing and maintaining stemness or, on the other hand, initiating a differentiation program. The reprogramming of somatic cells provides a new opportunity to study the contribution of epigenetics to differentiation. A mature cell can be converted into a pluripotent state by three experimental approaches: somatic nuclear transfer into enucleated oocytes, the in vitro application of a defined set of transcription factors creating iPS cells, or fusing ES cells with somatic cells to generate heterokaryons and hybrids [47]. Epigenetic rearrangements are observed independently of the technique [48,49]. In fact, there is evidence that HDAC inhibitors and DNA demethylating agents are useful for enhancing iPS reprogramming [50,51]. A prerequisite in reprogramming of iPS from somatic cells is that some stemness-related promoters become demethylated. How might this demethylation be achieved? It could be done through a DNA repair mechanism [52,53] or by the recent discovery of TET proteins, a group of enzymes that convert methylated 5-methylcytosine to 5-hydroxymethylcytosine [54]. Although experimental models for reprogramming have generated a considerable amount of information, many questions remain. How different is the epigenetic regulation of pluripotent and multipotent cells? Does CpG methylation underpin self-renewal in adult stem cells, as it does in ES cells? Do epigenetic marks define the lineage potential of an adult stem cell? Is it possible to revert the differentiation program by manipulating the epigenome? How safe is this reversion? The recent discovery that nearly one quarter of all methylation identified in ES cells was found in a non-CG context [55] suggests that the genomic context must also be addressed. Do ES cells use a different methylation mechanism for gene regulation? Furthermore, long-term in vitro culture of adult stem cells, a prerequisite for large-scale expansion previous to implantation with therapeutic purposes, showed specific alterations of CpG island methylatyion [56]. As a consequence, it is necessary to optimize and standardize the experimental protocols used for in vitro expansion which minimize epigenetic-related instability. In conclusion, although manipulation of epigenetic activity might be an interesting means of generating populations of specific cell types, additional epigenetic research on the understanding of stem cell biology must be done before they can be used as differentiation agents in stem cell-based therapies.
Abbreviations
5-ADC: 5-aza-2'-deoxycytidine; ASC: adipose-derived stem cell; BM-MSC: mesenchymal stem cell from bone marrow; ES: embryonic stem; HCP: high CpG promoter; HDAC: histone deacetylase; iPS: induced pluripotent stem; LCP: low CpG promoter; MSC: mesenchymal stem cell; TSA: trichostatin A.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
María Berdasco, Email: mberdasco@idibell.cat.
Manel Esteller, Email: mesteller@idibell.cat.
Acknowledgements
This work was supported by EU FP6 ESTOOLS LSHG-CT-2006-018739, SAF2007-00027-65134, Consolider CSD2006-49, the Lilly Foundation, the Dr. Josef Steiner Cancer Research Foundation, and European Research Council Advanced Grant EPINORC.
References
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, Gonzalez R, Crook J, Davidson B, Schulz TC, Robins A, Khanna A, Sartipy P, Hyllner J, Vanguri P, Savant-Bhonsale S, Smith AK, Chakravarti A, Maitra A, Rao M, Barker DL, Loring JF, Fan JB. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AL, Jacobsen BM, Reiner AH, Andersen IS, Collas P. Promoter DNA methylation patterns of differentiated cells are largely programmed at the progenitor stage. Mol Biol Cell. 2010;21:2066–2077. doi: 10.1091/mbc.E10-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Imao Y, Nishino K, Hattori N, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Barrand S, Collas P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem Biophys Res Commun. 2010;391:762–767. doi: 10.1016/j.bbrc.2009.11.134. [DOI] [PubMed] [Google Scholar]
- Barrand S, Andersen IS, Collas P. Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes. Biochem Biophys Res Commun. 2010;401:611–617. doi: 10.1016/j.bbrc.2010.09.116. [DOI] [PubMed] [Google Scholar]
- Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–2295. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansranjavin T, Krehl S, Mueller T, Mueller LP, Schmoll HJ, Dammann RH. The role of promoter CpG methylation in the epigenetic control of stem cell related genes during differentiation. Cell Cycle. 2009;8:916–924. doi: 10.4161/cc.8.6.7934. [DOI] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Grunau C, Hindermann W, Rosenthal A. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum Mol Genet. 2000;9:2651–2663. doi: 10.1093/hmg/9.18.2651. [DOI] [PubMed] [Google Scholar]
- Imamura T, Ohgane J, Ito S, Ogawa T, Hattori N, Tanaka S, Shiota K. CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. Genomics. 2001;76:117–125. doi: 10.1006/geno.2001.6607. [DOI] [PubMed] [Google Scholar]
- Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis Rheum. 2009;60:1416–1426. doi: 10.1002/art.24472. [DOI] [PubMed] [Google Scholar]
- Kang MI, Kim HS, Jung YC, Kim YH, Hong SJ, Kim MK, Baek KH, Kim CC, Rhyu MG. Transitional CpG methylation between promoters and retroelements of tissue-specific genes during human mesenchymal cell differentiation. J Cell Biochem. 2007;102:224–239. doi: 10.1002/jcb.21291. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Kogo Y, Ohgane J, Hattori N, Yagi S, Tanaka S, Shiota K. Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochem Biophys Res Commun. 2008;366:360–366. doi: 10.1016/j.bbrc.2007.11.137. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Shi YY, Aalami OO, Chou YF, Mari C, Thomas R, Quarto N, Contag CH, Wu B, Longaker MT. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat Biotechnol. 2004;22:560–567. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- Roelandt P, Pauwelyn KA, Sancho-Bru P, Subramanian K, Ordovas L, Vanuytsel K, Geraerts M, Firpo M, De Vos R, Fevery J, Nevens F, Hu WS, Verfaillie CM. Human embryonic and rat adult stem cells with primitive endoderm-like phenotype can be fated to definitive endoderm, and finally heaptocyte-like cells. PLoS One. 2010;5:e12101. doi: 10.1371/journal.pone.0012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AP, Foraker JE, Ylostalo J, Prockop DJ. Human stem/progenitor cells from bone marrow enhance glial differentiation of rat neural stem cells: a role for transforming growth factor β and Notch signaling. Stem Cells Dev. 2011;20:289–300. doi: 10.1089/scd.2009.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S, Eichler H, Stoeve J, Klüter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- Sørensen AL, Timoskainen S, West FD, Vekterud K, Boquest AC, Ahrlund-Richter L, Stice SL, Collas P. Lineage-specific promoter DNA methylation patterns segregate adult progenitor cell types. Stem Cells Dev. 2010;19:1257–1266. doi: 10.1089/scd.2009.0309. [DOI] [PubMed] [Google Scholar]
- Boquest AC, Noer A, Sørensen AL, Vekterud K, Collas P. CpG methylation profiles of endothelial cell-specific gene promoter regions in adipose tissue stem cells suggest limited differentiation potential toward the endothelial cell lineage. Stem Cells. 2007;25:852–861. doi: 10.1634/stemcells.2006-0428. [DOI] [PubMed] [Google Scholar]
- Hupkes M, van Someren EP, Middelkamp SH, Piek E, van Zoelen EJ, Dechering KJ. DNA methylation restricts spontaneous multi-lineage differentiation of mesenchymal progenitor cells, but is stable during growth factor-induced terminal differentiation. Biochim Biophys Acta. 2011;1813:839–849. doi: 10.1016/j.bbamcr.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Rangappa S, Fen C, Lee EH, Bongso A, Sim EK. Transformation of adult mesenchymal stem cells isolated from the fatty tissue into cardiomyocytes. Ann Thorac Surg. 2003;75:775–779. doi: 10.1016/s0003-4975(02)04568-x. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lee YM, Kim MJ, Choi JY, Park EK, Kim SY, Lee SP, Yang JS, Kim DS. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol Cells. 2006;22:182–188. [PubMed] [Google Scholar]
- Zhou GS, Zhang XL, Wu JP, Zhang RP, Xiang LX, Dai LC, Shao JZ. 5-Azacytidine facilitates osteogenic gene expression and differentiation of mesenchymal stem cells by alteration in DNA methylation. Cytotechnology. 2009;60:11–12. doi: 10.1007/s10616-009-9203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serafi AT, Oreffo RO, Roach HI. Epigenetic modifiers influence lineage commitment of human bone marrow stromal cells: differential effects of 5-aza-deoxycytidine and trichostatin A. Differentiation. 2011;81(1):35–41. doi: 10.1016/j.diff.2010.09.183. [DOI] [PubMed] [Google Scholar]
- Alexanian AR. Epigenetic modifiers promote efficient generation of neurallike cells from bone marrow-derived mesenchymal cells grown in neural environment. J Cell Biochem. 2007;100:362–371. doi: 10.1002/jcb.21029. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, Alaminos M, Cheung NK, Rahman N, Esteller M. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JM, Hackanson B, Lübbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2010;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K. Use of adult marrow mesenchymal stem cells for regeneration of cardiomyocytes. Bone Marrow Transplant. 2003;32(Suppl 1):S25–27. doi: 10.1038/sj.bmt.1703940. [DOI] [PubMed] [Google Scholar]
- Liu J, Sandoval J, Doh ST, Cai L, López-Rodas G, Casaccia P. Epigenetic modifiers are necessary but not sufficient for reprogramming non-myelinating cells into myelin gene-expressing cells. PLoS One. 2010;5:e13023. doi: 10.1371/journal.pone.0013023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SC, Yoon J, Shim WJ, Ro YM, Lim DS. 5-azacytidine induces cardiac differentiation of P19 embryonic stem cells. Exp Mol Med. 2004;36:515–523. doi: 10.1038/emm.2004.66. [DOI] [PubMed] [Google Scholar]
- Makino S, Fukuda K, Miyoshi S, Konishi F, Kodama H, Pan J, Sano M, Takahashi T, Hori S, Abe H, Hata J, Umezawa A, Ogawa S. Cardiomyocytes can be generated from marrow stromal cells in vitro. J Clin Invest. 1999;103:697–705. doi: 10.1172/JCI5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Chen WM, Hsu KH, Kuo CD, Hung SC. Sodium butyrate activates ERK to regulate differentiation of mesenchymal stem cells. Biochem Biophys Res Commun. 2007;355:913–918. doi: 10.1016/j.bbrc.2007.02.057. [DOI] [PubMed] [Google Scholar]
- Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniyan V, Boddeke E, Bakels R, Küst B, Kooistra S, Veneman A, Copray S. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Qian H, Zhang X, Zhu W, Yan Y, Ye S, Peng X, Li W, Xu Z, Sun L, Xu W. 5-Azacytidine induces cardiac differentiation of human umbilical cord derived mesenchymal stem cells by activating extracellular regulated kinase (ERK) Stem Cells Dev. 2011. in press . [DOI] [PMC free article] [PubMed]
- Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- Yoon BS, Yoo SJ, Lee JE, You S, Lee HT, Yoon HS. Enhanced differentiation of human embryonic stem cells into cardiomyocytes by combining hanging drop culture and 5-azacytidine treatment. Differentiation. 2006;74:149–159. doi: 10.1111/j.1432-0436.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Bacanamwo M. DNA methyltransferase inhibition induces mouse embryonic stem cell differentiation into endothelial cells. Exp Cell Res. 2010;316:172–180. doi: 10.1016/j.yexcr.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T, Hasegawa K. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- Hosseinkhani M, Hasegawa K, Ono K, Kawamura T, Takaya T, Morimoto T, Wada H, Shimatsu A, Prat SG, Suemori H, Nakatsuji N, Kita T. Trichostatin A induces myocardial differentiation of monkey ES cells. Biochem Biophys Res Commun. 2007;356:386–391. doi: 10.1016/j.bbrc.2007.02.151. [DOI] [PubMed] [Google Scholar]