Abstract
The focus of the present review is on the extent to which epigenetic alterations influence the development of systemic lupus erythematosus. Lupus is a systemic autoimmune disease characterized by the production of autoantibodies directed at nuclear self-antigens. A DNA methylation defect in CD4+ T cells has long been observed in idiopathic and drug-induced lupus. Recent studies utilizing high-throughput technologies have further characterized the nature of the DNA methylation defect in lupus CD4+ T cells. Emerging evidence in the literature is revealing an increasingly interconnected network of epigenetic dysregulation in lupus. Recent reports describe variable expression of a number of regulatory microRNAs in lupus CD4+ T cells, some of which govern the expression of DNA methyltransferase 1. While studies to date have revealed a significant role for epigenetic defects in the pathogenesis of lupus, the causal nature of epigenetic variation in lupus remains elusive. Future longitudinal epigenetic studies in lupus are therefore needed.
Introduction
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of autoantibodies directed against nuclear self-antigens. The role of genetic variation has been extensively investigated in lupus, revealing polymorphisms throughout the genome correlated with disease [1-3]. Although studies investigating common genetic variants in lupus have revealed a number of susceptibility loci, the cumulative effect size of these loci accounts for a small fraction of disease heritability, approximately 15 to 20% [4]. The inability of present genetic association studies to comprehensively account for disease heritability has led many to postulate that the missing heritability of complex human disease may broadly reside in epigenetic mechanisms [5].
Epigenetics refers to heritable modifications that regulate gene expression without changes to DNA sequence [6]. Common epigenetic mechanisms include DNA methylation and histone modification. MicroRNAs are also involved in epigenetic regulation. The most widely researched epigenetic modification to date is DNA methylation, which is known to have diverse regulatory function, governing gene expression patterns in a cell-and tissue- specific manner. Broadly speaking, DNA methylation is a negative regulator of gene expression whereby higher levels of promoter and CpG island methylation are typically observed to correlate with lower levels of gene expression. Aberrant DNA methylation and gene expression are observed in a number of conditions and diseases, including cancer, neuro-psychiatric disorders, and autoimmunity.
DNA methylation patterns are established during development and maintained throughout the life course by a class of enzymes known as DNA methyltransferases [7]. Further, widespread epigenomic differences are observed throughout cellular development [8,9]. Once established, patterns of DNA methylation are heritably preserved from parent to daughter cells during replication by DNA methyltransferase 1 (DNMT)1. DNMT1 associates with replication fork complex proteins where it transfers methylation patterns from the template strand to the incipient copy.
Histone proteins act to spatially organize chromatin and determine the extent to which chromatin is accessible for active transcription. Covalent modification of histone proteins represents another facet of epigenetic variation through which gene expression and chromatin structure are regulated. Methylation and acetylation of specific histone lysine residues act to regulate the transcriptional state of chromatin in cis. Locus-specific chromatin architecture varies between cell types, as genes are differentially expressed among different cells at specified times.
Epigenetic regulation acts to program gene expression patterns in the various cell types that comprise the tissues and organ systems of the human body. In effect, epigenetic modification facilitates the differentiation of diverse cell types from static genomic DNA. The possibility for epigenetic variation underlying the disease process of lupus is intriguing as patients experience periods of calm punctuated by disease flares. Because epigenetic states vary with time and between cell types, it is in the context of disease activity that epigenetic variation in lupus is likely to stand in sharpest relief against the epigenome of normal tissues.
Epigenetic regulation in normal lymphocytes
The human immune system is composed of a number of cell types that serve specific functions in establishing and maintaining cell-mediated and humoral immunity. The diversity of lymphocyte populations that comprise the human immune system is established and maintained through epigenetic modification. Distinct and highly conserved epigenetic profiles of DNA methylation are observed in normal B and T lymphocytes [10,11]. While DNA methylation patterns are broadly conserved within cell types, widespread differences are observed between lymphocyte subtypes. DNMT1 function and DNA methylation patterns are essential in ensuring T cell differentiation and function [12]. DNA methylation and histone modifications reinforce repressive and permissive chromatin states at key regulatory loci in CD4+ T cells lineages [13,14].
Demethylation and changes in chromatin architecture at transcription factor and cytokine genes accompany effect or cell differentiation in response to cytokine signaling among lymphocyte populations. Chromatin remodeling of the IL17 locus is observed upon differentiation of Th17 cells [15]. Likewise, DNA demethylation and histone acetylation of the IFNG (IFN-γ) and the IL4-IL5-IL13 genetic loci, upon Th1 and Th2 differentiation, respectively, is mediated by T cell lineage -specific transcription factors following T-cell receptor signaling [16]. Indeed, Th2-specific chromatin remodeling and demethylation takes place in a locus control region that flanks a number of Th2 cytokines [17]. Further, these epigenetic changes are accompanied by changes in long-range interactions and chromatin architecture at cytokine gene loci under Th1 and Th2 polarizing conditions [18].
Drug-induced lupus points to epigenetic dysregulation in autoimmunity
While approximately 90% of lupus cases are idiopathic, prolonged exposure to certain medications is known to induce lupus. It was eventually discovered that procainamide and hydralazine, medications that can cause lupus, inhibit DNA methylation in T cells [19]. It was further shown that in vitro treatment of Th2 cells with procainamide or 5-azacytidine (a DNA methylation inhibitor) leads to their autoreactivity, and upon adoptive transfer these cells are capable of inducing autoimmunity in mice [20].
A better understanding of the mechanisms through which drug-induced lupus occurs has provided insight into the epigenetic mechanisms and pathways involved in idiopathic lupus. Global hypomethylation of genomic DNA is observed in CD4+ T cells of lupus patients [21]. This decline is attributable to a reduction in methyl-transferase activity. Procainamide acts as a competitive inhibitor of DNMT1 [22]. Hydralazine reduces DNA methylation via inhibiting extracellular signal-regulated kinase (ERK) pathway signaling [23], which regulates the expression of DNMT1 [24,25]. The ERK pathway further ensures proper immune cell development and function [26].
Reduced ERK pathway signaling in lupus CD4+ T cells
An ERK pathway signaling defect has been observed in lupus CD4+ T cells [27,28] and has been attributed to originate from decreased signaling through protein kinase C delta in lupus CD4+ T cells [29]. Decreased protein kinase C signaling may further account for defective T cell activation in lupus [30]. Additionally, mice lacking the protein kinase C delta gene display germinal center formation in the absence of stimulation and develop autoimmunity [31].
A reduction of ERK pathway signaling leading to reduced expression of DNMT1 has been shown to lead to T cell autoreactivity. Mice that express a dominant-negative form of the ERK pathway component MEK show reduced ERK pathway signaling and reduced expression of DNMT1. This leads to overexpression of methylation-sensitive genes, anti-double stranded DNA antibody production, and activation of IFN-regulated signaling pathways similar to lupus patients [32].
Sex-chromosome complement and skewed X chromosome inactivation in lupus
Lupus is a sexually dimorphic autoimmune disease with a 9:1 female bias, a fact that makes sex chromosome complement and skewed X chromosome inactivation interesting candidates for investigating the origins of the disease. Under normal circumstances, females have only one X chromosome from which genes are actively transcribed, while the additional X chromosome remains transcriptionally inactive through epigenetic repression. As previously stated, widespread demethylation of genomic DNA is observed in lupus CD4+ T cells, and this, it is thought, might lead to the reactivation of methylation-sensitive genes on the normally inactive X chromosome. An example of the reactivation of a gene on the inactive X chromosome in lupus CD4+ T cells is that of CD40 ligand (CD40LG). CD40LG is a B-cell co-stimulatory molecule that, upon binding with CD40, promotes B cell activation, plasma cell differentiation, immunoglobulin production and antibody class switching [33,34]. Overexpression of CD40LG is observed in lupus CD4+ and CD8+ T cells as well as lupus B cells [35]. CD4+ T cells from women but not men overexpress CD40LG when treated with 5-azacytidine, while the overexpression of autosomal methylation-sensitive genes is comparable between the sexes [36]. CD40LG-trans-fected normal human T cells lead to autologous B cell activation and plasma cell differentiation in co-culture assays [37]. Further, T cells from women but not men increase B cell IgG production when treated with DNA methylation inhibitors, which can be inhibited by anti-CD40LG antibody [37].
There is evidence for skewed X chromosome inactivation in a number of other autoimmune diseases apart from lupus, including autoimmune thyroiditis and scleroderma, many of which also have a strong female bias [38]. Further evidence for the role of sex chromosome complement in the pathogenesis of lupus comes from Klinefelter's syndrome (47, XXY), a condition in which phenotypic male individuals possess an additional X chromosome. It was found that lupus is approximately 14 times more frequent among men with Klinefelter's syndrome than it is among (46, XY) men, a rate similar to that observed among women [39].
Aberrant methylation and gene expression in lupus CD4+ T cells
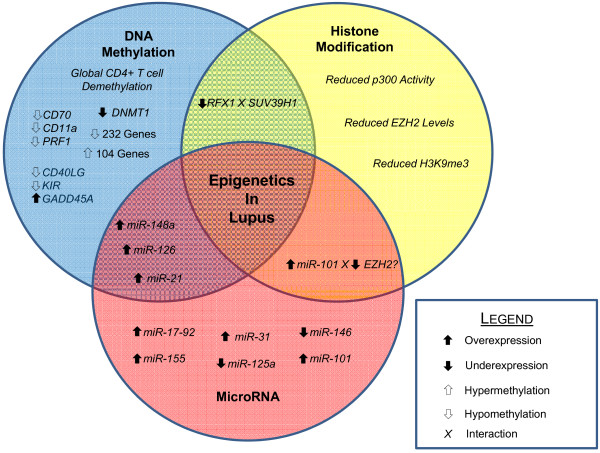
Global reduction of DNA methylation is a hallmark of CD4+ T cells in lupus (Figure 1). Several autosomal genes are known to be hypomethylated in lupus CD4+ T cells. Many of these genes encode proteins involved in processes related to immune function and inflammation. It is thought that the overexpression of methylation-sensitive genes leads to lupus T cell autoreactivity and subsequent induction of autoreactive B cell immunoglobulin production.
Figure 1.
Epigenetics of lupus CD4+ T cells. A broad DNA methylation defect is observed in lupus CD4+ T cells. Reduced expression of DNA methyltransferase 1 (DNMT1) is observed to lead to the hypomethylation and overexpression of several genes in lupus CD4+ T cells. Specifically, hypomethylation of promoter regions of CD70, CD11a, PRF1, CD40LG (CD40 ligand), and killer immunoglobulin-like receptor (KIR) genes leads to their overexpression and subsequent induction of T-cell autoreactivity. Further, overexpression of GADD45A (growth arrest and DNA-damage-inducible 45 alpha) contributes to decreased DNA methylation in lupus CD4+ T cells. There is overlap between the DNA methylation defect in lupus and other epigenetic mechanisms. Overexpression of miR-148a and miR-126 affect DNA methylation levels by directly targeting DNMT1 transcripts. miR-21 overexpression also affects DNA methylation; however, this microRNA indirectly regulates DNMT1 expression through ERK pathway signaling. Variable expression of other individual regulatory microRNAs also contributes to the disease process. Overexpression of miR-17-92 in murine T lymphocytes is observed and may affect lymphocyte stability. miRNA-155 is overexpressed in CD4+ T cells and contributes to reduced stability of regulatory T cell populations. miR-31 negatively regulates the expression of Foxp3 and has been observed to be overexpressed in murine splenocytes. Underexpression of miR-146 leads to increased type I IFN signaling in lupus CD4+ T cells. Reduced expression of miR-125a leads to increased expression of the inflammatory chemokine RANTES by reduced targeting of KLF13 transcripts. Overexpression of miR-101 has been reported in murine T lymphocytes. miR-101 has also been reported to negatively regulate EZH2, a histone methyltransferase involved in epigenetic repression. Aberrant patterns of histone modification are also observed in lupus CD4+ T cells. Reduced levels of histone 3 lysine 9 trimethylation (H3K9me3) are observed as well as reduced p300 histone acetyltransferase activity. Further, EZH2 levels are reduced in lupus CD4+ T cells. Reduced levels of the transcription factor RFX1 lead to loss of local epigenetic repression characterized by reduced levels of DNA methylation and H3K9me3 via the histone methyltransferase SUV39H1.
Lymphocyte function-associated antigen 1 (LFA-1) is a multi-unit, integrin family protein involved in lymphocyte adhesion that is composed of the products of two genes, CD11a and CD18. DNA methylation and local chromatin architecture regulate the tissue-specific expression of CD11a [40]. Promoter hypomethylation of the ITGAL (CD11a) gene is observed in lupus CD4+ T cells [41]. This leads to overexpression of LFA-1 among an autoreactive T-cell subset. Further, ectopic expression of CD18 recapitulates the effects of treatment with any of the multiple DNA methylation inhibitors in inducing T-cell autoreactivity [42]. LFA-1 overexpression is a driver of autoreactivity directly linked to reduced levels of DNA methylation.
CD70 (TNFSF7) is a membrane bound, B-cell co-stimulatory protein involved in regulating B-cell immuno-globulin production. CD70 is overexpressed in lupus CD4+ T cells and T cells treated with DNA methylation inhibitors, and this overexpression stimulates B cell IgG production in vitro [43]. Indeed, the promoter sequence of CD70 that demethylates upon treating normal CD4+ T cells with DNA methylation inhibitors is similarly demethylated in lupus CD4+ T cells [44]. Reduced methylation of a CpG island in the CD70 promoter was also observed to correspond with higher CD70 expression and reduced DNMT1 mRNA levels in MRL/lpr mice at 16 weeks when autoimmunity is established [45]. Overexpression of CD70 in response to reduced promoter methylation is also observed in patients with subacute cutaneous lupus erythematosus. Increased expression was observed at both the mRNA and protein level by flow cytometry, and although the overall proportion of CD70-expressing cells was unchanged, the mean florescent intensity was greater among subacute cutaneous lupus erythematosus patient samples [46].
Many transcription factors are known to influence local chromatin architecture through specific interaction with DNA and histone methyltransferases. Decreased levels of RFX1 at CD11a and CD70 promoters leads to decreased methylation and increased expression of both of these genes in lupus CD4+ T cells [47]. Moreover, this change in DNA methylation and expression is reinforced by concomitant reductions in histone 3 lysine 9 trimethylation (H3K9me3) at these promoters in a RFX1-SUV39H1 interaction-dependent manner [48].
A role for the Gadd45A (growth arrest and DNA-damage-inducible 45 alpha) gene in autoimmunity has long been suggested. Initial knock-out studies of Gadd45a reported by Salvador and colleagues revealed a protective role for the gene product, as knockout Gadd45A-/- mice grew to develop a lupus-like systemic autoimmune disease [49]. More recently, transfection of HEK293 cells with a Gadd45A overexpression vector was shown to result in both site-specific and global DNA demethylation. Further, increased expression of Gadd45A following exposure to ultraviolet light was shown to lead to reduced genomic DNA methylation, a trend partially counteracted by knockdown of Gadd45A [50]. These findings were recapitulated in recent work in CD4+ T cells in which overexpression of Gadd45A and exposure to UV-B light radiation led to increased expression and promoter demethylation of CD70 and CD11a [51].
Perforin is a protein encoded by the PRF1 gene, expressed primarily by CD8+ T lymphocytes and natural killer cells. Perforin acts to promote target celllysis by promoting the formation of pores in the target cell membrane. Initially, overexpression of perforin in T cells was observed in response to treatment with DNA methylation inhibitors. Promoter demethylation and perforin overexpression were accompanied by changes in chromatin architecture as evidenced by the gain of a DNAse hypersensitive site upstream of the PRF1 transcription start site [52]. Further, this trend of PRF1 promoter demethylation leading to increased perforin expression was then confirmed in lupus CD4+ T cells, where increased perforin was observed via flow cytometry in CD4+ T cells of lupus patients with active disease [53].
Killer immunoglobulin-like receptors (KIRs) comprise a diverse set of polymorphic genes involved in HLA type I stimulatory and inhibitory signals in natural killer cells and CD4+ CD28- T cells [54]. Expression of KIR genes is regulated by methylation of their promoter regions, and treatment of T cells with 5-azacytidine leads to activation of a number KIR genes [55]. Interestingly, overexpression of KIR genes is also observed in lupus CD4+ T cells, and this overexpression is seen to correspond to disease activity [56].
DNA methylation in lupus B cells
Several genes are observed to be differentially regulated in lupus B cells. Surface expression of CD5, which is important in maintaining B cell anergy [57], is known to be reduced in lupus B cells. IL-6 has been observed to alter CD5 expression levels in lupus B cells by inhibiting DNA methylation [58]. Increased exposure to IL-6 leads to ERK pathway inhibition, decreased DNMT1 levels, and increased expression of CD5-1EB, the cytoplasmic form of CD5, which in turn limits the expression of CD5 on the cell surface. This trend was observed in lupus B cells and confirmed among B cells from healthy controls exposed to increased levels of IL-6 or treated with DNA methylation inhibitors [58].
Widespread DNA methylation differences between lupus patients and healthy controls
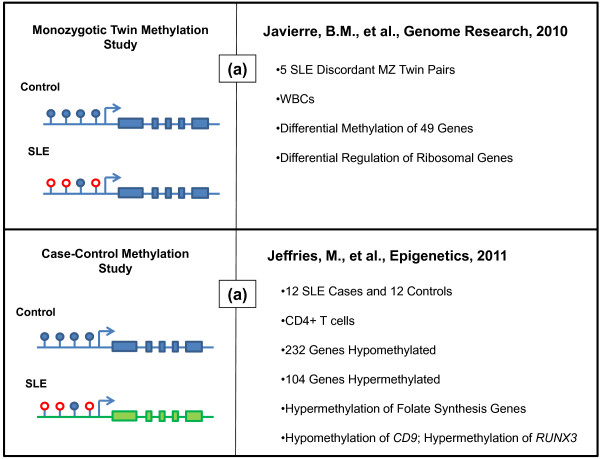
Studies in disease-discordant monozygotic twins are useful when investigating epigenetic aspects of complex human disease. Monozygotic twin studies enable the control of many of the confounding influences of genetic variation, although there is evidence for copy number variation among monozygotic twins [59], while focusing on disease-relevant, inter-individual epigenetic variation. Monozygotic twins display significant life course variation in epigenetic modifications, and this variation, it is thought, may drive disease pathogenesis [60]. A recent twin study in lupus revealed significant epigenetic variation in white blood cells between disease-discordant monozygotic twins [61]. The study reported and validated a number of genes found to possess promoter methylation differences. Specifically, 49 genes were found to be hypomethylated in lupus patients, highlighting ontologies relevant to autoimmunity. In addition, variable methylation of ribsomal genes was reported whereby 18S and 28S ribosomal subunit genes are hypomethylated in lupus individuals of discordant pairs and hypomethylated in both individuals of concordant lupus twin pairs examined.
A recent case-control methylation study utilizing high-throughput methylation arrays identified a number of differentially methylated genes and pathways in lupus CD4+ T cells. The methylation status of over 27,000 CpG sites in promoter regions of nearly 15,000 genes was collected and analyzed for 12 female lupus cases and 12 controls. In the study, Jeffries and colleagues report hypomethylation of 236 CpG sites and hypermethylation of another 105 (representing 232 and 104 genes, respectively). Among hypomethylated genes, ontology analysis highlighted genes involved in connective tissue development, including ADAMTS1, ALX4, CD9, ESRRA, FGF8, HOXA13, HOXD11, MMP9, MSX1, PDGFRA, and SOX5 [62]. Hypermethylation of genes involved in folate biosynthesis, a pathway related to maintenance of DNA methylation, was observed [62]. Interestingly, RUNX3, which belongs to a transcription factor family involved in mediating cell proliferation signals [63], was found to be hypermethylated in lupus CD4+ T cells. Protein interaction network analysis also revealed differential methylation of several apoptosis-associated genes, and genes involved in cell growth, tissue development, and cell division. This study also provided data to support a potential role for some specific DNA methylation changes as novel biomarkers for lupus disease activity, with the caveat that these need to be first validated and then examined in the setting of a clinical trial before being considered of any clinical benefit (Figure 2).
Figure 2.
Genome-wide methylation studies in lupus. (a) Differences in promoter DNA methylation were investigated in the context of identical genetic background (that is, discordant monozygotic (MZ) twins). Javierre and colleagues [61] performed a genome-wide methylation scan investigating epigenetic differences in five pairs of monozygotic twins discordant for lupus. The study investigated these differences in white blood cells (WBCs) of patients and their related controls. The study reported variable methylation of 49 of the 807 genes investigated. Pathway analysis of these genes reveals putative functions in immune response, cell activation, and cell proliferation. The study further identifies hypomethylation and overexpression of ribosomal genes, which may correspond to increased ribosomal-autoantibody formation. (b) Jeffries and colleagues [62] investigated the methylation status of over 27,000 individual CpG dinucleotides located in promoter regions of nearly 15,000 genes. Twelve lupus cases of varying disease activity and healthy control individuals were included in the study. The study reports hypomethylation of CpG dinucleotides in 232 genes and the hypermethylation of 104 genes. Pathway analysis reveals genes involved in folate biosynthesis, a pathway related to maintenance of DNA methylation. Further, modifications of individual genes observed to be dysregulated included the hypomethylation of CD9, encoding a known activator of T cell signaling, and hypermethylation of RUNX3, which encodes a transcription factor that mediates profileration signals in lymphocytes. In both of the diagrams on the left side of each panel, empty red circles represent unmethylated CpG dinucleotides, whereas solid blue circles represent methylated CpG sites. In the lower panel, the differences in color of the gene body indicate the presence of genetic heterogeneity.
Variable regulation of microRNAs in lupus
MicroRNAs represent a post-transcriptional regulatory mechanism whereby single-stranded RNAs 20 to 24 bp in length homologously bind to mRNA transcripts, effectively blocking their subsequent translation [64]. MicroRNA regulatory networks work to fine-tune immune function. Further, differential regulation of microRNAs appears to play a role in the disease process of lupus.
MiR-146, which plays a role in negatively regulating innate immune responses [65], has been observed to be intrinsically underexpressed in lupus CD4+ T cells. Reduced expression of miR-146 leads to induction of type I interferon signaling, and miR-146 expression levels negatively correlate with disease activity. Interestingly, IFN signatures of lupus patient peripheral blood mononuclear cells were alleviated upon transfection with a miR-146 expression vector [66]. Zhao and colleagues observed targeting of the 3' UTR of the KLF13 gene by miR-125a in a dose-dependent manner. Underexpression of miR-125a was also reported in lupus CD4+ T cells leading to increased levels of the inflammatory chemokine RANTES [67]. Normalization of miR-125a expression in CD4+ T cells of lupus patients resulted in reduced RANTES levels.
Investigation of microRNA patterns in three models of murine lupus reveals common microRNA pathways. The study investigated common defects in regulatory microRNAs among splenic T and B lymphocytes in B6/lpr, MRL/lpr, and NZB/W mice when compared with their respective age-matched control mice. Dai and colleagues report a number of regulatory microRNAs, including miR-155, miR-150, miR-182-96-183, miR-31, miR-127, and miR-379, to be significantly increased in splenic B and T cells of MRL/lpr mice. Significantly, upregulation of miR-146, miR101a, and miR-17-92 in MRL/lpr mice was observed exclusively among T lymphocytes [68]. Mice with increased miR-17-92 expression display uncontrolled expansion and autoimmunity characterized by reductions in the proapoptotic protein Bim and the tumor suppressor PTEN (phosphatase and tensin homolog) [69]. Interestingly, genomic loss of miR-101 has been associated with overexpression of the repressive histone methyltransferase EZH2 in human prostate cancer cells [70]. EZH2, the histone methyltransferase that mediates H3K27me3, functionally associates with DNMT1 [71]. Overexpression of miR-101 might contribute to the loss of repression observed in lupus CD4+ T cells. Moreover, downregulation of EZH2 in CD4+ T cells has been reported in lupus patients with active disease [72].
A systematic, microarray-based investigation of microRNA expression in Epstein-Barr virus-transformed B cell lines of lupus patients with nephritis and controls in Caucasian and African-American populations observed reproducible overexpression of a number of microRNAs across ethnicities [73]. This study also included a discordant monozygotic twin pair in which the broader microRNA expression patterns were partially conserved.
More recently, analysis of microRNA expression profiles in lupus peripheral blood mononuclear cells compared to controls revealed the differential expression of 27 microRNAs out of 365 analyzed [74]. Importantly, the levels of miR-21, miR-25, miR-106b and miR-148b correlated with disease activity in lupus patients, while miR-196a and miR-379 expression inversely correlated with disease activity, suggesting a potential role for microRNA profiling as disease biomarker in lupus [74].
A recent comprehensive review on the role of micro-RNA in the pathogenesis of lupus and other autoimmune disease has been recently published [75].
Crosstalk between microRNAs and DNA methylation in lupus
Targeting of DNA methyltransferases by regulatory microRNAs is observed in both development and disease. MicroRNAs appear to play a critical role in regulation of de novo methylation in mouse embryonic stem cells [76]. Targeting of DNMT3a and DNMT3b by miR-29 family members is observed in human lung cancer [77]. Two recent studies demonstrate downregulation of DNMT1 as a result of variable expression of regulatory microRNAs in lupus.
Pan and colleagues [78] demonstrated a connection between aberrant regulation of certain microRNAs and reduced expression of DNMT1. A genome-wide micro-RNA scan in both lupus patients and MRL/lpr lupusprone mice revealed increased expression of both miR-21 and miR-148a. miR-21 targets and inhibits RSGRP1, a signaling molecule involved in the ERK signaling pathway, through which it is able to indirectly downregulate DNMT1 expression. MiR-148a, however, directly targets the DNMT1 transcript.
MiR-126 is among a number of microRNAs observed to be variably regulated in lupus CD4+ T cells. Zhao and colleagues [79] report overexpression of miR-126 in lupus CD4+ T cells, which correlates with decreased DNMT1 expression. They further demonstrate that miR-126 targets a 3' UTR region in DNMT1 and reduces DNMT1 protein expression. Furthermore, ectopic overexpression of miR-126 in CD4+ T cells from healthy donors led to hypomethylation and overexpression of the methylation sensitive genes CD11a and CD70 and induction of T cell autoreactivity [79].
Variable histone protein modification in lupus
Covalent modification of histone protein represents another layer of epigenetic regulation. Acetylation of histone tails leads to an open conformation of chromatin that is permissive to gene expression. The acetyltrans-ferase activity of p300 in B cells is crucial in preventing autoimmunity. Mice with a B cell-specific knock-in mutant version of p300 that lacks only histone acetyl-transferase activity develop a lupus-like autoimmune disease characterized by splenomegaly, nephritis, and vasculitis [80]. Reduced global levels of H3K9me3 and histone 3 acetylation have been reported in lupus CD4+ T cells, and differences in the activity of histone acetyl-transferases have been observed in lupus CD4+ T cells [72].
Sullivan and colleagues [81] initially reported variability in histone 4 acetylation at the tumor necrosis factor alpha (TNF-α) locus in lupus monocytes. This same group further studied histone 4 acetylation at the genome level in lupus monocytes where they found hyperacetylation and overexpression of several genes with modest overlap with the acetylation and expression patterns of IFN-α treated cells [82]. These findings point to widespread changes in chromatin architecture in lupus monocytes.
The altered epigenetic state of regulatory T cell populations in murine lupus
Foxp3+ CD25high regulatory T cells broadly serve to suppress autoimmunity. Expression of the transcription factor Foxp3 regulates the differentiation of the regulatory T cell lineage [83]. Mice that lack Foxp3 display a paucity of regulatory T cells and subsequently develop fatal autoimmunity [84]. Epigenetic modifications regulate the emergence and durability of regulatory T cells. Demethylation of the Foxp3 promoter accompanies the emergence of the regulatory T cell lineage in both mice and humans [85,86]. Foxp3 expression is also governed post-transcriptionally by microRNAs. Specifically, miR-31 is a negative regulator of Foxp3 expression, while miR-21 positively regulates Foxp3 [87].
Regulatory T cells of lupus patients with active disease are reduced in quantity commensurate to disease severity and display decreased suppressive function [88,89]. Divekar and colleagues [90] have recently reported a regulatory T cell defect of murine lupus whereby regulatory T cells exhibit reduced suppressive potential in MRL/lpr mice that is further characterized by decreased dicer expression and increased miR-155 expression. Interestingly, miR-155 plays a critical role in the regulation of germinal center formation and is critical in maintaining proper immune function [91,92]. Divekar and colleagues suggest that increased expression of miR-155 may account for the regulatory T cell defect observed in MRL/lpr mice. These findings further coincide with previous reports in which dicer deficiency among regulatory T cells led to diminished regulatory T cell populations and autoimmune pathology [93,94].
Epigenetic variation in lupus: cause or consequence?
This review offers clear evidence of the involvement of epigenetic variation in lupus. The studies mentioned herein describe widespread lupus-associated epigenetic variation, yet association and causation are not interchangeable terms. Many questions regarding the causal nature of epigenetic variation in lupus remain to be answered. At what stage of disease are epigenetic manifestations first present? In what cell type(s) do epigenetic perturbations first appear in lupus? Does epigenetic variation merely reflect prior canalization of disease, or are these modifications ultimately causal? Is disease-associated epigenetic variation a proximate or an ultimate cause of disease?
The causal nature of epigenetic variation in lupus is a complicated question. In our view, large longitudinal cohort studies that will allow the epigenome to be examined before and after disease will be crucial to answer this question. Of course, these types of studies will be complicated to design, and samples from specific cell types will have to be maintained and subsequently examined. The evidence that inhibiting T cell ERK path-way signaling using a mouse model results in decreased DNMT1 expression, overexpression of methylation sensitive genes similar to lupus patients, the development of anti-double stranded DNA antibody, and an interferon signature-like gene expression profile represents the strongest argument for a causal role of DNA methylation in lupus to date [32]. Recent studies have shown that epigenetic variation in lupus occurs along multiple axes. Unraveling the causal web of lupus epigenetics has become an increasingly complicated task as recent studies have revealed a complex, interconnected network of epigenetic regulation that leads to altered gene expression in lupus. These studies pose new questions about the nature of complexity of regulatory events involved in lupus pathogenesis. Future epigenetic studies investigating the origins of complex human disease will therefore need to incorporate multiple-level systems modeling and complexity science to uncover disease etiology in complex and dynamic biologic systems.
Determining the environmental-epigenetic interactions that contribute to lupus is important in understanding the ultimate cause of lupus. There is much reference made to environmental factors in the lupus epigenetics literature, yet these typically refer to a few examples or are invoked as an unknown explanatory variable. With a few notable exceptions, the specific mechanisms through which environmental triggers operate remain largely uncharacterized to date. Further characterization of the putative environmental-epigenetic tipping point between active and inactive disease is therefore needed.
Conclusion
Studies to date have revealed a substantial epigenetic component of disease in lupus, particularly in T cells (Table 1, Figure 1), yet ultimately the causal nature of disease associated epigenetic variation remains ambiguous. Although the etiology of lupus is indeterminate, some generalizations about the origins of lupus can be made: first, there is a significant genetic component to the disease; second, known genetic variation does not comprehensively describe the origin, development, and even the heritability of the disease; third, epigenetic variation correlates with variable expression of a number of genes in pathways involved in disease pathogenesis; and fourth, lupus is the result of complex interactions between genetic susceptibility, environmental exposure, and epimutation. The path forward in understanding lupus is not merely in the clear delineation of the various components of disease but in understanding emergent phenotypes that result from their interaction over time.
Table 1.
Epigenetic perturbations observed in lupus T cells
| Gene/pathway | Epigenetic defect | Cell type | Consequence | Reference |
|---|---|---|---|---|
| ERK pathway | Defective signaling | Human and murine T cells | Reduced DNMT1 expression and reduced T cell DNA methylation | [23,32] |
| PRF1 | Hypomethylation | CD4+ T cells | Increased perforin expression | [52,53] |
| ITGAL | Hypomethylation | CD4+ T cells | Increased CD11a expression | [40,41] |
| CD40LG | Hypomethylation | CD4+ T cells | B-cell hyperactivation | [36] |
| KIR2DL4 (KIR) | Hypomethylation | CD4+ T cells | Increased KIR expression | [55,56] |
| TNFSF7 | Hypomethylation | CD4+ T cells | Increased CD70 expression | [43-45] |
| GADD45A | Overexpression | CD4+ T cells | Decreased DNA methylation | [51] |
| Folate biosynthesis | Hypermethylation | CD4+ T cells | Impaired DNA methylation? | [62] |
| RUNX3 | Hypermethylation | CD4+ T cells | Impaired T cell proliferation? | [62] |
| CD9 | Hypomethylation | CD4+ T cells | Increased T cell activation? | [62] |
| MMP9 | Hypomethylation | CD4+ T cells | ? | [62] |
| PDGFRA | Hypomethylation | CD4+ T cells | ? | [62] |
| SOX5 | Hypomethylation | CD4+ T cells | ? | [62] |
| miR-126 | Overexpression | CD4+ T cells | Downregulation of DNMT1 | [79] |
| miR-146 | Underexpression | CD4+ T cells; murine T splenocytes | Increased type I IFN | [66,68] |
| miR-17-92 | Overexpression | Murine T splenocytes | Lymphocyte stability | [68] |
| miR-155 | Overexpression | CD4+ T cells; CD4+ CD25+ T cells | Reduced Treg cell stability | [90] |
| miR-101a | Overexpression | Murine T splenocytes | Reduced EZH2? | [68] |
| miR-31 | Overexpression | Regulatory T cells; murine T splenocytes | Repression of FOXP3 | [62,87] |
| miR-21 | Overexpression | CD4+ T cells | Targets ERK pathway leading to lower DNMT1 | [78] |
| miR-125a | Underexpression | CD4+ T cells | Targets KLF13 leading to increased RANTES | [67] |
| miR-148a | Overexpression | CD4+ T cells | Downregulates DNMT1 | [78] |
| RFX1 | Underexpression | CD4+ T cells | Reduced H3K9me3 and DNA methylation | [47,48] |
CD40LG, CD40 ligand; DNMT, DNA methyltransferase; ERK, extracellular signal-regulated kinase; GADD45A, growth arrest and DNA-damage-inducible 45 alpha; H3K9me3, histone 3 lysine 9 trimethylation; KIR, killer immunoglobulin-like receptor; Treg, regulatory T cell.
Abbreviations
CD40LG: CD40 ligand; DNMT: DNA methyltransferase; ERK: extracellular signal-regulated kinase; GADD45A: growth arrest and DNA-damage-inducible 45 alpha; H3K9me3: histone 3 lysine 9 trimethylation; IFN: interferon; IL: interleukin; KIR: killer immunoglobulin-like receptor; LFA-1: lymphocyte function-associated antigen 1; UTR: untranslated region.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Travis Hughes, Email: Travis-Hughes@omrf.org.
Amr H Sawalha, Email: amr-sawalha@ouhsc.edu.
References
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, González-Escribano MF, Martin J, Abderrahim H, Alarcón-Riquelme ME. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapää-Dahlqvist S, Petri M, Manzi S, Seldin MF, Rönnblom L, Syvänen AC, Criswell LA, Gregersen PK, Behrens TW. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010;11:554–560. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci USA. 2007;104:17052–17057. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med. 2006;203:1493–1505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–3035. [PubMed] [Google Scholar]
- Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- Scheinbart LS, Johnson MA, Gross LA, Edelstein SR, Richardson BC. Procainamide inhibits DNA methyltransferase in a human T cell line. J Rheumatol. 1991;18:530–534. [PubMed] [Google Scholar]
- Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- Rouleau J, MacLeod AR, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem. 1995;270:1595–1601. doi: 10.1074/jbc.270.4.1595. [DOI] [PubMed] [Google Scholar]
- Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- Kammer GM, Perl A, Richardson BC, Tsokos GC. Abnormal T cell signal transduction in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1139–1154. doi: 10.1002/art.10192. [DOI] [PubMed] [Google Scholar]
- Cedeño S, Cifarelli DF, Blasini AM, Paris M, Placeres F, Alonso G, Rodriguez MA. Defective activity of ERK-1 and ERK-2 mitogen-activated protein kinases in peripheral blood T lymphocytes from patients with systemic lupus erythematosus: potential role of altered coupling of Ras guanine nucleotide exchange factor hSos to adapter protein Grb2 in lupus T cells. Clin Immunol. 2003;106:41–49. doi: 10.1016/S1521-6616(02)00052-9. [DOI] [PubMed] [Google Scholar]
- Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- Tada Y, Nagasawa K, Yamauchi Y, Tsukamoto H, Niho Y. A defect in the protein kinase C system in T cells from patients with systemic lupus erythematosus. Clin Immunol Immunopathol. 1991;60:220–231. doi: 10.1016/0090-1229(91)90065-I. [DOI] [PubMed] [Google Scholar]
- Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, Nakayama KI. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferonregulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal IS, Foellmer HG, Grewal KD, Xu J, Hardardottir F, Baron JL, Janeway CA Jr, Flavell RA. Requirement for CD40 ligand in costimulation induction, T cell activation, and experimental allergic encephalomyelitis. Science. 1996;273:1864–1867. doi: 10.1126/science.273.5283.1864. [DOI] [PubMed] [Google Scholar]
- Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–431. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Desai-Mehta A, Lu L, Ramsey-Goldman R, Datta SK. Hyperexpression of CD40 ligand by B and T cells in human lupus and its role in pathogenic autoantibody production. J Clin Invest. 1996;97:2063–2073. doi: 10.1172/JCI118643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yuan J, Pan Y, Fei Y, Qiu X, Hu N, Luo Y, Lei W, Li Y, Long H, Sawalha AH, Richardson B, Lu Q. T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus. Clin Immunol. 2009;132:362–370. doi: 10.1016/j.clim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbalkan Z, Bagişlar S, Kiraz S, Akyerli CB, Ozer HT, Yavuz S, Birlik AM, Calgüneri M, Ozçelik T. Skewed X chromosome inactivation in blood cells of women with scleroderma. Arthritis Rheum. 2005;52:1564–1570. doi: 10.1002/art.21026. [DOI] [PubMed] [Google Scholar]
- Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, Reveille JD, Alarcón GS, Vilá LM, Reid J, Harris B, Li S, Kelly JA, Harley JB. Klinefelter's syndrome (47, XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99:4503–8. doi: 10.1182/blood.V99.12.4503. [DOI] [PubMed] [Google Scholar]
- Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- Richardson B, Powers D, Hooper F, Yung RL, O'Rourke K. Lymphocyte function-associated antigen 1 overexpression and T cell autoreactivity. Arthritis Rheum. 1994;37:1363–1372. doi: 10.1002/art.1780370915. [DOI] [PubMed] [Google Scholar]
- Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, Richardson B. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupusinducing drugs. J Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur J Immunol. 2007;37:1407–1413. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhao M, Lu Q. Demethylation of promoter regulatory elements contributes to CD70 overexpression in CD4+ T cells from patients with subacute cutaneous lupus erythematosus. Clin Exp Dermatol. 2010;35:425–430. doi: 10.1111/j.1365-2230.2009.03611.x. [DOI] [PubMed] [Google Scholar]
- Zhao M, Sun Y, Gao F, Wu X, Tang J, Yin H, Luo Y, Richardson B, Lu Q. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun. 2010;35:58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Zhao M, Wu X, Zhang Q, Luo S, Liang G, Su Y, Tan Y, Lu Q. RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Res Ther. 2010;12:R227. doi: 10.1186/ar3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador JM, Hollander MC, Nguyen AT, Kopp JB, Barisoni L, Moore JK, Ashwell JD, Fornace AJ Jr. Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity. 2002;16:499–508. doi: 10.1016/S1074-7613(02)00302-3. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, Döderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhao M, Yin H, Gao F, Wu X, Luo Y, Zhao S, Zhang X, Su Y, Hu N, Long H, Richardson B, Lu Q. Overexpression of the growth arrest and DNA damageinduced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62:1438–47. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr Opin Immunol. 2004;16:626–33. doi: 10.1016/j.coi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Liu Y, Wu A, Yarlagadda S, Gorelik GJ, Kaplan MJ, Hewagama A, Hinderer RC, Strickland FM, Richardson BC. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J Immunol. 2009;183:3481–3487. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippen KL, Tze LE, Behrens TW. CD5 maintains tolerance in anergic B cells. J Exp Med. 2000;191:883–890. doi: 10.1084/jem.191.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaud S, Le Dantec C, Jousse-Joulin S, Hanrotel-Saliou C, Saraux A, Mageed RA, Youinou P, Renaudineau Y. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. 2009;182:5623–5632. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, Diaz de Ståhl T, Menzel U, Sandgren J, von Tell D, Poplawski A, Crowley M, Crasto C, Partridge EC, Tiwari H, Allison DB, Komorowski J, van Ommen GJ, Boomsma DI, Pedersen NL, den Dunnen JT, Wirdefeldt K, Dumanski JP. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genomewide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, Luo X, Huang X, Li J, Chen S, Shen N. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, Ahmed SA. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010;5:e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W, Zhang G, Zhou Y, Su Y, Lu Q. Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol. 2008;35:804–810. [PubMed] [Google Scholar]
- Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, Bruner GR, Harley JB, Ojwang JO. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, Iliopoulos D, Boumpas DT. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70:1496–1506. doi: 10.1136/ard.2010.139857. [DOI] [PubMed] [Google Scholar]
- Ceribelli A, Yao B, Dominguez-Gutierrez PR, Nahid MA, Satoh M, Chan EK. MicroRNAs in systemic rheumatic diseases. Arthritis Res Ther. 2011;13:229. doi: 10.1186/ar3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, Yin H, Lu Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- Forster N, Gallinat S, Jablonska J, Weiss S, Elsässer HP, Lutz W. p300 protein acetyltransferase activity suppresses systemic lupus erythematosus-like autoimmune disease in mice. J Immunol. 2007;178:6941–6948. doi: 10.4049/jimmunol.178.11.6941. [DOI] [PubMed] [Google Scholar]
- Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. 2007;123:74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010;11:124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothé F, Simion A, Akl H, Mourtada M, El Rifai M, Burny A, Romero P, Martiat P, Badran B. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, Nochy D, Debré P, Piette JC, Gorochov G. Global natural regulatory T cell depletion in active systemic lupus erythematosus. J Immunol. 2005;175:8392–8400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007;178:2579–2588. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- Divekar AA, Dubey S, Gangalum PR, Singh RR. Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J Immunol. 2011;186:924–930. doi: 10.4049/jimmunol.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]